The orientation of stem cell divisions is maintained beyond one cell cycle thanks to microtubule polymerization and apical centrosome positioning.
Abstract
The mechanisms that maintain the orientation of cortical polarity and asymmetric division unchanged in consecutive mitoses in Drosophila melanogaster neuroblasts (NBs) are unknown. By studying the effect of transient microtubule depolymerization and centrosome mutant conditions, we have found that such orientation memory requires both the centrosome-organized interphase aster and centrosome-independent functions. We have also found that the span of such memory is limited to the last mitosis. Furthermore, the orientation of the NB axis of polarity can be reset to any angle with respect to the surrounding tissue and is, therefore, cell autonomous.
Introduction
Asymmetric division of Drosophila melanogaster neuroblasts (NBs), a neural stem cell population, results in the generation of two unequally fated daughter cells. One daughter, the ganglion mother cell (GMC), enters a differentiation pathway, whereas the other daughter is a self-renewed NB. During the asymmetric division of NBs, orientation of the mitotic spindle relies on the apicobasal polarity axis (Gonzalez, 2007; Chia et al., 2008; Doe, 2008; Knoblich, 2008; Siller and Doe, 2009). NB cortical polarization involves the apical localization of the Par (partitioning defective) and Pins (partner of Insc [Inscuteable]) complexes. The Par complex, which includes Baz (Bazooka; the fly homologue of Caenorhabditis elegans Par-3), Par-6, and atypical PKC (Wodarz et al., 1999; Petronczki and Knoblich, 2001; Rolls et al., 2003), directs the basal localization of cell-fate determinants such as Pros (Prospero), Brat (Brain Tumor), and Numb through their adaptor proteins Mira (Miranda) and Pon (partner of Numb; Doe et al., 1991; Ikeshima-Kataoka et al., 1997; Li et al., 1997; Shen et al., 1997; Lu et al., 1998; Schober et al., 1999; Izumi et al., 2004; Betschinger et al., 2006; Lee et al., 2006b; Caussinus and Hirth, 2007). The Pins complex includes Pins, the heterotrimeric G protein subunit Gαi, and Mud (Mushroom body defect; Parmentier et al., 2000; Schaefer et al., 2000; Yu et al., 2003; Izumi et al., 2004, 2006; Bowman et al., 2006; Siller et al., 2006) and seems to be mainly involved in aligning the spindle along the apicobasal axis. Pins and Gαi are also involved in the control of unequal daughter cell size (Cai et al., 2003; Fuse et al., 2003; Yu et al., 2003; Izumi et al., 2004). Both complexes contain known auxiliary modulators (Chia et al., 2008; Doe, 2008; Knoblich, 2008). Organization of the basal cortex is also dependent on the activity of Dlg (Discs large) and Lgl (Lethal [2] giant larvae; Ohshiro et al., 2000; Peng et al., 2000; Betschinger et al., 2003). These two proteins have long been known for their function as tumor suppressors (Gateff, 1978). Some of the proteins of the aforementioned apical and basal complexes also have tumor suppressor functions (Caussinus and Gonzalez, 2005; Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006a,b; Doe, 2008; Januschke and Gonzalez, 2008).
In both embryonic and larval NBs, the axis of cortical polarity remains roughly unchanged through successive rounds of cell division, and the side of GMC delivery is fixed (Ito and Hotta, 1992; Yu et al., 2006; Rebollo et al., 2009). NBs delaminate from the neuroectoderm during embryogenesis (Lu et al., 2000; Egger et al., 2008). Delaminating NBs inherit the apical cortex of the epithelium, which contains the Par complex and is stabilized by the expression of the NB-specific protein Insc. Insc also mediates the assembly of the Pins complex into the apical cortex and controls spindle alignment (Lu et al., 1998; Siegrist and Doe, 2005). This model of apical cortex polarization seems to explain well how apicobasal polarity is established and maintained during the first round of asymmetric cell division that follows delamination (Parmentier et al., 2000; Schaefer et al., 2000; Yu et al., 2000). However, the Par and Pins complexes are dismantled at mitosis exit, and Baz cortical localization, the earliest sign of cortical polarity, only starts at the end of the following interphase (Siller et al., 2005; Lee et al., 2006a; Rebollo et al., 2007; Rusan and Peifer, 2007). Therefore, it is unclear how the orientation of its first apicobasal polarity axis is memorized and reestablished at almost invariant positions through consecutive cell cycles (Yu et al., 2006).
Live imaging studies of larval NBs labeled with centriolar reporters have shown that a few minutes after cytokinesis, soon after the splitting of the centriolar signal (Rebollo et al., 2007; Rusan and Peifer, 2007), only one of them retains pericentriolar material, thus becoming the major microtubule-organizing center (MTOC) of the cell. This active centrosome positions itself in close contact with the region of the cortex where the apical crescent was localized during mitosis, which is indeed the same in which the apical crescent will form in the next mitosis (Rebollo et al., 2007; Rusan and Peifer, 2007). Thus, for most of the cell cycle, the microtubule cytoskeleton is organized from a centrosome that is bound to the presumptive apical cortex, its localization accurately predicting the position of the next apical crescent much earlier than the onset of asymmetric localization of any of the known markers of cortical polarity. The same process takes place in embryonic NBs (Rebollo et al., 2009), with the exception of the first cell cycle when delamination occurs, in which spindles assemble orthogonally to the polarity axis and later rotate to align with it (Kaltschmidt et al., 2000).
It has been proposed that the microtubule aster organized by the apical centrosome of the NB could contribute to pass on polarity information from one cell cycle to the next (Rebollo et al., 2007; Rusan and Peifer, 2007). However, the well-established fact that assembly of the cortical crescents does not require microtubules (Knoblich et al., 1995; Kraut et al., 1996; Broadus and Doe, 1997; Siegrist and Doe, 2005) appears to contradict this hypothesis.
Results
Microtubule depolymerization erases the memory of cortical polarity orientation in larval NBs
To assess the possible contribution of microtubules to maintaining the orientation of apicobasal polarity and asymmetric mitosis in larval NBs, we first performed time-lapse recordings of these cells in the presence of the microtubule-depolymerizing drug colcemid. As reported previously (Rebollo et al., 2007; Rusan and Peifer, 2007), during interphase, NBs contain one major microtubule aster organized by the centrosome that is localized near the region of the cortex where the next apical crescent will be assembled (Fig. 1 A, t0, arrow). The other centrosome is highly motile, has little, if any, pericentriolar material, and cannot organize microtubules (Fig. 1 A, t0, arrowhead). The addition of colcemid at a final concentration of 50 µM results in the depolymerization of most of the microtubule network of the NB and the consequent dispersion of the tubulin-GFP reporter, which becomes a diffuse cloud over the cytoplasm before nuclear envelope breakdown (NEB; Fig. 1 A, t1) and through the entire cell after NEB (Fig. 1 A, t2). Microtubule depolymerization also causes the release of the cortex-bound centrosome (Fig. 1 A, t1 and t2, arrowheads), showing that such a link is microtubule dependent (Video 1). Under these conditions, NBs enter mitosis but are arrested by the spindle assembly checkpoint at c-metaphase, the metaphase-like state induced by microtubule depolymerization (Karess, 2005).
Figure 1.
Microtubule depolymerization severely compromises the fidelity of cortical crescent position in larval NBs. (A–G) Still images from time-lapse recordings of larval NBs before (t0) and after (t1 and t2) treatment with colcemid. NBs are shown oriented with their apical side up in all t0 panels. Fluorescent reporter constructs are indicated to the left of each row. (A–F) Yellow and green asterisks mark pre- and posttreatment crescent position, respectively. Arrows mark apical centrosomes; white arrowheads refer to the basal centrosome in t0 and motile centrosomes in t1 and t2. (A) Before colcemid treatment, the apical centrosome nucleates an aster, and the basal shows migratory behavior (t0). When microtubules are depolymerized, the apical centrosome detaches from the cortex, and both centrosomes localize randomly (t1 and t2; Video 1). (B–D) In the control division, metaphase crescents and position of centrosomes reflect the polarity axes (t0; yellow lines). Behavior of Dlg (B), Pins (C) and Baz (D; Video 2) crescents upon colcemid treatment is shown. Loss of microtubules renders both centrosomes motile (t1). The place of crescent formation becomes unpredictable, suggesting a change in polarity axis orientation that bears no apparent relation with centrosome position at the time of crescent assembly (t2; green lines). (E and F) Yellow and green lines represent pre- and posttreatment cortical polarity orientation, respectively. (E) Basal crescent positioning responds similarly to colcemid treatment (Video 2). (F) Alignment of apical with basal crescents is unperturbed in colcemid-arrested cells despite occurring at an ectopic position (t0 and t2; yellow and green arrowheads, highlighting the limits of the basal crescent, before and after microtubule depolymerization, respectively). Depolymerizing microtubules can prolong interphase, but crescent formation occurred always closely preceding or at the time of NEB, as in controls (not depicted). (G) A crescent assembled before colcemid treatment (t0, yellow asterisk, arrow [apical centrosome], and arrowhead [basal centrosome]) does not change position when microtubules are depolymerized (visible with the loss of centrosome anchoring; t1 and t2, arrowheads). (H and I) Plot of the angle of crescent position in two successive cycles in control cells (yellow) and before and after colcemid treatment (green). Time is shown in hours:minutes:seconds. Bar, 10 µm.
We then determined the effect that these same microtubule-depolymerizing conditions might have on apical crescent assembly. To this end, we followed the behavior of GFP-Dlg, GFP-Baz, and YFP-Pins, three core components of different complexes which localize at the apical cortex at mitosis (Chia et al., 2008; Doe, 2008; Knoblich, 2008). We first followed NBs for one cell cycle in the absence of colcemid to identify the region of the cortex where apical crescents normally form (Fig. 1, B–D, t0, yellow asterisks) and then added 50 µM colcemid to the culture to monitor the position of apical crescent assembly without microtubules (Fig. 1, B–D, t1). As previously reported, apical crescents did assemble in the presence of colcemid (Knoblich et al., 1995; Kraut et al., 1996; Broadus and Doe, 1997). However, unlike crescents assembled under normal microtubule dynamics, which form at an angle between −14° and 24° (n = 20) with respect to the previous (Fig. 1 H, yellow sector), the position of crescents formed in the presence of colcemid was widely scattered (−150° to 140°; n = 23) with respect to where they were before microtubules were depolymerized (Fig. 1, B–D [t2, green asterisks] and H [green asterisks]; and Video 2).
Similar results were obtained regarding the effect of microtubule depolymerization on the localization of basal crescents. Using Mira-GFP (Fig. 1 E, t0) and GFP-Pon as reporters, we found that the addition of colcemid significantly distorted the memory of the site of basal crescent assembly (Fig. 1 E, t1 and t2, green asterisk), which was scattered over an arc ranging from −175° to 138° (n = 10; Fig. 1 I, green asterisks), which is much wider than that observed under normal microtubule dynamics (−13° to 21°; n = 10; Fig. 1 I, yellow sector). Simultaneous imaging of apical (YFP-Pins) and basal (GFP-Pon) crescents under microtubule-depolymerizing conditions showed that they still assembled aligned to each other so that the cortical accumulation of YFP-Pins, which occurs first (Fig. 1 F, t1, green asterisk), can be used to unequivocally predict the place of assembly of the GFP-Pon crescent diametrically opposed to it (Fig. 1 F, t2, green arrowheads). This observation confirms previous results showing that the mechanisms that specify the positioning of the basal protein complexes opposite to the apical cortex are not microtubule dependent (Broadus and Doe, 1997). We also observed that once assembled, crescent position was not affected by microtubule depolymerization (Fig. 1 G).
From these observations, we conclude that retention of the orientation of the cortical polarity axis in successive cell cycles is microtubule dependent. However, notably, a certain bias toward the control orientation can be observed in the population of colcemid-treated NBs, suggesting that either microtubule-independent functions also contribute to the memory of cortical polarity orientation or microtubule depolymerization was not fully achieved in some of these cells. We also conclude that under microtubule-depolymerizing conditions, the entire cell cortex is competent for the assembly of apical crescents that, like those formed under normal conditions, appear shortly before NEB and direct the localization of basal cortical complexes.
The ectopic cortical polarity axis established under microtubule depolymerization conditions drives self-renewing asymmetric division once microtubule dynamics are restored
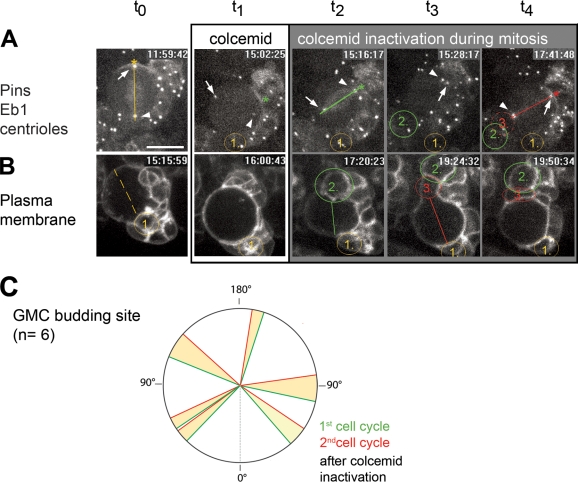
We then wondered whether the ectopic orientation of cortical polarity brought about by microtubule depolymerization might affect the orientation of cell division. To address this question, we treated cells with colcemid as described previously and then delivered a UV pulse to inactivate the drug and restore microtubule dynamics (Theurkauf and Hazelrigg, 1998).
In a first set of experiments, microtubule dynamics were restored after the cells had entered mitosis and established a new cortical polarity axis (Fig. 2 A). Colcemid treatment in these cells lasted for 71 ± 15 min (n = 18). In NBs expressing tubulin and centriole-GFP reporters, the effect of the UV pulse became immediately apparent by the growth of microtubule asters over the two centrosomes (Fig. 2 A, t2, arrowheads), the assembly of the mitotic spindle (Fig. 2 A, t3), and the completion of cytokinesis, which delivered a new GMC (Fig. 2 A, t4, green circle 2). All of these processes appeared to proceed as they did in control untreated cells, except for a major difference: the ectopic site of GMC delivery that, in extreme cases like the NB shown in Fig. 2 A (t4, green circle 2), can be almost diametrically opposed to where the previous GMC was delivered before colcemid treatment (Fig. 2 A, t0, yellow circle 1; and Video 3). A quantified view of this phenotype is shown in Fig. 2 E. Under normal conditions, taking as 0° the place of delivery of a GMC, successive GMCs were produced nearly on top of each other, clustered within an arc ranging from −25° to 24° (n = 54; Fig. 2 E, yellow sector). After transient microtubule depolymerization, the place of delivery of the GMCs with respect to their previous sibling was much wider, ranging from −168° to 119° (n = 12; Fig. 2 E, green circles). Colabeling with microtubule reporters and cortical polarity markers showed that in all cells examined (n = 13), immediately after colcemid inactivation, the newly assembled spindle rotated to align with the cortical polarity axis so that the ectopically positioned basal cortex was duly segregated to the GMC (Fig. 2 B). These observations show that the ectopic cortical polarity axis assembled in the absence of microtubules is capable of driving self-renewing asymmetric division once microtubule dynamics are restored. They also show that transient microtubule depolymerization can reorient NB asymmetric division at almost any angle with respect to the surrounding tissue.
Figure 2.
Transient loss of microtubules establishes ectopic cortical polarity that drives cell division once microtubule dynamics are restored. (A–D) Still images from time-lapse recordings of larval NBs dividing twice. Fluorescent reporter constructs are indicated to the left of each row. White arrows mark apical centrosomes; white arrowheads refer to motile interphase centrosomes and to the basal centrosome in cells in metaphase. GMCs are highlighted by circles and are colored and numbered according to birth order (yellow, first; and green, second). (A–C) Yellow and green lines refer to pre- and posttreatment division orientation, respectively, in the transient microtubule depolymerization experiments followed by colcemid inactivation during (A and B) or before (C) mitosis. (A) The first GMC is delivered basally opposing the apical aster (t0). Microtubule depolymerization mispositions the centrosomes (t1). After microtubule polymerization is restored (t2), the cell enters metaphase (t3) and divides asymmetrically, delivering the second GMC ectopically (t4). Then one centrosome organizes an aster at the ectopic apical pole, whereas the other is down-regulated and motile (colcemid added, 19:06:12; effect detectable, 19:20:22; UV pulse delivered, 20:13:17; and total time exposed, 67 min; Video 3). (B) Transient colcemid treatment produces an ectopic basal cortex (t0–t3; yellow and green arrowheads outline pre- and posttreatment crescents, respectively). Upon colcemid inactivation, the spindle, reflected by the centrosomes at the spindle poles, aligns with this crescent (t2) that is segregated to the ectopically delivered GMC (t3 and t4; colcemid added, 13:35:43; effect detectable, 14:00:43; UV pulse delivered, 14:51:54; and total time exposed, 76 min). (C) The control division delivers the GMC basally (t0). 25 min later, the apical centrosome has detached, yet microtubules remain detectable (t1 and t2, red arrow). When the UV pulse is delivered shortly thereafter, before entry into mitosis, microtubules regrow only over the ectopically localized apical centrosome (t3), and once assembled, the spindle rotates, and the following division occurs at the pretreatment orientation (t4; colcemid added,16:00:46; effect detectable, 16:09:12; UV pulse delivered, 16:32:25; and total exposure time, 32 min; Video 4). (D) Apical crescents form before (t0, yellow asterisk) and after the addition of Taxol (t4, green asterisk) at roughly the same sector of the cortex (Video 5). (E) Plot of GMC budding site variations induced by transient microtubule depolymerization (microtubule dynamics restored after entry into mitosis, green; two consecutive control divisions, yellow). (F) Plot of variations of the place of apical crescent assembly after the addition of Taxol (green) compared with two successive control divisions (yellow). Time is shown in hours:minutes:seconds. Bar, 10 µm.
In a second set of experiments, we assessed the effect of restoring microtubule dynamics while the NBs were still in interphase, before cortical polarity was established (Fig. 2 C). Colcemid treatment in these cells lasted for 42 ± 12 min (n = 8). One of these NBs behaved as described in the previous paragraph, dividing along an ectopic axis. However, in the remaining seven, the former apical centrosome moved back to the region of the cortex where it was before microtubules were depolymerized, the site of apical crescent assembly did not change, and the axis of division was maintained (Fig. 2 C, t0 and t4; and Video 4). Therefore, under these conditions, the cortex was capable of retaining polarity information even though the apical centrosome was temporarily removed from the cortex and unable to organize an aster. Such a short-term memory effect might reflect a microtubule-independent property of the cortex itself or it might simply be a consequence of microtubules that remained stable after colcemid was added. Interestingly, a remnant population of subcortical apical microtubules was observed in some of these cells until shortly before drug inactivation was performed (Fig. 2 C, t2, red arrow). The effect of Taxol on the orientation of polarity in larval NBs is also consistent with this interpretation. Taxol treatment resulted in chemically stabilized microtubules that could not depolymerize, thus providing a means to perturb the cytoskeleton that is totally different from the effect of microtubule-depolymerizing drugs. In larval NBs, Taxol brought about distinctively large asters dislodged from the apical cortex, where, nonetheless, some microtubules remained stabilized. Taxol treatment had no effect in the orientation of cortical polarity (Fig. 2, D and F), showing that as long as a stable population of microtubules remains linked to the apical cortex, polarity orientation can be maintained (Video 5). Whatever its nature, the transient polarity memory observed is damaged if microtubule depolymerization conditions persist at the time of entry into mitosis (Fig. 1 H).
Transient microtubule depolymerization permanently resets the orientation of asymmetric cell division in larval NBs
We have shown that transient microtubule depolymerization can result in NBs that have undergone self-renewing asymmetric division in two different orientations: one is the natural orientation that took place repeatedly before microtubules were depolymerized; the second is the orientation in which the NB divided once microtubule dynamics were restored. Therefore, we wondered what the orientation would be if these cells were allowed to proceed for one more cycle under normal microtubule dynamics. The next orientation could (a) be randomized once more, strongly arguing that the signals that fix polarity orientation were irreversibly damaged by microtubule depolymerization, (b) go back to where it always was before microtubules were depolymerized, a result which would strongly suggest that signals that are upstream of microtubules and remain stable after transient microtubule depolymerization specify polarity orientation, or (c) coincide with the last, ectopic orientation, thus showing that at each cell cycle, the orientation of NB self-renewing asymmetric division is set to match the orientation in the previous cycle, whatever its angle and regardless of the position of the NB with respect to the surrounding cells.
One example of the results of these experiments is shown in Fig. 3 A. As shown before (Broadus and Doe, 1997), microtubule depolymerization resulted in an ectopic apical crescent (Fig. 3 A, t1, green asterisk). Upon recovery of microtubule dynamics, the cell divided along the newly established axis (Fig. 3 A, t2, green line), delivering an ectopic GMC (Fig. 3 A, t3, green circle 2). Importantly, the sites of apical crescent assembly and GMC delivery in the next cell cycle coincided with the previous (Fig. 3 A, t4, red asterisk and red circle 3). In a second example, the two GMCs delivered after transient microtubule depolymerization (Fig. 3 B, t4, green circle 2 and red circle 3) could be seen on top of each other nearly 170° away from the GMC delivered before colcemid was added (Fig. 3 B, t0, yellow circle 1; and Video 6). A plot showing the extent to which the division axis of the second cell cycle after colcemid inactivation was oriented like the first is shown in Fig. 3 C. Despite the relatively small sample size, owed to the technical difficulties of this experiment, it is clear that although the site of delivery of the first (green) GMC was almost random with respect to the control (0°), the second (red) GMC was in all cases as close to the first as successive GMCs were to each other in untreated control brains (Fig. 2 E, yellow sector). These results demonstrate that the microtubule-dependent memory effect that maintains polarity orientation in larval NBs reads only the orientation of the last cell cycle.
Figure 3.
Ectopic division orientations induced by transient microtubule depolymerization are permanently kept once microtubule dynamics are restored. (A and B) Still images from time-lapse recordings of larval NBs under normal microtubule dynamics (t0), after microtubule depolymerization by colcemid (t1), and after colcemid inactivation (t2–t4). Fluorescent reporter constructs are indicated to the left of each row. Arrows mark apical centrosomes; arrowheads refer to motile interphase centrosomes and to the basal centrosome in cells in metaphase. Colored circles highlight GMCs, and colored lines highlight the division axis. Dashed circles indicate the position where GMCs will be delivered. Each are colored according to birth order (yellow, first; green, second; and red, third). (A) An NB dividing three times (colcemid added, 13:55:46; effect detectable, 14:11:46; UV pulse delivered, 15:05:17; and total time exposed, 70 min). The orientation of the apical crescent and the spindle reflect the polarity axis (t0, yellow line). Under microtubule depolymerization conditions, the apical crescent forms ectopically (t1, green asterisk). After the drug is inactivated, the newly assembled spindle reorients to align with the ectopic crescent (t2, green asterisk/line). The same ectopic orientation is kept in the next cell cycle so that the third GMC is delivered on top of the second, away from the first (t4, red asterisk/line). In this NB, the original apical centrosome ended up in the GMC, and the other remained in the NB (t1 and t2). (B) An NB dividing three times (colcemid added, 15:34:41; UV pulse delivered, 16:45:23; and total time exposed to colcemid, 71 min; Video 6). (t0) The dashed line indicates the control polarity axis, as judged by the place of the most recent GMC. The cell rounds up upon colcemid treatment and upon release from colcemid arrest, divides asymmetrically with an ectopic orientation (t1 and t2, green line). This orientation is kept in the next mitosis, which delivers the third GMC on top of the second, diametrically opposed to the first GMC (t3 and t4). (C) Quantification of GMC budding sites plotting the variation (yellow sector) between the first and second cell division orientation (green and red, respectively) with respect to the control division (dashed line, 0°). Time is shown in hours:minutes:seconds. Bar, 10 µm.
In some of the cells in which we were able to unequivocally trace both centrosomes during the course of the transient microtubule depolymerization experiments, we found that the centrosome that initially organized the apical aster and was fated to remain in the NB (Fig. 3 A, t0, arrow) could end up at the spindle pole facing the ectopic basal side (Fig. 3 A, t2, arrow) and therefore be inherited by the ectopic GMC. Consistently, in this cell, the centrosome originally destined for the GMC ended up within the NB (Fig. 3 A, t2, arrowhead). Therefore, the structural and functional differences between the two NB centrosomes do not necessarily dictate their fates, which can be switched by transient microtubule depolymerization. However, we do not know whether such fate switch has any long-term developmental consequences.
Larval NBs without a stable interphase aster do not accurately memorize the orientation of cortical polarity and cell division
To further test the possible contribution of the interphase aster to defining the axis of NB self-renewing asymmetric division, we first determined the effect of mutants in dsas-4, which result in the absence of centrioles and centrosomes (Basto et al., 2006). In agreement with a previous study (Basto et al., 2006), about one fourth of the dsas-4 mutant NBs that we monitored failed asymmetric cell division. We focused our attention on the remaining three quarters. Fig. 4 A shows a wild-type NB in an MARCM (mosaic analysis with a repressible cell marker) clone (Lee and Luo, 1999). Mitosis orientation in three consecutive cell cycles changed very little in this cell (Fig. 4 A, t0–t3, yellow, green, and red lines; and Video 7). In contrast, in the NB within the MARCM dsas-4 clone shown in Fig. 4 B, the orientation of cortical polarity and cell division changed by 90° from one mitosis to the next (Fig. 4 B, t0–t3, yellow and green asterisks [orientation of cortical polarity] and lines [orientation of cell division]; and Video 8). In the dsas-4 NBs shown in Fig. 4 (C and D), changes in orientation occurred consistently in one direction, resulting in a net error of ∼90° over the course of a few cell cycles (Fig. 4, C and D, yellow, red, green, and white lines). The observed scattering of the sites of apical crescent assembly (−65° to 93°; n = 16) and GMC delivery (−90° to 128°; n = 24) in two consecutive cycles in dsas-4 NBs is plotted in Fig. 4 (E and F).
Figure 4.
The loss of centrioles severely compromises the fidelity of polarity orientation in larval NBs. (A–D) Still images from time-lapse recordings of MARCM clones labeled by the expression of nuclear GFP (NLS-GFP [nlsGFP]). Genotypes and fluorescent reporters are indicated to the left of each row. Colored asterisks, lines, and circles mark the position of apical crescents, the division axes, and small daughter cells, respectively, in successive cell cycles (yellow, first; green, second; red, third; and white, fourth). Dashed circles indicate the position where GMCs will be delivered. (A) NB in a control wild-type (wt) clone dividing three times. Polarity orientation, as judged by apical centrosome position (arrows) and small daughter cell budding sites, changes very little from one cycle to the next (t0–t3). The resulting GMCs are clustered (t3; Video 7). (B–D) dsas-4 mutant NB clones. (B) The position of apical crescents and the place of daughter cell delivery, which appear to be aligned, vary by nearly 90° between two successive cell cycles (t0 and t2; Video 8). (C) Projections made to reflect small daughter cell budding sites in successive rounds of division, which are scattered around the NB cortex in four successive divisions (t0–t3). (D) Apical crescent positions scatter similarly over three consecutive cell cycles (t0–t3). (E) Plot of apical crescent position variations between two consecutive cell cycles of control dsas-4/+ NBs (yellow) and homozygous dsas-4 mutant NBs (green) from mosaic brains. (F) Plot of daughter cell budding site variations between two consecutive cell cycles of control dsas-4/+ NBs (yellow) and homozygous dsas-4 mutant NBs (green). Time is shown in hours:minutes:seconds. Bar,10 µm.
Similar results were obtained with asl3/Df(3R)1577, asl, a severe loss of function condition in which astral microtubules are virtually absent (Bonaccorsi et al., 1998; Varmark et al., 2007). Unlike control cells, which displayed a prominent interphase apical aster and divided consistently in the same orientation in consecutive divisions (Fig. 5 A, t0–t4), hemizygous asl3 NBs showed no sign of centrosome-organized asters and divided at changing angles at each cell cycle (Fig. 5 B, t0–t4; and Video 9). A plot showing the actual changes observed in the direction of division in two consecutive cell cycles in a sample of 15 cells is shown in Fig. 5 C. The observed scattering of daughter cell budding sites in hemizygous asl3 NBs is comparable with that observed in dsas-4 NBs. A previous study based on five asl2 homozygous NBs reported more subtle shifts (Rusan and Peifer, 2007). Differences in sample size and in the leaky MTOC activity of asl alleles (Raff, 2001; Basto et al., 2006; Varmark et al., 2007) might account for this disagreement.
Figure 5.
Mutation in asl affects the fidelity of polarity orientation in larval NBs. (A and B) Still images from time-lapse recordings of larval NBs expressing tubulin-GFP. Division axes and daughter cells are highlighted by color-coded lines and circles, respectively, for successive divisions (yellow, first; and green, second). The orientation of each division was judged by the position of the midbody (arrowheads). (A) In control NBs, the orientation of successive divisions varies little, and the interphase aster (t2, asterisk) coincides with the location of the apical spindle pole of the previous division (t0 and t1) and allows predicting the position of the apical spindle pole in the next division (t4). (B) asl mutant NB dividing twice (Video 9). Interphase asters are absent (t2), and variations in the orientation of the division axis were observed: in contrast to controls, daughter cells resulting from two consecutive divisions were born, in this case, separated by several daughter cell diameters (t1 and t4). (C) Plot of daughter cell bud site variations in asl mutant NBs (green) compared with control NBs (yellow). Time is shown in hours:minutes:seconds. Bar,10 µm.
A previous study has shown that in larval NBs, loss of Pins function results in unstable apical asters that are lost as interphase progresses (Rebollo et al., 2007). Therefore, we decided to determine the effect that pins mutants might have in the memory of self-renewing asymmetric division orientation in Drosophila larval NBs. The case of one of the pinsP62/pinsP89 NBs that we recorded is shown in Fig. 6 A. In this cell, the site of GMC delivery rotated ∼100° between the first (Fig. 6 A, t0, yellow circle 1) and second (Fig. 6 A, t1, green circle 2) mitoses and a further 80° between the second and third (Fig. 6 A, t2, red circle 3; and Video 10). Thus, altogether, the division axis of this cell rotated ∼180° over the course of three consecutive divisions. A plot showing the orientation offset observed between two consecutive divisions (n = 6) is shown in Fig. 6 E. Our attempts to plot the orientation of apical crescents in pinsP62/pinsP89 NBs expressing Baz-GFP failed because the Baz-GFP signal was always (n = 20) extraordinarily faint and short lasted and could not be reliably detected in successive divisions of the same NB. This difficulty was to be expected because Pins is a core component of the apical complex and is required for apical crescent stability (Parmentier et al., 2000; Schaefer et al., 2000; Yu et al., 2000). In the few asymmetric mitoses in which a Baz-GFP crescent was detected (unpublished data), the crescent was always aligned with the spindle axis revealed by the position of the two centrosomes (Asl-YFP; n = 5).
Figure 6.
Probing the mechanism that controls the fidelity of polarity orientation in larval NBs. (A–D) Still images from time-lapse recordings of larval NBs. Fluorescent reporters are indicated to the left of each row. Division axes, daughter cells, and apical crescents are highlighted by color-coded lines, circles, and asterisks, respectively, for successive divisions (yellow, first; green, second; and red, third). (A) An NB mutant for pins. This cell, which lacks interphase asters, divides three times, producing daughters that are scattered over an arc of ∼180° (t0 and t2; Video 10). (B) An NB mutant for polo. Two inactive centrosomes (arrowheads) are visible in interphase (t1). (C) An NB expressing Khc-73 RNAi. The arrow in t1 points to the stable microtubule-nucleating apical centrosome. (D) An NB expressing Par-1–GFP. The arrow in t1 points to the apical aster during interphase. (E–H) plots of apical crescents or daughter cell budding site variations in NBs mutant for pins (E) or polo (F) or expressing Khc-73 (G) or Par-1–GFP (H). Green indicates mutant NBs and yellow indicates control (worniu-Gal4 UASCherryTubulin) NBs. Time is shown in hours:minutes:seconds. Bars,10 µm.
Finally, because Polo has been shown to specifically localize on the centrosome that remains apical and organizes the interphase aster (Rusan and Peifer, 2007), we decided to test the effect of loss of polo function on polarity orientation memory. Polo regulates a wide range of functions, including cell cycle progression, centrosome maturation, cytokinesis, and NB cortical polarity. Consequently, polo loss of function conditions are highly pleiotropic (Carmena et al., 1998; Glover, 2005; Wang et al., 2007; Chia et al., 2008). Therefore, we focused our attention on polo1 cells that divided asymmetrically, organized well-defined Dlg-GFP crescents, and were not obviously polyploid. First, we found that in polo1 NBs, the aster was lost soon after mitosis (not depicted) and none of the two centrosomes remained cortex bound in interphase (Fig. 6 B, t1, arrowheads). Consistently, we observed a significant apical crescent orientation offset in successive mitoses (Fig. 6 B, t0 [yellow asterisk] and t2 [green asterisk]), ranging from −38° to 68°, in these cells (n = 14).
Altogether, these observations are consistent with the centrosome-organized interphase aster having a role in keeping the memory of self-renewing asymmetric division orientation in Drosophila larval NBs. However, intriguingly, the loss of polarity orientation memory caused by colcemid treatment is greater than that caused by any of the mutants that we have studied (Fig. 1, H and I; Fig. 2 E; Fig. 4, E and F; and Fig. 5 C). These results, particularly those regarding dsas-4 NBs in which the total absence of centrioles rules out the possibility of a leaky MTOC activity, show that noncentrosomal microtubule-dependent functions also contribute to polarity orientation memory in these cells.
Interestingly, 5 out of the 15 cells with impaired centrosome function that we were able to record for more than two cycles showed consistent clockwise or anticlockwise displacement of polarity orientation. Most likely, these cases simply reflect a normal distribution of random events rather than a real tendency to rotate in one direction. However, random as they might be, these cases make the point that observations limited to two consecutive cycles, like those shown in Figs. 4 (E and F), 5 C, and 6 E, do not reveal the full extent to which orientation can be affected over a few cell cycles and, indeed, over the lifespan of a cell that typically undergoes dozens of mitoses.
Our initial attempts to identify the genes that mediate the memory of polarity orientation have been, so far, negative. We have focused on Khc-73 and Par-1. Khc-73 coimmunoprecipitates with Dlg and is required for microtubule-dependent Pins/Gαi/Dlg crescent formation during metaphase (Siegrist and Doe, 2005). Khc-73 RNAi impairs alignment between the metaphase spindle and the cortical polarity axis in 30-35% of embryonic NBs (Siegrist and Doe, 2005). Par-1, like atypical PKC, phosphorylates Baz, which is dephosphorylated by PP2A (protein phosphatase 2A; Krahn et al., 2009). In embryonic NBs, loss of PP2A function or Par-1 overexpression can result in a complete reversal of cortical polarity (Krahn et al., 2009). Therefore, we decided to check the effect of Khc-73 RNAi (Fig. 6 C) and Par-1 overexpression (Fig. 6 D) in the memory of polarity orientation in larval NBs. We found that in both cases, GMC budding sites in successive cell cycles were as tightly clustered as in wild-type NBs (Fig. 6, G and H). However, negative as they are, these results must be taken with great caution.
Discussion
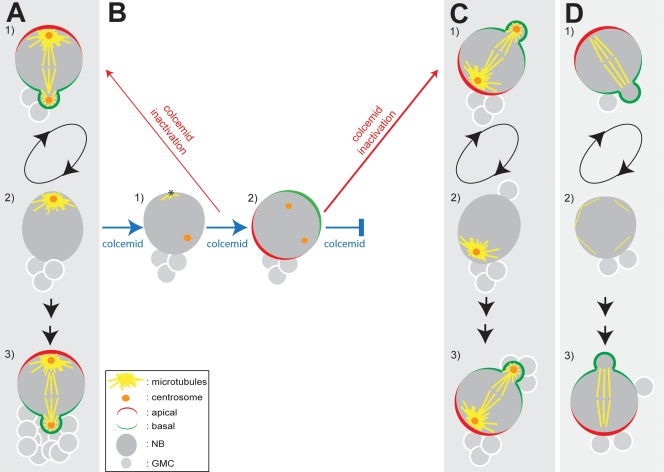
A distinct feature of self-renewing asymmetric division in Drosophila NBs is that its orientation remains roughly unchanged through successive rounds of cell division, and consecutive GMC siblings are tightly clustered (Fig. 7 A). The bases for such orientation control are unknown.
Figure 7.
The role of the microtubule network in the orientation of asymmetric cell division in Drosophila NBs. (A) Under normal conditions, the apical position of the main MTOC is maintained through the cell cycle and passed on from mother to daughter NB (1 and 2), polarity orientation is retained in consecutive mitoses, and the small differentiating cells (GMCs) are delivered in a cluster (3). (B and C) Under microtubule depolymerization conditions, the interphase aster is disassembled, and the cortical attachment of the apical centrosome is lost. If microtubule dynamics are restored during interphase, the memory of cortical polarity orientation is often unaffected (B, 1, asterisk). However, if microtubule-depolymerizing conditions are kept until the cell enters mitosis, the orientation of cortical polarity is randomized (B, 2), and once microtubule dynamics are restored, asymmetric cell division takes place along the new, ectopic axis of cortical polarity (C, 1). This situation results in the ectopic delivery of a GMC, the relocation of the interphase aster of the NB (C, 2), and the resetting of the axes of cortical polarity and asymmetric cell division orientation to the new ectopic orientation (C, 3). (D) In different mutant conditions that affect the assembly or the stability of the interphase asters, the memory of polarity orientation is partially lost (1 and 2) and daughter cells are not clustered (3).
Polarity in Drosophila NBs is most conspicuous at anaphase–telophase, when specific markers tag the apical and basal sides of the cortex, the spindle is oriented apicobasally, and the outlines of the two unequally sized daughters start to be apparent (Fig. 7 A, 1). All of these cues are lost when cytokinesis cleaves the GMC away, the spindle disassembles, and none of the known cortical polarity markers remain asymmetrically localized at the cell cortex. However, polarity information could be passed on to the next cycle by the position of the apical centrosome that stays in the apical side of the renewed NB, near the nucleus, facing and close to the side of the cortex where the apical crescent was last localized (Fig. 7 A, 2). Indeed, the cortical localization of the large aster organized by this centrosome during interphase accurately predicts the site of apical crescent assembly and with it the axis of cortical polarity and cell division of the next mitosis (Fig. 7 A, 3). Our experiments aimed at establishing whether the potential polarity cue provided by the position of the apical centrosome actually contributes to define the orientation of self-renewing asymmetric division in larval NBs.
We found that when NBs are kept throughout interphase under microtubule-depolymerizing conditions that disassemble the apical aster, cortical polarity is established ectopically (Fig. 7 B, 2). This result shows that microtubules are required for the memory of cortical polarity orientation. The ectopic site of apical crescent assembly in these cells bears no relation with the position of the asterless centrosomes. Moreover, if normal microtubule dynamics resume, the aster organized by one of the centrosomes moves toward the apical crescent (Fig. 7 C, 1), presumably by the Pins–Dlg–Mud–Khc-73C pathways (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). Thus, in mitosis, the ectopic apical crescent governs the position of the centrosome/aster. However, as these NBs divide and enter the next cell cycle, the new position of the centrosome/aster during interphase (Fig. 7 C, 2) labels the site where the apical crescent will form at mitosis onset (Fig. 7 C, 3). This observation suggests that aster to cortex signaling during interphase contributes to define the site of apical crescent assembly and with it the orientation of NB polarity in the next mitosis.
The possible role of the interphase aster in defining NB polarity orientation is further substantiated by the phenotype of mutants without centrosomes (dsas-4), with centrosomes that have none or very little MTOC activity (asl), or with unstable interphase asters (pins and polo). In these mutants, cell division orientation memory is impaired (Fig. 7 D, 1–3) and successive GMC siblings are not clustered as in wild-type brains (Fig. 7, A [3] and D [3]). However, importantly, the extent of loss of polarity orientation memory caused by colcemid is greater than that caused by centrosome loss, strongly suggesting that noncentrosomal microtubule-dependent functions also contribute to such memory. The critical phase of microtubule to cortex signaling seems to take place late in interphase because orientation memory is maintained in a majority of cells in which transient microtubule depolymerization stops before mitosis onset (Fig. 7 B, 1). Signaling could come from the convergence of the microtubule minus ends on such major MTOC, the interaction with the cortex of the plus ends of astral microtubules, the centrosome itself, or, indeed, any combination of these. Probably because of the proximity of the large aster, NBs often have a slightly elongated appearance and a pointed apical side (Rebollo et al., 2007, 2009), which is likely to reflect tension discontinuities that could also mediate signaling. Precedence for centrosome to cortex signaling has been reported in C. elegans (Cowan and Hyman, 2004; Tsai and Ahringer, 2007). The molecular nature of such signaling remains unknown.
After transient microtubule depolymerization, NBs divide asymmetrically, delivering a GMC at the ectopic basal side (Fig. 7 C, 1), showing that the ectopic cortical polarity axis assembled under microtubule-depolymerizing conditions is able once microtubule dynamics are restored, to drive what resembles normal self-renewing NB mitosis, including daughter cell size asymmetry. Cell size differences between the NBs and their GMC daughters are thought to be controlled by a distance-dependent effect of the apical crescent of Gαi that results in spindle asymmetry and the shifting of the cytokinesis furrow toward the GMC (Cai et al., 2003; Fuse et al., 2003; Yu et al., 2003; Izumi et al., 2004). Consistent with this model, it is well documented that spindle misorientation with respect to the axis of cortical polarity affects daughter cell size asymmetry to the extent that a 90° shift, which positions the spindle poles equidistant to the apical cortex, results in equally sized daughters (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006; Rebollo et al., 2007; Rusan and Peifer, 2007). This model predicts that cell size asymmetry should not be compromised if both the spindle and the cortical polarity axis rotated coordinately and remained aligned. Our results confirm this prediction.
Our results also reveal that, somehow counterintuitively, the orientation of the axis of cortical polarity is reset at each cell cycle to match the orientation in the last. They also show that cortical polarity can be oriented at different angles regardless of the position of the NB with respect to the surrounding cells, strongly suggesting that cortical polarity orientation is controlled in a cell-autonomous manner.
Materials and methods
Fly strains
The following strains were used: Asterless-YFP, asl3 and Df(3R)1577, asl (Varmark et al., 2007); tubulin-GFP (Rebollo and González, 2004); GFP-Dlg and GFP-Baz fusions were obtained in protein-trap screens, Flytrap (Kelso et al., 2004), and are therefore under the control of the corresponding natural promoter and thus likely to be expressed at levels that are similar to those of the normal protein (Morin et al., 2001); pinsP62 and pinsP89 (Yu et al., 2000); YFP-Pins (provided by Y. Bellaiche, Institute Curie, Paris, France) is under the control of the polyubiquitin promoter and recombined to the pinsP62 allele (David et al., 2005); GFP-Pon (a gift from J. Knoblich, Institute of Molecular Biotechnology GmbH, Vienna, Austria) artificially enters the nucleus during interphase, allowing to reveal the basal NB cortex at mitosis; worniu-Gal4 (provided by C. Doe, University of Oregon, Eugene, OR; Albertson et al., 2004); UAS–Khc-73 RNAi (Siegrist and Doe, 2005); Eb1-GFP (Rebollo et al., 2007); PLC-γ pleckstrin homology domain–GFP (provided by F. Pichaud, University College London, London, England, UK; Pinal et al., 2006); Mira-GFP (Mollinari, 1997); FRT 82B dsas-4S2214 (provided by J. Raff, University of Oxford, Oxford, England, UK; Stevens et al., 2007); UAS–Par-1 N1S GFP (provided by D. St. Johnston, University of Cambridge, Cambridge, England, UK; Doerflinger et al., 2006); UASCherryTubulin (provided by M. Peifer, University of North Carolina at Chapel Hill, Chapel Hill, NC; Rusan and Peifer, 2007); polo1 (Tavares et al., 1996); and FRT 82B ry (Bloomington Stock Center). Mosaic larval brain analysis was performed using the MARCM technique (Lee and Luo, 1999). Homozygous mutant clones were marked by the expression of NLS-GFP (Shiga et al., 1996). Flies were kept under standard conditions at 25°C.
Time-lapse recording
Brain explants from L3 larvae, 60–80 h after larval hatching, were prepared for time-lapse recordings as described previously (Siller et al., 2005) using the clot method (Forer and Pickett-Heaps, 1998; Rebollo et al., 2007). Tissue culture dishes with coverglass bottoms (FluoroDish; World Precision Instruments) were used for mounting the brains in the clot with the ventral side facing the cover glass and covered with Schneider’s Drosophila medium with l-glutamine (Biological Industries) and supplemented with fat body from wild-type L3 larvae. NBs of the central brain were sampled. Images were acquired at 22°C on a spinning disc confocal system (Andor Technology) using an inverted microscope (IX71; Olympus) equipped with an electron-multiplying charge-coupled device camera (iXON DU-897E-#BV-500; Andor Technology) using a 60× NA 1.42 oil Plan-Apochromat objective. 30–40 z sections were taken at 0.5–0.7-µm intervals. z stacks were recorded every 30–60 s. Control NBs expressing tubulin-GFP and Asl-YFP were able to undergo up to a maximum of six consecutive cell cycles under these conditions. z stacks were projected, dynamic range was adjusted, and avi files were generated using IQ (Andor Technology). Applying a Gaussian blur (radius: 1) was used to reduced image noise (ImageJ 1.42q; National Institutes of Health). Videos were annotated and compressed using After Effects 7.0 (Adobe). Thanks to its optimal signal to noise ratio, the Asl-YFP reporter unequivocally identifies centrioles. In all figures, we have labeled the centrioles within the NB of interest to tell them apart from others that are in sections above or below but appear to be within that NB in z projections.
Drug treatment
Colcemid (EMD) was added to the medium to a final concentration of 50 µM. Colcemid effects were readily visible ∼15 min after addition of the drug. Colcemid was inactivated by changing to fresh, colcemid-free medium, followed by one to three 5-s pulses of the microscope’s UV light. This treatment had no effect on NB polarity in control brains. Taxol (Paclitaxel; Sigma-Aldrich) was added to the medium to a final concentration of 20 µM. Quantifications of polarity orientation were made on z projections. Therefore, observed changes were limited to the X–Y plane. Polarity axes were defined as the straight line that intersects the center of the crescent (for apical crescent position) or the center of the daughter cell (for daughter cell budding site) with the center of the NBs.
Online supplemental material
Video 1 shows that apical centrosome attachment during interphase requires microtubules. Video 2 demonstrates that the location of cortical crescent assembly is not memorized from one cell cycle to the next when microtubules are depolymerized. Video 3 shows that transient microtubule depolymerization can change the orientation of NB division. Video 4 reveals that transient microtubule depolymerization during interphase does not always change NB division orientation. Video 5 shows that the location of cortical crescent assembly is memorized from one cell cycle to the next in the presence of Taxol. Video 6 shows that an ectopic division axis induced by transient microtubule depolymerization is kept in subsequent cycles. Video 7 shows that the offspring of wild-type NBs are clustered at the basal pole. Videos 8 and 9 show polarity orientation defects in dsas-4 mutant NBs and in asl hemizygous NBs, respectively. Video 10 shows that division orientation errors can be accumulative over successive divisions in pins mutant NBs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905024/DC1.
Acknowledgments
We thank Y. Bellaiche, C. Doe, D. St. Johnston, J. Knoblich, M. Peifer, F. Pichaud, J. Raff, the Carnegie Flytrap, and the Bloomington Stock Center for fly strains, S. Llamazares, E. Rebollo, and P. Sampaio for help and advise, and M. Llamazares for proofreading.
Work in our laboratory is supported by European Union and Spanish grants: ONCASYM-037398 FP6, BFU2006-05813, SGR2005 Generalitat de Catalunya, and Consolider-Ingenio 2010 CENTROSOME_3D CSD2006-23. J. Januschke was partially funded by an Association pour la recherche sur le Cancer fellowship.
Footnotes
Abbreviations used in this paper:
- Baz
- Bazooka
- Dlg
- Discs large
- GMC
- ganglion mother cell
- Insc
- Inscuteable
- MARCM
- mosaic analysis with a repressible cell marker
- Mira
- Miranda
- MTOC
- microtubule-organizing center
- NB
- neuroblast
- NEB
- nuclear envelope breakdown
- Par
- partitioning defective
- Pins
- partner of Insc
- Pon
- partner of Numb
References
- Albertson R., Chabu C., Sheehan A., Doe C.Q. 2004. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J. Cell Sci. 117:6061–6070 10.1242/jcs.01525 [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. 2006. Flies without centrioles. Cell. 125:1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bello B., Reichert H., Hirth F. 2006. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 133:2639–2648 10.1242/dev.02429 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 422:326–330 10.1038/nature01486 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. 2006. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 124:1241–1253 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M.G., Gatti M. 1998. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J. Cell Biol. 142:751–761 10.1083/jcb.142.3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.K., Neumüller R.A., Novatchkova M., Du Q., Knoblich J.A. 2006. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell. 10:731–742 10.1016/j.devcel.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Broadus J., Doe C.Q. 1997. Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr. Biol. 7:827–835 10.1016/S0960-9822(06)00370-8 [DOI] [PubMed] [Google Scholar]
- Cai Y., Yu F., Lin S., Chia W., Yang X. 2003. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell. 112:51–62 10.1016/S0092-8674(02)01170-4 [DOI] [PubMed] [Google Scholar]
- Carmena M., Riparbelli M.G., Minestrini G., Tavares A.M., Adams R., Callaini G., Glover D.M. 1998. Drosophila polo kinase is required for cytokinesis. J. Cell Biol. 143:659–671 10.1083/jcb.143.3.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E., Gonzalez C. 2005. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37:1125–1129 10.1038/ng1632 [DOI] [PubMed] [Google Scholar]
- Caussinus E., Hirth F. 2007. Asymmetric stem cell division in development and cancer. Prog. Mol. Subcell. Biol. 45:205–225 10.1007/978-3-540-69161-7_9 [DOI] [PubMed] [Google Scholar]
- Chia W., Somers W.G., Wang H. 2008. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J. Cell Biol. 180:267–272 10.1083/jcb.200708159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C.R., Hyman A.A. 2004. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 431:92–96 10.1038/nature02825 [DOI] [PubMed] [Google Scholar]
- David N.B., Martin C.A., Segalen M., Rosenfeld F., Schweisguth F., Bellaïche Y. 2005. Drosophila Ric-8 regulates Galphai cortical localization to promote Galphai-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat. Cell Biol. 7:1083–1090 10.1038/ncb1319 [DOI] [PubMed] [Google Scholar]
- Doe C.Q. 2008. Neural stem cells: balancing self-renewal with differentiation. Development. 135:1575–1587 10.1242/dev.014977 [DOI] [PubMed] [Google Scholar]
- Doe C.Q., Chu-LaGraff Q., Wright D.M., Scott M.P. 1991. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 65:451–464 10.1016/0092-8674(91)90463-9 [DOI] [PubMed] [Google Scholar]
- Doerflinger H., Benton R., Torres I.L., Zwart M.F., St Johnston D. 2006. Drosophila anterior-posterior polarity requires actin-dependent PAR-1 recruitment to the oocyte posterior. Curr. Biol. 16:1090–1095 10.1016/j.cub.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Egger B., Chell J.M., Brand A.H. 2008. Insights into neural stem cell biology from flies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:39–56 10.1098/rstb.2006.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forer A., Pickett-Heaps J.D. 1998. Cytochalasin D and latrunculin affect chromosome behaviour during meiosis in crane-fly spermatocytes. Chromosome Res. 6:533–549 10.1023/A:1009224322399 [DOI] [PubMed] [Google Scholar]
- Fuse N., Hisata K., Katzen A.L., Matsuzaki F. 2003. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr. Biol. 13:947–954 10.1016/S0960-9822(03)00334-8 [DOI] [PubMed] [Google Scholar]
- Gateff E. 1978. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 200:1448–1459 10.1126/science.96525 [DOI] [PubMed] [Google Scholar]
- Glover D.M. 2005. Polo kinase and progression through M phase in Drosophila: a perspective from the spindle poles. Oncogene. 24:230–237 10.1038/sj.onc.1208279 [DOI] [PubMed] [Google Scholar]
- Gonzalez C. 2007. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8:462–472 10.1038/nrg2103 [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J.B., Nabeshima Y., Doe C.Q., Matsuzaki F. 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 390:625–629 10.1038/37641 [DOI] [PubMed] [Google Scholar]
- Ito K., Hotta Y. 1992. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 149:134–148 10.1016/0012-1606(92)90270-Q [DOI] [PubMed] [Google Scholar]
- Izumi Y., Ohta N., Itoh-Furuya A., Fuse N., Matsuzaki F. 2004. Differential functions of G protein and Baz–aPKC signaling pathways in Drosophila neuroblast asymmetric division. J. Cell Biol. 164:729–738 10.1083/jcb.200309162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y., Ohta N., Hisata K., Raabe T., Matsuzaki F. 2006. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8:586–593 10.1038/ncb1409 [DOI] [PubMed] [Google Scholar]
- Januschke J., Gonzalez C. 2008. Drosophila asymmetric division, polarity and cancer. Oncogene. 27:6994–7002 10.1038/onc.2008.349 [DOI] [PubMed] [Google Scholar]
- Kaltschmidt J.A., Davidson C.M., Brown N.H., Brand A.H. 2000. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat. Cell Biol. 2:7–12 10.1038/71323 [DOI] [PubMed] [Google Scholar]
- Karess R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392 10.1016/j.tcb.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Kelso R.J., Buszczak M., Quiñones A.T., Castiblanco C., Mazzalupo S., Cooley L. 2004. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32:D418–D420 10.1093/nar/gkh014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Knoblich J.A., Jan L.Y., Jan Y.N. 1995. Asymmetric segregation of Numb and Prospero during cell division. Nature. 377:624–627 10.1038/377624a0 [DOI] [PubMed] [Google Scholar]
- Krahn M.P., Egger-Adam D., Wodarz A. 2009. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev. Cell. 16:901–908 10.1016/j.devcel.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Kraut R., Chia W., Jan L.Y., Jan Y.N., Knoblich J.A. 1996. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 383:50–55 10.1038/383050a0 [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Robinson K.J., Doe C.Q. 2006a. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 439:594–598 10.1038/nature04299 [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Wilkinson B.D., Siegrist S.E., Wharton R.P., Doe C.Q. 2006b. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 10:441–449 10.1016/j.devcel.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22:451–461 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Li P., Yang X., Wasser M., Cai Y., Chia W. 1997. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 90:437–447 10.1016/S0092-8674(00)80504-8 [DOI] [PubMed] [Google Scholar]
- Lu B., Rothenberg M., Jan L.Y., Jan Y.N. 1998. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 95:225–235 10.1016/S0092-8674(00)81753-5 [DOI] [PubMed] [Google Scholar]
- Lu B., Jan L., Jan Y.N. 2000. Control of cell divisions in the nervous system: symmetry and asymmetry. Annu. Rev. Neurosci. 23:531–556 10.1146/annurev.neuro.23.1.531 [DOI] [PubMed] [Google Scholar]
- Mollinari C. 1997. Molecular characterization of a new component of the centrosome in Drosophila melanogaster. PhD thesis. Ruprecht Karls Universität, Heidelberg, Germany: 219 pp [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W. 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 98:15050–15055 10.1073/pnas.261408198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshiro T., Yagami T., Zhang C., Matsuzaki F. 2000. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 408:593–596 10.1038/35046087 [DOI] [PubMed] [Google Scholar]
- Parmentier M.L., Woods D., Greig S., Phan P.G., Radovic A., Bryant P., O’Kane C.J. 2000. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 20:RC84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C.Y., Manning L., Albertson R., Doe C.Q. 2000. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 408:596–600 10.1038/35046094 [DOI] [PubMed] [Google Scholar]
- Petronczki M., Knoblich J.A. 2001. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3:43–49 10.1038/35050550 [DOI] [PubMed] [Google Scholar]
- Pinal N., Goberdhan D.C., Collinson L., Fujita Y., Cox I.M., Wilson C., Pichaud F. 2006. Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr. Biol. 16:140–149 10.1016/j.cub.2005.11.068 [DOI] [PubMed] [Google Scholar]
- Raff J.W. 2001. Centrosomes: Central no more? Curr. Biol. 11:R159–R161 10.1016/S0960-9822(01)00082-3 [DOI] [PubMed] [Google Scholar]
- Rebollo E., González C. 2004. Time-lapse imaging of male meiosis by phase-contrast and fluorescence microscopy. Methods Mol. Biol. 247:77–87 [DOI] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., González C. 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 12:467–474 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Rebollo E., Roldán M., Gonzalez C. 2009. Spindle alignment is achieved without rotation after the first cell cycle in Drosophila embryonic neuroblasts. Development. 136:3393–3397 10.1242/dev.041822 [DOI] [PubMed] [Google Scholar]
- Rolls M.M., Albertson R., Shih H.P., Lee C.Y., Doe C.Q. 2003. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163:1089–1098 10.1083/jcb.200306079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N.M., Peifer M. 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177:13–20 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko A., Shevchenko A., Knoblich J.A. 2000. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10:353–362 10.1016/S0960-9822(00)00401-2 [DOI] [PubMed] [Google Scholar]
- Schober M., Schaefer M., Knoblich J.A. 1999. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 402:548–551 10.1038/990135 [DOI] [PubMed] [Google Scholar]
- Shen C.P., Jan L.Y., Jan Y.N. 1997. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 90:449–458 10.1016/S0092-8674(00)80505-X [DOI] [PubMed] [Google Scholar]
- Shiga Y., Tanaka-Matakatsu M., Hayashi S. 1996. A nuclear GFP/beta-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev. Growth Differ. 38:99–106 10.1046/j.1440-169X.1996.00012.x [DOI] [Google Scholar]
- Siegrist S.E., Doe C.Q. 2005. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 123:1323–1335 10.1016/j.cell.2005.09.043 [DOI] [PubMed] [Google Scholar]
- Siller K.H., Doe C.Q. 2009. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11:365–374 10.1038/ncb0409-365 [DOI] [PubMed] [Google Scholar]
- Siller K.H., Serr M., Steward R., Hays T.S., Doe C.Q. 2005. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell. 16:5127–5140 10.1091/mbc.E05-04-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Cabernard C., Doe C.Q. 2006. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8:594–600 10.1038/ncb1412 [DOI] [PubMed] [Google Scholar]
- Stevens N.R., Raposo A.A., Basto R., St Johnston D., Raff J.W. 2007. From stem cell to embryo without centrioles. Curr. Biol. 17:1498–1503 10.1016/j.cub.2007.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares A.A., Glover D.M., Sunkel C.E. 1996. The conserved mitotic kinase polo is regulated by phosphorylation and has preferred microtubule-associated substrates in Drosophila embryo extracts. EMBO J. 15:4873–4883 [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W.E., Hazelrigg T.I. 1998. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multi-step anterior localization pathway. Development. 125:3655–3666 [DOI] [PubMed] [Google Scholar]
- Tsai M.C., Ahringer J. 2007. Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J. Cell Biol. 179:397–402 10.1083/jcb.200708101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmark H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H., Gonzalez C. 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17:1735–1745 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Wang H., Ouyang Y., Somers W.G., Chia W., Lu B. 2007. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 449:96–100 10.1038/nature06056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., Knust E. 1999. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 402:544–547 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X., Chia W. 2000. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 100:399–409 10.1016/S0092-8674(00)80676-5 [DOI] [PubMed] [Google Scholar]
- Yu F., Cai Y., Kaushik R., Yang X., Chia W. 2003. Distinct roles of Gαi and Gβ13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 162:623–633 10.1083/jcb.200303174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Kuo C.T., Jan Y.N. 2006. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 51:13–20 10.1016/j.neuron.2006.06.016 [DOI] [PubMed] [Google Scholar]