Defective attenuation of BMP signaling causes synapses to overgrow in Drosophila Ema mutants due to impaired endosomal maturation.
Abstract
Endosomal maturation is critical for accurate and efficient cargo transport through endosomal compartments. Here we identify a mutation of the novel Drosophila gene, ema (endosomal maturation defective) in a screen for abnormal synaptic overgrowth and defective protein trafficking. Ema is an endosomal membrane protein required for trafficking of fluid-phase and receptor-mediated endocytic cargos. In the ema mutant, enlarged endosomal compartments accumulate as endosomal maturation fails, with early and late endosomes unable to progress into mature degradative late endosomes and lysosomes. Defective endosomal down-regulation of BMP signaling is responsible for the abnormal synaptic overgrowth. Ema binds to and genetically interacts with Vps16A, a component of the class C Vps–HOPS complex that promotes endosomal maturation. The human orthologue of ema, Clec16A, is a candidate susceptibility locus for autoimmune disorders, and its expression rescues the Drosophila mutant demonstrating conserved function. Characterizing this novel gene family identifies a new component of the endosomal pathway and provides insights into class C Vps–HOPS complex function.
Introduction
Endosomes mediate membrane trafficking as they transit among the trans-Golgi network, the plasma membrane, and lysosomes (Gruenberg and Stenmark, 2004; Maxfield and McGraw, 2004; Bonifacino and Rojas, 2006; Luzio et al., 2007). The endosomal system is essential for normal development and physiology, and impaired endosomal function contributes to many human diseases including cancers, neurological diseases, and immune disorders (Jeyakumar et al., 2005; Huizing et al., 2008; Mosesson et al., 2008). Elegant genetic studies in yeast have identified molecular constituents of the endosomal compartment, and subsequent biochemical analysis and in vitro reconstitution experiments have elucidated important aspects of molecular trafficking through this compartment.
Trafficking through the endosomal compartment involves a series of membrane fusion and maturation steps that are tightly controlled by a complex network of molecules. Early endosomes grow via homotypic fusion and mature to late endosomes by Rab conversion via the exchange of Rab5 for Rab7 (Rink et al., 2005). Known as a Rab effector as well as a tethering complex, the class C Vps–HOPS complex plays important roles in the membrane fusion and maturation (Nickerson et al., 2009). The complex consists of four core subunits, Vps11, Vps18 (also known as Dor), Vps16, and Vps33 (also known as Car), as well as two accessory subunits, Vps39 and Vps41 for the HOPS complex or Vps3 and Vps8 for the CORVET complex (Seals et al., 2000; Peplowska et al., 2007). Yeast class C Vps genes were originally identified based on their roles in protein sorting to the vacuole, the yeast equivalent of the lysosome, and have been subsequently characterized as a tethering complex for vacuolar fusion (Banta et al., 1988; Sato et al., 2000; Seals et al., 2000; Wurmser et al., 2000). Orthologues for the class C Vps–HOPS complex from multicellular organisms participate in many aspects of endosomal trafficking including homo-/heterotypic fusion of endosomes and lysosomes, maturation of endosomes via Rab conversion, biogenesis of lysosome-related organelles, and maturation of autophagosomes and phagosomes (Sevrioukov et al., 1999; Poupon et al., 2003; Sriram et al., 2003; Richardson et al., 2004; Pulipparacharuvil et al., 2005; Rink et al., 2005; Kinchen et al., 2008; Liang et al., 2008). However, the molecular mechanisms by which the class C Vps–HOPS complex participates in such diverse functions remain largely unexplored.
Endosomal trafficking of BMP receptors regulates developmental plasticity at the Drosophila neuromuscular junction (NMJ). In a forward genetic screen for abnormal synaptic development and defective protein trafficking, we identified the novel Drosophila gene ema. ema is a member of a highly conserved gene family in multicellular eukaryotes whose cellular and molecular function has not been characterized. The human ema orthologue Clec16A (KIAA0350) has been identified as a strong candidate susceptibility locus for a variety of autoimmune disorders including type 1 diabetes (T1D) and multiple sclerosis (Hakonarson et al., 2007; International Multiple Sclerosis Genetics Consortium, 2007; Todd et al., 2007; Wellcome Trust Case Control Consortium, 2007). In this study, we demonstrate that Ema is required for endosomal function and maturation, and that in its absence defective endosomal down-regulation of BMP signaling leads to abnormal synaptic terminal overgrowth. The mammalian orthologue of ema functionally substitutes for the Drosophila gene, demonstrating conservation of function. Finally, we show that Ema is an endosomal membrane protein that functions with the class C Vps–HOPS complex to promote endosomal maturation.
Results
Mutation of the highly conserved Drosophila gene ema leads to synaptic overgrowth
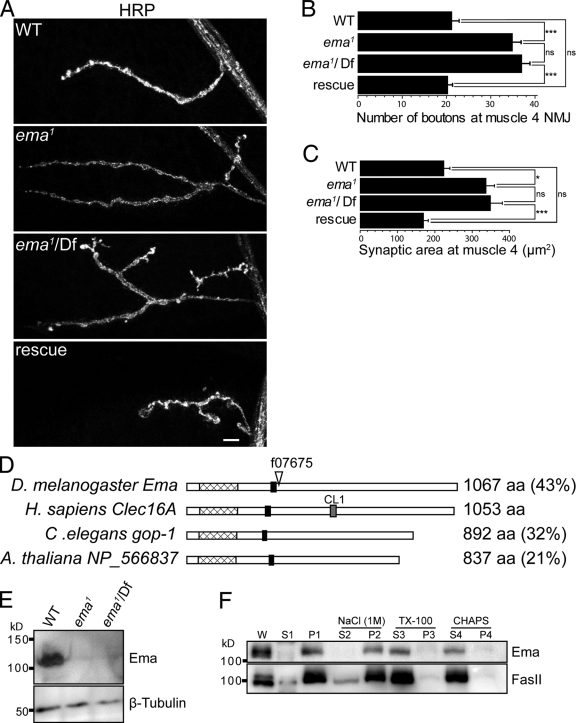
The Drosophila NMJ is a popular model system for identifying mechanisms controlling developmental synaptic plasticity (Collins and DiAntonio, 2007). To identify new genes regulating synaptic terminal growth, we performed a forward genetic screen for mutants with abnormal NMJ morphology. We screened a collection of ∼1,500 lines in which a transposable element is inserted in or near a gene on the third chromosome (Thibault et al., 2004). Lines that survived to third instar larvae were dissected and stained for the presynaptic active zone protein Bruchpilot (Wagh et al., 2006) and the essential postsynaptic glutamate receptor DGluRIII (Marrus et al., 2004). In the Drosophila forward genetic screen, a mutant for ema was identified based on synaptic terminal overgrowth at the NMJ (Fig. 1, A–C). The ema mutant f07675 (renamed ema1) is pupal lethal and has a transposable element inserted within a coding exon of the previously uncharacterized gene CG12753 (Fig. 1 D; Thibault et al., 2004). We name CG12753 ema (endosomal maturation defective) based on the mutant phenotype described below. Animals homozygous for ema1 have an approximately twofold increase in both the number of synaptic boutons and the synaptic area when compared with wild type (Fig. 1, B and C). In addition, trans-heterozygous mutants of ema1 in combination with a genetically unrelated deficiency for the ema locus, Df(3R)Exel7330 (Parks et al., 2004), show a very similar phenotype. This indicates that the phenotype maps to the insertion, and because the phenotype is quantitatively no worse over the deficiency, that the mutant behaves as a genetic null.
Figure 1.
Identification of the ema mutant. (A) Confocal images of larval NMJs from muscle 4 stained with an antibody to the neuronal membrane marker HRP reveal synaptic terminal overgrowth in the ema1 and ema1/Df mutants that is rescued by the neuronal expression of ema. WT (wild type) = Canton S; ema1 = CG12753 f07675; ema1/Df = CG12753 f07675/Df(3R)Exel7330; and rescue = ema1/Df; Elav-Gal4/UAS-EmaGFP. Bar, 10 µm. (B and C) Quantification is presented for the (B) number of synaptic boutons and (C) synaptic area (µm2) at muscle 4 for the genotypes shown in A; n > 10 for all genotypes. Data represent mean ± SEM. ANOVA analysis. *, P < 0.05; ***, P < 0.001; ns, not significant. (D) Schematic diagram of Ema orthologues highlighting the single putative transmembrane domain (black boxes), the highly conserved uncharacterized domain (pfam09758; hatched boxes), the C_type_lectin_1 domain signature (PS00615) in hClec16A (gray box), the percentage of sequence identity of each orthologue to human Clec16A, and the piggyBac insertion f07675 (∇) in the ema mutant. (E) Representative Western blots of third instar larval lysate from wild type, ema1, and ema1/Df probed for Ema and tubulin reveals the absence of Ema protein in the ema mutants. (F) Representative Western blots of Ema and the integral membrane protein FasII from fractionated wild-type cell lysates. Wild-type adult heads were homogenized (W) in a homogenization buffer (10 mM Hepes, pH 7.4, and 100 mM NaCl) and fractionated via centrifugation into soluble (S1) and insoluble (P1) membrane fractions. The P1 pellet was treated with either 1 M NaCl to extract peripherally associated proteins and then fractionated into soluble (S2) and insoluble (P2) fractions, or the nonionic detergent Triton X-100 (2%) and fractionated into soluble (S3) and insoluble (P3) fractions, or with the zwitterionic detergent CHAPS (4%) and fractionated into soluble (S4) and insoluble (P4) fractions. Both Ema and FasII are detergent extractable as predicted for integral membrane proteins.
Highly conserved ema orthologues are present in multicellular organisms including human (CLEC16A), mouse (KIAA0350), C. elegans (GOP-1), and Arabidopsis (NCBI Protein database accession no. NP_566837), but not in unicellular yeast, suggesting an important function for multicellularity. The molecular and cellular functions of this gene family have not been described; however, a number of genome-wide association studies have consistently identified the human orthologue Clec16A as a strong candidate susceptibility locus for autoimmune disorders (Hakonarson et al., 2007; International Multiple Sclerosis Genetics Consortium, 2007; Todd et al., 2007; Wellcome Trust Case Control Consortium, 2007). The ema orthologues are predicted to encode an ∼1,000 amino acid protein with a single predicted transmembrane domain near the N terminus (Fig. 1 D). Domain profile and signature analysis highlights a highly conserved uncharacterized domain (pfam09758) at the N-terminal region in all the ema orthologues (Marchler-Bauer et al., 2009). A putative sugar binding C-type lectin domain signature (PS00615) is predicted in mammalian Clec16A orthologues, but is not conserved in the orthologues from flies, worms, or plants (Hakonarson et al., 2007; Todd et al., 2007).
To test Ema expression, we generated a polyclonal antibody against a C-terminal fragment of Ema and identified an ∼110-kD band from lysates of wild-type third instar larvae on immunoblots that is absent in the ema mutants (Fig. 1 E). The absence of detectable protein is consistent with the genetic data suggesting that ema1 is a null allele. We performed cell fractionation and extraction to test the prediction that Ema is an integral membrane protein. We find that Ema is enriched in the membrane fraction and is not extracted by 1M NaCl, but, like the known membrane protein FasII, is solubilized from the membrane by the detergents Triton X-100 and CHAPS (Fig. 1 F). These findings are consistent with the prediction that Ema is an integral membrane protein.
To test whether the ema1 phenotypes are due to the loss of ema function, we performed genetic rescue experiments. We generated transgenic flies containing the ema gene under the control of the UAS promoter (UAS-ema). Expression of ema rescues the pupal lethality, the synaptic phenotype at the NMJ, and phenotypes described below (Fig. 1, A–C). Hence, loss of ema function is responsible for these phenotypes.
ema mutants have a dramatically enlarged endosomal compartment
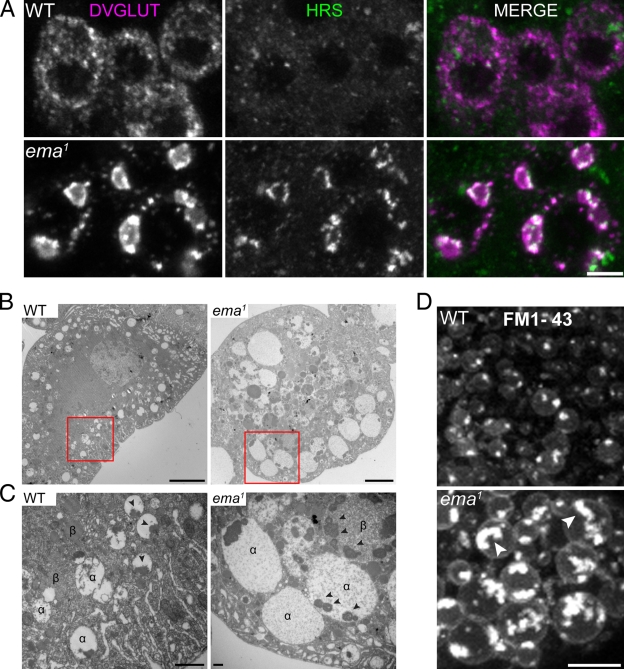
While examining the ema mutants, we observed large aggregates of the Drosophila vesicular glutamate transporter (DVGLUT; Daniels et al., 2004) in the motoneuron cell bodies (Fig. 2 A; Fig. S1, A and B). To identify the nature of the aggregates, we co-stained for DVGLUT and markers of cellular organelles. We observed that the endosomal sorting complex required for transport (ESCRT) protein Hrs (also known as Vps27; Lloyd et al., 2002) and the class C Vps–HOPS complex proteins Dor and Vps16A (Sevrioukov et al., 1999; Pulipparacharuvil et al., 2005) associate with the DVGLUT aggregates, whereas markers of ER, Golgi, and late endosome/lysosomes do not (Fig. 2 A; Fig. S2). These data suggest that the DVGLUT aggregates are present in an abnormally expanded endosomal compartment.
Figure 2.
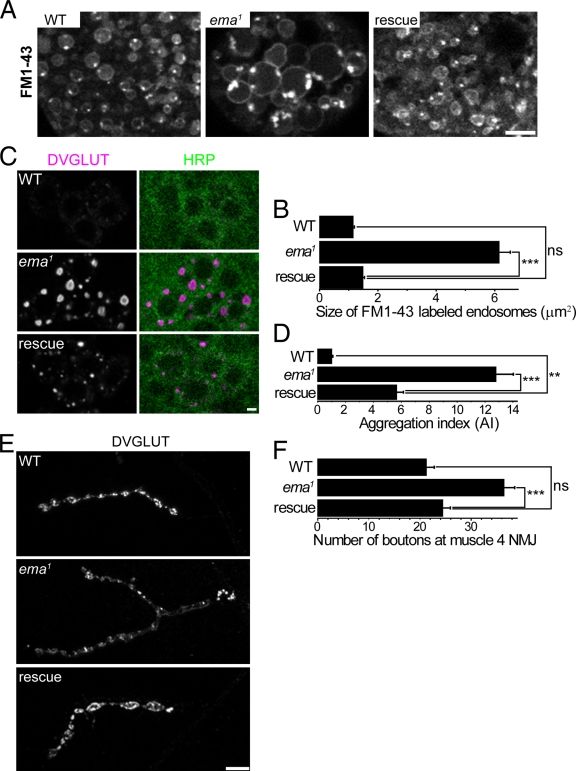
The endosomal compartment is enlarged in the ema mutant. (A) Colocalization between DVGLUT aggregates and the endosomal ESCRT protein Hrs. Representative confocal images of motor neuron cell bodies stained for DVGLUT (magenta) and Hrs (green). Bar, 5 µm. (B) Transmission electron micrograph of third instar larval garland cells. Bar, 5 µm. (C) High magnification of insets from B. Electron-lucent α and electron-dense β vacuoles and intraluminal electron-dense granular aggregates (arrowheads) are indicated. Bar, 500 nm. (D) Representative live confocal images of garland cells labeled with the lipophilic styryl dye FM1-43. Brightly labeled lipophilic contents are indicated within the enlarged endosomal compartment in the mutant (arrowheads). Bar, 5 µm.
To investigate endosomal structure and function, we performed transmission electron microscopy (TEM) in the Drosophila nephrocyte garland cell, a favorite in vivo model for studies of the endocytic pathway (Kosaka and Ikeda, 1983; Weavers et al., 2009). Wild-type garland cells display extensive tubulo-vesicular and vacuolar profiles of endosomal compartments, including electron-lucent α vacuoles akin to early endosomes in the cell cortex, and electron-dense β vacuoles that represent late endosomes more centrally (Fig. 2 B; Kosaka and Ikeda, 1983). Strikingly, ema mutant garland cells are filled with very large α and β vacuoles, many of which are 5–10 µm in diameter, whereas the largest vacuoles in wild type are ∼2 µm in diameter. In wild type, both types of vacuoles usually contain a single aggregate of electron-dense granular material that appears to be attached to the limiting membrane (Fig. 2 C, see arrowheads; Kosaka and Ikeda, 1983). However, the large vacuolar compartments in the mutant contain numerous electron-dense granular structures (Fig. 2 C, arrowheads), suggesting they arise from homo-/heterotypic fusions of α and β vacuoles. Despite these enlarged vacuolar profiles, other subcellular organelles including two nuclei, ER, and Golgi are present in ema mutant garland cells, although their organization is often distorted by neighboring enlarged vacuoles (unpublished data).
To address directly whether the large vacuolar profiles in the mutant are endosomes, we performed live labeling of the endosomal compartment with the lipophilic membrane tracer FM1-43. After a 5-min pulse of FM1-43 dye, vesicular and vacuolar profiles are extensively labeled in wild-type garland cells (Fig. 2 D). The size of the labeled endosomes increases as they advance toward the center of the cell, but rarely exceeds 2 µm in diameter. In the ema, FM1-43 dye labels the limiting membrane and the intraluminal contents of very large vacuoles (Fig. 2 D; average area of FM1-43–labeled endosomes (μm2) = 0.91 ± 0.04 in wild type vs. 6.14 ± 0.65 in mutants; Student t test, P ≪ 0.001), which likely represent the enlarged vacuolar profiles observed in the electron micrographs. Collectively, the immunocytochemistry in motoneurons and the ultrastructural and live imaging analysis in garland cells demonstrate that ema mutants have a dramatically enlarged endosomal compartment.
Ema localizes to endosomes
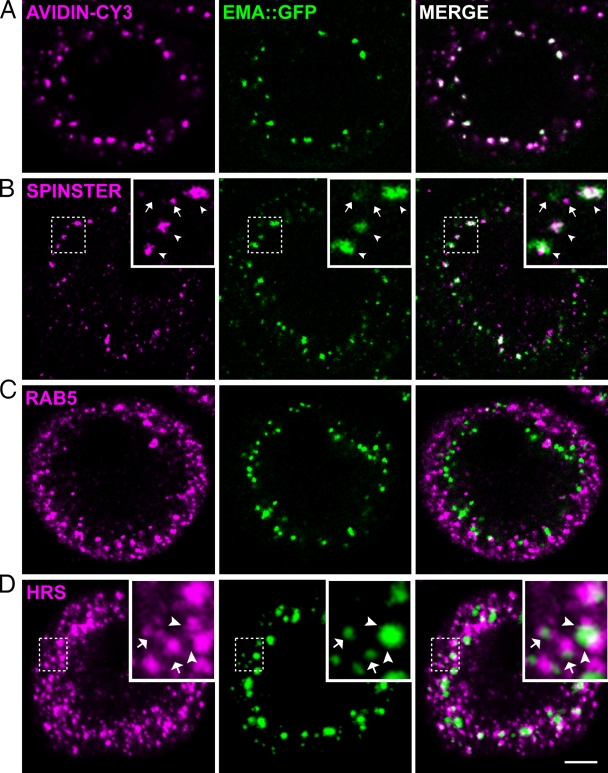
To begin investigating how ema is required for the normal organization of the endosomal compartment, we examined its subcellular localization. Although we have generated α-Ema antibodies that work on immunoblots, we were unable to generate antisera that recognize Ema for immunocytochemical studies. As an alternative, we generated an Ema::GFP fusion protein that rescues all the ema mutant phenotypes without any apparent adverse effects (rescue data in Fig. 1, A–C; see Fig. 4, A and B; Fig. 5, D and E; Fig. S1). When expressed in wild-type garland cells with the ubiquitous daughterless-Gal4 driver, Ema::GFP localizes to puncta (Fig. 3), which are often larger and brighter in the perinuclear region than in the cortical area. To investigate whether the Ema puncta represent endosomal structures, we tested whether they colocalize with endosome-specific probes. We first labeled the endosomal compartment with a 5-min pulse of the fluid-phase endocytic tracer avidin-Cy3 followed by an additional 5-min chase. Ema::GFP puncta colocalize with avidin-Cy3–labeled endosomes, demonstrating that Ema localizes to functional endosomes (Fig. 3 A).
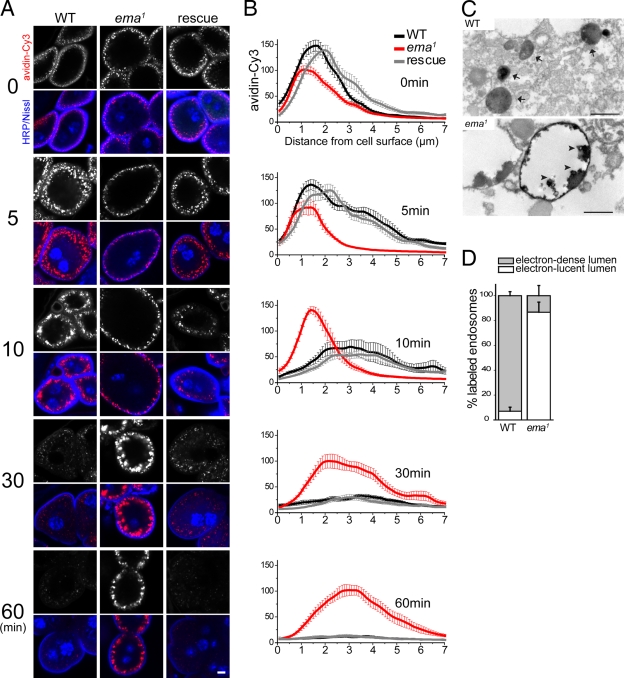
Figure 4.
Defective endosomal trafficking in the ema mutant. (A and B) Endosomal trafficking of the fluorescent endocytic tracer avidin-Cy3. Garland cells were incubated with avidin-Cy3 for 5 min and chased for the indicated time period (min). (A) Representative confocal images of garland cells traced with avidin-Cy3 (red) and stained for the plasma membrane marker HRP (blue) and nuclear marker Nissl (blue). Bar, 5 µm. (B) Line-depth intensity plot of avidin-Cy3 tracer. (Black) wild type (Canton S), (red) ema1, and (gray) rescue (ema1; Actin5C-Gal4/UAS-EmaGFP). n > 10 for all genotypes at all chase times. Data represent mean ± SEM. (C and D) Ultrastructural analysis of endosomal trafficking of the endocytic tracer HRP in garland cells. (C) Representative transmission electron micrograph of HRP-labeled endosomes, with electron-dense lumen in wild type (arrow) and electron-lucent lumen in mutant. Electron-dense HRP tracers highlight the limiting membrane and intraluminal protrusions (arrowheads) in mutant. Bar, 500 nm. (D) Quantification of HRP-labeled endosomes. n = 191 for 8 wild-type cells; n = 16 for 6 mutant garland cells. Data represent mean ± SEM.
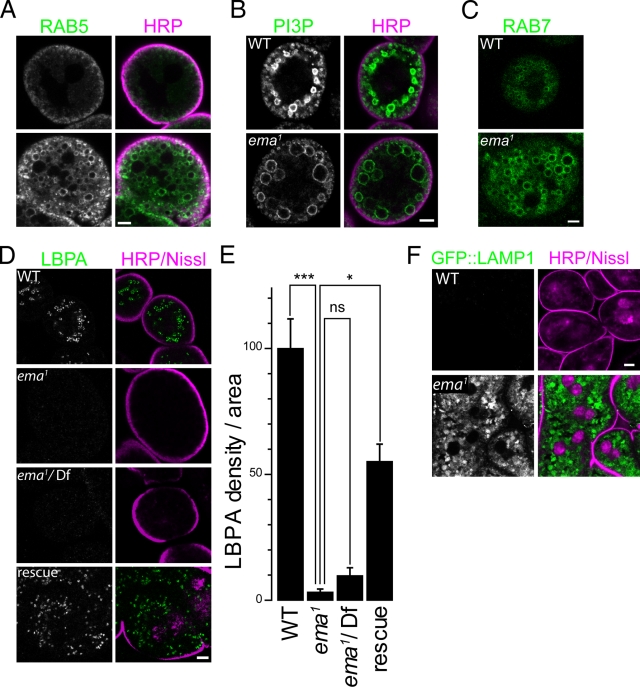
Figure 5.
Endosomal maturation is disrupted in the ema mutant. (A–D and F) Representative confocal images of garland cells labeled for (A) the early endosomal Rab5::GFP fusion protein, (B) the early endosomal phospholipid PI3P by a FYVE::GFP fusion protein, (C) the late endosomal Rab7::GFP fusion protein, (D) the late endosomal and lysosomal phospholipid LBPA by a monoclonal α-LBPA antibody, and (F) the GFP::LAMP1 fusion protein. Cells were co-labeled for plasma membrane (HRP/magenta) and/or nuclei (Nissl/magenta) except in C, where live imaging was taken for Rab7::YFP. Rescue genotype in D is ema1; actin5C-Gal4, UAS-Ema::GFP. Bar, 5 µm. (E) Quantification of LBPA level in garland cells for the genotypes in D. Total LBPA density per cell area was measured. Data represent mean ± SEM. ANOVA analysis. *, P < 0.05; ***, P < 0.001; ns, not significant.
Figure 3.
Ema localizes to endosomes. Representative single confocal sections of third instar larval garland cells labeled for Ema::GFP fusion protein (green) and endosomal markers (magenta), (A) the endocytic tracer avidin-Cy3 (pulse for 5 min and chase for 5 min), (B) the late endosomal and lysosomal protein Spinster, (C) the early endosomal protein Rab5, and (D) the early endosomal ESCRT protein Hrs. In insets of B, Ema puncta colocalize (arrowheads) or do not colocalize (arrows) with Spinster puncta. Insets of D depict partial overlapping (arrowheads) and directly adjacent (arrows) distributions between Ema and Hrs puncta. In D, the Ema channel (green) was enhanced by adjusting gamma output to visualize weak Ema puncta. Bar, 5 µm.
To investigate in which endosomal compartment Ema localizes, we compared the distribution of Ema::GFP with three different stage-specific endosomal proteins: the early endosomal protein Rab5, the MVB ESCRT protein Hrs, and the late endosomal/lysosomal protein Spinster. Many of the larger Ema::GFP puncta deeper in the cell do colocalize with the late endosomal/lysosomal protein Spinster (Fig. 3 B, arrowheads in insets), indicating that some Ema localizes to a late endosomal compartment. In contrast, Ema::GFP puncta do not colocalize with Rab5-positive early endosomes that preferentially localize to a more subcortical region of the cell (Fig. 3 C). Ema::GFP puncta also do not directly colocalize with Hrs; however, Hrs and Ema::GFP puncta are usually directly adjacent (Fig. 3 D, arrows in insets) and partially overlapping (Fig. 3 D, arrowheads in insets). These findings indicate that Ema is an endosomal protein, and so the defects in the endosomal compartment observed in the ema mutant are likely due to a direct effect of ema on endosomal function.
ema mutants display severe defects in endosomal trafficking
To test whether ema is required for endosomal function, we examined the trafficking of the fluid-phase endocytic tracer avidin-Cy3 in garland cells from wild type, the ema mutant, and the ema mutant rescued by the wild-type ema transgene (Fig. 4, A and B). After a pulse of avidin-Cy3 and a chase with fresh media, we examined the distribution of avidin-Cy3 by measuring the average density of the tracer across a fixed-width line from the surface to the center of the cell (Fig. 4 B). During the pulse, tracer is internalized and concentrates in bright puncta within 2 µm of the cell surface in all the genotypes, although internalization is slightly less efficient in mutant cells (Fig. 4 B, 0 min). In wild-type and rescued cells, the tracer advances and spreads throughout the entire cortical area during the first 5-min chase, which is reflected as multiple peaks beyond 2 µm from the cell surface in the line-depth intensity profile. After a 10-min chase, most tracer in wild-type and rescued cells leaves the subcortical area and appears as large and diffuse spots around 3 µm from the surface. During this time, wild-type and rescued cells lose much of the tracer, suggesting that the avidin-Cy3 has either been degraded or recycled. In contrast, in ema mutant garland cells the avidin-Cy3 is still concentrated in a very narrow subcortical area with its main intensity peak within 2 µm from the surface after a 10-min chase, indicating early endosomal trafficking is greatly retarded. Indeed, 30 min of chase are required for the tracer in the mutant cells to access to large puncta around 3 µm from the surface, which is accessible within 10 min of chase in the wild-type cell. After a 30-min chase in wild-type or rescued cells, what little tracer that is still visible has reached the perinuclear region, and by 60 min almost all tracer has been degraded or released from the cell (Fig. 4, A and B). In stark contrast, the large spots of avidin-Cy3 observed in mutant garland cells persist without a significant loss of intensity for 2 h (unpublished data). The persistence of signal suggests that the tracer is either irreversibly trapped or dramatically slowed in its progression. These functional studies demonstrate that ema is required for the trafficking and processing of internalized avidin-Cy3 tracer through the endosomal pathway.
To investigate the morphological details of endosomal trafficking, we followed internalized horseradish peroxidase (HRP) at the ultrastructural level via transmission electron microscopy. Most HRP-labeled endosomes in wild-type garland cells are small (∼100 nm in diameter; Fig. 4 C, see arrows in wild type) and distribute throughout the cell (not depicted). In the majority of the labeled endosomes (∼90% of total), the HRP reaction product fills the luminal space (electron-dense lumen; Fig. 4 D). In the ema mutant, none of the extremely enlarged endosomes (>5 µm in diameter) are detectably labeled by HRP (not depicted), indicating they are inaccessible by the endocytic tracer. Such strong resistance to the access of the endocytic tracer is consistent with the delayed kinetics in the early phase of endosomal trafficking of the avidin-Cy3 tracer (Fig. 4, A and B; see 5- and 10-min chase). In the mutant cell, there are a few HRP-labeled endosomes that are relatively large (∼500 nm; Fig. 4 C) and exclusively located cortically (not depicted). Furthermore, HRP reaction product in most labeled endosomes (∼90% of total) from mutant cells does not fill the luminal space as in wild type (electron-lucent lumen; Fig. 4 D), but instead is concentrated along the limiting membrane and in intraluminal clusters, which often extend from the limiting membrane (Fig. 4 D, arrowheads in the mutant). Taken together, inaccessibility of the HRP tracer to the enlarged endosomes and abnormal distribution of HRP tracer within endosomes in the mutant garland cells indicate that ema is important for trafficking and processing of molecular cargo through endosomal compartments.
Ema is required for endosomal maturation
Enlarged endosomal compartments often reflect functional changes in endosomal trafficking and maturation, which can be investigated by analyzing their association with endosomal Rabs and phospholipid species (Stenmark et al., 1994; Lloyd et al., 2002; Rink et al., 2005; Piper and Katzmann, 2007; Zoncu et al., 2009). To investigate the roles of Ema for endosomal maturation, therefore, we examined the steady-state distribution of such molecules in the garland cell. The early endosomal protein Rab5::GFP (Zhang et al., 2007) mainly concentrates in the subcortical area near the plasma membrane. In contrast, in the mutant Rab5::GFP labels numerous enlarged endosomes beyond the cortical area, suggesting an expansion of early endosomal compartments (Fig. 5 A). The early endosomal phospholipid PI3P was visualized via its binding to the FYVE::GFP fusion protein (Bökel et al., 2006). When expressed in wild-type garland cells, FYVE::GFP preferentially localizes in two regions: small vesicles in the subcortical area and large vesicles deeper into the cell (Fig. 5 B). In the ema mutant, two similar groups of endosomes are labeled with FYVE::GFP. The small endosomes near the plasma membrane of mutant garland cell are comparable to those in wild type; however, the structures present deeper in the cell are much enlarged. The intense labeling of enlarged endosomes with FYVE::GFP indicates that some of the huge vacuoles present in the mutant garland cells represent relatively early endosomes. Next, we examined the late endosomal compartment by expressing Rab7::YFP fusion protein (Zhang et al., 2007). Live confocal imaging highlights the enrichment of Rab7::YFP–positive late endosomes at the deep area of wild-type garland cells (Fig. 5 C). The size and distribution of these endosomes is comparable to the large FYVE::GFP–labeled vesicles. In mutant garland cells, Rab7::YFP is associated with enlarged endosomes predominantly located in the cell cortex, indicating that some enlarged vesicles in the ema mutant have characteristics of late endosomes. To examine the distribution of mature late endosomes and lysosomes, we examined the localization of a unique late endosomal/lysosomal lipid LBPA (Kolter and Sandhoff, 2005). In wild-type garland cells, numerous bright LBPA puncta are distributed in the perinuclear region of the cell where late endosomes and lysosomes reside (Fig. 5 D). Surprisingly, mutant garland cells are completely devoid of LBPA and LBPA-containing structures (Fig. 5, D and E). The absence of LBPA is rescued by the transgenic expression of ema, demonstrating that ema is required for the maturation of PI3P- and Rab7-containing endosomes into LBPA-containing endosomes. To investigate whether this block in late endosomal maturation disrupts the formation or function of degradative late endosomes, we expressed a GFP::LAMP1 fusion protein that is a substrate for lysosomal degradation (Pulipparacharuvil et al., 2005). When expressed in wild-type garland cells, GFP::LAMP1 is hardly detectable (Fig. 5 F). In contrast, the density of GFP::LAMP1 is increased more than eightfold in the mutant and accumulates in the enlarged endosomes, demonstrating that the endosomes have not matured into degradative late endosomes (Fig. 5 F, average GFP::LAMP1 density per area [a.u.] = 3.9 ± 0.5 in wild type vs. 33.6 ± 2.6 in mutant; Student’s t test, P ≪ 0.001). Collectively, the compartmental identity of endosomes in the ema mutant is dramatically perturbed in that enlarged endosomes retain both early and late endosomal characteristics. Furthermore, the absence of LBPA and the accumulation of the degradation reporter GFP::LAMP1 indicate that ema is critical for the maturation of endosomal compartments into degradative late endosomes and lysosomes.
Defective endosomal down-regulation of BMP signaling is responsible for synaptic overgrowth at the ema mutant NMJs
The endolysosomal pathway regulates the levels and distribution of membrane receptors (Seto et al., 2002), which are marked for internalization by the addition of ubiquitin (Hicke and Dunn, 2003). To determine if ema regulates receptor trafficking, we examined the distribution of ubiquitinated proteins and the trafficking of the receptor tyrosine kinase EGFR, a well-characterized endolysosomal receptor cargo (Felder et al., 1990). In the ema mutant, the levels of both anti-conjugated ubiquitin staining as well as GFP-tagged EGF receptor are dramatically increased in motoneuron cell bodies and partially colocalize with DVGLUT aggregates (Fig. 6, A and B), suggesting the endolysosomal traffic and degradation of membrane receptor proteins is disrupted.
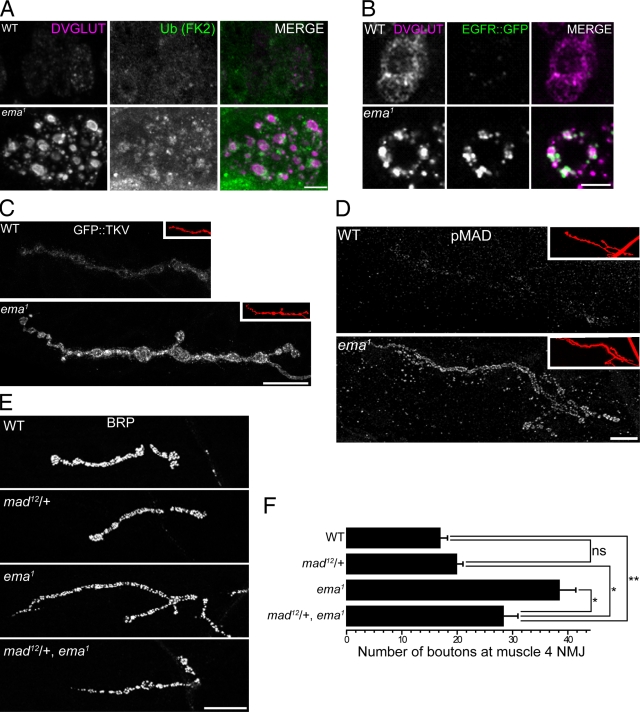
Figure 6.
Up-regulated BMP signaling promotes synaptic overgrowth in the ema mutant. (A and B) Endolysosomal trafficking of membrane receptor cargos in motoneurons. Representative confocal images of motoneuron cell bodies labeled for (A) ubiquitinated proteins by a monoclonal α-FK2 antibody (green) or (B) EGFR::GFP fusion protein (green) and for DVGLUT protein (magenta). Bar, 5 µm. (C and D) Distribution of BMP signaling components at the NMJs. Representative confocal images of muscle 4 type 1b NMJs labeled for (C) neuronally expressed GFP::TKV fusion protein and (D) endogenous pMAD protein. In insets, the NMJs are visualized by the neuronal membrane marker HRP (red). Bar, 20 µm. (E and F) A mad mutation dominantly suppresses the synaptic overgrowth in ema. (E) Representative confocal images of wild type (WT), a heterozygous mad mutant (mad12/+), ema1, and mad12/+, ema1 NMJs labeled for the presynaptic protein BRP. Bar, 20 µm. (F) Quantification of number of synaptic boutons at the muscle 4 NMJs for the genotypes in E. n > 10 for all genotypes. Data represent mean ± SEM. ANOVA analysis. *, P < 0.05; **, P < 0.01; ns, not significant.
To test for functional consequences of defective endolysosomal receptor trafficking, we examined the BMP pathway at the Drosophila NMJ where the endosomal regulation of the BMP receptor thick veins (Tkv) is required for normal synaptic terminal growth (Sweeney and Davis, 2002; O’Connor-Giles et al., 2008). In the ema mutant, there is a twofold increase in synaptic terminal growth at the NMJs (Fig. 1, A–C), consistent with increased BMP signaling. At wild-type NMJs, GFP::Tkv is distributed as tiny, dim speckles throughout synaptic boutons, whereas in the ema mutant GFP::Tkv is often clustered as bright puncta and its density per synaptic area is approximately doubled (Fig. 6 C; average GFP::TKV density per NMJ [a.u.] = 9.7 ± 0.8 in wild type vs. 17.4 ± 1.3 in mutant; Student t test, P ≪ 0.001). This is consistent with a defect in the endolysosomal down-regulation of the Tkv receptor. To test the efficacy of endogenous BMP signaling, we examined the distribution of phosphorylated MAD (pMAD), the immediate downstream target of activated BMP receptors. In the ema mutant, there is a more than fourfold increase in pMAD level compared with wild type (Fig. 6 D; average pMAD density per NMJ [normalized a.u.] = 1.0 ± 0.2 in wild type vs. 4.6 ± 0.2 in mutant; Student t test, P ≪ 0.001). Hence, endogenous BMP signaling is significantly up-regulated at the NMJs of ema mutants. To investigate if activation of BMP signaling is responsible for the NMJ overgrowth in the ema mutant, we performed genetic interaction studies between ema and the essential downstream BMP signaling component mad. Removing one copy of mad does not lead to a decrease in synaptic bouton number in wild-type larvae; however, in the ema mutant, removing one copy of mad leads to a significant decrease in synaptic bouton number (Fig. 6, E and F). The dosage-sensitive interactions between mad and ema, in conjunction with the defects in Tkv trafficking and signaling, are all consistent with the model that synaptic terminal overgrowth is due to the inability of the ema mutant to down-regulate and inactivate BMP receptors. Collectively, these data indicate that ema is required for the endosomal trafficking of membrane receptors and, in the case of Tkv, that this regulates NMJ development.
Ema orthologues are functionally conserved
The human ema orthologue Clec16A is a recently identified candidate susceptibility locus for a number of autoimmune disorders (Hakonarson et al., 2007; International Multiple Sclerosis Genetics Consortium, 2007; Todd et al., 2007; Wellcome Trust Case Control Consortium, 2007). The high level of sequence conservation between the Drosophila and human orthologues (Fig. 1 D) suggests conserved function. To test this hypothesis, we asked if CLEC16A can substitute for ema and rescue the Drosophila ema mutant phenotypes. We generated transgenic flies in which the human gene is expressed under the control of the yeast UAS promoter (UAS-CLEC16A). Expression of CLEC16A is sufficient to rescue the lethality of the ema mutant (80.5 ± 10.1% survival rate). In fact, this rescue is comparable to that obtained by expressing the Drosophila gene (142.2 ± 8.4% survival rate). Hence, the essential function of ema orthologues is conserved from Drosophila to humans.
To investigate whether CLEC16A functions in endosomal trafficking, we assessed the ability of human CLEC16A to rescue the endosomal phenotypes in garland cells and motoneurons from the ema mutant. Expression of CLEC16A in ema mutant garland cells leads to a significant reduction in the size of endosomes (Fig. 7, A and B). When expressed in motoneurons, CLEC16A leads to a significant decrease in the size and intensity of DVGLUT aggregates (Fig. 7, C and D). In addition, neuronal expression of CLEC16A fully rescues NMJ overgrowth in the ema mutant (Fig. 7, E and F). These findings suggest that ema and CLEC16A play a conserved role in endosomal trafficking and maturation, and imply that key interaction partners must also be conserved across species.
Figure 7.
Human Clec16A rescues ema. (A and B) Human Clec16A rescue of enlarged endosomes in the ema mutant garland cells. (A) Representative confocal images of live FM1-43–labeled endosomes from wild-type (WT), ema1, and rescue (ema1; actin5C-Gal4, UAS-GFP::hClec16A) garland cells. (B) Quantification of the size of live FM1-43–labeled endosomes for the genotypes in A. n > 200 for all genotypes. (C and D) Human Clec16A rescue of DVGLUT aggregates in the ema mutant motoneurons. (C) Representative confocal images of wild-type (WT), ema1, and rescue (BG380-Gal4, UAS-GFP::hClec16A; ema1) motoneuron cell bodies labeled for DVGLUT protein (magenta) and the neuronal membrane marker HRP (green). Bar, 2 µm. (D) Quantification of DVGLUT aggregation for the genotypes in C. Aggregation index (AI) of a motor neuron = total DVGLUT density × average size of DVGLUT aggregates in that neuron. n > 30 for all genotypes. (E and F) Human Clec16A rescue of synaptic overgrowth at the ema mutant NMJs. (E) Representative confocal images of wild-type (WT), ema1, and rescue (BG380-Gal4, UAS-GFP::hClec16A; ema1) type Ib NMJs at the muscle 4 labeled for DVGLUT protein. The ema1 image was taken at a higher gain for clarity. Bar, 10 µm. (F) Quantification of number of type Ib synaptic boutons at muscle 4 for the genotypes in E. n > 10 for all genotypes. For B, D, and F, data represent mean ± SEM. ANOVA analysis. **, P < 0.01; ***, P < 0.001; ns, not significant.
Ema interacts with the class C Vps–HOPS complex
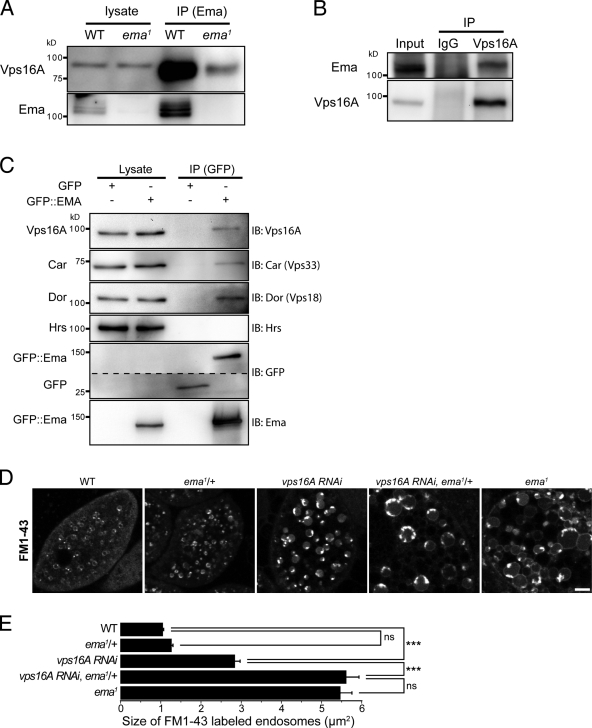
To investigate the molecular function of Ema, we performed a yeast two-hybrid screen for binding partners of Ema from a Drosophila embryonic library. Using the N-terminal region of Ema (aa 1–285) harboring the highly conserved uncharacterized domain pfam09758 (Fig. 1 D), we isolated the C-terminal region (aa 492–883) of the endosomal protein Vps16A (unpublished data). Vps16A is a subunit of the class C Vps–HOPS complex that is a tethering complex during vacuolar fusion in yeast (Sato et al., 2000; Wurmser et al., 2000) and that promotes maturation of endosomes in higher eukaryotes (Pulipparacharuvil et al., 2005; Rink et al., 2005; Kinchen et al., 2008; Liang et al., 2008). Reciprocal coimmunoprecipitation experiments demonstrate that endogenous Ema and Vps16A proteins bind in vivo (Fig. 8, A and B). Class C Vps subunits bind in a functional complex (Sevrioukov et al., 1999; Sato et al., 2000; Seals et al., 2000; Wurmser et al., 2000; Pulipparacharuvil et al., 2005; Peplowska et al., 2007), and we find that transgenic Ema binds in vivo to the class C Vps–HOPS complex subunits Vps16A, Car (Vps33A), and Dor (Vps18), but does not bind the ESCRT protein Hrs (Fig. 8 C).
Figure 8.
Ema interacts with the class C Vps–HOPS complex. (A and B) Reciprocal coimmunoprecipitation between endogenous Ema and Vps16A proteins. (A) Western blots for Vps16A and Ema of the immunocomplex isolated with an α-Ema antibody from whole-cell lysates of wild-type and ema mutant third instar larvae. (B) Western blots of Ema and Vps16A of the immunocomplex isolated with either the α-Vps16A antibody or control IgG from whole-cell lysates of wild-type adult fly heads. (C) Coimmunoprecipitation between Ema::GFP fusion protein and endogenous proteins of the class C Vps–HOPS complex. Western blots for Vps16A, Car, Dor, Hrs, Ema, and GFP of the immunocomplex isolated with an α-GFP antibody from whole-cell lysates of third instar larvae expressing either Ema::GFP fusion protein or GFP alone. (D and E) Genetic interaction of ema and vps16A for enlarged endosomes. (D) Representative confocal images of live FM1-43–labeled endosomes of wild-type (WT), a heterozygote ema mutant (ema1/+), Vps16A knockdown (vps16A RNAi/+; daGal4/+), a Vps16A knockdown in the heterozygote ema mutant (vps16A RNAi/+; daGal4/+; ema1/+), and ema1 garland cells. Bar, 5 µm. (E) Quantification of the size of live FM1-43–labeled endosomes (µm2) for the genotypes in D. n > 500 endosomes from at least 10 cells for all genotypes. Data represent mean ± SEM. ANOVA analysis. ***, P < 0.001; ns, not significant.
In Drosophila mutants of Vps16A and other class C Vps mutants, defective endolysosomal trafficking and maturation results in the accumulation of a lysosomal substrate GFP::LAMP1 fusion protein (Sevrioukov et al., 1999; Pulipparacharuvil et al., 2005). Similarly, GFP::LAMP1 accumulates in ema mutant garland cells (Fig. 5 F), suggesting that Ema and the class C Vps complex may function in a common trafficking pathway. The Drosophila mutants of class C Vps–HOPS genes also exhibit enlarged endolysosomal compartments (Sevrioukov et al., 1999; Sriram et al., 2003) and knockdown of Vps16A in mammalian culture system results in the enlargement of endosomal compartments due to defective endosomal maturation (Rink et al., 2005). We find that the expression of a Vps16A RNAi construct (Pulipparacharuvil et al., 2005) in the garland cell system leads to enlarged endosomes twice as big as those in wild type (Fig. 8, D and E). Hence, a partial loss of vps16A gives a similar but quantitatively weaker phenotype than loss of ema. Genes that function together in a pathway can show dosage-sensitive genetic interactions, so we tested whether loss of a single copy of ema would enhance the vps16A phenotype. Removing a single copy of ema has no effect on the size of endosomes in an otherwise wild-type background. However, when vps16A is inhibited, then removing a single copy of ema leads to an additional twofold increase in the size of endosomes compared with inhibition of vps16A alone (Fig. 8, D and E). Hence, ema dominantly enhances the vps16A knockdown phenotype. Collectively, the physical and genetic interactions between ema and vps16A suggest that they cooperate to promote endosomal maturation.
Discussion
Here we demonstrate that Ema is an evolutionarily conserved endosomal membrane protein that interacts with the class C Vps–HOPS complex to promote maturation of endosomes. In its absence there is an accumulation of large endosomal intermediates, membrane trafficking is disrupted, and lysosomal degradation is inhibited. Down-regulation of signaling receptors is impaired, and up-regulated BMP signaling induces synaptic overgrowth at the ema mutant NMJs. In addition, we show that human Clec16A can substitute for Drosophila ema, suggesting that members of this gene family play a conserved function in endosomal maturation. The human ema orthologue Clec16A is a strong candidate susceptibility locus for autoimmune disorders including type 1 diabetes and multiple sclerosis (Hakonarson et al., 2007; International Multiple Sclerosis Genetics Consortium, 2007; Todd et al., 2007; Wellcome Trust Case Control Consortium, 2007). Therefore, our analysis of this understudied gene family sets the stage for testing the role of human Clec16A in the regulation of endosomal functions critical for immunity and self-tolerance.
Ema and the class C Vps–HOPS complex
The class C Vps–HOPS complex is well conserved from yeast to multicellular organisms where it promotes endosomal membrane fusion (Sato et al., 2000; Seals et al., 2000; Wurmser et al., 2000; Poupon et al., 2003; Richardson et al., 2004). The absence of an Ema orthologue in yeast puts some constraints on potential roles of Ema for the function of the class C Vps–HOPS complex. Localization of Ema on a subset of endosomes (Fig. 3) suggests that Ema could promote membrane fusion between endosomes and specific membrane compartments such as lysosomes and Golgi-derived vesicles. In that scenario, the expansion of endosomes in the ema mutant would stem from improper fusion and subsequently impaired processing of endosomal cargo, as shown for the defective trafficking and processing of GFP::LAMP1 and endocytic cargos in the ema mutant. An alternative explanation of the functional interaction between Ema and the class C Vps–HOPS complex comes from studies on the orthologues of the complex in multicellular organisms where the class C Vps–HOPS complex promotes compartmental maturation. Such observations suggest that specific endosomal compartments may contain compartment-specific proteins that facilitate the function of the class C Vps–HOPS complex. Our characterization of Ema as a transmembrane protein localizing to a subset of endosomal intermediates is consistent with such a role for Ema.
How might Ema promote class C Vps–HOPS complex function? Although its precise molecular function is unknown, it is attractive to speculate that Ema may function with the class C Vps–HOPS complex in the process of Rab conversion. During endosomal maturation there is a replacement of Rab5 by Rab7 in a conversion process that was proposed to require the class C Vps–HOPS complex (Rink et al., 2005). Many of the phenotypes associated with disrupting this Rab conversion process match those described here for loss of ema. For example, endosomal trafficking is dramatically slowed in mammalian cells expressing the GTPase-deficient Rab5Q88L mutant and in cells in which the class C Vps–HOPS complex genes Vps39 and Vps16 are knocked down (Rink et al., 2005). We demonstrate a similar delay in endosomal trafficking in the ema mutant (Fig. 4, A and B). In addition, delayed Rab conversion leads to enlarged endosomal compartments with early and late endosomal characteristics as described here for the ema mutant cells (Fig. 5). Rink et al. (2005) proposed that the class C Vps–HOPS complex participates in a rate-limiting step in the transition of Rabs on endosomes. Given the functional and physical interaction between Ema and the complex (Fig. 8) and the localization of Ema to endosomes that are internal to the Rab5 endosomes (Fig. 3), Ema could promote such a rate-limiting step in the conversion process. Although additional models for Ema function are possible, it is clear that Ema plays an important role in multicellular organisms in conjunction with the class C Vps–HOPS complex to promote endosomal maturation.
Ema and receptor trafficking
Receptor-mediated endocytosis and subsequent trafficking through the endosomal pathway can modulate the efficacy of signaling through membrane receptors. Signaling can be attenuated by targeting active receptors for degradation in the lysosome, or potentiated by trafficking receptors to signaling endosomes (Seto et al., 2002). As such, defects in the endosomal system can have profound effects on receptor-mediated signaling. Ubiquitination of membrane receptors targets these proteins for endolysosomal trafficking. In the ema mutant, there is an accumulation of ubiquitinated proteins to the same compartment where the ESCRT protein Hrs accumulates, consistent with defects in receptor trafficking. In addition, levels of the EGF receptor in the motoneurons and in the garland cells (unpublished data) are increased in the mutant, suggesting a defect in the degradation of receptors. Finally, levels of the type I BMP receptor Tkv, a membrane serine kinase, are increased at the NMJ.
The inability to down-regulate BMP signaling at the NMJ has important consequences for synaptic terminal development. BMP signaling drives growth of the synaptic terminal, and mutants with defects in the endosomal trafficking of BMP receptors display NMJ overgrowth (Sweeney and Davis, 2002; Korolchuk et al., 2007; Wang et al., 2007; O’Connor-Giles et al., 2008). In the ema mutant, there is a more than fourfold increase in the levels of phospho-MAD at the NMJ compared with wild type, demonstrating that signaling through the endogenous BMP pathway is potentiated. In combination with the increase in Tkv levels, these data show that ema is required to limit BMP signaling. We identified the ema mutant because it shows synaptic overgrowth. This overgrowth is sensitive to the dose of MAD, hence synaptic overgrowth in the ema mutant is likely due to excessive BMP signaling, suggesting that in the mutant Tkv is retained in a signaling endosome (Bökel et al., 2006). Collectively, these data support the model that Ema restrains synaptic growth during development by inhibiting BMP signaling via the endolysosomal degradation of the Tkv receptor. A role for ema in the regulation of receptor trafficking suggests that ema may participate in other receptor-mediated developmental processes. Indeed, ema mutant clones in the wing have developmental defects that are consistent with effects on multiple signaling pathways (Fig. S3).
The endosomal function of ema and autoimmunity
Many genome-wide association studies have identified the human ema orthologue Clec16A as a candidate susceptibility factor in type I diabetes and other autoimmune disorders (International Multiple Sclerosis Genetics Consortium, 2007; Hakonarson et al., 2007; Todd et al., 2007; Wellcome Trust Case Control Consortium, 2007; Rubio et al., 2008; Skinningsrud et al., 2008; Awata et al., 2009; Wu et al., 2009; Zoledziewska et al., 2009). However, how Clec16A confers susceptibility to autoimmune diseases remains unknown. Because human Clec16A can rescue the endosomal defects of the Drosophila ema mutant, we propose that the fundamental function for members of this gene family is to promote endosomal maturation. Such a function suggests at least two mechanisms by which Clec16A may confer susceptibility to autoimmune disorders. First, Clec16A could participate in the endosomal regulation of immune receptor signaling pathways and restrain the aberrant activation and proliferation of autoreactive T lymphocytes. Consistent with this hypothesis, association studies link type 1 diabetes to a long list of T cell–signaling components including PTPN22, CTLA4, IL2RA, and PTPN2 (Ounissi-Benkalha and Polychronakos, 2008). Second, Clec16A may be involved in the biogenesis and/or function of the MHC class II compartment (MIIC), a lysosome-related endosomal compartment that is involved in establishing self-tolerance during the development of naive T cells in the thymus (Rocha and Neefjes, 2008; van Niel et al., 2008). Consistent with this possibility, there is a strong association between HLA class II alleles and susceptibility to type 1 diabetes and other autoimmune disorders (Suri et al., 2008; International Multiple Sclerosis Genetics Consortium (IMSGC), 2009). Ultimately, understanding the function of Clec16A for autoimmunity will require analyzing its function in mammalian cells and relating the genetic changes in susceptible patients to the expression or function of Clec16A.
In summary, we have demonstrated that ema is a member of a novel and conserved gene family that encodes an endosomal membrane protein that functions with the class C Vps–HOPS complex to promote endosomal maturation and function. These findings provide insights into the mechanism of endosomal trafficking in higher eukaryotes and the potential role of this pathway in development and disease.
Materials and methods
Sequence analysis of ema orthologues
Sequence identity (%) to human Clec16A was analyzed via pairwise sequence alignment using ClustalW2 and transmembrane domains were predicted using TMpred. NCBI Protein database accession numbers of analyzed ema orthologues: NP_056041.1 (H. sapiens), NP_650542.1 (D. melanogaster), NP_498117 (C. elegans), and NP_566837 (A. thaliana).
Molecular biology
A Drosophila cDNA clone (RE60631) for ema (CG12753) was obtained from Berkeley Drosophila Genome Project (Berkeley, CA). To generate UAS-GFP::EMA, the ema cDNA was cloned into pUAST-EGFP using NdeI and XhoI. To generate UAS-hClec16A, a human cDNA clone (I.M.A.G.E Clone ID: 5727832) for Clec16A (KIAA0350) was obtained from Invitrogen and cloned into pUAST-EGFP using BglII and XhoI–SalI. Transgenic fly lines were generated using standard techniques. To generate the His-tagged EMA-C fusion protein construct, the ORF of EMA (aa 704–998) was cloned into pQE30 (QIAGEN) using BamHI and SalI. To make the bait construct pBGKT7-EMA_N in the yeast two-hybrid screening, the ORF of EMA (aa 1–285) was PCR amplified and cloned into pBGKT7 (BD) using NdeI and BamHI.
Yeast two-hybrid screen
Yeast two-hybrid screening was performed using Matchmaker Gal4 two-hybrid system 3 (Takara Bio, Inc.) according to the manufacturer’s instructions. In brief, ∼5 × 106 independent clones were produced in a library-scale sequential transformation of AH109 with the bait construct pBGKT7-EMA_N (aa 1–285) and a Drosophila embryo Matchmaker cDNA Library (Takara Bio, Inc.). Transformants were selected and isolated on high-stringency SD/-Ade/-His/-Leu/-Trp/X-α-Gal plates. Plasmids isolated from positive clones were sequenced and transformed into AH109 either alone to test for autonomous false positive activity or with the bait construct to confirm their interaction. No autonomous positive activity either from the bait construct or the isolated Vps16A clone was detected.
Fly stocks and genetics
Drosophila were cultured using standard techniques. Crosses were kept at 25°C on standard Drosophila medium. ema1 (f07675) (Thibault et al., 2004), Df(3R)Exel7330 (Parks et al., 2004) and mad1-2 (Wiersdorff et al., 1996) flies were obtained from the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN). We have confirmed the piggyBac insertion f07675 at 3R: 12,265,747 (corresponding at 369 aa in the ema ORF) by sequence analysis of PCR-amplified flanking genomic regions. Canton S was used as wild-type control, unless otherwise indicated. UAS-Vps16A RNAi and UAS-GFP::Lamp1 flies were provided by Dr. H. Kramer (University of Texas Southwestern Medical Center, Dallas, TX; Pulipparacharuvil et al., 2005). UAS-myc::FYVE::GFP and UAS-GFP::TKV flies were provided by Dr. M. González-Gaitán (University of Geneva, Geneva, Switzerland; Bökel et al., 2006). BG380-Gal4 was provided by Dr. V. Budnik (University of Massachusetts Medical School, Worcester, MA; Budnik et al., 1996). UAS-EGFR::GFP fly was provided by Dr. J.B. Duffy (Indiana University, Bloomington, IN). FRT82B (Xu and Rubin, 1993), UAS-GFP::RAB5 (Zhang et al., 2007), UAS-YFP::RAB7 (Zhang et al., 2007), elav-Gal4 (Yao and White, 1994), daughterless (da)-Gal4 (Wodarz et al., 1995), and actin5C-Gal4 (Ito et al., 1997) flies were obtained from the Bloomington Drosophila Stock Center. hs-flp, tub-GFP; FRT82B, tub-Gal80 fly was provided by Dr. C. Micchelli (Washington University in St. Louis, St. Louis, MO).
Immunocytochemistry
Wandering third instar larvae were dissected in ice-cold PBS and fixed with Bouin’s solution for 5 min. Blocking and antibody incubation were performed in PBS containing 0.1% Triton X-100. For pMAD and Rab5 staining, larvae were fixed in 4% PFA in PBS for 30 min. For LBPA staining, specimens were fixed in 4% PFA in PBS for 1 h at 4°C, permeabilized in PBS containing 0.1% Triton X-100 for 10 min at 4°C, and the remaining staining procedures were performed in PBS. Antibodies were used at the following dilutions. α-DVGLUT (Daniels et al., 2004) at 1:10,000; α-HRS-FL (Lloyd et al., 2002) at 1:1,000; α-Spinster (Sweeney and Davis, 2002) at 1:250; α-pMAD (Persson et al., 1998) at 1:1,000; α-Rab5 (Wucherpfennig et al., 2003) at 1:50; α-LBPA (Echelon) at 1:50; α-BRP (Developmental Studies Hybridoma Bank, Iowa City, IA; Wagh et al., 2006) at 1:250; α-GFP (3E6; Invitrogen) at 1:1,000; α-Vps16A (Pulipparacharuvil et al., 2005) at 1:1,000; α-DOR (Sevrioukov et al., 1999) at 1:1,000; α-LAVA LAMP (Sisson et al., 2000) at 1:250; α-GM130 (Nakamura et al., 1995) at 1:800; α-KDEL receptor (Abcam) at 1:1,000; NeuroTrace 640/660 deep-red fluorescent Nissl (Invitrogen) at 1:500; Cy3-/Cy5-conjugated α-HRP (Jackson Immunoresearch Laboratories, Inc.) at 1:1,000; and Alexa 488–/Cy3-conjugated secondary antibodies (Jackson Immunoresearch Laboratories, Inc.) at 1:1,000. After staining, specimens were equilibrated in 70% glycerol in PBS and mounted with VectaShield (Vector Laboratories).
Generation of polyclonal α-EMA-C antibodies, Western blot, and coimmunoprecipitation
His-tagged EMA-C fusion protein was expressed and purified using a Ni-NTA column (QIAGEN) according to the manufacturer’s instructions. Polyclonal antibodies against His-tagged EMA-C were generated in guinea pigs (Cocalico Biologicals) and rabbits (YenZym Antibodies). For Western blot of endogenous Ema protein, wandering third instar larvae were dissected in ice-cold PBS and lysed in SDS sample buffer. Lysates were run on 6% SDS-PAGE gels according to standard procedures. The blot was probed with guinea pig α-Ema antibody at 1:1,000, then stripped and reprobed with α-β-tubulin (Developmental Studies Hybridoma Bank) at 1:500. HRP-conjugated secondary antibodies (Jackson Immunoresearch Laboratories, Inc.) were used at 1:10,000. For coimmunoprecipitation experiments, third instar larvae or adult fly heads were homogenized in 1 ml of ice-cold lysis buffer (10 mM Hepes, pH 7.4, 100 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail [Roche]). After centrifugation at 13,000 rpm for 10 min at 4°C, the supernatant was precleared with 100 µl of protein A–Sepharose beads (Invitrogen). The precleared lysate was then incubated with 5 µl of guinea pig α-Ema antibody, 10 µl of mouse α-GFP (3E6; Invitrogen), or 5 µl of rabbit α-Vps16A antibody for 1 h at 4°C and then with 50 µl of protein A–Sepharose beads for an additional 1 h at 4°C. Immunocomplexes were washed in the lysis buffer three times and dissolved in SDS sample buffer. Blots were probed with rabbit or guinea pig α-Ema antibodies at 1:1,000; rabbit α-Vps16A antibody (Pulipparacharuvil et al., 2005) at 1:1,000; rabbit α-Car antibody at 1:1,500; guinea pig α-Dor antibody at 1:1,000; mouse α-GFP antibody (11E5; Invitrogen) at 1:500; or guinea pig α-Hrs antibody (Lloyd et al., 2002) at 1:10,000.
Live FM1-43 labeling in garland cells
Third instar larvae were dissected in ice-cold PBS, equilibrated in the Drosophila Schneider medium (Invitrogen) for 5 min at room temperature, incubated in 10 µM of FM1-43 (Invitrogen) for 5 min at room temperature, and rinsed thoroughly. Garland cells were dissected out and transferred in Schneider medium. Live confocal imaging was performed at room temperature.
Avidin-Cy3 uptake and trafficking in garland cells
Third instar larvae were dissected in ice-cold PBS, equilibrated in the Schneider medium for 5 min at room temperature, pulsed with 10 µg/ml of Extravidin-Cy3 (Sigma-Aldrich) at room temperature for 5 min, rinsed thoroughly, and chased in Schneider medium for the indicated chase period at room temperature. Then, samples were fixed in 4% PFA in PBS for 10 min at room temperature and processed for staining with Cy5-conjugated α-HRP antibody and 640/660 deep-red fluorescent Nissl. For each chase period, all genotypes of animals were simultaneously processed in the same dish throughout all procedures.
HRP uptake and trafficking in garland cells
Third instar larvae were dissected in ice-cold PBS, equilibrated in a HL-3 buffer at room temperature, incubated in 0.7% HRP (Sigma-Aldrich grade VI) for 5 min at room temperature, rinsed thoroughly, and chased for 10 min at room temperature. Samples were fixed for 1 h at 4°C in 2% paraformaldehyde, 2% glutaraldehyde, and 1% tannic acid in 0.1 M cacodylic acid buffer, pH 7.2, and processed for DAB reaction as described previously (Kosaka and Ikeda, 1983). Garland cells were dissected out and processed for EM as described below.
Rescue of ema lethality
The fly Ema (UAS-GFP::Ema) and human hClec16A (UAS-GFP::hClec16A) transgenes were ubiquitously expressed by daughterless-Gal4 in the ema mutant. Survival rate (%) is the percentage of observed/expected number of progeny. Three independent crosses were performed for each rescue experiment. Data represent mean ± SEM (%).
Electron microscopy
As described previously (Daniels et al., 2004), samples were fixed for 1 h at 4°C in 2% paraformaldehyde, 2% glutaraldehyde, and 1% tannic acid in 0.1 M cacodylic acid buffer, pH 7.2. Then, the samples were postfixed in 1% OsO4 in 0.1 M cacodylic acid buffer, pH 7.2, for 1 h at room temperature, stained en bloc with 1% uranyl acetate, dehydrated in a grade series of ethanol and propylene oxide, and embedded in epon resin (Electron Microscopy Sciences). Blocks were sectioned in an ultramicrotome (RMC Products) at ∼70-nm thickness with a Delaware Diamond knife and post-stained for 1 h in Reynolds lead citrate and uranyl acetate. Electron micrographs were taken on a transmission electron microscope (H-7500; Hitashi).
Imaging and analysis
Confocal images were acquired with a confocal microscope (model C1; Nikon) and accompanying EZ-C1 software using argon (excitation at 488 nm) and HeNe (excitation at 543 and 633 nm) lasers and a 60x Plan-Apochromat NA 1.4 objective (Nikon) at room temperature. Samples for each experiment were processed using the same confocal gain setting, unless otherwise specified in figure legends. All quantifications were performed while blinded to genotype. ImageJ (National Institutes of Health, Bethesda, MD) was used for all quantitative manipulation and analysis of images. In the analysis of bouton number, synaptic area and synaptic levels of DVGLUT, pMAD, and TKV::GFP, confocal sections of similar sizes of muscle 4 on A3/A4 hemisegments were maximally projected. In the analysis of DVGLUT aggregation index (A.I.), the product of average area and average intensity of DVGLUT aggregates was derived from individual motoneurons and pooled together. Garland cells were identified according to bright HRP staining, two nuclei, and location near the esophagus and proventriculus. All chase periods of images in avidin-Cy3 uptake and trafficking experiments were acquired at the same gain setting at which the avidin-Cy3 in mutant garland cells at a 30-min chase period was minimally saturated. For the line-plot density analysis of avidin-Cy3 tracer in garland cells, images were taken at the plane where either nucleus was clearly seen. A line density profile was measured along the longest axis from cell surface to the center of the cell. The line width was set as 50 px (5.3 µm) because it is wide enough to average multiple tracers while minimizing interference from neighboring tracers moving in different directions. All images were processed with Photoshop CS3 and Illustrator CS3 software (Adobe).
Statistics
Results are represented as mean ± SEM. Statistical tests were performed with Origin 7.0 software (Origin Laboratory). Groups of means were compared using one-way ANOVA, and comparisons between two means were performed using Student’s t test. *, P < 0.05; **, P < 0. 01; ***, P < 0.001; ns, not significant.
GenBank accession number
The GenBank accession no. for the ema cDNA sequence is NM_001038969.1.
Online supplemental material
Fig. S1 shows defective DVGLUT trafficking in the ema mutant motoneurons. Fig. S2 shows how DVGLUT aggregates associate with the endosomal proteins DOR and Vps16A in the ema mutant motoneurons. Fig. S3 shows MARCM analysis of ema in the Drosophila wing. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200911126/DC1.
Supplementary Material
Acknowledgments
We thank H. Kramer, M. González-Gaitán, H.J. Bellen, G.W. Davis, P. ten Dijke, K. Moses, G. Warren, V. Budnik, J.C. Sisson, D.E. Krantz, K.E. Zinsmaier, J.B. Duffy, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for antibodies and flies. We thank P.I. Hanson, C. Micchelli, and T.J. Baranski for advice and discussion on the project and for comments on the manuscript. We thank S. Kornfeld for comments on the manuscript. We thank the Inner Ear Research Core Center at Washington University for use of their electron microscopy facility and H. Wynder for help with EM sample preparation. We thank X. Sun and S.L. Johnson for excellent technical assistance, and members of the laboratory for critical discussions.
This work was supported by the Juvenile Diabetes Research Foundation and the National Institutes of Health (DA 020812) to A. DiAntonio.
Footnotes
Abbreviations used in this paper:
- DVGLUT
- Drosophila vesicular glutamate transporter
- ESCRT
- endosomal sorting complex required for transport
- NMJ
- neuromuscular junction
- Tkv
- thick veins
References
- Awata T., Kawasaki E., Tanaka S., Ikegami H., Maruyama T., Shimada A., Nakanishi K., Kobayashi T., Iizuka H., Uga M., et al. ; Japanese Study Group on Type 1 Diabetes Genetics. 2009. Association of type 1 diabetes with two Loci on 12q13 and 16p13 and the influence coexisting thyroid autoimmunity in Japanese. J. Clin. Endocrinol. Metab. 94:231–235. 10.1210/jc.2008-0718 [DOI] [PubMed] [Google Scholar]
- Banta L.M., Robinson J.S., Klionsky D.J., Emr S.D.. 1988. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107:1369–1383. 10.1083/jcb.107.4.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bökel C., Schwabedissen A., Entchev E., Renaud O., González-Gaitán M.. 2006. Sara endosomes and the maintenance of Dpp signaling levels across mitosis. Science. 314:1135–1139. 10.1126/science.1132524 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Rojas R.. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 7:568–579. 10.1038/nrm1985 [DOI] [PubMed] [Google Scholar]
- Budnik V., Koh Y.H., Guan B., Hartmann B., Hough C., Woods D., Gorczyca M.. 1996. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron. 17:627–640. 10.1016/S0896-6273(00)80196-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A., DiAntonio A.. 2007. Synaptic development: insights from Drosophila. Curr. Opin. Neurobiol. 17:35–42. 10.1016/j.conb.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Daniels R.W., Collins C.A., Gelfand M.V., Dant J., Brooks E.S., Krantz D.E., DiAntonio A.. 2004. Increased expression of the Drosophila vesicular glutamate transporter leads to excess glutamate release and a compensatory decrease in quantal content. J. Neurosci. 24:10466–10474. 10.1523/JNEUROSCI.3001-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S., Miller K., Moehren G., Ullrich A., Schlessinger J., Hopkins C.R.. 1990. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 61:623–634. 10.1016/0092-8674(90)90474-S [DOI] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H.. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317–323. 10.1038/nrm1360 [DOI] [PubMed] [Google Scholar]
- Hakonarson H., Grant S.F., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C., et al. 2007. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 448:591–594. 10.1038/nature06010 [DOI] [PubMed] [Google Scholar]
- Hicke L., Dunn R.. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141–172. 10.1146/annurev.cellbio.19.110701.154617 [DOI] [PubMed] [Google Scholar]
- Huizing M., Helip-Wooley A., Westbroek W., Gunay-Aygun M., Gahl W.A.. 2008. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu. Rev. Genomics Hum. Genet. 9:359–386. 10.1146/annurev.genom.9.081307.164303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium. 2007. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 357:851–862. 10.1056/NEJMoa073493 [DOI] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium (IMSGC). 2009. The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 10:11–14. 10.1038/gene.2008.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D.. 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 124:761–771. [DOI] [PubMed] [Google Scholar]
- Jeyakumar M., Dwek R.A., Butters T.D., Platt F.M.. 2005. Storage solutions: treating lysosomal disorders of the brain. Nat. Rev. Neurosci. 6:713–725. [DOI] [PubMed] [Google Scholar]
- Kinchen J.M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C.D., Hengartner M.O., Ravichandran K.S.. 2008. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10:556–566. 10.1038/ncb1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter T., Sandhoff K.. 2005. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 21:81–103. 10.1146/annurev.cellbio.21.122303.120013 [DOI] [PubMed] [Google Scholar]
- Korolchuk V.I., Schütz M.M., Gómez-Llorente C., Rocha J., Lansu N.R., Collins S.M., Wairkar Y.P., Robinson I.M., O’Kane C.J.. 2007. Drosophila Vps35 function is necessary for normal endocytic trafficking and actin cytoskeleton organisation. J. Cell Sci. 120:4367–4376. 10.1242/jcs.012336 [DOI] [PubMed] [Google Scholar]
- Kosaka T., Ikeda K.. 1983. Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J. Cell Biol. 97:499–507. 10.1083/jcb.97.2.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U.. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10:776–787. 10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd T.E., Atkinson R., Wu M.N., Zhou Y., Pennetta G., Bellen H.J.. 2002. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 108:261–269. 10.1016/S0092-8674(02)00611-6 [DOI] [PubMed] [Google Scholar]
- Luzio J.P., Pryor P.R., Bright N.A.. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632. 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37(Database issue):D205–D210. 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrus S.B., Portman S.L., Allen M.J., Moffat K.G., DiAntonio A.. 2004. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J. Neurosci. 24:1406–1415. 10.1523/JNEUROSCI.1575-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F.R., McGraw T.E.. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- Mosesson Y., Mills G.B., Yarden Y.. 2008. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer. 8:835–850. 10.1038/nrc2521 [DOI] [PubMed] [Google Scholar]
- Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T.E., Warren G.. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715–1726. 10.1083/jcb.131.6.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D.P., Brett C.L., Merz A.J.. 2009. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 21:543–551. 10.1016/j.ceb.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor-Giles K.M., Ho L.L., Ganetzky B.. 2008. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 58:507–518. 10.1016/j.neuron.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounissi-Benkalha H., Polychronakos C.. 2008. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol. Med. 14:268–275. 10.1016/j.molmed.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Peplowska K., Markgraf D.F., Ostrowicz C.W., Bange G., Ungermann C.. 2007. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell. 12:739–750. 10.1016/j.devcel.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Persson U., Izumi H., Souchelnytskyi S., Itoh S., Grimsby S., Engström U., Heldin C.H., Funa K., ten Dijke P.. 1998. The L45 loop in type I receptors for TGF-beta family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 434:83–87. 10.1016/S0014-5793(98)00954-5 [DOI] [PubMed] [Google Scholar]
- Piper R.C., Katzmann D.J.. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519–547. 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupon V., Stewart A., Gray S.R., Piper R.C., Luzio J.P.. 2003. The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles. Mol. Biol. Cell. 14:4015–4027. 10.1091/mbc.E03-01-0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S., Akbar M.A., Ray S., Sevrioukov E.A., Haberman A.S., Rohrer J., Krämer H.. 2005. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 118:3663–3673. 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Richardson S.C., Winistorfer S.C., Poupon V., Luzio J.P., Piper R.C.. 2004. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell. 15:1197–1210. 10.1091/mbc.E03-06-0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Rocha N., Neefjes J.. 2008. MHC class II molecules on the move for successful antigen presentation. EMBO J. 27:1–5. 10.1038/sj.emboj.7601945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio J.P., Stankovich J., Field J., Tubridy N., Marriott M., Chapman C., Bahlo M., Perera D., Johnson L.J., Tait B.D., et al. 2008. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun. 9:624–630. 10.1038/gene.2008.59 [DOI] [PubMed] [Google Scholar]
- Sato T.K., Rehling P., Peterson M.R., Emr S.D.. 2000. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell. 6:661–671. 10.1016/S1097-2765(00)00064-2 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Eitzen G., Margolis N., Wickner W.T., Price A.. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 97:9402–9407. 10.1073/pnas.97.17.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E.S., Bellen H.J., Lloyd T.E.. 2002. When cell biology meets development: endocytic regulation of signaling pathways. Genes Dev. 16:1314–1336. 10.1101/gad.989602 [DOI] [PubMed] [Google Scholar]
- Sevrioukov E.A., He J.P., Moghrabi N., Sunio A., Krämer H.. 1999. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell. 4:479–486. 10.1016/S1097-2765(00)80199-9 [DOI] [PubMed] [Google Scholar]
- Sisson J.C., Field C., Ventura R., Royou A., Sullivan W.. 2000. Lava lamp, a novel peripheral Golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. 151:905–918. 10.1083/jcb.151.4.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinningsrud B., Husebye E.S., Pearce S.H., McDonald D.O., Brandal K., Wolff A.B., Løvås K., Egeland T., Undlien D.E.. 2008. Polymorphisms in CLEC16A and CIITA at 16p13 are associated with primary adrenal insufficiency. J. Clin. Endocrinol. Metab. 93:3310–3317. [DOI] [PubMed] [Google Scholar]
- Sriram V., Krishnan K.S., Mayor S.. 2003. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J. Cell Biol. 161:593–607. 10.1083/jcb.200210166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Parton R.G., Steele-Mortimer O., Lütcke A., Gruenberg J., Zerial M.. 1994. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri A., Levisetti M.G., Unanue E.R.. 2008. Do the peptide-binding properties of diabetogenic class II molecules explain autoreactivity? Curr. Opin. Immunol. 20:105–110. 10.1016/j.coi.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney S.T., Davis G.W.. 2002. Unrestricted synaptic growth in spinster-a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron. 36:403–416. 10.1016/S0896-6273(02)01014-0 [DOI] [PubMed] [Google Scholar]
- Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287. 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Todd J.A., Walker N.M., Cooper J.D., Smyth D.J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S.F., Payne F., et al. Genetics of Type 1 Diabetes in Finland; Wellcome Trust Case Control Consortium. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39:857–864. 10.1038/ng2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G., Wubbolts R., Stoorvogel W.. 2008. Endosomal sorting of MHC class II determines antigen presentation by dendritic cells. Curr. Opin. Cell Biol. 20:437–444. 10.1016/j.ceb.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Wagh D.A., Rasse T.M., Asan E., Hofbauer A., Schwenkert I., Dürrbeck H., Buchner S., Dabauvalle M.C., Schmidt M., Qin G., et al. 2006. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 49:833–844. 10.1016/j.neuron.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Wang X., Shaw W.R., Tsang H.T., Reid E., O’Kane C.J.. 2007. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat. Neurosci. 10:177–185. 10.1038/nn1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weavers H., Prieto-Sánchez S., Grawe F., Garcia-López A., Artero R., Wilsch-Bräuninger M., Ruiz-Gómez M., Skaer H., Denholm B.. 2009. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature. 457:322–326. 10.1038/nature07526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 447:661–678. 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersdorff V., Lecuit T., Cohen S.M., Mlodzik M.. 1996. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development. 122:2153–2162. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Hinz U., Engelbert M., Knust E.. 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 82:67–76. 10.1016/0092-8674(95)90053-5 [DOI] [PubMed] [Google Scholar]
- Wu X., Zhu X., Wang X., Ma J., Zhu S., Li J., Liu Y.. 2009. Intron polymorphism in the KIAA0350 gene is reproducibly associated with susceptibility to type 1 diabetes (T1D) in the Han Chinese population. Clin. Endocrinol. (Oxf.). 71:46–49. 10.1111/j.1365-2265.2008.03437.x [DOI] [PubMed] [Google Scholar]
- Wucherpfennig T., Wilsch-Bräuninger M., González-Gaitán M.. 2003. Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J. Cell Biol. 161:609–624. 10.1083/jcb.200211087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A.E., Sato T.K., Emr S.D.. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151:551–562. 10.1083/jcb.151.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Rubin G.M.. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237. [DOI] [PubMed] [Google Scholar]
- Yao K.M., White K.. 1994. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J. Neurochem. 63:41–51. [DOI] [PubMed] [Google Scholar]
- Zhang J., Schulze K.L., Hiesinger P.R., Suyama K., Wang S., Fish M., Acar M., Hoskins R.A., Bellen H.J., Scott M.P.. 2007. Thirty-one flavors of Drosophila rab proteins. Genetics. 176:1307–1322. 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoledziewska M., Costa G., Pitzalis M., Cocco E., Melis C., Moi L., Zavattari P., Murru R., Lampis R., Morelli L., et al. 2009. Variation within the CLEC16A gene shows consistent disease association with both multiple sclerosis and type 1 diabetes in Sardinia. Genes Immun. 10:15–17. 10.1038/gene.2008.84 [DOI] [PubMed] [Google Scholar]
- Zoncu R., Perera R.M., Balkin D.M., Pirruccello M., Toomre D., De Camilli P.. 2009. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 136:1110–1121. 10.1016/j.cell.2009.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.