Abstract
Protein therapeutics made up of artificially combined proteins or protein domains, so called fusion proteins, are a novel and growing class of biopharmaceuticals. We have studied abatacept (Orencia®), a fusion protein that is constructed of a modified IgG Fc domain and the soluble part of the T-cell receptor CTLA-4. In accelerated degradation studies conducted at at 40 °C, a pH shift from 7.5 to 6.0 yields significantly faster aggregation kinetics, as measured by size-exclusion chromatography. To understand how the fusion domains and their interactions contribute to this result, we considered aggregation in light of the modified Lumry-Eyring reaction pathway. Protein conformational stabilities against chaotropes and temperature were measured. The structural consequences of these perturbations were observed by a variety of experimental techniques, including differential scanning calorimetry, circular dichroism, and intrinsic fluorescence. Abatacept’s colloidal stability was studied by measuring zeta potentials and osmotic second virial coefficients, as well as by modeling electrostatic potentials on the protein’s surface. The domains of abatacept exhibit different conformational stabilities that are highly pH dependent, whereas abatacept was weakly colloidally unstable at pH 6 or pH 7.5. These results are ascribed to conformational instability of the CTLA-4 and CH2 domains, which unfold to form a molten globule-like structure that is aggregation-prone. We suggest the instability against aggregation is determined by the least stable domains.
Protein aggregation is currently a topic of widespread interest, both because of its implication in a number of human disease states (more than 50 diseases are associated with protein misfolding/aggregation, including Alzheimers’, Parkinson’s and Huntington’s diseases (1)) and because of the challenges that it presents to the biotherapeutic industry (2). Aggregates within therapeutic protein formulations are a safety concern because of their potential for immunogenicity and altered efficacy in patients (3). Aggregation of therapeutic proteins is costly, requiring additional recovery and purification steps, reducing production yields and shortening shelf life (4, 5).
Recent theoretical and experimental advances have lead to a greater understanding of the mechanisms by which proteins may aggregate (4, 6–9). Using the framework of the Lumry-Eyring model (10), protein aggregation has been explained as requiring an initial (often reversible) unimolecular conformational change to an aggregation-competent, partially unfolded species, which then goes on to react with other protein molecules to form an aggregate, that can continue to grow (6).
In the Lumry-Eyring framework, the observed reaction orders depend on which of the various steps that is rate-limiting. In one limiting case, a first-order dependence on protein concentration is observed if the rate of unfolding to form the aggregation-competent species is slow. Alternatively, second-order kinetics may be observed if the association step in the second part of the reaction is rate limiting. In solutions where the protein is colloidally stable (e.g., at pH values removed from the isoelectric point, pI), k2 may be greatly reduced. Typically, neither conformational stability nor colloidal stability dominate, and the observed reaction order is found to be concentration-dependent and variable between the those expected for the limiting cases.
For proteins that are composed of multiple domains, the interpretation of conformational stability is more complicated. In such cases, unfolding is frequently a multi-step process, and individual domains may have different conformational stabilities and kinetics (11–13), and presumably different colloidal stabilities (14). If a partially unfolded state is involved in the aggregation reaction, we expect that the stability of the least stable domain will dominate the conformational stability contributions to aggregation. In one case, mutations lowering only the stability of the CH2 domain of a therapeutic antibody resulted in increased aggregation rates (15). Solution conditions increasing the surface tension (e.g. adding citrate and sucrose) stabilized the CH2 domain and lowered the aggregation rate (16). In another case, an fusion protein constructed from human growth hormone and human albumin had its agitation-induced aggregation rate decreased when the domain with the lower stability, the growth hormone part, was stabilized by non-ionic surfactants (17).
Therapeutic fusion proteins are a type of multidomain protein of particular current interest as biotherapeutics (18). Fusion proteins are constructed by splicing two or more proteins or protein domains to obtain new non-natural polypeptides with combined functionalities of the parent proteins. Therapeutic fusion proteins may exhibit advantages such as connecting desired, but previously non-coupled functions; increasing half-life, and increasing biological activity (18, 19).
In naturally-occuring multidomain proteins such as antibodies, the protein structure is stabilized by both inter- and intradomain interactions (20–22). For example, complete (IgG1) antibodies often show conformational stabilities that are intermediate between those of the CH2 and CH3 subdomains of isolated Fc domains, and close to the stability of the isolated Fab domains (23–26). In contrast, because artificial fusion proteins have not co-evolved, we expect that interdomain interactions might not contribute significantly to overall conformational stability, and might even serve to destabilize such a construct. Likewise, we expect that the overall colloidal stability of an artificial fusion protein might be lower than that of a naturally-occurring multidomain protein such as an antibody.
To investigate the conformational and colloidal stability and the domain influence on aggregation in a fusion protein system, we have studied abatacept (Orencia®, Bristol-Myers Squibb, New York, USA), an FDA-approved therapeutic fusion protein for treatment of Rheumatoid Arthritis. Abatacept is a fusion protein consisting of the soluble extra-cellular domain of human CTLA-4 (cytotoxic T lymphocyte-associated molecule-4) receptor that is expressed by activated T cells fused to a modified version of the Fc domain of human immunoglobulin G1, IgG1 (27). Abatacept is a first-in-class drug for selective costimulation modulators, designed to block a key costimulatory signal (CD80/CD86) required for T-cell activation (28).
Structurally, abatacept is a glycosylated fusion protein with a MALDI-MS molecular weight of 92,300 Da and it is a homodimer of two homologous polypeptide chains of 357 amino acids each (for amino acid sequence and structural model see Figure 1 and Figure 2, respectively). An intermolecular disulphide bond between the CTLA-4 domains of the fusion protein links the two chains. Three cysteines and one proline in the hinge-region of the Fc domain are mutated to serines, thus abolishing the native covalent Fc-chain connection as well as its complement-dependent cytotoxicity (CDC) or antibody-dependent cellular cytotoxicity (ADCC) properties (29). In addition, the CTLA-4 domain and the Fc domain each have 4 intramolecular disulphide bridges. In the dimer there are 4 N-linked glycolysation sites in the CTLA-4 domain and 2 in the Fc domain. Potentially there is an O-linked glycosylation on the P139S mutation site in Fc domain (29). Heterogenous glycosylation gives a pI of 4.5–5.5. Abatacept has 8 tryptophan residues, all exclusively located in the Fc domain, and 30 tyrosine residues distributed over both domains.
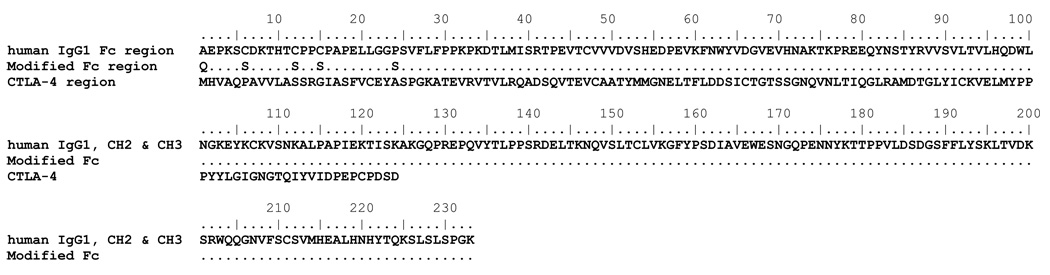
Figure 1.
Amino acid sequences of the CTLA-4 and the modified Fc regions of abatacept and sequence comparasion to the Fc region of human IgG1 (starting at amino acid 125) from the PDB file 1HZH. Dots indicate identical amino acids.
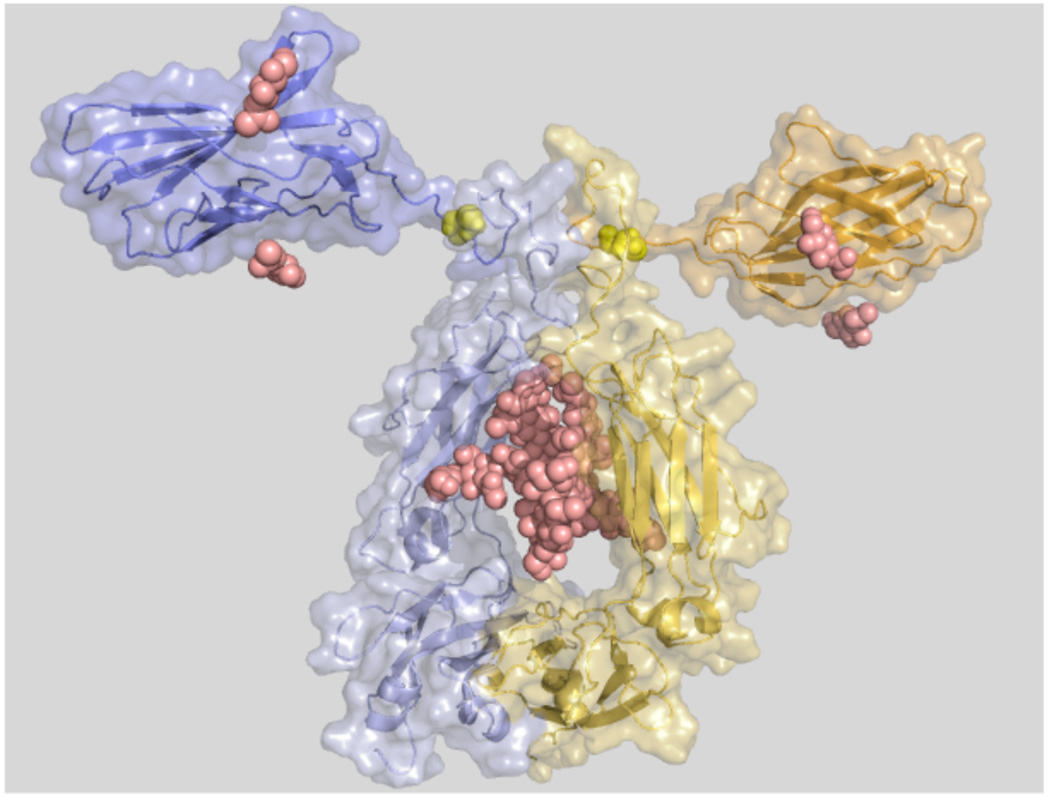
Figure 2.
Structural model of abatacept. Blue and yellow cartoon and surface representations for each chain. Small domains (top) are the extracellular soluble domains of human CTLA-4. Large domains are the CH2 and CH3 domains in the modified Fc part of human IgG1. Yellow spheres are Cys120 in CTLA-4 that makes a disulphide connection between the chains. Sugar residues are represented as pink spheres. The amino acid sequence of one chain of abatacept was sent to the Robetta server (http://robetta.bakerlab.org/) for structure prediction. Robetta models were aligned in PyMOL (http://pymol.org/) with the heavy chains of IgG1 from 1HZH to build a dimer. To create the final dimer model, the CTLA-4 domain predicted by Robetta was replaced by the actual crystal structure of CTLA-4 (PDB id 1I8L) by aligning the crystal structure with each of the modeled domains in the dimer model. Graphics were made in PyMOL.
To probe the roles of abatacept’s two domains in determining its overall solution stability, we conducted “accelerated degradation studies” to determine the stability of abatacept against aggregation at elevated storage temperatures (40 °C) and in various solution conditions (pH 6.0 and 7.5). We then used a variety of physical and spectroscopic techniques to elucidate the contributions of the two domains to that stability. In order to probe the conformational stability of abatacept and its domains, chaotrope- and thermally-induced structural perturbations were monitored using absorption and fluorescence spectroscopy, circular dichroism (CD), differential scanning calorimetry (DSC), and ANS binding. Colloidal stability was measured by static light scattering (SLS) and the zeta potential.
Materials and Methods
All chemicals and reagents were of highest laboratory purity and obtained from Fisher Scientific (Pittsburg, PA) and Sigma-Aldrich (St Louis, MO) unless otherwise indicated. Abatacept (CTLA4-Ig - Orencia®, Bristol Myers Squibb, New York, USA) was purchased (University of Colorado at Boulder Wardenburg Pharmacy) as a lyophilized powder in vials containing 250 mg protein, 500 mg maltose, 17.2 mg monobasic sodium phosphate, and 14.1 mg NaCl. CTLA-4 extracellular soluble domain (further only called CTLA-4) expressed in E. coli was donated by Barofold Inc. (Boulder, CO).
Dialysis and Sample Preparation
One vial of lyophilized abatacept formulation was dissolved in 10 mL USP water. Exipients were removed by extensive dialysis against millipore water for >10h at room temperature with at least two water changes. The protein was further dialyzed into a buffer using slide-a-lyzer dialysis cassettes (MWCO 10,000 Da, Thermo-Fisher) with 2–3 buffer exchanges over >10h at room temperature. Purity and no significant protein loss were confirmed by SDS-PAGE and UV absorption. Protein stock solutions at protein concentrations of typically 20–25 mg/mL were stored at 4 °C and showed no aggregation (samples contain <2% dimer fraction that does not change over time) over more than three months as monitored by analytical ultracentrifugation and SEC-HPLC. Samples were filtered through a 0.22 µm PVDF syringe filter (Whatman) prior to experiments.
The ionization heat of sodium phosphate buffer is small, and thus, the pH of phosphate buffer is almost constant with rising temperature. Due to this and the similarity to the therapeutic formulation we used 10 mM mono/dibasic sodium phosphate, 25 mM NaCl, at pH 6 or 7.5. All buffers were filtered through a 0.22 µm PVDF syringe filter (Whatman) and degassed prior use.
Protein Concentration Determination
UV absorbance was measured using a Lambda 35 UV/VIS spectrophotometer (Perkin-Elmer, Waltham, MA). The extinction coefficient (ε280) for abatacept was calculated to be 1.0 mL mg−1 cm−1 by the +/− GdnHCl method by Pace et al. (30), which also was cross-correlated with the amount lyophilized protein in the vials. The extinction coefficient (ε280) for E. coli-expressed CTLA-4 was 0.6 mL mg−1 cm−1.
Accelerated Degradation Studies
For the reaction rate studies, a 5 mg/mL solution of abatacept in 10 mM sodium phosphate, 25 mM sodium chloride, 0.1 g/L sodium azide, pH 6 or pH 7.5, was aliquoted into 100 µL samples in microcentrifuge tubes (0.6 mL, Fisher). Samples were then incubated at room temperature (ca. 22 °C), or 40 °C. Three samples per condition were removed at each time point, centrifuged for five minutes to remove insoluble aggregates and analyzed on a Beckman Coulter System Gold 126 HPLC (Fullerton, CA) equipped with a with a Beckman Coulter 166 Detector, a Waters 717 autosampler (Milford, MA), and a TSK-Gel 3000SWXL size exclusion column with guard column (Tosoh Biosciences, Montgomeryville, PA). Run time was 40 minutes with a flow rate of 0.6 mL/min. Sample injection volumes were 40 µL and the mobile phase was 100 mM sodium phosphate, 300 mM sodium chloride, 0.1 g/L sodium azide, pH 7. Data were analyzed using Thermo Galactic GRAMS AI software (v. 7) and the concentration of remaining monomor was calculated according to the method outlined earlier (31, 32).
The initial rate of aggregation, measured at the starting protein concentration of 5 mg/mL, was calculated from triplicate measurements at 5 separate time points. These time points were chosen so that there was less than 10% conversion of the initial monomer to dimers and other multimers. At this limited conversion, all multimers were soluble and the observed total peak areas (monomer/dimer/multimer for pH 6 and monomer/dimer for pH 7.5) were equal to the initial peak area for the starting monomer (within error). No lag phase was observed before aggregation began.
Deglycosylation of Abatacept
N-linked deglycoslylation was performed with 1 unit PNGase F per 100 µg protein in 1 % n-octyl p-D-glucopyranoside (OG). Samples were incubated at 37 °C between 2 and 24h. Deglycosylation was monitored by isoelectric focusing (IEF) PAGE, MALDI-MS and ES-MS. After 5h incubation with enzyme, no further changes in IEF mobility were observed.
Chaotrope-induced Unfolding of Abatacept
Samples containing 0–6 M guanidine hydrochloride (GdnHCl) or 0–9.75 M urea were mixed from stock solutions of 17 mg/mL abatacept, buffer with 7 M GdnHCl or 10 M urea, and buffer. All stock solutions were at pH 6 or 7.5. Identical samples were mixed without protein for background measurements. Protein and background samples were incubated overnight at 4 °C before measurements. Reversibility was monitored by incubating a stock solution of 4.5 mg/mL abatacept in 8 M GdnHCl overnight at 4 °C. This stock solution was mixed with 8 M GdnHCl solution and buffer in proportions to yield samples in the range from 0.25 – 6.25 M GdnHCl. Those samples were further incubated at 4 °C overnight before fluorescence measurements at 25 °C.
Fluorescence Spectroscopy
Sample concentrations were 0.1 mg/mL abatacept in sodium phosphate buffer at pH 6 or 7.5. The protein solution was placed in a 5 mm pathlength square quartz cuvette thermostated at 25 °C on an Aminco Bowman Series 2 fluorescence-spectrophotometer (SLM Aminco, Urbana, IL). Samples were excited at 280 or 295 nm and emission was collected between 305 and 400 nm with excitation and emission slits were 4 nm and scan rate 50 nm/min. Background correction was performed, total fluorescence intensity and center of spectral mass (CSM) were calculated as:
| (1) |
where Fi is the fluorescence intensity at wave number νi, ΣFi is the spectral integral in the spectrum range. Triplicate samples were collected and fitting was performed as described below.
Fourth Derivative UV Absorption Spectroscopy
Fourth derivative UV absorption (4DUV) spectroscopy as a function of GdnHCl was carried out at 25 °C at pH 6 and pH 7.5 with samples prepared as above with a protein concentration of 0.5 mg/mL. Absorbance spectra between 260 and 305 nm were recorded in steps of 0.1 nm as a function of denaturant (averaging 3 scans per sample) using a Lambda 35 UV/VIS spectrophotometer (Perkin-Elmer, Waltham, MA). 4DUV spectra were determined as described (33). Transitions between spectral forms were quantified within the 275–290 nm range, typical of Tyr effects, by cumulative difference amplitude (CDA) as reported (34).
ANS Fluorescence
Freshly made 8-anilino-1-naphtalene-sulfonic acid (ANS) stock solutions (final concentration 100 µM) were mixed with protein and GdnHCl stock solutions to yield samples containing 0.1 mg/mL abatacept (ca. 1 µM), 100 µM ANS, and various concentrations of GdnHCl. Samples containing only ANS and GdnHCl were also prepared as background blanks for each point. Excitation wavelength was 350 nm and the emission wave length range was 400–600 nm with 4 nm slit widths. ANS fluorescence intensity was plotted at 480 nm as a function of GdnHCl after background spectra were subtracted from protein spectra.
Circular Dichroism
Far-UV circular dichroism (CD) spectra were obtained using a JASCO J-820 spectropolarimeter with a JASCO PTC-343 Peltier type thermostatted cell holder. Quartz cuvettes with 0.1 mm or 1 mm path length were used for spectra recorded between 195 and 250 nm. Near-UV CD spectra were recorded between 260 and 340 nm using a 10 × 10 mm cuvette. Abatacept and CTLA-4 concentrations were <24 µM and <2 µM, respectively, for far-UV CD and ~30 µM for abatacept for near-UV CD. The following settings were used: temperature, 10 °C; resolution, 1 nm; bandwidth, 1 nm; sensitivity, 20 mdeg; response time, 8 sec; accumulation, 6; and scan rate, 50 nm/min. Baseline spectra were recorded with pure buffer in the cuvette and subtracted from the spectra for protein solutions. Data are reported as molar ellipticity and was determined as:
| (2) |
where c is the protein concentration (mg/mL), l is the path length of the cell (cm), θλ is the measured ellipticity at wavelength λ (mdeg), Mw is the molecular mass of the protein (g/mol), and n is the number of residues.
Thermally-induced Unfolding of Abatacept
Fluorescence Spectroscopy
Thermally-induced unfolding of abatacept was performed on a Photon Technology International (PTI) spectrofluorometer (Lawrenceville, NJ) equipped with a turreted four-position Peltier-controlled cell holder and a xenon lamp. Sample concentrations were 0.1 mg/mL in sodium phosphate buffer at pH 6 and 7.5. The protein solution was placed in a 10-mm pathlength triangular quartz cuvette with a Teflon® cap. Tryptophan fluorescence was excited at 295 nm. Emission was collected between 305 and 400 nm with 4 nm slit widths and a scan rate of 50 nm/min. Thermally-induced denaturation was followed by a scan every 2.5 or 5 °C between 10–90 °C. After background correction, total fluorescence intensity was integrated and center of spectral mass were calculated as above. Triplicate samples were collected and fitting was performed as described below.
Circular Dichroism
Thermal melts were acquired for samples in phosphate buffer at pH 6 and pH 7.5 by monitoring the CD signal at 217 nm over the temperature range from 10 to 90 °C at intervals of 1 °C with a scanning rate of 1 °C/min; bandwidth, 2 nm; sensitivity, 20 mdeg; response time, 16 sec. Blank buffer scans were subtracted from protein scans and data was transformed to molar ellipticity as above. Fitting was performed as described below.
Differential Scanning Calorimetry
Calorimetric experiments were performed on a VP-DSC (Microcal LLC, Northampton, MA) with an active cell volume of 0.5 mL at 1.5 atm over-pressure using scanning rates of 15, 30, 60 or 90 °C/min. Dialysed and degassed samples with a protein concentration of 0.5–11 mg/mL were monitored from 5–115 °C in phosphate buffer. All transition midpoint temperatures are apparent as the system was not fully reversible under any experimental conditions used. Specifically, the post-transition baseline did not reach a steady baseline level and there was no or only minor transition/excess enthalpy on repetitive runs on the same sample. Visual inspection of each sample following heat denaturation showed clear solutions for phosphate-buffered solutions. Buffer scans were subtracted from the sample scans followed by normalization for protein concentration and baseline correction using standard procedures in Origin v7.0 (Microcal LLC, Northampton, MA). Resulting apparent excess heat capacity thermograms were non-lineary fitted to a non-2 state model with 2 or 3 transitions in Origin v7.0.
Activation energies for abatacept structural transitions
Different scanning rates (v = 15, 30, 60 and 90 °C/hour) were employed to estimate the activation energy (Ea) for each thermally-induced structural transition of abatacept, using the approach described by Sanchez-Ruiz (35), where activation energies were estimated from the slope of a plot of ln(ν/Tm) vs. 1/Tm using eq. 3:
| (3) |
where A is a pre-exponential factor the Arrhenius equation, Ea is the activation energy, R is the gas constant, ν is defined as the scan rate, and Tm is the mid point transition temperature.
Data Analysis of Unfolding Curves
The CD, fluorescence and absorbance denaturation data were analyzed using the software Kaleidagraph (Synergy Software, Reading, PA), Sigmaplot 11 (Systat Software Inc., San Jose, CA), or Origin v7.0 (Microcal LLC, Northampton, MA). Denaturation data for each transition were fitted to two-state folding model using a non-linear least square method to determine thermodynamic parameters (36, 37):
| (4) |
where Y0 is the response signal, kN, bN, kU, and bU are the intercepts and the slopes of the baselines of the native and the unfolded states (or more specifically, the intermediate state in the case of the first transition’s end state and the second transition’s start state), respectively, and are assumed to be linear, ΔGNU is the free energy of unfolding, R is the molar gas constant, and T is temperature. The free energy expressions for the different perturbations are outlined below:
Chaotrope-induced Unfolding
| (5) |
where [D] is the chaotrope concentration; ΔGNU(H2O) is the unfolding free energy in buffer without denaturant; m is the influence of denaturant concentration on the stability and is related to the change in surface area and heat capacity change (38).
Thermally-induced Unfolding
| (6) |
where Tm is the apparent transition midpoint, and ΔHNU is the enthalpy difference between the two states at Tm. ΔCp is the heat capacity difference between unfolded and native protein. The Tm values are not sensitive to the value of ΔCp.
Light Scattering
Static light scattering was used to estimate the second virial coefficient (SVC), representing overall attractive or repulsive solution specific interactions, as a measure of the collodial stability. Samples consisted of protein concentrations ranging 0.5 – 5 mg/mL in 10 mM sodium phosphate, 25 mM NaCl, pH 6 or pH 7.5 and the temperature was controlled at 24 °C. An Electro-Optics laser model 1145AP (Hsintien City, Taiwan), a Brookhaven Instruments goniometer and cascade photodiode detector model BI-200SM and BI-APD (Holtsville, NY) respectively, were used to determine the excess Rayleigh ratios at a 90° angle (scattering due to protein only) to the incident 633 nm light beam. The relationship used to determine the osmotic second virial coefficient is given here and is derived from the virial expansion of the ideal osmotic pressure equation (39).
| (7) |
Where Mw is the mass-averaged molecular weight, R90 is the excess Rayleigh ratio at 90° (39) and the optical constant K is described by:
| (8) |
where n0 is refractive index of the solvent, dn/dc is the refractive index increment and λ is the wavelength of the incident beam. A refractive index increment of 0.185 mL/g, estimated from a weight averaged contribution from the protein and carbohydrate portions of the monoclonal antibody, was used for all light scattering analysis (40).
Dynamic Light Scattering
For the dynamic light scattering a Malvern Zetasizer Nano ZS (Malvern, UK) was used. Protein concentration was 5 mg/mL in 10 mM sodium phosphate, 25 mM NaCl, pH 6 or pH 7.5. The resulting correlation functions were used to determine the diffusion coefficients from which the hydrodynamic diameters were calculated by the Stokes equation. The temperature for all light scattering measurements was controlled at 25 °C. The hydrodynamic diameter of abatacept determined by DLS (10.2 ± 0.2 nm) did not vary significantly with the solvent conditions used in this study.
Zeta Potentials
A Malvern Zetasizer Nano ZS (Malvern, UK) was used to measure the electrophoretic mobility of the antibody via laser Doppler velocimetry. The zeta potential is calculated from Henry’s equation using the Smoluchoski approximation which is valid for ionic strengths above 1 mM (41).
| (9) |
Where µe is the electrophoretic mobility, ε is the dielectric constant or permittivity of the solution, ks is a model-based constant which from the Smoluchoski approximation is 1.5 and ζ is the zeta potential. 5 mg/mL of abatacept in 10 mM sodium phosphate, 25 mM NaCl, pH 6 or pH 7.5, was used for all samples. The measurement was repeated on three samples at 25 °C and the errors are reported as the standard deviation.
The effective charge of an equivalent sphere can be estimated via the linearized Poisson-Boltzman equation, also referred to as the Debye-Hückel approximation, and is given by eq. 10 (42).
| (10) |
Where Z is the effective charge, rp is the effective sphere radius, κ is the inverse electric double layer thickness or inverse Debye length and e is the elementary charge.
The electrostatic contribution to the osmotic second virial coefficient, also referred to as the Donnan term can be calculated using eq. 11 (43).
| (11) |
Here SVCelectrostatic is the electrostatic component of the second virial coefficient, Mw is the actual molecular weight of the protein, ρs is the solvent density and mions is the molal concentration of ions.
Results
Aggregation Reaction Rates from Accelerated Degradation Studies of Abatacept
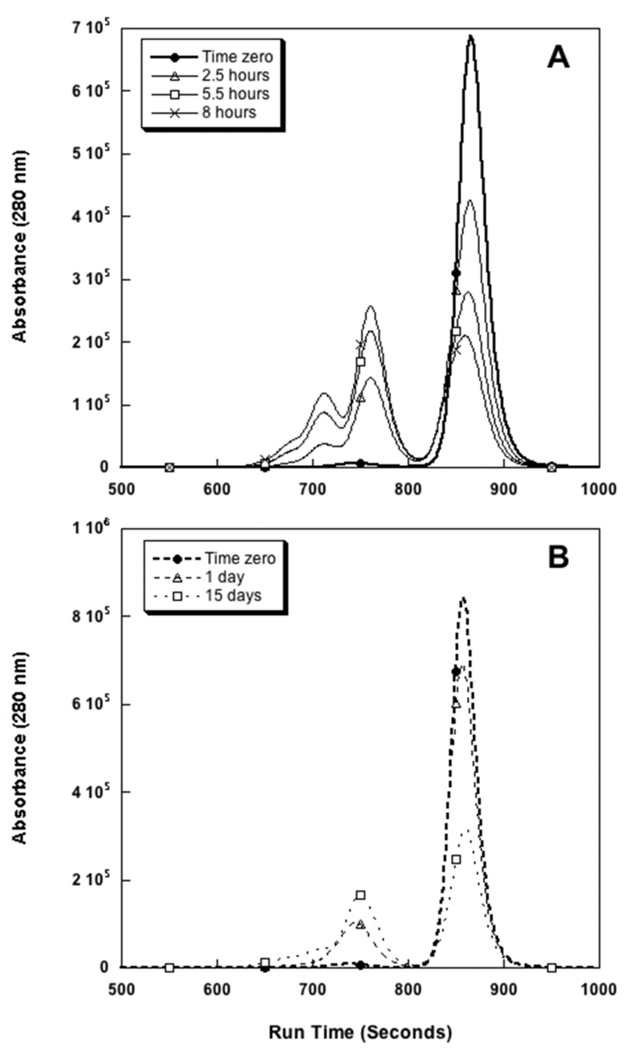
Aggregation reaction rates were measured after incubation at 40 °C at either pH 6 or pH 7.5. Figures 3A and 3B correspond to the 40 °C incubation experiments at pH 6 and pH 7.5, respectively. From these chromatograms it is apparent that the aggregation of abatacept proceeds much more rapidly at the lower pH. After only 8 hours of incubation at pH 6, 40 °C, half of the monomer has associated into higher order oligomers. For pH 7.5, 15 days of incubation was needed to see the same decrease in the monomer peak. The rate of monomer loss (subsequently referred to as the aggregation rate) is two orders of magnitude faster at pH 6 than at pH 7.5 (800 µg/(mL h) at pH 6 versus 6 µg/(mL h) at pH 7.5). At room temperature (ca. 23 °C), less than 1 % loss of monomer was seen over the course of one month at pH 7.5 and less than 2 % of initial monomer was lost over the same period at pH 6.0 (data not shown). A fraction of dimer corresponding to less than 2% of the total protein is present after dissolving the lyophilized protein.
Figure 3.
Accelerated stability studies of abatacept at 40 °C followed by SEC-HPLC at A. pH 6 (solid) and B. pH 7.5 (dashed). Ca. 50 % monomer loss is reached after 8h and 15 days at pH 6 and pH 7.5, respectively.
In studies conducted at 40 °C, the majority of higher order complexes are soluble dimers and trimers, as seen by SEC-HPLC and cross-validated with sedimentation velocity analytical ultracentrifugation (data not shown, see supplemental results for more details). Reducing and non-reducing polyacrylamide gel electrophoresis show that the aggregates are non-covalent (data not shown).
Reversibility of Abatacept Aggregates
The reversibility of abatacept aggregation was briefly examined. Samples were incubated at 40 °C in pH 6 buffer for 5.5 hours. Half of the samples were analyzed immediately following incubation while the remaining samples were stored at 4 °C for 24 hours prior to analysis. The sample stored at 4 °C prior to analysis shows an increase in the monomer peak area of ca. 18 %, compared to samples analyzed immediately, as well as a decrease in the oligomeric peaks (Supplementary figure 1). This indicates the aggregates may be at least partially reversible.
Conformation of Abatacept and E.coli expressed CTLA-4 at pH 6 and 7.5
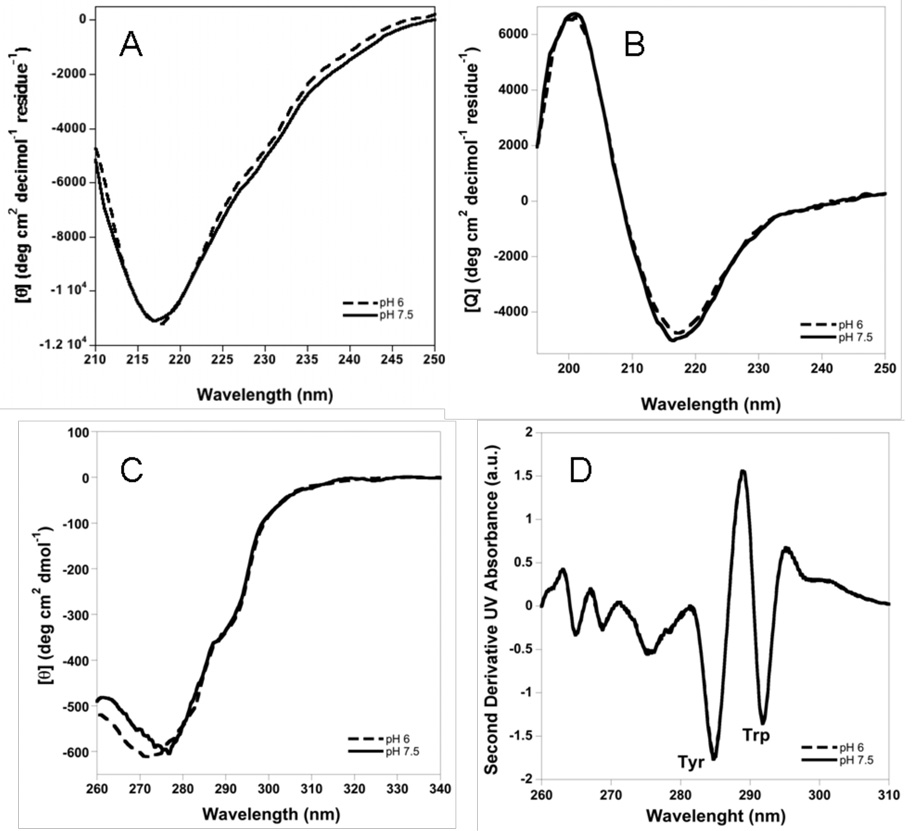
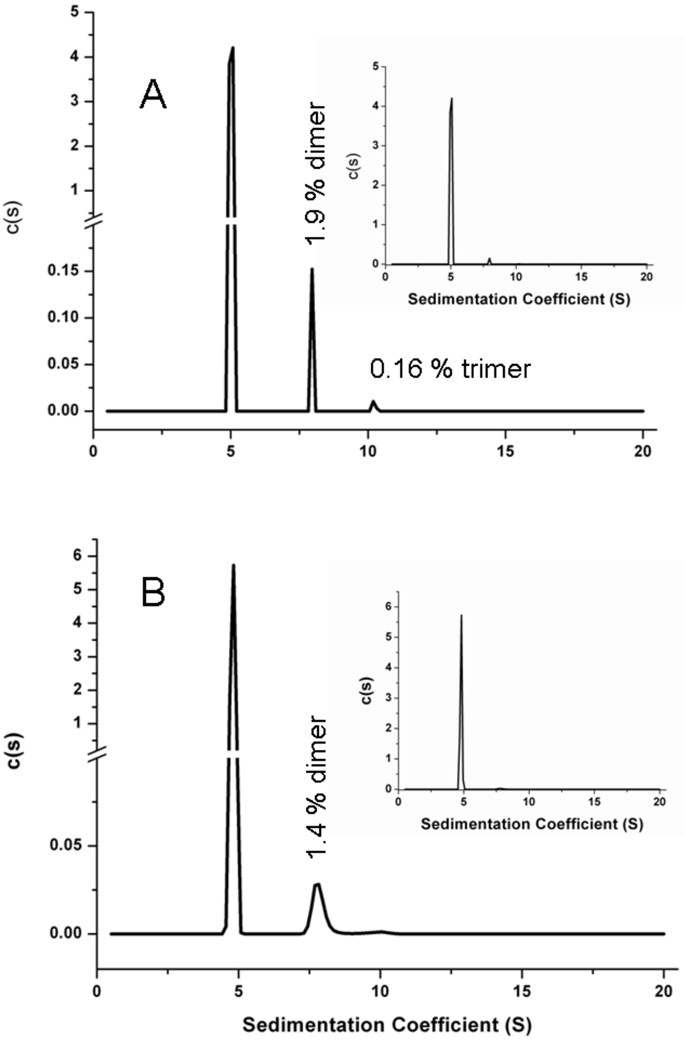
Structural features of abatacept at pH 6 and 7.5 were monitored using circular dichroism (CD), absorbance, SEC-HPLC, gels, and SV-AUC. Far-UV CD spectra at 10 °C at pH 6 and 7.5 are essentially identical and show a single minimum at 217 nm, a signature typical for β-sheet-rich proteins (Figure 4A). Near-UV CD spectra at 10 °C show differences below 280 nm, with a minimum of ca. 272 nm at pH 6 and ca. 277 nm at pH 7.5 (Figure 4C), whereas second derivative UV absorbance spectra at 25 °C at pH 6 and 7.5 completely overlap (Figure 4D). This implies that abatacept has a very similar secondary structure at the two pH conditions tested with some differences in the environment around aromatic residues. Additionally, SEC-HPLC, SDS and native PAGE, and SV-AUC studies conducted at 4 °C or 20 °C at pH 6 or pH 7.5 show that abatacept is monomeric under the these solution conditions with <2% dimer present (Figure 5).
Figure 4.
A. Far-UV CD of abatacept, B.Far-UV CD of CTLA-4, C. Near-UV CD of abatacept. All at 10 °C. D. Second derivative UV absorbance spectra overlapping at both pH’s at 20 ° C. Protein in 10 mM sodium phosphate buffer with 25 mM NaCl at pH 6 (dashed line or filled circles) and 7.5 (solid line or open circles)
Figure 5.
Sedimentation velocity analytical centrifugation distributions of 1.3 mg/mL abatacept in 10 mM sodium phosphate, 25 mM NaCl, at pH 6 (A) and pH 7.5 (B) at 20 °C. Inserts show c(s) distributions in full scale.
Using non-reducing SDS-PAGE and SV-AUC, E. coli-expressed CTLA-4 is shown to be >99% monomeric (data not shown). The CTLA-4 domain shows a similar far-UV CD spectrum to abatacept with a minimum of 217 nm and the characteristics of a beta-sheet protein (Figure 4B).
Assignment of Unfolding Transitions of Abatacept
We observe two major structural transitions for abatacept upon perturbation with chaotrope or temperature (see below for more details), which we anticipate are due to unfolding of the individual Fc and CTLA-4 polypeptides that comprise the domains of the abatacept fusion protein. In order to assign these transitions, we first measured the unfolding properties of separated, individual domains. To that end, we attempted to cleave the linking region between the Fc and CTLA-4 domains of abatacept using papain (44) and thrombin (45). However, application of papain resulted in additional, undesired fragmentation of abatacept, whereas thrombin did not cleave the molecule with any significant yield, in spite of previous reports that thrombin exhibits proteolytic activity against the EPK sequence found in the hinge region of abatacept that links the two domains (45, 46).
In another approach, we used CTLA-4 that was produced in E. coli to measure the Tm for thermal unfolding of CTLA-4. The observed Tm, 58 °C at pH 7.5 (far-UV CD), was close to that of the first thermal transition observed for abatacept. Because CTLA-4 produced in E. coli is not glycosylated, we also measured the Tm’s for unfolding of abatacept after treating the protein with PNGase F to deglycosylate the molecule. Deglycosylation resulted in only minor decreases (ca. 2.5 and 1 °C for the first and second transition, respectively) in the observed Tm values. Depending on solution conditions and heating rates, Tm values for Fc domains in intact antibodies are generally in the range of 80–84 °C, with the CH3 domain generally more stable than the CH2 domain (24, 25, 47), which corresponds well to the second transition in abatacept (83 °C at a scan rate of 60 °C/h).
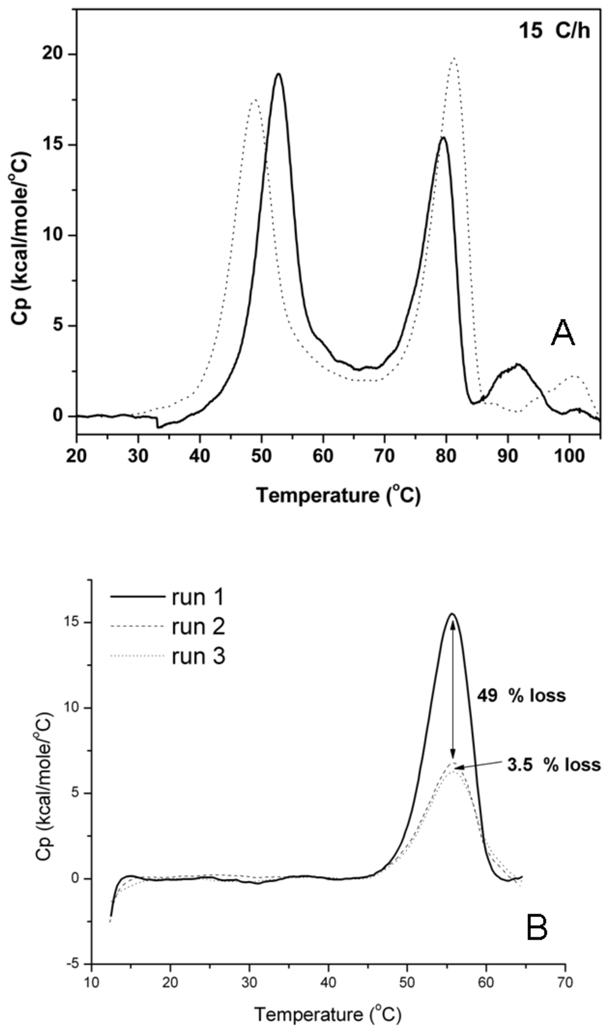
DSC scans to 60 °C (a temperature between the first and second transitions), followed by cooling and reheating to 60 °C show 50 % recovery of peak area measured in the first scan. Further scans on the same sample results in close to 100 % recovery of the peak area seen in the second scan (Figure 8B). This indicates that the low-temperature transition is a composite of one irreversible and one highly reversible population with equal changes in heat capacity upon unfolding. In an earlier study (48), a DSC thermogram of a Fc domain with a sequence similar to that in abatacept shows a Tm for the CH2-domain of ca. 60 °C, close to the first transition measured in our study. These results imply that the thermal unfolding transitions of the CTLA-4 and the CH2 domains likely overlap to give the first observed transition, whereas unfolding of the CH3 domain is responsible for the second transition. Therefore, we assign the lower temperature transition to the CTLA-4 and CH2 domains and the high-temperature transition to the CH3 domain of abatacept.
Figure 8.
A. DSC thermograms between 20–105 °C of abatacept in sodium phosphate buffer at pH 6 (dotted) and pH 7.5 (solid). B. Refolding of first thermal transition between 15–65 °C of abatacept in 10 mM sodium phosphate, 25 mM NaCl, at pH 7.5. First heat-cool-heat cycle yields 51% reversibility (solid to dashed) and second cycle yields 96.5% reversibility (dashed to dotted).
Chaotrope-induced Unfolding of Abatacept
Fluorescence Spectroscopic Analysis of the Fc-domain
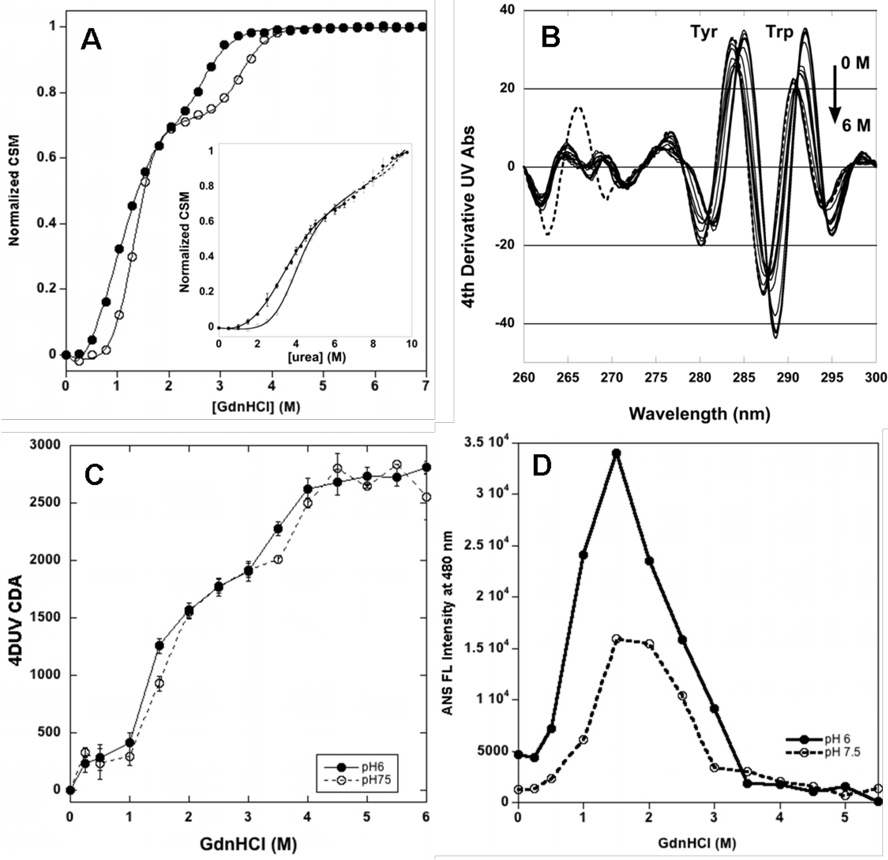
GdnHCl- and urea-induced denaturation monitored by intrinsic fluorescence of tryptophan residues in the Fc domain of abatacept at 25 °C yields two transitions at pH 6 and pH 7.5 (Figure 6A and B). The first transition is assigned to overlapping CTLA-4 and CH2 domain unfolding and the second transition is assigned to CH3 domain unfolding (for more details see the Discussion).
Figure 6.
A. GdnHCl denaturation of abatacept at 25 C at pH 6 (filled circles) and pH 7.5 (open circles) monitored by fluorescence. Insert: Fluorescence scans in 0–6 M GdnHCl at pH 6 at 25 °C, between 310–390 nm, ex wl 295 nm. B.. Denaturation curves in 0–9.75 M urea of ca 2 mM abatacept at 25 °C at pH 6 (filled circles) and pH 7.5 (open circles) in 10 mM sodium phosphate buffer and 25 mM NaCl. Insert: Fluorescence scans at pH 6 between 310–390 nm, ex wl 295 nm. C. Denaturation curve in 0–6 M GdnHCl of ca 5 mM abatacept at 25 °C at pH 6 (filled circles)and pH 7.5 (open circles) in 10 mM sodium phosphate buffer and 25 mM NaCl. Insert: 4d UV scans at pH 6 between 260–310 nm. D. ANS binding to abatacept (ca 100:1 molar ratio) at pH 6 (filled circles) and pH 7.5 (open circles) in 0–5.5 M GdnHCl at 25 °C in 10 mM sodium phosphate buffer with 25 mM NaCl.
Fitting a two-state model to each transition in GdnHCl (Figure 6A) yields the unfolding free energy and a midpoint of transition (Cm) for the first and second transition that is shown in Table 1. For the first transition the m values are −9.0 +/− 0.6 and −12.1 +/− 1.0 kJ/mol at pH 6 and 7.5, respectively. The second transitions have m values of −11.6 +/− 0.5 and −12.4 +/− 0.2 kJ/mol at pH 6 and 7.5, respectively.
Table 1.
Parameters for chaotrope-and thermally-induced unfolding at pH 6 and pH 7.5 monitored by intrinsic fluorescence.
| pH 6 | pH 7.5 | ||
|---|---|---|---|
| GdnHCl | ΔG1, kJ/mol | 7.1±0.6 | 15.6±1.2 |
| 25 °C | ΔG2, kJ/mol | 33.1±1.8 | 44.3±0.9 |
| Cm1, mol/l | 0.8±0.1 | 1.3±0.2 | |
| Cm2, mol/l | 2.9±0.2 | 3.6±0.1 | |
| Urea | ΔG1, kJ/mol | 6.8±1.3 | 14.5±1.3 |
| 25 °C | ΔG2, kJ/mol | 47.3±3.9 | nd |
| Cm1, mol/l | 3.1±0.2 | 3.8±0.2 | |
| Cm2, mol/l | 8.6±0.3 | nd | |
| Temperature | Tm1 , °C | 47.7±0.1 | 51.6±0.3 |
| Tm2 , °C | 80.9±0.2 | 78.5±0.2 | |
nd = not determined due to short baseline
Urea solutions at 25 °C for abatacept also yield two unfolding transitions at pH 6 and pH 7.5 (Figure 6B). Fitting a two-state model to each transition yields free energy of unfolding and Cm that are shown in Table 1. Abatacept is more stable towards urea than GdnHCl. Unfolding m values for the first transition at pH 6 and 7.5 are −2.2 +/− 0.4 and −3.7 +/− 0.4 kJ/mol, respectively. The second transition yields an m value of −5.5 +/− 0.4 kJ/mol at pH 6, but at pH 7.5 this transition cannot be reliably analyzed because unfolding is not complete even in saturated urea solutions.
The CSM values for the native and unfolded state of abatacept in both GdnHCl and urea solutions are independent of pH and are located at 338 and 351 nm, respectively. The CSM for free trypthophan is typically around 350 nm, thus it appears that the aromatic residues of abatacept are fully solvent-exposed in the final state. An intermediate state with a CSM around 346 nm is evident at GdnHCl concentrations around 2 M, and similarily at urea concentrations of 5.5 M.
Reversibility studies of the GdnHCl-induced unfolding transitions show a two-state transition with a Cm of ca. 2 M for the refolding; a value in between the two unfolding transitions (Supplementary figure 2). The CSM observed after dilution from 7 M GdnHCl to 0.25 M GdnHCl is 340 nm, compared to 338 nm observed at the same GdnHCl concentrations during unfolding. The change in apparent number of folding steps and shift in CSM during refolding compared to unfolding experiments indicates partial reversibility of the unfolding transition.
4DUV Spectroscopy
A blue shift and decreased intensity in 4DUV spectral signal is observed for the peaks arising from tryptophan residues during GdnHCl denaturation (Figure 6C insert), whereas peaks arising from tyrosine first decrease in intensity and then increase. GdnHCl also causes a blue shift of the peaks from tyrosine absorbance. Blue shifts suggest an increase in the polarity of the aromatic environment as the protein unfolds. As tyrosine residues are fairly evenly distributed in abatacept, the tyrosine-optimized derivative analysis likely reflects the “global” unfolding of the protein. The spectral changes in the fourth derivative data were expressed as the cumulative difference amplitude (CDA) (34). Figure 6C shows a three-state transition at both pH 6 and 7.5. The GdnHCl concentrations at the midpoints of the first and second transition observed at pH 6 are 1.3 M and 3.5M, respectively, and 1.4 M and 3.9 M respectively at pH 7.5.
ANS Fluorescence
ANS fluorescence was followed to probe changes in exposure of hydrophobic regions upon denaturation, but charge interactions can modify the fluorescence/binding behavior of ANS (49, 50). In GdnHCl at 25 °C, ANS interacts significantly with abatacept between 0.5 and 3 M GdnHCl; this region correlates with the first transition and the intermediate area. The ANS fluoresence intensity is more pronounced at pH 6 compared to pH 7.5, with almost a two-fold higher maxiumum intensity at 480 nm and a slightly wider fluorescence range (Figure 6D).
Thermally-induced Unfolding of Abatacept
The thermal stability of abatacept in solution was determined using DSC, far-UV CD, and intrinsic fluoresence at pH 6 and 7.5. Fluorescence (Figure 7A) and CD (Figure 7B) show two distinct transitions upon heating. DSC exhibits two main transitions around 50 and 80 °C, with additional transitions detectable at higher temperatures (T > 90 °C) that probably are aggregation-related. All transitions are scan rate- and pH-dependent (Figure 8A and Table 2). Thermal denaturation is essentially 100% irreversible (Supplementary figure 3).
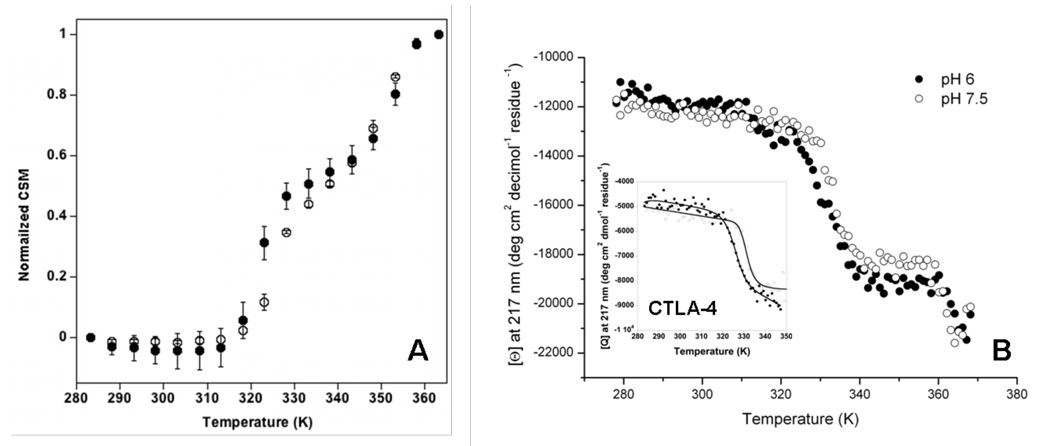
Figure 7.
A. FL CSM of thermal denaturation of ca 2 mM abatacept at pH 6 (filled circles) and pH 7.5 (open circles). Insert: Thermal FL scans 10–90 °C at pH 6, between 310–390 nm, ex wl 295 nm. B. Far-UV CD melt followed between 10 –95 °C at 217 nm for ca, 3 mM abatacept and ca. 4 mM CTLA-4 (insert) at pH 6 (filled circles) and pH 7.5 (open circles) in 10 mM sodium phosphate buffer and 25 mM NaCl.
Table 2.
DSC transition midpoint temperatures at different scan rates and transition activation energies at pH 6 and pH 7.5.
| (Tm in °C) | pH 6 | pH 7.5 | ||||
|---|---|---|---|---|---|---|
| Scanrate (°C/h) |
Tm1 | Tm2 | Tm3 | Tm1 | Tm2 | Tm3 |
| 15 | 48.8 | 81.2 | 100.2 | 52.7 | 79.6 | 91.6 |
| 30 | 50.5 | 82.2 | 100.5 | 54.2 | 81.1 | 93.8 |
| 60 | 52.6 | 84.9 | - | 55.8 | 83.0 | 95.5 |
| 90 | 54 | 85.9 | - | 57.1 | 84.3 | - |
| Ea** | 297±13 | 391±14 | -* | 361±19 | 391±20 | 422±18 |
| kJ/mol | ||||||
a third transition is not detected the higher scan rates at pH 6, precluding estimation of the activation energy
standard errors from fitting
dH<dHv for transition 3 → aggregation. Aggregation kinetics faster at pH 6 than pH 7.5 as Tm is less scan rate dependent at pH 6 compared to pH 7.5 (see Supplementary table 2).
Fluorescence Spectroscopic Monitoring of Thermally-induced Unfolding in the Fc-domain of Abatacept
Thermally-induced unfolding of abatacept monitored from 10 to 90 °C in sodium phosphate buffer exhibits two transitions in solutions at both pH 6 and pH 7.5 (Figure 7A). The transitions observed are likely unfolding of the CH2 and CH3 domains as CTLA-4 does not contain any Trp. Fitting a two-state model to each transition individually yields transition midpoint temperatures (Tm) that are shown in Table 1. The CSM for the unfolded state (90 °C) is around 346 nm and 348 nm for solutions at pH 6.0 and pH 7.5, respectively. An intermediate state observed at 62 °C has a CSM around 343.5 nm. It should be noted that both the unfolded and intermediate state observed at elevated temperatures have different CSM values compared to those measured in urea or GdnHCl solutions, indicate less solvent exposure of aromatics at elevated temperature than in chaotrope solutions.
Far-UV CD Monitoring of Thermally-induced Unfolding in Abatacept
Thermally-induced unfolding followed by far-UV CD at 217 nm at temperatures from 10 to 95 °C in sodium phosphate buffer yields two transitions at both pH 6 and pH 7.5 (Figure 7B), with a decreasing CD signal with increasing temperature, typical for beta-sheet proteins. We expect to observe the unfolding of all domains as far-UV CD monitors the protein backbone. As we only can observe two transitions CTLA-4 unfolding is overlapping with the CH2 domain unfolding in the first transition (see Discussion). CD measurements coupled with heat capacity values determined from DSC experiments were used to yield Tm values for the the first transition of 59.5 +/− 0.7 °C for pH 6, and 60.4 +/− 0.4 °C at pH 7.5. The second transition has a Tm = 89.4 +/− 4.6 °C for pH 6, and 87.7 +/− 2.4 °C at pH 7.5. The posttransitional baseline is short and noisy due to the proximity to the lower boiling point due to altitude (ca. 94–95 °C at 1600 m above sea level).
Differential Scanning Calorimetric Analysis of Abatacept Unfolding
Thermally-induced unfolding monitored by calorimetry shows two major transitions and additional scan rate- and pH-dependent transitions (Figure 8A). Tm values at different scan rates are tabulated in Table 2. As observed in fluorescence and CD studies, the second transition has higher thermal stability at pH 6 compared to that at pH 7.5. In contrast, the first transition is more stable at pH 7.5. The Tm’s are intermediate between those obtained by fluorescence and CD. Unfolding is irreversible after full unfolding, but if heating is stopped after the first transition (ca. 60 °C) and the sample is cooled to 10 °C, a subsequent heating to 60 °C shows a 50% recovery of the excess heat capacity observed during the first run, indicating partial reversibility. Interestingly, a third scan gives 96.5% reversibility relative to the second scan (Figure 8B). The Tm’s were not affected by varying abatacept concentrations from 1–11 mg/mL at either pH (Supplementary figure 4). Lower van’t Hoff enthalpies compared to the calorimetric enthalpies in the two first transitions for all scan rates indicate that those transitions are not two-state unfolding reactions. For the third transition, the van’t Hoff enthalpies are higher compared to the calorimetric enthalpies, consistent with aggregation reactions being responsible for the observed excess heat capacity (Supplementary table 1).
Heat Capacity Changes during Thermally-induced Unfolding of Abatacept
At pH 6, complete abatacept unfolding, CTLA-4/CH2 domain unfolding, and CH3 domain unfolding yielded heat capacity changes of 5.2, 3.1, and 2.1 kcal/mol/K, respectively. At pH 7.5 those heat capacity changes were 8.0, 5.7, and 2.3 kcal/mol/K, respectively (Supplementary figure 3). The heat capacity change for unfolding of abatacept was estimated to 8.4 kcal/mol/K based on the primary sequence (51), close to the experimental value at pH 7.5.
Scan rate Dependence and Activation Energies
Tm values increase with increasing scan rates (Table 2). The activation energies for each transition are tabulated in Table 2. Activation energies were pH-dependent for the lower-temperature transition. At both pH’s, activation energies were smaller for the lower-temperature transition.
Thermally-induced Unfolding in Deglycosylated Abatacept and CTLA-4
Thermally-induced unfolding of abatacept deglycosylated with PNGase F and E. coli-expressed CTLA-4 (thus aglycosylated) were followed by DSC, far-UV CD or intrinsic fluorescence. For deglycosylated abatacept in pH 7.5 sodium phosphate buffer, the first transition midpoint temperature is lowered by around 2 °C compared to that in complete abatacept, whereas the second transition midpoint temperature is affected by less than 1 °C compared to the abatacept (Supplementary figures 5 and 6). The denaturation is irreversible and the solution is opaque afterwards indicating the presence of insoluble aggregates.
The thermal stability of the CTLA-4 covalent dimer in sodium phosphate buffer was monitored using DSC and far-UV CD. Melting followed by far-UV CD at 217 nm shows one single transition with a midpoint transition temperature of 52.3 and 58.4 °C at pH 6 and 7.5, respectively (Figure 7B insert). One single transition with a Tm of 54.4 °C is also seen using DSC at pH 8 in 20 mM Tris, 200 mM NaCl, and 10 % glycerol (Supplementary figure 7). The thermal denaturation is irreversible.
Colloidal Stability
Zeta Potentials and Second Virial Coefficient from Light Scattering (SVC)
The osmotic second virial coefficients are negative at both pH 6 and pH 7.5, indicating net pair-wise attraction between abatacept molecules (Table 3). By measuring the zeta potential from the electrophoretic mobility of abatacept, the electrostatic contribution to its solution behavior could be measured at pH 6 and 7.5. As seen in Table 3, pH 6 solution conditions show slightly lower negative zeta potential compared to pH 7.5 conditions. Both values are well below the arbitrary limit for colloidal stability of +/− 30 mV (52). The Debye-Huckel approximation (eq. 10) yields estimated effective charges at both pH conditions that are negative and only separated by one charge unit. At pH 6, the theoretical and the effective charge are very close, but the effective charge at pH 7.5 is ca. 4-fold less negative than the theoretical charge. This effective charge measured at the diffuse layer boundary or slipping plane contributes to the intermolecular interactions and is reflected in the estimate of the electrostatic contribution to the second virial coefficient (Table 3). At both pH 6 and pH 7.5 this contribution is insignificant and the net attractive second virial coefficient in sodium phosphate buffer can be explained by both the low protein surface charge and electrostatic screening of the remaining charges.
Table 3.
Second virial coefficient (SVC) from light scattering and zeta potential from laser doppler velocimetry for abatacept at pH 6 and pH 7.5 in 10 mM sodium phosphate, 25 mM NaCl (ionic strength = 75 mM), at 25 °C.
| Light Scattering | Laser Doppler Velocimetry | ||||
|---|---|---|---|---|---|
| SVCel × 10−6 | |||||
| pH | SVC × 10−4 | Mw x103 | Zeta potential | (mL mol g−2) | Effective charge(Z)/Theoretical charge |
| (mL mol g−2) | (g mol −1) | (mV) | (% of SVC) | ||
| 6 | −6.5±3.7 | 78.9±2.7 | −9.3±1 | 2.8 (0.4 %) | −2.6/−2 |
| 7.5 | −6.0±1.9 | 68.6±3.0 | −12.8±1.2 | 5.2 (0.9 %) | −3.5/−14.8 |
Discussion
Aggregation Profile of Abatacept/ Lumry-Eyring Framework
During incubation studies at 40 °C, abatacept aggregated much more rapidly in solutions at pH 6.0 than at pH 7.5. Based on the Lumry-Eyring theory (10), we expect that the increased aggregation rate seen at pH 6.0 vs. pH 7.5 could be due to decreased conformational stability of abatacept at the lower pH, or more attractive intermolecular interactions (decreased colloidal stability), or both.
Aggregation generally does not require complete unfolding, but rather proceeds via partially unfolded intermediates (6, 8). In support of this expectation, we observed that abatacept is susceptible to aggregation at moderately elevated temperature (40 °C), even though complete unfolding of the molecule requires relatively high concentrations of chaotrope (>9 M urea), elevated temperatures (>90 °C). At temperatures from 10–40 °C, no differences in abatacept structure can be detected in solutions at pH 6.0 vs. 7.5 using far-UV CD or second derivative UV absorbance spectroscopy, suggesting that the majority of the protein remains in the native state ensemble under both pH conditions.
Molten globule-like intermediate in the Lumry-Eyring scheme
If a molten globule intermediate is formed as a protein unfolds, techniques that monitor secondary vs. tertiary structural changes often show different onset points for the unfolding transition, with tertiary structure losses preceeding disappearance of secondary structures (53, 54). Under identical solution conditions, the apparent midpoints for thermally-induced unfolding transitions of abatacept detected using various spectroscopic techniques increase in the order fluorescence < DSC < far-UV CD (Table 1). This indicates an intermediate state during the melting where the secondary structure, probed by far-UV CD, is more stable than the tertiary structure that is reflected in the tryptophan environment as probed by fluorescence. Calorimetry measurements, which follow overall thermal processes during unfolding, show transition temperatures in between those detected by fluorescence or far-UV CD.
ANS fluorescence as a function of GdnHCl is consistent with formation of an intermediate state with greater exposed hydrophobic surface area that is populated at moderate GdnHCl concentrations (e.g., 0.5–3.5 M). ANS fluorescence is more pronounced at pH 6, suggesting that the intermediate state has increased solvent-accessible hydrophobic regions at pH 6 than at pH 7.5.
A likely model for an aggregation-prone abatacept intermediate is a hybrid of a molten globule-like CTLA-4 and CH2 domains and an intact CH3 domain. If a partially unfolded state is involved in the aggregation reaction, we expect that the stability of the least stable domain will dominate the conformational stability contributions to aggregation (13, 15). In the case of abatacept, the CTLA-4 and CH2 domains are less stable than the CH3 domain at both solution conditions tested. Using chaotropes to perturb the structure of abatacept, we found that the free energy of unfolding of the CTLA-4/CH2 domains decreases by approximately a factor of 2 (15.6 to 7.1 kJ/mol) as the pH decreases from 7.5 to 6.0. Thus, a contributing factor to increased aggregation rates at pH 6 may be the greater instability of the CTLA-4/CH2. In contrast, the stability of the CH3 domain decreases by 25% (44.3 to 33.1 kJ/mol) upon decreasing pH from 7.5 to 6.0, and is always significantly higher than that of the CTLA-4/CH2.
In addition, the apparent activation energy for domain unfolding is pH-dependent for the CTLA-4/CH2 domains, but not for the CH3 domain (Table 2), suggesting that the pH sensitivity in rates of aggregation is related to pH-induced changes in the stability of the CTLA-4/CH2 domains rather than in the CH3 region. Furthermore, the theoretical heat capacity change of unfolding per residue for CTLA-4/CH2 domains is larger than that for the CH3 domain, suggesting that unfolding increases solvent exposure of hydrophobic residues to a greater degree for CTLA-4 compared to the Fc domain of abatacept (55). Thus, unfolded or partly folded CTLA-4/CH2 domains would be expected to be more prone to aggregation via hydrophobic interactions than the CH3 domain. All those contributions would result in a larger fraction partly unfolded aggregation-prone species at pH 6 compared to pH 7.5 at 40 °C.
Colloidal Stability
The second step in the general Lumry-Eyring framework is a collisional reaction step, which is expected to be influenced by the intermolecular forces between abatacept molecules. Typically, these forces are dominated by electrostatic interactions (56). Overall, the measured pI for abatacept is 4.5–5.5, based on isoeletric focusing. Thus, we would expect that the intermolecular electrostatic interactions would be repulsive, since abatacept should carry a negative charge at both pH 6.0 and 7.5. Indeed, this is borne out by the zeta potential values for abatacept (−9.3 and −12.8 mV at pH 6.0 and 7.5, respectively). However, for colloidal stability, the absolute value of the zeta potential should be greater than 30 mV (52, 56), an empirical value that separates low-charged surfaces from high-charged surfaces, so abatacept appears to be colloidally unstable at both solution pH’s, but only slightly more so at pH 6.
Interestingly, the pI’s of the individual domains (calculated based on amino acid sequence, but not taking potential counterion binding into account) are near 7.2 and 4.2 for the Fc and CTLA-4 domains, respectively, compared to the calculated pI of 5.9 for the whole molecule. The calculated pI’s are 8.9 and 6.5 for the CH2 and CH3 Fc subdomains, respectively. Thus, whereas at pH 7.5 the charge calculated based on amino acid sequence on both domains will be negative (−14.2 for CTLA-4, −0.6 for Fc, but +2.3 for CH2, and −3 for CH3), at pH 6.0 the CTLA-4 domain is expected to be negatively charged (−12) and the Fc domain positively charged (+10), with +8 for CH2, and +2.2 for CH3 (supplementary information). These opposite charges would be expected to increase domain-domain intramolecular attractive interactions at pH 6.0, and also to increase intermolecular attraction via increased dipole-dipole interactions. The observation of pH-dependent dimer and trimer populations as a result of accelerated stability studies may originate from the different domain charge distributions at the two pH conditions.
The SVC values for abatacept do not differ significantly at the two pH values studied, consistent with the measured zeta potentials. Although the SVC values under both conditions are negative, reflecting attractive protein-protein pairwise interactions, the similarity of the SVC values suggests that differences in colloidal instabilities at pH 6.0 and 7.5 are not responsible for the large differences in rates of aggregation at the two pH’s. This conclusion is further substantiated by the fact that at room temperature, well below temperatures where the thermal transitions are observed, abatacept does not aggregate appreciably over at least 30 days when stored at either pH. Rapid aggregation did not allow SVC determination at elevated temperatures.
Based on the calculated charge distribution on abatacept at pH 6.0, strong electrostatic contributions to the SVC might be expected. However, based on zeta potential measurements made in solution, the electrostatic contribution to the virial coefficient is insignificant. This may arise from both counterion binding that reduces the effective surface charges of abatacept at the two pH conditions and from electrostatic screening of remaining charges at the ionic strength tested (75 mM). The relatively small net attractive pair-wise interactions may be explained by van der Waals attractions, and/or by attractive dipolar interactions because of the opposite charges of various domains. Interestingly, the SVC in pH 6 and pH 7.5 solutions containing 50 mM Tris, 25 mM MES, 25 mM acetic acid, and 25 mM NaCl (a solution with the same, but pH-independent, ionic strength of 75 mM) instead of phosphate, is positive at both pH conditions, implying repulsive net pair-wise interactions. This could indicate that binding of phosphate ions to abatacept has an important role in making abatacept self-interactions attractive, but is beyond the scope of the current study.
Conclusion
At 40 °C, abatacept shows strikingly different propensity to aggregate at pH 6.0 compared to pH 7.5. This propensity correlates with the pH effect on conformational stability of the domains of abatacept. In contrast, colloidal stability of abatacept is relatively insensitive to the pH’s studied. This suggests that partial unfolding of the least stable domains dominates abatacept’s aggregation propensity.
Because protein aggregation generally proceeds through partially unfolded intermediates, it is likely that the least stable domain of a multi-domain protein will predominately determine the aggregation propensity of the protein. In the case of abatacept, the CTLA-4/CH2 domains appear to be much less stable than the more stable CH3 domain, and thus likely are responsible for abatacept aggregation under accelerated aggregation conditions.
Intact antibodies have generally a good temperature tolerence, as exemplified by the lack of any negative effect on the biological activity after the use of heat treatment at 60 °C for 10 h (pasteurization) of intravenous Ig-preparations for virial clearance (57). In comparison with native IgG proteins, wherein interdomain interactions presumably have evolved to provide mutual stabilization, fusion proteins may lack such stabilizing interdomain stabilization. This is the case for abatacept: the unfolding transitions show minimal cooperativity between domains. This has been seen in other artificial fusion proteins as well (58). In contrast, many natural multidomain proteins such as antibodies show cooperative transitions that are apparent two-state (21).
As with most randomly introduced mutations, fusion of two or more foreign proteins or domains would be expected only rarely to create stabilizing, cooperative interactions. There may be a vast potential to increase the stability and modulate aggregation properties of artificial fusion proteins by optimizing inter- and intramolecular domain-domain interactions by not only adjusting formulation conditions, but by using protein engineering approaches.
Supplementary Material
Acknowledgements
LaToya Jones Braun’s lab at School of Pharmacy and the Biophysical Core Center at University of Colorado in Denver are acknowledged for instrument access. Ingemar Andre at David Baker’s lab at University of Washington, Seattle, is gratefully thanked for helpful discussions concerning structural modelling with Robetta and PyMOL. Barofold is gratefully thanked for donating CTLA-4. Malvern Inc. is acknowledged for help with zeta potential and DLS measurements. Joan Torrent and Reinhard Lange, INSERM, Montpellier, France, are gratefully thanked for providing software for CDA calculations, as well as for helpful comments on the manuscript.
Abbreviations
- CD
circular dichroism
- CH2
constant heavy chain domain 2
- CH3
constant heavy chain domain 3
- CSM
center of spectral mass
- CTLA-4
cytotoxic T lymphocyte-associated molecule-4
- DLS
dynamic light scattering
- DSC
differential scanning calorimetry
- ES-MS
electrospray-mass spectroscopy
- Fc
fragment crystallizable
- GdnHCl
guanidine hydrochloride
- HPLC
high-pressure liquid chromatography
- IEF
isoelectric focusing
- MALDI-MS
matrix-assisted laser desorption/ionization mass spectroscopy
- MWCO
molecular weight cut-off
- PAGE
polyacrylamide gel electrophoresis
- PDB
protein data bank
- RPM
revolutions per minute
- SASA
solven accessible surface area
- SEC
size-exclusion chromatography
- SV-AUC
sedimentation velocity analytical ultracentrifugation
- SVC
second virial coefficient
- UV
ultraviolet.
Footnotes
Funding was provided by NIH Grant R01 EB006006.
Supporting Information Available
Further calorimetric, AUC, charge distribution calculations, deglycosylation stability and reversibility data for abatacept and CTLA-4 is presented in figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Uversky V, Fink A, editors. Protein Misfolding, Aggregation and Conformational Diseases. Vol. 1–2. Kluwer Academic/Plenum; 2006. [Google Scholar]
- 2.Wang W. Protein aggregation and its inhibition in biopharmaceutics. International Journal of Pharmaceutics. 2005;289:1–30. doi: 10.1016/j.ijpharm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg A. Effects of Protein Aggregates: An Immunologic Perspective. AAPS Journal. 2006;8:501–507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regina M, Murphy BSK. Protein Misfolding and Aggregation. Biotechnology Progress. 2007;23:548–552. doi: 10.1021/bp060374h. [DOI] [PubMed] [Google Scholar]
- 5.Cromwell M, Hilario E, Jacobson F. Protein Aggregation and Bioprocessing. AAPS Journal. 2006;8:572–579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts CJ. Nonnative protein aggregation kinetics. Biotechnology and Bioengineering. 2007;98:927–938. doi: 10.1002/bit.21627. [DOI] [PubMed] [Google Scholar]
- 7.Andrews JM, Roberts CJ. A Lumry-Eyring Nucleated Polymerization Model of Protein Aggregation Kinetics: 1. Aggregation with Pre-Equilibrated Unfolding. J. Phys. Chem. B. 2007;111:7897–7913. doi: 10.1021/jp070212j. [DOI] [PubMed] [Google Scholar]
- 8.Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical Stability of Proteins in Aqueous Solution: Mechanism and Driving Forces in Nonnative Protein Aggregation. Pharmaceutical Research. 2003;20:1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- 9.Mahler HC, Friess W, Grauschopf U, Kiese S. Protein aggregation: Pathways, induction factors and analysis. J Pharm Sci published online Sept 29. 2008 doi: 10.1002/jps.21566. [DOI] [PubMed] [Google Scholar]
- 10.Lumry R, Eyring H. Conformation changes of proteins. Journal of Physical Chemistry. 1954;58:110–120. [Google Scholar]
- 11.Batey S, Clarke J. Apparent cooperativity in the folding of multidomain proteins depends on the relative rates of folding of the constituent domains. Proceedings of the National Academy of Sciences. 2006;103:18113–18118. doi: 10.1073/pnas.0604580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randles LG, Batey S, Steward A, Clarke J. Distinguishing Specific and Nonspecific Interdomain Interactions in Multidomain Proteins. Biophys. J. 2008;94:622–628. doi: 10.1529/biophysj.107.119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azuaga AI, Dobson CM, Mateo PL, Conejero-Lara F. Unfolding and aggregation during the thermal denaturation of streptokinase. European Journal of Biochemistry. 2002;269:4121–4133. doi: 10.1046/j.1432-1033.2002.03107.x. [DOI] [PubMed] [Google Scholar]
- 14.Chi EY, Krishnan S, Kendrick BS, Chang BS, Carpenter JF, Randolph TW. Roles of conformational stability and colloidal stability in the aggregation of recombinant human granulocyte colony-stimulating factor. Protein Sci. 2003;12:903–913. doi: 10.1110/ps.0235703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oganesyan V, Damschroder MM, Leach W, Wu H, Dall'Acqua WF. Structural characterization of a mutated, ADCC-enhanced human Fc fragment. Molecular Immunology. 2008;45:1872–1882. doi: 10.1016/j.molimm.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 16.Salinas B. Chemical and Biological Engineering. Boulder: University of Colorado; 2009. Phase behaviour and opalescence of concentrated antibody formulations. [Google Scholar]
- 17.Chou DK, Krishnamurthy R, Randolph TW, Carpenter JF, Manning MC. Effects of Tween 20 and Tween 80 on the stability of Albutropin during agitation. Journal of Pharmaceutical Sciences. 2005;94:1368–1381. doi: 10.1002/jps.20365. [DOI] [PubMed] [Google Scholar]
- 18.Chamow SM, Ashkenazi A, editors. Antibody Fusion Proteins. 1 ed. New York: Wiley-Liss Inc.; 1999. [Google Scholar]
- 19.Teillaud J-L. Engineering of monoclonal antibodies and antibody-based fusion proteins: successes and challenges. Expert Opinion on Biological Therapy. 2005;5:S15–S27. doi: 10.1517/14712598.5.1.s15. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Singh S, Zeng DL, King K, Nema S. Antibody structure, instability, and formulation. Journal of Pharmaceutical Sciences. 2007;96:1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- 21.Honegger A. Engineering Antibodies for Stability and Efficient Folding. In: Chernajovsky Y, Nissim A, editors. Therapeutic Antibodies. SpringerLink: 2008. pp. 47–68. [DOI] [PubMed] [Google Scholar]
- 22.Daugherty AL, Mrsny RJ. Formulation and delivery issues for monoclonal antibody therapeutics. Advanced Drug Delivery Reviews. 2006;58:686–706. doi: 10.1016/j.addr.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Ionescu RM, Vlasak J, Price C, Kirchmeier M. Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. Journal of Pharmaceutical Sciences. 2008;97:1414–1426. doi: 10.1002/jps.21104. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Bulseco G-G, Sun J. Effect of posttranslational modifications on the thermal stability of a recombinant monoclonal antibody. Immunology Letters. 2006;106:144–153. doi: 10.1016/j.imlet.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Demarest SJ, Glaser SM. Antibody therapeutics, antibody engineering, and the merits of protein stability. Current Opinion in Drug Discovery and Development. 2008;11:675–687. [PubMed] [Google Scholar]
- 26.Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochemical and Biophysical Research Communications. 2007;355:751–757. doi: 10.1016/j.bbrc.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 27.Linsley P, Brady W, Urnes M, Grosmaire L, Damle N, Ledbetter J. CTLA-4 is a second receptor for the B cell activation antigen B7. J Exp Med. 1991;174:561–569. doi: 10.1084/jem.174.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreland L, Bate G, Kirkpatrick P. Abatacept. Nat Rev Drug Discov. 2006;5:185–186. doi: 10.1038/nrd1989. [DOI] [PubMed] [Google Scholar]
- 29.Davis PM, Abraham R, Xu L, Nadler SG, Suchard SJ. Abatacept binds to the Fc receptor CD64 but does not mediate complement-dependent cytotoxicity or antibody-dependent cellular cytotoxicity. J Rheumatol. 2007;34:2204–2210. [PubMed] [Google Scholar]
- 30.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Shouvik R, Jones LS, Krishnan S, Kerwin BA, Chang BS, Manning MC, Randolph TW, Carpenter JF. Mechanism for benzyl alcohol-induced aggregation of recombinant human interleukin-1 receptor antagonist in aqueous solution. Journal of Pharmaceutical Sciences. 2004;93:3076–3089. doi: 10.1002/jps.20219. [DOI] [PubMed] [Google Scholar]
- 32.Krishnan S, Chi EY, Webb JN, Chang BS, Shan D, Goldenberg M, Manning MC, Randolph TW, Carpenter JF. Aggregation of Granulocyte Colony Stimulating Factor under Physiological Conditions: Characterization and Thermodynamic Inhibition. Biochemistry. 2002;41:6422–6431. doi: 10.1021/bi012006m. [DOI] [PubMed] [Google Scholar]
- 33.Lange R, Frank J, Saldana JL, Balny C. Fourth derivative UV-spectroscopy of proteins under high pressure .1. Factors affecting the fourth derivative spectrum of the aromatic amino acids. European Biophysics Journal with Biophysics Letters. 1996;24:277–283. [Google Scholar]
- 34.Torrent J, Connelly JP, Coll MG, Ribo M, Lange R, Vilanova M. Pressure versus Heat-Induced Unfolding of Ribonuclease A: The Case of Hydrophobic Interactions within a Chain-Folding Initiation Site. Biochemistry. 1999;38:15952–15961. doi: 10.1021/bi991460b. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Ruiz JM. Theoretical analysis of Lumry-Eyring models in differential scanning calorimetry. Biophysical Journal. 1992;61:921–935. doi: 10.1016/S0006-3495(92)81899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 37.Webb JN, Webb SD, Cleland JL, Carpenter JF, Randolph TW. Partial molar volume, surface area, and hydration changes for equilibrium unfolding and formation of aggregation transition state: High-pressure and cosolute studies on recombinant human IFN-gamma. Proceedings of the National Academy of Sciences. 2001;98:7259–7264. doi: 10.1073/pnas.131194798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kratochvil P. Classical Light Scattering for Polymer Solutions. Amsterdam, Netherlands: Elsevier; 1987. [Google Scholar]
- 40.Arakawa T, Wen J. Determination of Carbohydrate Contents from Excess Light Scattering. Analytical Biochemistry. 2001;299:158–161. doi: 10.1006/abio.2001.5432. [DOI] [PubMed] [Google Scholar]
- 41.Faude A, Zacher D, Muller E, Bottinger H. Fast determination of conditions for maximum dynamic capacity in cation-exchange chromatography of human monoclonal antibodies. Journal of Chromatography A. 2007;1161:29–35. doi: 10.1016/j.chroma.2007.03.114. [DOI] [PubMed] [Google Scholar]
- 42.Chun M-S, Lee I. Rigorous estimation of effective protein charge from experimental electrophoretic mobilities for proteomics analysis using microchip electrophoresis. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2008;318:191–198. [Google Scholar]
- 43.Asthagiri D, Paliwal A, Abras D, Lenhoff AM, Paulaitis ME. A Consistent Experimental and Modeling Approach to Light-Scattering Studies of Protein-Protein Interactions in Solution. 2005;88:3300–3309. doi: 10.1529/biophysj.104.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao S-h, Putnam FW. The Cleavage of Human ggr-Globulin by Papain. J. Biol. Chem. 1961;236:122–135. [PubMed] [Google Scholar]
- 45.Linsley PS, Nadler SG, Bajorath Jr, Peach R, Leung HT, Rogers J, Bradshaw J, Stebbins M, Leytze G, Brady W, Malacko AR, Marquardt H, Shaw S-Y. Binding Stoichiometry of the Cytotoxic T Lymphocyte-associated Molecule-4 (CTLA-4) J. Biol. Chem. 1995;270:15417–15424. doi: 10.1074/jbc.270.25.15417. [DOI] [PubMed] [Google Scholar]
- 46.Muszbek L, Laki K. Cleavage of Actin by Thrombin. Proceedings of the National Academy of Sciences. 1974;71:2208–2211. doi: 10.1073/pnas.71.6.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghirlando R, Lund J, Goodall M, Jefferis R. Glycosylation of human IgG-Fc: influences on structure revealed by differential scanning micro-calorimetry. Immunol Lett. 1999;68:47–52. doi: 10.1016/s0165-2478(99)00029-2. [DOI] [PubMed] [Google Scholar]
- 48.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL. Structure and Stability Changes of Human IgG1 Fc as a Consequence of Methionine Oxidation. Biochemistry. 2008;47:5088–5100. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]
- 49.Matulis D, Lovrien R. 1-Anilino-8-Naphthalene Sulfonate Anion-Protein Binding Depends Primarily on Ion Pair Formation. 1998;74:422–429. doi: 10.1016/S0006-3495(98)77799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasymov OK, Glasgow BJ. ANS fluorescence: Potential to augment the identification of the external binding sites of proteins. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2007;1774:403–411. doi: 10.1016/j.bbapap.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edelhoch H, Osborne JJ. The thermodynamic basis of the stability of proteins, nucleic acids, and membranes. Adv Protein Chem. 1976;30:183–250. doi: 10.1016/s0065-3233(08)60480-5. [DOI] [PubMed] [Google Scholar]
- 52.ASTM Standard. D 4187-82 Test Methods for Zeta Potential of Colloids in Water and Waste Water. 1985. [Google Scholar]
- 53.Ohgushi M, Wada A. Liquid-like state of side chains at the intermediate stage of protein denaturation. Adv Biophys. 1984;18:75–90. doi: 10.1016/0065-227x(84)90007-8. [DOI] [PubMed] [Google Scholar]
- 54.Ptitsyn O. Molten globule and protein folding. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 55.Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proceedings of the National Academy of Sciences. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Everett DH. Basic Principles of Colloid Science. Cambridge, UK: The Royal Society of Chemistry; 1988. [Google Scholar]
- 57.Bridonneau P, Marcilly H, Vernois-Martin M, Goigoux P, Bourdel V, Laulan A, Deramoudt FX, Desmadril M, Sitbon M, Basuyaux B, Steinbuch M, Schmitthaeusler R. Liquid Pasteurization of an Immunoglobulin Preparation without Stabilizer: Effects on its Biological and Biochemical Properties. Vox Sanguinis. 1996;70:203–209. doi: 10.1111/j.1423-0410.1996.tb01327.x. [DOI] [PubMed] [Google Scholar]
- 58.Souillac PO. Biophysical characterization of insoluble aggregates of a multi-domain protein: An insight into the role of the various domains. Journal of Pharmaceutical Sciences. 2005;94:2069–2083. doi: 10.1002/jps.20423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.