Abstract
Life on earth is dependent on sulphur (S) and nitrogen (N). In plants, the second step in the reduction of sulphate and nitrate are mediated by the enzymes sulphite and nitrite reductases, which contain the iron (Fe)-containing siroheme as a cofactor. It is synthesized from the tetrapyrrole primogenitor uroporphyrinogen III in the plastids via three enzymatic reactions, methylation, oxidation and ferrochelatation. Without siroheme biosynthesis, there would be no life on earth. Limitations in siroheme should have an enormous effect on the S- and Nmetabolism, plant growth, development, fitness and reproduction, biotic and abiotic stresses including growth under S, N and Fe limitations, and the response to pathogens and beneficial interaction partners. Furthermore, the vast majority of redoxreactions in plants depend on S-components, and S-containing compounds are also involved in the detoxification of heavy metals and other chemical toxins. Disturbance of siroheme biosynthesis may cause the accumulation of light-sensitive intermediates and reactive oxygen species, which are harmful, or they can function as signaling molecules and participate in interorganellar signaling processes. This review highlights the role of siroheme in these scenarios.
Key words: iron, nitrogen, plant/microbe interaction, redox, siroheme, sulphur
Introduction
Life on earth is absolutely dependent on sulphur (S) and nitrogen (N). In plants, S is mainly taken up from the soil as sulphate, the oxidized form of S, before reduction and metabolism into S-containing compounds.1–3 Animals are unable to reduce sulphate and thus require S-containing amino acids (such as cysteine and methionine) or proteins as diet. Therefore, sulphate assimilation by plants is essential for all life on earth. The presence of S in many redox mediators also highlights its importance for signaling processes.4,5 Likewise, plants recruit N mainly from the soil as nitrate, which is reduced to ammonium before integration into N-containing compounds. N supply is a limiting factor for plant growth and ultimately for the production of food for heterotrophic organisms (reviewed in refs. 6–9).
The second steps in the reduction of sulphate and nitrate are mediated by the enzymes sulphite and nitrite reductases (SiR, NiR). Higher plant SiRs and NiRs contain siroheme as prosthetic group which is central to the catalytic activity of the higher plant enzymes10 and catalyze the six electron reduction of sulphite and nitrite, respectively. Thus, assimilation of all inorganic S and the majority of N in the biosphere depend on the availability of siroheme and without siroheme, there would be no reduced S for the synthesis of the amino acids cysteine and methionine and for the biogenesis of Fe-S centers. Interestingly, both SiRs and NiRs also contain a Fe-S cofactor.11
Biosynthesis of Siroheme
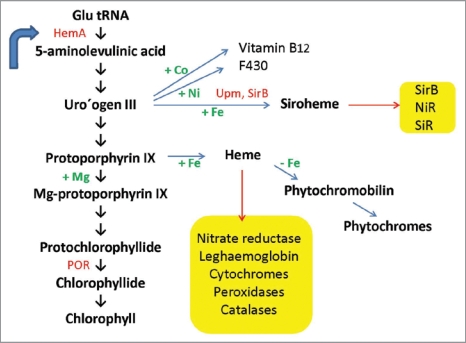
In higher plants tetrapyrrole synthesis occurs in plastids,12,13 where it is initiated by the reduction of the glutamyl moiety of glutamyl-tRNA to glutamate-1-semialdehyde. Intermolecular transamination of glutamate semialdehyde generates 5-aminolevulinic acid (ALA), which is then transformed into the first macrocyclic intermediate of the pathway, uroporphyrinogen III, by the enzymes ALA dehydratase, porphobilinogen deaminase, and uroporphyrinogen III synthase.14–17 ALA dehydratase converts two molecules of ALA to porphobilinogen (PBG) and PBG deaminase and uroporphyrinogen III cosynthase condense four molecules of PBG and inverse the ring D to form uroporphyrinogen III (Fig. 1). It represents the first branch point in the pathway, as methylation of this intermediate directs it toward siroheme synthesis, whereas decarboxylation steers it toward heme and chlorophyll synthesis.18
Figure 1.
Tetrapyrrole biosynthesis pathway leading to chlorophylls, siroheme and phytochromes. Crucial enzymes are in red: HemA, Glu tRNA synthase, POR, protochlorophyllide oxidoreductase. Chelexed ions are in green. Yellow boxes: Enzymes and subtrates, which require siroheme and heme, respectively. Blue error: feedback regulation of HemA enzyme activity.
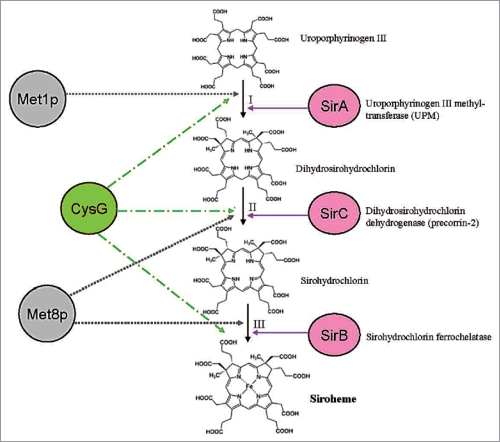
Siroheme biosynthesis from uroporphyrinogen III is mediated by three enzymatic reactions (Fig. 2): (1) Two methylation steps, (2) oxidation and (3) ferrochelatation. Methylation in rings A and B by uroporphyrinogen III methyltransferase (Upm, At5g40850, acc. no. in Arabidopsis) forms first precorrin-1 (first methylation step) and then precorrin-2 (second metylation step) for siroheme synthesis, while decarboxylation by uroporphyrinogen III decarboxylase generates coproporphyrinogen III leading to chlorophyll and heme biosynthesis. UPM requires S-adenosyl-L-methionine as a methyl donor and has been characterized from a wide variety of species including higher plants.8,13,19 Precorrin-2 is further dehydrogenated to sirohydrochlorin and Fe is inserted to the center of the tetrapyrrole to form siroheme by sirohydrochlorin ferrochelatase (SirB; At1g50170, acc. no. in Arabidopsis). A gene/enzyme for the second step is not yet known in plants (Fig. 2). Interestingly, not only SiR and NiR, but also SirB contain 2Fe-2S centers.11
Figure 2.
Siroheme biosynthesis from uroporphyrinogen III . The plant, yeast and bacterial enzymes are indicated. For details, see text.
In yeast, the first step in this pathway is catalyzed by an enzyme called Met1p, which shows no homology to UPM from higher plants (Fig. 2). The last two steps are catalyzed by a single bifunctional enzyme called Met8p,20 which houses both dehydrogenase and chelatase functionalities within the same active site (reviewed in ref. 21; Fig. 2). However, no orthologs of Met8p are found in higher plants.
In some bacteria, the transformation of dihydrosirohydrochlorin into siroheme is catalyzed by two separate enzymes called SirC (dihydrosirohydrochlorin dehydrogenase) and SirB.22,23 The latter inserts ferrous Fe into sirohydrochlorin to give siroheme. Interestingly, CysG, a homodimeric enzyme of 50-kDa subunits from bacteria such as S. enterica serovar typhimurium and Escherichia coli, catalyzes all reactions.15,24,25
Little is known about the regulation of siroheme biosynthesis in higher plants. As expected, knock-out lines for the two known plant enzymes UPM and SirB are lethal, thus only manipulation of the upm and sirb mRNA levels in plants will allow the analysis of these enzymes by genetic approaches.
Several evolutionary related chelatases insert various ions into the tetrapyrrole skeleton (reviewed in ref. 26; Fig. 3). Many of the SirB protein sequence of both higher plants and bacteria are closely related to CbiXs, cobalt chelatases from bacteria.27 They are responsible for the chelation of Co2+ into sirohydrochlorin and important for vitamin B12 biosynthesis. CbiX often contains a C-terminal histidine-rich region that may be important for metal delivery and/or storage, and may also contain an Fe-S center. Both are found in a wide range of bacteria. This subgroup also contains single domain proteins from archaea and bacteria which may represent the ancestral form of ferro- and cobaltchelatases. Thus, the nuclear-encoded sirb gene of higher plants is of prokaryotic origin. The gene became part of the eukryotic cell after the stable establishment of the endosymbiosis and must have been shifted from the plastid genome to the nucleus.
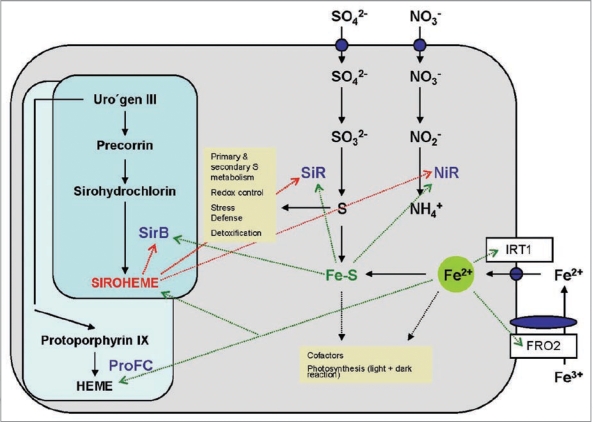
Figure 3.
Role of Fe, Fe-S clusters and siroheme in the metabolism of a plant cell. The synthesis of siroheme and heme from uroporphyrinogen III and the uptake and assimilation of S, N and Fe is shown. Dashed lines show the requirement of these compounds as cofactors and their involvement in biological pathways (yellow boxes). ProFC, protochlorophyllide ferrochelatase; IR T1, inducible iron transporter 1, FRO2, ferric-chelate reductase 2.
Uroporphyrinogen III is a Light Sensitive Component
Interestingly, the tetrapyrrole primogenitor uroporphyrinogen III is the first light-sensitive compound in the tetrapyrrole biosynthesis pathway. Accumulation of uroporphyrin III, a nonenzymatic oxidation product of uroporphyrinogen III, is likely to cause photodamage and to initiate ROS formation in the plastids and the cells. The tetrapyrolle biosynthesis pathway in higher plants is highly regulated through metabolic feedback inhibition of Glu tRNA reductase (HemA) by heme and Pchlide presumably via the FLU protein.28,29 HemA responsible for the synthesis of ALA is located upstream of UPM that methylates uroporphyrinogen III, and thus regulates ALA and siroheme biosynthesis. The feedback regulation ensures that the light-sensitive intermediates do not accumulate in the chloroplasts, when they are not utilized for siroheme, chlorophyll or heme biosynthesis. A sophisticated control system must ensure that the pathways obtain sufficient substrate to fulfil their functions under a given situation. Manipulation of the siroheme biosynthesis pathway may result in the accumulation of uroporphyrinogen III that is oxidised to highly photoactive uroporphyrin III leading to tetrapyrrolemediated type II photosensitization reaction that generates 1O2 (singlet oxygen).30–32 Furthermore, elevated levels of ROS activate the stimulation of the scavenging system and might have consequences for the redox status in the cell. ROS formation in the plastids and activation of the scavenging systems might influence the redox status of cytoplasm/nucleus. Due to severe metabolic and redox changes, stress and defense related genes are activated (cf. below). Since all enzymes of the scavenging systems are nuclear-encoded, disturbance in the tetrapyrrole biosynthesis pathway and ROS production in the plastids might influence the retrograde signaling from the plastids to the nucleus.33
Crucial Metabolic Pathway Depending on Siroheme
Primary S and N metabolism.
Sulphate is taken up from the soil by the roots via high affinity sulphate uptake systems (Fig. 3). Reduction to sulphite by the sulphate reductase and to sulphide via a 6 electron-transfer reaction by SiR occurs in the plastids, before the reduced S is integrated into cysteine and methionine. The S-containing amino acids are then distributed to all other compartments of the cell, where they can be integrated into proteins or used for the synthesis of other S-containing compounds, such as redox-active peptides or heavy metal-complexing metallothioneins. Many enzymes of S assimilation are subjected to complex regulations via transcriptional, post-transcriptional and translational processes, control of the enzyme activities and feedback processes.34
The vast majority of reduced N in the plants derives from nitrate assimilation (Fig. 3), while legumes in association with rhizobia can also form nodules and fix N from the air (cf. below). Nitrate is taken up by the roots by a number of high- and low-affinity transporters (Fig. 3) and reduced in the cytoplasm to nitrite by nitrate reductase. Further reduction of nitrite to ammonium occurs in the plastids via a 6-electron transfer reaction mediated by NiR (Fig. 3). In photosynthetic organisms, the electrons for the reduction of nitrite to ammonium derive from ferredoxin of photosynthetic electron transport chain. Besides siroheme, SiR and NiR contain Fe and Fe-S clusters as cofactors, which suggests that Fe homeostasis in the cell might play a regulatory role in the control of sulphate and nitrate assimilation (Fig. 3).
Recently, the transcription factor SLIM1 has been identified as a central transcriptional regulator of the primary and secondary S metabolism in Arabidopsis.35 Furthermore, a forward screen identified SLIM1 as an essential component in the beneficial interaction between P. indica and Arabidopsis (Sherameti et al. unpublished). In the yeast-two hybrid system, SLIM1 also forms dimers with MYB72, a transcription factor that is required for induced systemic resistance (ISR) in Arabidopsis.36,37 If this holds true for the in vivo situation, SLIM1 is directly involved in the plant/microbe interaction (reviewed in ref.38). It is conceivable, at least for Brassicaceae, that the S metabolism is controlled by beneficial microbes for two reasons: to strengthen plant performance by a better supply of S, and to promote the S-dependent defense machinery for better protection against pathogens and/or to maintain a balanced growth of the beneficial microbe in the host (cf. below).
Redox regulation, detoxification of the cell.
One of the primary mechanisms of a cell is to maintain an environmental-dependent balance of the oxidized and reduced form of glutathione to maintain the cellular redox state.39,40 Glutathione biosynthesis is a key component of plant stress responses and counteracts oxidative damage. The synthesis of glutathione occurs in two ATP-dependent steps: glutamate-cysteine ligase catalyzes the formation of γ-glutamylcysteine from cysteine and Glu, the rate limiting step in the pathway. Glutathione synthase adds Gly to γ-glutamylcysteine to yield glutathione. The reduced form of glutathione provides a substrate for multiple cellular reactions that yield oxidized glutathione, in which two molecules are linked by a disulphide bridge. Regulation of the glutathione pool is complex. It occurs at the transcriptional level for enzymes responsible for synthesis and utilization of glutathione. It is also regulated post-translationally by enzyme modifications, the availability of substrates and feedback loops. Glutathione is also the substrate for metallothioneins, which chelate and detoxify excess heavy metals in the cell.
Extreme temperatures, drought, pathogen attacks, or chemicals/ toxins induce the production of toxic levels of ROS.40–44 Detoxification of ROS, xenobiotics and heavy metals requires gluthatione which acts as an antioxidant and plays a key role in the ascorbate-glutathione cycle.45–50 Changes in redox status also occur during immune responses in different organisms. Redox changes regulate the conformation of NPR1, a master regulator of salicylic acid-mediated defense genes.51–53 NPR1 is sequestered in the cytoplasm as an oligomer through intermolecular disulphide bonds. S-nitrosylation of NPR1 facilitates its oligomerization, whereas salicylic acid-induced NPR1 oligomer-to-monomer reaction is catalyzed by thioredoxins53 and the transition of cytoplasmic NPR1 to the nucleus. Thus, oxidative stress and H2O2 production results in the oxidation of redox sensors which needs to be counteracted by the plant. In contrast, beneficial microbes establish reducing conditions in the cell.54,55
Secondary S metabolism.
Besides the requirement of S in the primary metabolism, it also plays an important role in the biosynthesis of glucosinolates. In Brassicaceae, up to 30% of the S can be incorporated into glucosinolates, S-rich plant metabolites of the order Brassicales that function in the defense of plants against pests and pathogens.56–58 It appears that different S-containing antimicrobial substances of the secondary S-metabolism contribute to the resistance against necrotrophic and biotropic fungi. A role of glucosinolates in beneficial plant/microbe interactions has also been postulated.59 Their breakdown products might restrict hyphal growth, which is necessary to maintain the interaction in a beneficial stage. Glucosinolates are also important as flavor components, cancer-prevention agents, and crop biofumigants. Glucosinolate accumulation depends intimately on the S status of the entire plant. S fertilisation usually led to an increase in glucosinolate content ranging from 25% to more than 50-fold. The effect was greater on glucosinolates derived from methionine than from tryptophan. This is regulated by extensive gene transcription and under the control of SLIM1. In S-deficient plants, there is a general downregulation of glucosinolate biosynthetic genes which accompanies an upregulation of genes controlling S uptake and assimilation. Glucosinolates may be considered a potential source of S for other metabolic processes under low-S conditions, since increased breakdown of glucosinolates has been reported under S deficiency. However, the pathway for S mobilisation from glucosinolates has not been determined.56,60 The breakdown of indolic glucosinolates to form auxin in roots under S-deficient conditions may help stimulate root formation for S uptake.60
Camalexin is probably the best characterized phytoalexin from Arabidopsis which is induced by a large variety of plant pathogens. It is substituted with S- and N-containing side chains, which again highlights the importance of siroheme for plant/microbe interactions. Both biotrophic and necrotrophic plant pathogens as well as beneficial microbes induce camalexin, and this includes bacteria, viruses and fungi. Camalexin can be induced in shoots and roots61 and the major inducers are ROS, since also abiotic factors generating ROS (such as heavy metals, UV-B light, ROS, chemicals, etc.) activate camalexin biosynthesis. Besides ROS, salicylic acid signaling and the redox state of glutathione are important for the induction. Although the formation of camalexin in response to many biotic and abiotic substances is well documented, we are only at the beginning to understand the function of camalexin in plant defense.
Fe Metabolism
Proton transporting ATPases in the plasma membrane of Fe-deficient roots increase the solubility of Fe3+ hydroxides by generating a slightly acidic pH in the apoplast. The lower pH stimulates the activity of the plasma membrane-bound Fe3+ chelate reductase FRO2 which transfers electrons from intracellular NADH to extracellular Fe3+. This enzyme activity is co-regulated with the Fe-regulated transporter IRT1 (Fig. 3). Within the cells, Fe is distributed into the different subcellular locations i.e., plastids, mitochondria, cytoplasm and vacuole.62 At the appropriate cellular compartment, it is bound to ferritin or chelatases that bring the Fe to the required positions, or it can be directly bound by the enzymes which require Fe as a cofactor. Considering the complexity of these processes and the central role of Fe for siroheme synthesis, S and N assimilation and as a cofactor for several enzymes, it is tempting to speculate that the Fe homeostasis in the cell or even in the organelles may play a crucial regulatory role in all S and N requiring metabolic processes.
Fe is also central for rhizobacteria-mediated N2-fixation in legumes, since leghaemoglobin and the nitrogenase contain huge amounts of Fe. While the Fe-containing heme cofactor of leghaemoglobin is synthesized by the bacterium, the apoprotein is of plant origin. Furthermore, the nitrogenase consists of two subunits: subunit 1 contains MoFe cofactors and subunit 2 Fe cofactors. Thus, the availability of huge amounts of Fe and the proper insertion into the proteins is a prerequisite for the function of a symbiotic nodule.
Fe homeostasis might also be important for Fe-S cluster biosynthesis in plant cells. Although the chemical structure of the Fe-S clusters is simple and the cluster can self-assemble under anaerobic conditions, under aerobic conditions, their biosynthesis requires dedicated proteins. The assembly process consists of three steps: (1) the release of S from cysteine, (2) co-assembly with Fe on a scaffold protein and (3) transfer of the nascent cluster to the target Fe-S protein.
Because of the unstable nature of Fe-S clusters, every cell and organelle contain Fe-S cluster assembly proteins. Fe-S cluster biosynthesis is quite old since all bacteria can synthesize them. Regulation and distribution of Fe-S clusters in the cells is complex.63 The majority of the Fe-S clusters in autotrophic, photosynthetic organisms is probably used for photosynthesis. For instance, the photosystem I complex contains 3 Fe-S clusters and represents one of the most abundant protein complexes in higher plants.64–68 Crucial enzymes involved in the dark reaction of photosynthesis contain also Fe-S centers.
Role in Organellar and Cellular Signaling
For many years intermediates of the chlorophyll biosynthesis have been proposed to play a crucial role in this signaling process.69 Although a direct involvement of chlorophyll intermediates appear to be less likely,70,71 impairments in chlorophyll biosynthesis still have an effect on the expression of nuclear genes for plastids proteins, for instance, following photodamaging effects and ROS production. Manipulation of siroheme biosynthesis might affect the accumulation of the light-sensitive uroporphyrinogen III (or other light-sensitive intermediates) if they are not used for chlorophyll and/or heme pathways. This might interfere with the feedback regulation from heme and protochlorophyllide to ALA. Microarray analyses will provide important information on the expression of the nuclear encoded genes for proteins of the metabolic and signaling pathways which are controlled by siroheme (Fe, N and S metabolism; Fe-S cluster biosynthesis; ROS scavenging systems; redox control; glucosinolate biosynthesis; defense responses; plastid → nucleus signaling; etc.). A comparative analyses of these data with those from photodamaged plastids, plastids which are impaired in specific signal processes or plastids which accumulate ROS due to photodamage will clearly demonstrate the role of heme and siroheme intermediates in the retrograde signaling.72
Open Questions
Considering the important role of siroheme in central cellular processes in plants, many questions remain unanswered:
What determines the distribution of uroporphyrinogen III between the two pathways? Determining physiological parameters such as photosynthetic activities or chlorophyll accumulation in wild-type and knock-down lines in the absence or presence of Fe-chelator α-α-dipyridyl needs to be performed to understand the competition between ferrochelatase and siroheme chelatase leading to synthesis of Fe-protoporphyrin IX and siroheme. Generating reduced uroporphyrinogen III pools in wild-type, knock-down and overexpressor lines in the presence of levulinic acid that inhibits uroporphyrinogen synthesis or in the presence of ALA in dark that causes high accumulation of the substrate for siroheme, heme and chlorophyll biosynthesis, will shine light on this regulation. The distribution of uroporphyrinogen III between the two pathways should be dependent on external parameters such as nutrient availability or other stress, or sensed by the Fe-S centers in SirB. Again, the presence of an Fe-S center in SirB and its unstability to reduction might play a role in branchpoint regulation. Redox-signals may allow or prevent protein-protein interactions, which can control the efficiencies of the two branches of the pathways. Since this cofactor is highly sensitive to external manipulations and redox agents, it might have an ion-sensing function. Therefore, plants impaired in upm and/or sirb might be highly sensitive to reduced Fe and/or S concentrations, since not only the biochemical pathways are impaired if the ions are limiting, but that the limitation might be perceived ahead of time by the plants. The Fe-S center in SirB could be involved in redox-sensing and/or the initiation of signaling pathways that regulate downstream processes. Finally, the enzyme catalysing the second step in the biosynthesis is not known in plants. Different biological disciplines are required to tackle these questions.
Acknowledgements
Supported by the DFG-INSA and DFG OE13/28-1.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10173
References
- 1.Leustek T, Martin MN, Bick JA, Davies JP. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:141–165. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- 2.Sato S, Soga T, Nishioka T, Tomita M. Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection. Plant J. 2004;40:151–163. doi: 10.1111/j.1365-313X.2004.02187.x. [DOI] [PubMed] [Google Scholar]
- 3.Kopriva S, Wiedemann G, Reski R. Sulphate assimilation in basal land plants–what does genomic sequencing tell us? Plant Biol. 2007;9:556–564. doi: 10.1055/s-2007-965430. [DOI] [PubMed] [Google Scholar]
- 4.Höfgen R, Kreft O, Willmitzer L, Hesse H. Manipulation of thiol contents in plants. Amino Acids. 2001;20:291–299. doi: 10.1007/s007260170045. [DOI] [PubMed] [Google Scholar]
- 5.Townsend DM, Tew KD, Tapiero H. Sulfur containing amino acids and human disease. Biomed Pharmacother. 2004;58:47–55. doi: 10.1016/j.biopha.2003.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawlor DW. Carbon and N assimilation in relation to yield: mechanisms are the key to understanding production systems. J Exp Bot. 2002;53:773–787. [PubMed] [Google Scholar]
- 7.Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. Nitrate transport and signalling. J Exp Bot. 2007;58:2297–2306. doi: 10.1093/jxb/erm066. [DOI] [PubMed] [Google Scholar]
- 8.Leustek T, Smith M, Murillo M, Singh DP, Smith AG, Woodcock SC, et al. Siroheme biosynthesis in higher plants. Analysis of an S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase from Arabidopsis thaliana. J Biol Chem. 1997;272:2744–2752. doi: 10.1074/jbc.272.5.2744. [DOI] [PubMed] [Google Scholar]
- 9.Lillo C. Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J. 2008;415:11–19. doi: 10.1042/BJ20081115. [DOI] [PubMed] [Google Scholar]
- 10.Crane BR, Siegel LM, Getzoff ED. Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange. Biochem. 1997;36:12101–12119. doi: 10.1021/bi971065q. [DOI] [PubMed] [Google Scholar]
- 11.Crane BR, Getzoff ED. The relationship between structure and function for the sulfite reductases. Curr Opin Struct Biol. 1996;6:744–756. doi: 10.1016/s0959-440x(96)80003-0. [DOI] [PubMed] [Google Scholar]
- 12.Cornah JE, Terry MJ, Smith AG. Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 2003;8:224–230. doi: 10.1016/S1360-1385(03)00064-5. [DOI] [PubMed] [Google Scholar]
- 13.Raux-Deery E, Leech HK, Nakrieko KA, McLean KJ, Munro AW, Heathcote P, et al. Identification and characterization of the terminal enzyme of siroheme biosynthesis from Arabidopsis thaliana: a plastidlocated sirohydrochlorin ferrochelatase containing a 2FE-2S center. J Biol Chem. 2005;280:4713–4721. doi: 10.1074/jbc.M411360200. [DOI] [PubMed] [Google Scholar]
- 14.Warren MJ, Scott AI. Tetrapyrrole assembly and modification into the ligands of biologically functional cofactors. Trends Biochem Sci. 1990;15:486–491. doi: 10.1016/0968-0004(90)90304-t. [DOI] [PubMed] [Google Scholar]
- 15.Warren MJ, Stolowich NJ, Santander PJ, Roessner CA, Sowa BA, Scott AI. Enzymatic synthesis of dihydrosirohydrochlorin (precorrin-2) and of a novel pyrrocorphin by uroporphyrinogen III methylase. FEBS Lett. 1990;261:76–80. doi: 10.1016/0014-5793(90)80640-5. [DOI] [PubMed] [Google Scholar]
- 16.Moser J, Schubert WD, Beier V, Bringemeier I, Jahn D, Heinz DW. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 2001;20:6583–6590. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schön A, Krupp G, Gough S, Berry-Lowe S, Kannangara CG, Söll D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature. 1986;322:281–284. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- 18.Phillips JD, Whitby FG, Kushner JP, Hill CP. Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase. EMBO J. 2003;22:6225–6233. doi: 10.1093/emboj/cdg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakakibara H, Takei K, Sugiyama T. Isolation and characterization of a cDNA that encodes maize uroporphyrinogen III methyltransferase, an enzyme involved in the synthesis of siroheme, which is prosthetic group of nitrite reductase. Plant J. 1996;10:883–892. doi: 10.1046/j.1365-313x.1996.10050883.x. [DOI] [PubMed] [Google Scholar]
- 20.Raux E, McVeigh T, Peters SE, Leustek T, Warren MJ. The role of Saccharomyces cerevisiae Met1p and Met8p in sirohaem and cobalamin biosynthesis. Biochem J. 1999;338:701–708. [PMC free article] [PubMed] [Google Scholar]
- 21.Schubert HL, Raux E, Brindley AA, Leech HK, Wilson KS, Hill CP, et al. The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase. EMBO J. 2002;21:2068–2075. doi: 10.1093/emboj/21.9.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raux E, Leech HK, Beck R, Schubert HL, Santander PJ, Roessner CA, et al. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin in Bacillus megaterium. Biochem J. 2003;370:505–516. doi: 10.1042/BJ20021443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson P, Hederstedt L. Organization of genes for tetrapyrrole biosynthesis in gram—positive bacteria. Microbiol. 1999;145:529–538. doi: 10.1099/13500872-145-3-529. [DOI] [PubMed] [Google Scholar]
- 24.Warren MJ, Roessner CA, Santander PJ, Scott AI. The Escherichia coli cysG gene encodes S-adenosylmethionine-dependent uroporphyrinogen III methylase. Biochem J. 1990;265:725–729. doi: 10.1042/bj2650725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren MJ, et al. Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem J. 1994;302:837–844. doi: 10.1042/bj3020837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert HL, Raux E, Wilson KS, Warren MJ. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38:10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- 27.Brindley AA, Raux E, Leech HK, Schubert HL, Warren MJ. A story of chelatase evolution: identification and characterization of a small 13–15-kDa “ancestral” cobaltochelatase (CbiXS) in the archaea. J Biol Chem. 2003;278:22388–22395. doi: 10.1074/jbc.M302468200. [DOI] [PubMed] [Google Scholar]
- 28.McCormac AC, Fischer A, Kumar AM, Söll D, Terry MJ. Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J. 2001;25:549–561. doi: 10.1046/j.1365-313x.2001.00986.x. [DOI] [PubMed] [Google Scholar]
- 29.Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004;40:957–967. doi: 10.1111/j.1365-313X.2004.02262.x. [DOI] [PubMed] [Google Scholar]
- 30.Tripathy BC, Chakraborty N. 5-Aminolevulinic acid induced photodynamic damage of the photosynthetic electron transport chain of cucumber (Cucumis sativus L) Cotyledons. Plant Physiol. 1991;96:761–767. doi: 10.1104/pp.96.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakraborty N, Tripathy BC. Involvement of singlet oxygen in photodynamic damage of isolated chloroplasts of cucumber (Cucumis sativus L.) cotyledons. Plant Physiol. 1992;98:7–11. doi: 10.1104/pp.98.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathy BC, Mohapatra A, Gupta I. Impairment of the photosyntheticparatus by oxidative stress induced by photosensitization reaction of protoporphyrin IX. Biochim Biophys Acta. 2007;1767:860–868. doi: 10.1016/j.bbabio.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Heiber I, Ströher E, Raatz B, Busse I, Kahmann U, Bevan MW, et al. The redox imbalanced mutants of Arabidopsis differentiate signaling pathways for redox regulation of chloroplast antioxidant enzymes. Plant Physiol. 2007;143:1774–1788. doi: 10.1104/pp.106.093328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirai MY, Saito K. Post-genomics approaches for the elucidation of plant adaptive mechanisms to sulphur deficiency. J Exp Bot. 2004;55:1871–1879. doi: 10.1093/jxb/erh184. [DOI] [PubMed] [Google Scholar]
- 35.Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Ent S, Verhagen BW, Van Doorn R, Bakker D, Verlaan MG, Pel MJ, et al. MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 2008;146:1293–1304. doi: 10.1104/pp.107.113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segarra G, Van der Ent S, Trillas I, Pieterse CM. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009;11:90–96. doi: 10.1111/j.1438-8677.2008.00162.x. [DOI] [PubMed] [Google Scholar]
- 38.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 39.Meister A. Glutathione metabolism. Methods Enzymol. 1995;252:26–30. doi: 10.1016/0076-6879(95)52005-8. [DOI] [PubMed] [Google Scholar]
- 40.Inzé D, van Montagu M. Oxidative stress in plants. Curr Opin Biotechnol. 1995;6:153–158. [Google Scholar]
- 41.Noctor G, Foyer CH. Ascorbate and glutathione: Keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 42.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer AJ, Hell R. Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res. 2005;86:435–457. doi: 10.1007/s11120-005-8425-1. [DOI] [PubMed] [Google Scholar]
- 44.Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F. Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007;49:159–172. doi: 10.1111/j.1365-313X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- 45.May MJ, Leaver CJ. Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 1993;103:621–627. doi: 10.1104/pp.103.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang C, Werner BL, Christensen EM, Oliver DJ. The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 2001;126:564–574. doi: 10.1104/pp.126.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer AJ, Fricker MD. Control of demand-driven biosynthesis of glutathione in green Arabidopsis suspension culture cells. Plant Physiol. 2002;130:1927–1937. doi: 10.1104/pp.008243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 49.Mullineaux PM, Rausch T. Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth Res. 2005;86:459–474. doi: 10.1007/s11120-005-8811-8. [DOI] [PubMed] [Google Scholar]
- 50.Hicks LM, Cahoon RE, Bonner ER, Rivard RS, Sheffield J, Jez JM. Thiol-based regulation of redoxactive glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell. 2007;19:2653–2661. doi: 10.1105/tpc.107.052597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somssich IE. Closing another gap in the plant SAR puzzle. Cell. 2003;113:815–816. doi: 10.1016/s0092-8674(03)00473-2. [DOI] [PubMed] [Google Scholar]
- 52.Pieterse CM, Van Loon LC. NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol. 2004;7:456–464. doi: 10.1016/j.pbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, et al. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baltruschat H, Fodor J, Harrach BD, Niemczyk E, Barna B, Gullner G, et al. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 2008;180:501–510. doi: 10.1111/j.1469-8137.2008.02583.x. [DOI] [PubMed] [Google Scholar]
- 55.Vadassery J, Tripathi S, Prasad R, Varma A, Oelmüller R. Monodehydroascorbate reductase 2 and dehydroascorbate reductase 5 are crucial for the mutualistic interaction between Piriformospora indica and Arabidopsis. J Plant Physiol. 2009;166:1263–1274. doi: 10.1016/j.jplph.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 56.Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 57.Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 58.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherameti I, Venus Y, Drzewiecki C, Tripathi S, Dan VM, Nitz I, et al. PYK10, a β-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J. 2008;50:1–17. doi: 10.1111/j.1365-313X.2008.03424.x. [DOI] [PubMed] [Google Scholar]
- 60.Falk KL, Tokuhisa JG, Gershenzon J. The effect of sulfur nutrition on plant glucosinolate content: physiology and molecular mechanisms. Plant Biol. 2007;9:573–581. doi: 10.1055/s-2007-965431. [DOI] [PubMed] [Google Scholar]
- 61.Bednarek P, Schneider B, Svatos A, Oldham NJ, Hahlbrock K. Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 2005;138:1058–1070. doi: 10.1104/pp.104.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeong J, Guerinot ML. Homing in on iron homeostasis in plants. Trends Plant Sci. 2009;14:280–285. doi: 10.1016/j.tplants.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009. p. e1000497. [DOI] [PMC free article] [PubMed]
- 64.Pilon M, Abdel-Ghany SE, Van Hoewyk D, Ye H, Pilon-Smits EA. Biogenesis of iron-sulfur cluster proteins in plastids. Genet Eng. 2006;27:101–117. doi: 10.1007/0-387-25856-6_7. [DOI] [PubMed] [Google Scholar]
- 65.Fillebeen C, Pantopoulos K. Redox control of iron regulatory proteins. Redox Rep. 2002;7:15–22. doi: 10.1179/135100002125000136. [DOI] [PubMed] [Google Scholar]
- 66.Eisenstein RS. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr. 2000;20:627–662. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 67.Kühn LC, Hentze MW. Coordination of cellular iron metabolism by post-transcriptional gene regulation. J Inorg Biochem. 1992;47:183–195. doi: 10.1016/0162-0134(92)84064-t. [DOI] [PubMed] [Google Scholar]
- 68.Hein P, Oelmüller R. Photosystem I and regulatory proteins for its biogenesis. Funct Plant Sci Biotech. 2007;1:106–111. [Google Scholar]
- 69.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 70.Moulin M, McCormac AC, Terry MJ, Smith AG. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA. 2008;105:15178–15183. doi: 10.1073/pnas.0803054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid- to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fey V, Wagner R, Braütigam K, Wirtz M, Hell R, Dietzmann A, et al. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem. 2005;280:5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]