Abstract
The vast quantities of antibiotics used in modern agriculture contaminate the environment and threaten human health. Recent studies have shown that crop plants grown in soil fertilized with manure from antibiotic-treated animals can accumulate antibiotic within the plant body, thus making them an additional antibiotic exposure route for consumers. Until recently, mechanisms of antibiotic entry and subcellular partitioning within plant cells were virtually unknown. We have uncovered and characterized a transporter gene in Arabidopsis thaliana, MAR1, which appears to control antibiotic entry into the chloroplast. Antibiotic resistance via MAR1 is specific to the aminoglycoside class, and is conferred by loss-of-function mutations, which is rather unusual, since most transporter-based antibiotic resistance is conferred by overexpression or gain-of-function mutations in efflux pumps with poor substrate specificity. Since MAR1 overexpression lines exhibit various iron starvation phenotypes, we propose that MAR1 transports an iron chelation molecule that is mimicked specifically by aminoglycoside antibiotics, and this facilitates their entry into the chloroplast. Knowledge about MAR1 enhances our understanding of how antibiotics might enter the plant cell, which may aid in the production of crop plants that are incapable of antibiotic accumulation, as well as further the development of new plant-based antibiotic resistance markers.
Key words: antibiotic, contamination, transport, import, chloroplast, membrane, iron, chelation, nicotianamine
Antibiotic Contamination of Crop Plants: An Emerging Public Health Problem
The amount of antibiotics used non-therapeutically in agriculture is estimated to be eight times greater than the amount used in all of human medicine,1 and accounts for about 70% of total antibiotic use in the United States.2 Until recently, major concerns about antibiotic use in agriculture have been related to their contamination of animal-based food products—such as milk and meat—and their pollution of the water supply via farm runoff. However, many antibiotics are not well absorbed in the animal gut and are excreted largely unchanged in manure,3,4 some retaining 75% of their activity after 2 years in soil.5 Despite this known presence and persistence, there are no guidelines on the presence of antibiotics in manure,6 and crop plants fertilized with antibiotic-contaminated manure are now emerging as additional antibiotic exposure routes. There is a growing body of work describing the ability of various plants to accumulate measurable levels of antibiotic after growth on contaminated soils,7–10 but unanswered questions remain about the molecular nature of this uptake, especially for hydrophilic antibiotics, which do not readily diffuse across membranes.
Antibiotic Resistance in Plants: A Poorly Studied Area
It is widely recognized that plants are sensitive to many antibiotics, and this fact has been exploited for the benefit of both basic and applied plant science to produce transgenic plants. Transgenic plant selection systems are often based on expression of bacterial genes, which typically provide resistance to antimicrobial compounds via enzymatic inactivation.11–13 Most of these enzymes are specific for one or a few particular aminoglycoside antibiotics, which generally target the translational machinery of prokaryotes. Because the eukaryotic organellar translational machinery is prokaryotic in nature, these antibiotics target chloroplast and mitochondrial translation in plants. Until now, endogenous, high-level resistance to aminoglycosides in plant lines has typically been found to be due to specific changes in organellar ribosomal subunits.14,15
Drug resistance in bacteria has been well studied, and is often conferred by expression of multidrug efflux transporters with low substrate specificity.16 These transporters encompass several protein families including ATP binding cassette (ABC) transporters, the major facilitator superfamily (MFS), the multidrug and toxic compounds efflux (MATE) family and others.17 However, in plants, there are only a few reports of antibiotic resistance that are based on overexpression of efflux transporters. In one study, it was shown that overexpression of an endogenous Arabidopsis thaliana ABC transporter gene, AtWBC19, confers kanamycin resistance in plants.18 Levels of resistance were similar to levels attained through expression of the bacterial neomycin phosphotransferase II (nptII), which is one of the most commonly used selectable markers in plants. The discovery and characterization of AtWBC19 has sparked the hope that plant-based antibiotic transport proteins may be promising new candidates for selectable markers.19
Since AtWBC19 is likely to be involved in antibiotic sequestration to the vacuole, it can be overexpressed for use as a marker. However, antibiotics must enter the cell in order to function, and a block of entry may also be sufficient to generate resistance. Once plant-endogenous antibiotic import proteins are uncovered and characterized, additional markers may be developed via RNAi-mediated downregulation of these proteins. Perhaps not surprisingly, movement of aminoglycoside antibiotics across the bacterial inner membrane involves energy-dependent transport,20,21 and recent work suggests that uptake of antibiotic into plants is also an energy-dependent process.22 Unfortunately, the specific plant transporter proteins capable of recognizing and importing antibiotics have remained unknown until now.
MAR1: A Gateway for Antibiotics into Plant Chloroplasts
We have recently uncovered and characterized a transport protein of Arabidopsis thaliana, MAR1, which we have shown is capable of transporting multiple aminoglycoside antibiotics.23 The original EMS mutant, mar1-1, which has a single amino acid change in a putative transmembrane domain of the protein, was resistant to the aminoglycosides kanamycin, tobramycin, gentamicin, streptomycin, amikacin and apramycin. However, mar1-1 showed no resistance to the non-aminoglycosides spectinomycin, chloramphenicol, lincomycin and tetracycline, or to aminoglycosides that inhibit both prokaryotic and eukaryotic translation (G418, hygromycin and paromomycin). Two independent T-DNA insertions in MAR1 were able to phenocopy the multiple resistance phenotype of mar1-1, while MAR1 overexpression lines were hypersensitive to aminoglycosides. Thus, MAR1 stands out as rather unusual in that it appears to recognize only one specific group of antibiotics, and resistance to these antibiotics is conferred by loss-of-function mutations.
Using a MAR1-YFP fusion protein, we went on to show that MAR1 localizes to the chloroplast, and is likely to be an inner membrane protein that allows entry of aminoglycoside antibiotics into the stroma, where they interfere with organellar (prokaryotic) translation.23 Thus, when MAR1 is disrupted, resistance is not seen to aminoglycosides that would interfere with cytoplasmic (eukaryotic) translation, as their entry into the cytoplasm is not barred. Evidence for the antibiotic transport functionality of MAR1 was uncovered using both yeast and isolated chloroplasts. Yeast expressing MAR1 cDNA were found to be hypersensitive to the aminoglycoside G418, but not to the non-aminoglycoside, chloramphenicol. Yeast expressing the mar1-1 mutant cDNA were also hypersensitive, but this hypersensitivity was intermediate between wild-type MAR1 yeast and empty vector controls.23 Thus, the mar1-1 mutant protein, with single amino acid change, may still have partial functionality.
We developed a novel assay to detect antibiotic in chloroplast extracts, and used this assay to measure antibiotic content of chloroplasts from mutant, wild-type and MAR1 overexpression lines. By spotting chloroplast lysates onto nitrocellulose membrane and using an antibody against gentamicin, we were able to show that lysates from mutant plants accumulated less gentamicin than wild-type, while overexpression lines hyper-accumulated gentamicin.23 Taken together, our data illustrate that MAR1 does, in fact, act to transport aminoglycoside antibiotics into the chloroplast.
MAR1 Transports Antibiotic Opportunistically
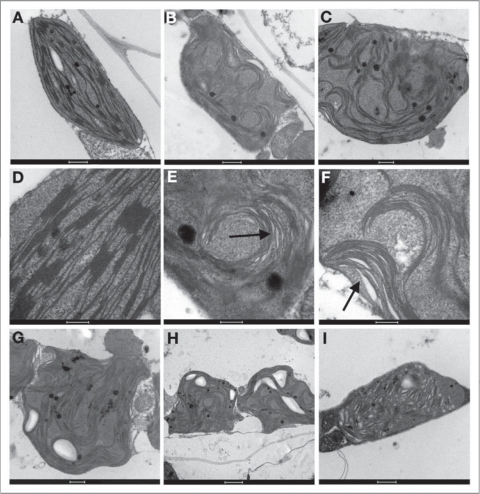
It is, of course, unlikely that evolutionary pressures would have selected for a means of entry for toxic antibiotics into plant chloroplasts. Thus, is it probable that the transport of antibiotics via MAR1 is opportunistic in nature. We have found that MAR1 overexpression lines are chlorotic, and this chlorosis can be rescued by iron supplementation. Additionally, MAR1 expression is downregulated under limiting iron conditions.23 We have therefore proposed that overexpression of MAR1 effectively creates a condition of iron limitation in the chloroplast. Since iron deficiency is often associated with alterations in chloroplast ultrastructure, we have recently used TEM to investigate the chloroplast ultrastructure of MAR1 overexpression lines (35S::MAR1). When compared to wild-type, we observed highly disorganized and misaligned thylakoid membranes, lack of proper grana stacking, and an overall distended and distorted shape in 35S::MAR1 chloroplasts (Fig. 1), all of which are symptoms of iron deficiency.24–26
Figure 1.
Chloroplast shape and ultrastructure in MAR 1 overexpression lines are distorted to varying degrees. TEM images of chloroplasts from Ler wild-type (A and D) and 35S::MAR1 (B, C and E-I). (E and F) are closeup images of (B and C), respectively. In (E and F), black arrows point to swollen lamellae. (I) illustrates swollen lamellae throughout, with no evidence of proper grana stacking. (G and H) illustrate gross shape distortion. Scale bars: (A-C), 500 nm; (D-F), 200 nm; (G and I), 500 nm; (H), 1 µm.
We have previously proposed that the iron limitation in MAR1 overexpressors could occur due to overaccumulation of an iron chelation molecule that aminoglycoside antibiotics are able to mimic structurally, thus obtaining entry into the chloroplast. Because aminoglycosides are known to mimic polyamines, and may exploit polyamine inward transport systems to gain entrance to cells,16 we have proposed that the elusive natural substrate of MAR1 may be the polyamine iron chelator, nicotianamine (NA). Mature MAR1 overexpression lines have a leaf chlorosis pattern that is opposite that of the NA-less chloronerva mutant of tomato (Lycopersicon esculentum)27—instead of interveinal chlorosis in young tissues, chlorosis arises in the midvein and in older tissues.23 We have hypothesized that this may be the result of a re-distribution of the cytoplasmic NA pool to the chloroplast, thus restricting NA from performing its role in phloem transport of iron and other metals.28
One of the MAR1 homologs in Arabidopsis, AtIREG1, was postulated to be involved in vessel loading of iron,29 and its downregulation in DwMYB2 overexpressors may be the cause of the disruption in iron translocation (from root to shoot) observed in these plants.30 Because iron is typically complexed with citrate in the xylem,31 we feel it is possible that AtIREG1 exports citrate (or an iron-citrate conjugate) from root cells into the vasculature. Thus, AtIREG1 may be playing a role similar to FRD3, which mediates citrate efflux into root vasculature,32 and is also downregulated in DwMYB2 overexpressors.30 More research is needed on AtIREG1 in order to explore this possibility. We postulate that the IREG proteins (AtIREG1, 2 and MAR1) may emerge as a family of transporters, distinct from the MATE and YSL families, which are also capable of transporting metal chelates or metal-chelate complexes.
Conclusions
Our work has begun to shed light on the possible mechanisms by which antibiotics—in this case, the particularly hydrophilic aminoglycosides—can cross membranes and reach their targets within plant cells. The MAR1 transporter is not solely dedicated to the transport of antibiotics, but rather, antibiotics are able to mimic the natural substrate of MAR1 to obtain entry into the chloroplast. It is interesting to note that many known drug and antibiotic efflux proteins in bacteria also have a more “conventional” function. For example, the E. coli cmr chloramphenicol efflux pump bears sequence similarity to several sugar transporters.33 The B. subtilis Blt drug transporter exports chloramphenicol and puromycin34 as well as polyamines.35 The mexAB/oprM multidrug efflux operon of P. aeruginosa is involved in efflux of tetracycline, chloramphenicol, several quinolones, and a range of β-lactams,36 but is also regulated by iron concentration, and proposed to be involved in the secretion of the iron chelator, pyoverdine, under conditions of iron starvation.37,38 Therefore, like these antibiotic export proteins, antibiotic importers may also be metabolite transporters that have been hijacked by antibiotic.
Our research on MAR1 has contributed to knowledge regarding the processes of antibiotic entry in plants, which is currently in its infancy. Further knowledge about antibiotic entry pathways could eventually enable the production of crop plants that are incapable of antibiotic accumulation, thus protecting consumers and keeping our food supply safe and healthy. This knowledge may also aid us in development of new plant-based molecular markers, and generally contributes to the understanding of how plants interact with the antibiotics they encounter, both in the laboratory and in the natural environment.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10142
References
- 1.Mellon M, Benbrook C, Benbrook K. Hogging it: Estimates of antimicrobial abuse in livestock. Cambridge, MA: Union of Concerned Scientists; 2001. [Google Scholar]
- 2.Florini K, Denison R, Stiffler T, Fitzgerald T, Goldburg R. Resistant bugs and antibiotic drugs: State and county estimates of antibiotics in agricultural feed and animal waste. Washington, DC: Environmental Defense; 2005. [Google Scholar]
- 3.Mackie R, Koike S, Krapac I, Chee-Sanford J, Maxwell S, Aminov R. Tetracycline residues and tetracycline resistance genes in groundwater impacted by swine production facilities. Anim Biotechnol. 2006;17:157–176. doi: 10.1080/10495390600956953. [DOI] [PubMed] [Google Scholar]
- 4.Sarmah A, Meyer M, Boxall A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006;65:725–759. doi: 10.1016/j.chemosphere.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Environmental Assessment for Apralan Premix for Swine, authors. Technical Report. Indianapolis: Elanco Products Company,; 1985. pp. 1–65. [Google Scholar]
- 6.Kumar K, Gupta S, Chander Y, Singh A. Antibiotic use in agriculture and its impact on the terrestrial environment. Adv Agron. 2005;87:1–54. [Google Scholar]
- 7.Migliore L, Cozzolino S, Flori M. Phytotoxicity to and uptake of enrofloxacin in crop plants. Chemosphere. 2003;52:1233–1244. doi: 10.1016/S0045-6535(03)00272-8. [DOI] [PubMed] [Google Scholar]
- 8.Kumar K, Gupta S, Baidoo S, Chander Y, Rosen C. Antibiotic uptake by plants from soil fertilized with animal manure. J Environ Qual. 2005;34:2082–2085. doi: 10.2134/jeq2005.0026. [DOI] [PubMed] [Google Scholar]
- 9.Boxall A, Johnson P, Smith E, Sinclair C, Stutt E, Levy L. Uptake of veterinary medicines from soils into plants. J Agr Food Chem. 2006;54:2288–2297. doi: 10.1021/jf053041t. [DOI] [PubMed] [Google Scholar]
- 10.Dolliver H, Kumar K, Gupta S. Sulfamethazine uptake by plants from manure-amended soil. J Environ Qual. 2007;36:1224–1230. doi: 10.2134/jeq2006.0266. [DOI] [PubMed] [Google Scholar]
- 11.Fraley R, Rogers S, Horsch R, Sanders P, Flick J, Adams S, et al. Expression of bacterial genes in plant cells. Proc Natl Acad Sci USA. 1983;80:4803–4807. doi: 10.1073/pnas.80.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayford M, Medford J, Hoffman N, Rogers S, Klee H. Development of a plant transformation selection system based on expression of genes encoding gentamicin acetyltransferases. Plant Physiol. 1988;86:1216–1222. doi: 10.1104/pp.86.4.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kavanagh TA, O’Driscoll KM, McCabe PF, Dix PJ. Mutations conferring lincomycin, spectinomycin, and streptomycin resistance in Solanum nigrum are located in three different chloroplast genes. Mol Gen Genet. 1994;242:675–680. doi: 10.1007/BF00283422. [DOI] [PubMed] [Google Scholar]
- 15.Rosellini D, LaFayette P, Barone P, Veronesi F, Parrott W. Kanamycin-resistant alfalfa has a point mutation in the 16S plastid rRNA. Plant Cell Rep. 2004;22:774–779. doi: 10.1007/s00299-004-0757-3. [DOI] [PubMed] [Google Scholar]
- 16.Van Bambeke F, Balzi E, Tulkens P. Antibiotic efflux pumps. Biochem Pharmacol. 2000;60:457–470. doi: 10.1016/s0006-2952(00)00291-4. [DOI] [PubMed] [Google Scholar]
- 17.Paulsen I. Multidrug efflux pumps and resistance: regulation and evolution. Curr Opin Microbiol. 2003;6:446–451. doi: 10.1016/j.mib.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Mentewab A, Stewart CN., Jr Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat Biotechnol. 2005;23:1177–1180. doi: 10.1038/nbt1134. [DOI] [PubMed] [Google Scholar]
- 19.Rea P. A farewell to bacterial ARMs? Nat Biotechnol. 2005;23:1085–1087. doi: 10.1038/nbt0905-1085. [DOI] [PubMed] [Google Scholar]
- 20.Taber H, Mueller J, Miller P, Arrow A. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987;51:439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao W, Warren M, Lee A, Mistry A, Lomovskaya O. MexXY-OprM efflux pump is required for antagonism of aminoglycosides by divalent cations in Pseudomonas aeruginosa. Antimicrob Agents Chemoth. 2001;45:2001–2007. doi: 10.1128/AAC.45.7.2001-2007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong W, Zhu Y, Liang Y, Zhang J, Smith F, Yang M. Uptake of oxytetracycline and its phytotoxicity to alfalfa (Medicago sativa L.) Environ Pollut. 2007;147:187–193. doi: 10.1016/j.envpol.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Conte S, Stevenson D, Furner I, Lloyd A. Multiple antibiotic resistance in Arabidopsis thaliana is conferred by mutations in a chloroplast-localized transport protein. Plant Physiol. 2009;151:559–573. doi: 10.1104/pp.109.143487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platt-Aloia K, Thomson W, Terry N. Changes in plastid ultrastructure during iron nutrition-mediated chloroplast development. Protoplasma. 1983;114:85–92. [Google Scholar]
- 25.Durrett T, Connolly E, Rogers E. Arabidopsis cpFtsY mutants exhibit pleiotropic defects including an inability to increase iron deficiency-inducible root Fe(III) chelate reductase activity. Plant J. 2006;47:467–479. doi: 10.1111/j.1365-313X.2006.02803.x. [DOI] [PubMed] [Google Scholar]
- 26.Graziano M, Beligni M, Lamattina L. Nitric oxide improves internal iron availability in plants. Plant Physiol. 2002;130:1852–1859. doi: 10.1104/pp.009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling H, Koch G, Baumlein H, Ganal M. Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA. 1999;96:7098–7103. doi: 10.1073/pnas.96.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Wiren N, Sukhbinder K, Suhkibar B, Briat J, Khodr H, Shioiri T, et al. Nicotianamine chelates both FeIII and FeII. Implications for metal transport in plants. Plant Physiol. 1999;119:1107–1114. doi: 10.1104/pp.119.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curie C, Briat J. Iron transport and signaling in plants. Annu Rev Plant Biol. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Wu X, Ling H, Yang W. Transgenic expression of DwMYB2 impairs iron transport from root to shoot in Arabidopsis thaliana. Cell Research. 2006;16:830–840. doi: 10.1038/sj.cr.7310099. [DOI] [PubMed] [Google Scholar]
- 31.Hell R, Stephan U. Iron uptake, trafficking and homeostasis in plants. Planta. 2003;216:541–551. doi: 10.1007/s00425-002-0920-4. [DOI] [PubMed] [Google Scholar]
- 32.Durrett T, Gassmann W, Rogers E. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144:197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsen I, Bakke I, Vader A, Olsvik O, El-Gewely M. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J Bacteriol. 1996;178:3188–3193. doi: 10.1128/jb.178.11.3188-3193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neyfakh A, Bidnenko V, Chen L. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jack D, Yang N, Saier M. The drug/metabolite transporter superfamily. Eur J Biochem. 2001;268:3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- 36.Paulsen I, Brown M, Skurray R. Proton-dependent multidrug eff lux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole K, Heinrichs D, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 38.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]