Abstract
The small ubiquitin modifier (SUMO) conjugation/deconjugation is an important regulatory progress in plant development and responses to abiotic stresses. However, much less is known about the roles of sumoylation in plant root development. Cytokinin and auxin play crucial roles in determining the balance between cell proliferation and cell differentiation in Arabidopsis roots. The SUMO E3 ligase AtMMS21 is a homologue of human NSE2/MMS21, which modulates DNA damage and DNA repair in human cells. This addendum summarizes our recent paper on the AtMMS21 mediating cytokinin signaling to regulate the root meristem cell proliferation. The mms21-1 roots had reduced responses to exogenous cytokinins and decreased expression of the cytokinin-induced genes ARR3, ARR4, ARR5 and ARR7, compared with the wild type. Furthermore, the expression of CRE1 and ARR1, which are both the receptor and positive regulator of cytokinin signaling, was also reduced in the mms21-1 mutant plants.
Key words: Arabidopsis thaliana, AtMMS21, cytokinin, root meristem, SUMO E3 ligase
All organisms use a variety of chemical modifiers for post-translational control of proteins that affect development, growth and homoeostasis. Ubiquitin is one such polypeptide that was first described to attach covalently to other proteins upon completion of their synthesis. SUMOs (small ubiquitin-like modifiers) are ubiquitin-like proteins with three-dimensional structures that are similar to ubiquitin.1 Similar to ubiquitination, sumoylation of substrates is catalyzed by a cascade of enzymes: the E1 SUMO-activating enzyme, the E2 conjugating enzyme and the E3 SUMO ligases.2 Sumoylation of target proteins in yeast and metazoans has been implicated in the regulation of innate immunity, cell cycle progression and mitosis, DNA repair, chromatin stability, cell division, nucleo-cytoplasmic trafficking, subnuclear targeting, ubiquitination antagonism and transcriptional regulation.3,4 PIAS/Siz proteins are SUMO E3 ligases that mediate the final step of SUMO conjugation. 2 Transcription factors are direct targets of SUMO conjugation mediated by PIAS/Siz proteins.4 The yeast SUMO E3 ligases Siz1 and Siz2 facilitate cell division at low temperatures.5 The yeast MMS21 and its human homologue of MMS21 contain an SP-RING domain that is related to the PIAS family of SUMO E3 ligases that autosumoylate, and are required for the prevention of DNA damage-induced apoptosis by facilitating DNA repair.6,7 SUMO conjugation/deconjugation in plants has been implicated in responses to heat shock, oxidative stress, hypoxia, phosphate limitation, ABA, flowering and pathogen defense.8–13
Recently, the Arabidopsis SUMO E3 ligase SIZ1 has been shown to participate in responses to phosphate starvation, salicylic acid-mediated signaling in plant pathogen defense, ABA signaling and basal thermotolerance, and in the development progress of flowering.11–16 Until now, the function of sumoylation in regulating the plant root development has not reported.
Here we demonstrated the role of Arabidopsis SUMO E3 ligase AtMMS21 in apical root meristem development. AtMMS21 null mutant mms21-1 exhibited severe short primary root phenotype caused by apical root meristem cell froliferation defect that linked to imparied expression of the cell division marker CYCB1:GUS. The mms21-1 roots had reduced sensitivity to exogenous cytokinins, and decreased expression of the cytokinins-induced genes ARR3, ARR4, ARR5 and ARR7. Interesting, the expression of AtMMS21 was downregulated obviously by exogenous cytokinin.17 We also detected that the expressions of the cytokinin receptor gene CRE1 and B-type ARRs gene ARR1 were decreased obviously in the mms21-1 loss-of-function mutant allete plants (Fig. 1). These results establish that AtMMS21 is invovled in cytokinin signaling in regualting root cell division and meristem development. Meanwhile, AtMMS21 (also named as HIGH PLOID2, HPY2) was considered to function as a repressor of endocycle onset in Arabidopsis meristems by regulating the cell cycle progression through the PLT-dependent auxin signaling pathway.18
Figure 1.
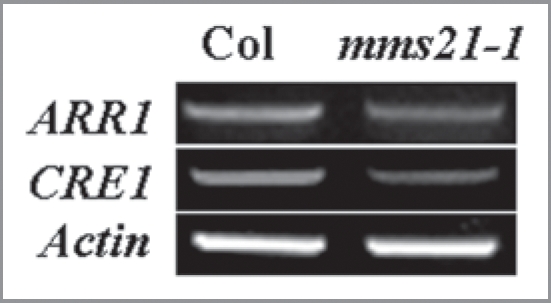
Expression of CRE1 and ARR1 genes were reduced in mms21-1 loss-of-function mutant plants.
Cytokinin and auxin play crucial roles in determining the balance between cell proliferation and cell differentiation in Arabidopsis roots. For meristem maintance and to enable continous root growth, the rate of cell differentiation must equal to the rate division in the root meristem. It has showed that this balance is the result of crosstalk between cytokinin and auxin through SHY2 gene, a member of the auxin repressor Aux/IAA gene family. 19 Auxin promote cell division in the root meristem and expansion/differentiation after cells have left the meristem.20 Cytokinin, on the other hand, counteracts auxin to promote cell expansion/differentiation at the root meristem.21,22 It has demonstrated that cytokinin does not interfere with specification of the QC and the stem cell function, nor with the overall division rate in the proximal meristem. Cytokinin affects primarily the rate of meristematic cell differentiation, resulting in shortening of the meristematic zone.21 The negative role of cytokinin on root growth has been proven by both exogenous cytokinin application and overexpression of the ISOPENTENYLTRANSFERASE (IPT) gene.23,24 Interestingly, the root meristem was reduced in the triple cytokinin receptor mutant ahk2, ahk3, ahk4 and multiple mutant in ahp members of the signal transduction cascade.25–28 Based on these phenotypes, it has supposed that the optimal cytokinin concentration is needed for the root meristem development.29 However, the specific function pathways of auxin and cytokinin and their interaction mechanisms in the root meristem are still unclear.
In wild-type roots, the transcription of AtMMS21 is decreased by cytokinin application. Furthermore, the mms21-1 loss-of-function mutant allete displays smaller root meristems, mimicking the effects of cytokinin application and auxin lack. Hence, application of exogenous cytokinin to the mms21-1 loss-of-function mutant allete did not trigger an obvious decrease in the root meristem size. Meanwhile, AtMMS21 can regulate positively the cytokinin receptor gene expression such as CRE1, and the ARRs genes expression such as ARR1, ARR3, ARR4, ARR5 and ARR7. These results indicate that the AtMMS21 protein is both necessary and sufficient for the cytokinin-mediated control of meristem size. Conversely, AtMMS21-mediated auxin signaling positively regulating the root meristem size by promoting the root meristem cell division. These regulation progresses sustaining the homeostasis between cytokinin and auxin signaling, and keeping the balance of root meristem cell division and differentiation. Based on these results, it can be speculated that AtMMS21 is a node component between cytokinin and auxin signaling to regulate the Arabidopsis root meristem development (Fig. 2).
Figure 2.
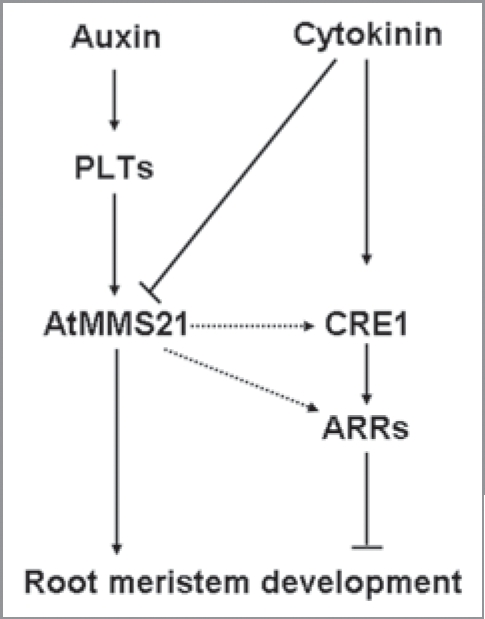
Proposed model of AtMM S21 function in root meristem development. AtMM S21 acts as a node element of cytokinin and auxin interaction. It plays role positively in the PLTs-mediated auxin signaling pathway to promote the root meristem cell division on one hand.18 AtMM S21 is inhibited by cytokinin to modulate the root meristem cell differenciation on the other hand.17 AtMM S21 also may stimulate cytokinin signaling by inducing the expression of CRE1 and ARRs genes to sustaining the homeostasis between cytokinin and auxin signaling in the progress of root meristem development.
Therefore, the SUMO E3 ligase AtMMS21 plays important roles in the root meristem maintance and growth. But the mechanisms of AtMMS21-mediated cytokinin signaling to regulate root meristem growth are yet to be established. Further researches on the identification of AtMMS21 target proteins, and their functions in cytokinin signaling and root meristem growth, are necessary to eluciate the mechanisms of AtMMS21-mediated sumoylation modulate the progress of root meristem growth and development.
Acknowledgements
This research was supported by the Program for New Century Excellent Talents in University (NCET-08-0644), the National Natural Science Foundation of China (30900789, 30770201) and the Natural Science Foundation of Guangdong (2007B020701005).
Addendum to: Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03992.x.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10158
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitinlike proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 4.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 6.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and −2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 9.Lois LM, Lima CD, Chua NH. Small ubiquitinlike modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell. 2003;15:1347–1359. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 14.Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 18.Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, et al. SUMO E3 Ligase HIGH PLOIDY2 Regulates Endocycle Onset and Meristem Maintenance in Arabidopsis. Plant Cell. 2009;21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 20.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 21.Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 22.Ruzicka K, Simaskova M, Duclercq J, Petrasek J, Zazimalova E, Simon S, et al. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA. 2009;106:4284–4289. doi: 10.1073/pnas.0900060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuderova A, Urbankova I, Valkova M, Malbeck J, Nemethova D, Hejatko J. Effects of conditional IPT-dependent cytokinin overproduction on root architecture of Arabidopsis seedlings. Plant Cell Physiol. 2008;49:570–582. doi: 10.1093/pcp/pcn029. [DOI] [PubMed] [Google Scholar]
- 24.Medford JI, Horgan R, El-Sawi Z, Klee HJ. Alterations of endogenous cytokinins in transgenic-plantsusing achimeric isopentenyl transferase gene. Plant Cell. 1989;1:403–413. doi: 10.1105/tpc.1.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18:3073–3087. doi: 10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riefler M, Novak O, Strnad M, Schmulling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development and cytokinin metabolism. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira FJ, Kieber JJ. Cytokinin signaling. Curr Opin Plant Biol. 2005;8:518–525. doi: 10.1016/j.pbi.2005.07.013. [DOI] [PubMed] [Google Scholar]


