Abstract
The RCD1 protein is a regulator of both developmental and stress responses in Arabidopsis thaliana and it interacts with several transcription factors. Its closest homolog, SRO1, seems to be dispensable for proper plant responses but the hardly viable phenotype of the rcd1 sro1 double mutant reveals that it encodes a functional protein that can partially compensate for the loss of RCD1 in the single rcd1 mutant. Both RCD1 and SRO1 contain a WWE domain, the catalytic core of poly(ADP-ribose) polymerases and a novel conserved domain termed RST which is also found in the transcription initiation complex component TAF4.
Here we summarize recent findings on the protein-protein interactions mediated by RCD1 and highlight the different functional possibilities that form the basis of our future experiments concerning the biochemical function of RCD1.
Key words: reactive oxygen species, signal transduction, RST, proteinprotein interaction, DREB2A
The Radical-induced Cell Death1 (RCD1) protein is a key regulator of several ROS-and abiotic stress-related responses in Arabidopsis thaliana. The rcd1 mutant plant displays several pleiotrophic phenotypes including ozone and salt sensitivity, UV-B and methyl viologen tolerance, early flowering and senescence. Additionally, it is an NO overproducer and has alterations in its responses to ethylene, salicylic acid and methyl jasmonate.1–5 The Arabidopsis genome encodes a close homolog of RCD1, called SRO1 (Similar to RCD1 One1). In contrast to rcd1, the sro1 single mutant displays only subtle phenotypes6 but the protein is necessary for plant development since the rcd1 sro1 double mutant is severely disturbed in its development.6,7
RCD1 seems to be involved in extensive protein-protein interactions: One of the earliest findings was reported by Lin and Heaton in 2001,8 when RCD1 was recovered in a yeast 2-hybrid (Y2H) screen with a viral movement protein from turnip crinkle virus. Katiyar-Agarwal and coworkers5 described its interaction with the cytoplasmic tail of Salt Overly Sensitive1 (SOS1), a well-known component of salt tolerance pathway. Furthermore, Miao and colleagues9 discussed a putative glutathione peroxidase3- RCD1 interaction and hypothesized that RCD1 could be a plant equivalent of the yeast redox-regulated transcription factor Yap1. Their idea was inspired by the earlier Y2H results by Belles-Boix and coworkers10 where two transcription factors, DREB2A and STO were found to interact with RCD1. Our group extended on these findings by doing a large-scale pair-wise Y2H interaction testing against the REGIA TF collection,11 and found a large number of RCD1-interacting TFs from several protein families, predominantly AP2/ERF, NAC and bHLH.7 All of the transcription factor interactions and the SOS1 interaction seemed to take place at the C-terminal end of the RCD1 protein.5,12
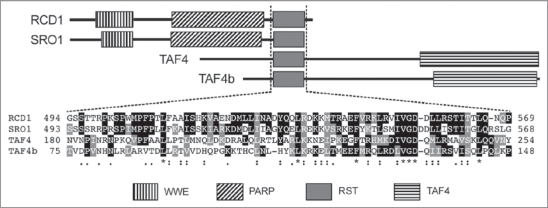
Both RCD1 and SRO1 contain three distinguishable domains: an N-terminal WWE domain which in Drosophila is known to mediate protein-protein interactions,13 and a poly(ADP-ribose) polymerase (PARP) catalytic domain that could be involved in protein modification reactions. The third conserved domain was recently identified in the C-terminus of RCD1 and named RST for RCD1-SRO-TAF4, since in addition to RCD1 and SRO1, it was found in four other homologs of these proteins (SRO2-SRO5) as well as in TBP-associated factor4,7 (TAF4; Fig. 1). The RST domain was present in RCD1-like proteins throughout the plant kingdom and consists of about 70 amino acids. The presence of the RST domain in TAF4s, which are involved in the assembly of the multimeric general transcription factor complex TFIID,14 indicated that this domain was likely to be involved in specifying protein-protein interactions. Furthermore, the large number of RCD1-TF interactions and similarity to a component of the transcription initiation complex hint that RCD1 might be involved in a protein complex that somehow regulates the function of the RCD1-interacting TFs.
Figure 1.
The structure of Arabidopsis RST domain-containing proteins RCD 1, SRO1, TA F4 and TA F4b and the amino acid sequence of the individual RST domains. The degree of amino acid conservation between the proteins within the RST domain is highlighted.
RCD1 and SRO1 are very similar on amino acid level (61% of the amino acids in the proteins are identical and 74% similar) and based on the double mutant phenotype SRO1 is a functional protein. However, the absence of a phenotype in the sro1 single mutant indicated that there must be some critical difference(s) between the two proteins. Do they differ in some key amino acids that confer specificity in interaction or are they differentially regulated on a post-translational level? In Y2H, RCD1 interacted with more transcription factors than SRO1, this suggests that the critical difference between the proteins might lie in the RST domain (Fig. 1). We are currently addressing these questions by creating domain swap constructs and targeted mutations to the proteins and assessing their functionality in mutant complementation and TF interaction experiments.
Biochemical activity of RCD1 or SRO1 has not been experimentally verified. Although their domain structure indicates a possible poly(ADP-ribose) polymerase activity, we are inclined to believe this is not the case (Vainonen JP, Overmyer K, Jaspers P, Kangasjärvi J, unpublished results). Instead, RCD1 could have a mechanistically similar activity or the domain could represent a necessary conserved structure without any enzymatic activity. Alternatively, the PARP domain could be a cofactor binding domain through which the function of the rest of the protein is regulated. This is an especially tempting scenario keeping in mind that the PARP domain is accompanied by two different putative protein-protein interaction domains, WWE and RST. Thus RCD1 or SRO1 could act as a scaffold protein and assemble TFs for their post-translational modification, relocalization or degradation.
Acknowledgements
The work was supported by the Academy of Finland Postdoctoral grants to K.O. (decision # 115034) and M.B. (decision # 108760) and by the Centre of Excellence program (2006–2011) to the laboratory of J.K. P.J. additionally acknowledges the Viikki Graduate School in Biosciences (VGSB).
Abbreviations
- RCD1
radical-induced cell death1
- ROS
reactive oxygen species
- NO
nitric oxide
- SRO1
similar to RCD1 one1
- Y2H
yeast two-hybrid
- REGIA
regulatory gene initiative in arabidopsis
- TF
transcription factor
- PARP
poly(ADP-ribose) polymerase
- RST
RCD1-SRO-TAF4
- TAF4
TBP-associated factor4
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10293
References
- 1.Ahlfors R, Brosché M, Kollist H, Kangasjärvi J. Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J. 2009;58:1–5. doi: 10.1111/j.1365-313X.2008.03756.x. [DOI] [PubMed] [Google Scholar]
- 2.Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, et al. Ozonesensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849–1862. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujibe T, Saji H, Arakawa K, Yabe N, Takeuchi Y, Yamamoto KT. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 2004;134:275–285. doi: 10.1104/pp.103.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahlfors R, Macioszek V, Rudd J, Brosché M, Schlichting R, Scheel D, Kangasjärvi J. Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone Exposure. Plant J. 2004;40:512–522. doi: 10.1111/j.1365-313X.2004.02229.x. [DOI] [PubMed] [Google Scholar]
- 5.Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu JK. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:18816–18821. doi: 10.1073/pnas.0604711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teotia S, Lamb RS. The paralogous genes RADICALINDUCED CELL DEATH1 AND SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol. 2009;151:180–198. doi: 10.1104/pp.109.142786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009;60:268–279. doi: 10.1111/j.1365-313X.2009.03951.x. [DOI] [PubMed] [Google Scholar]
- 8.Lin B, Heaton LA. An Arabidopsis thaliana protein interacts with a movement protein of Turnip crinkle virus in yeast cells and in vitro. J Gen Virol. 2001;82:1245–1251. doi: 10.1099/0022-1317-82-5-1245. [DOI] [PubMed] [Google Scholar]
- 9.Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell. 2006;18:2749–2766. doi: 10.1105/tpc.106.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belles-Boix E, Babiychuk E, Van Montagu M, Inzé D, Kushnir S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 2000;482:19–24. doi: 10.1016/s0014-5793(00)02016-0. [DOI] [PubMed] [Google Scholar]
- 11.Paz-Ares J. The REGIA Consortium. REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp Funct Genomics. 2002;3:102–108. doi: 10.1002/cfg.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahlfors R, Låang S, Overmyer K, Jaspers P, Brosché M, Tauriainen A, et al. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell. 2004;16:1925–1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zweifel ME, Leahy DJ, Barrick D. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure. 2005;13:1599–1611. doi: 10.1016/j.str.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 14.Marr MT. TAF4 takes flight. Proc Natl Acad Sci USA. 2009;106:1295–1296. doi: 10.1073/pnas.0812990106. [DOI] [PMC free article] [PubMed] [Google Scholar]