Maintenance of DNA integrity is critical for all cell types, but neurons are particularly sensitive to mutations in DNA repair genes, which lead to both abnormal development and neurodegeneration 1. We describe a previously unknown autosomal recessive disease characterized by microcephaly, early-onset, intractable seizures and developmental delay (MCSZ). Using genome-wide linkage analysis in consanguineous families, we mapped the disease locus to chromosome 19q13.33 and identified multiple mutations in PNKP that results in severe neurological disease, whereas a splicing mutation is associated with more moderate symptoms. Surprisingly, while patient cells are sensitive to radiation and other DNA damaging agents, no patient has yet developed cancer or immunodeficiency. Unlike other DNA repair defects that affect humans, PNKP mutations universally cause severe seizures. The neurological abnormalities in MCSZ patients may reflect a role for PNKP in several DNA repair pathways.
We identified the autosomal recessive disorder MCSZ with the following features: microcephaly, infantile onset seizures, developmental delay, and variable behavioral problems especially hyperactivity. MCSZ was observed in multiple pedigrees of Middle Eastern and European origin (Figure 1). The first three pedigrees were Arabic Palestinians living in Jordan and the USA. Three other families with similar manifestations were Arabic (Kingdom of Saudi Arabia), Turkish, and of mixed European ancestry (USA). We later found less severely affected patients from Family 7, which is also American of mixed European heritage (USA). Brain MRIs consistently show microcephaly with preserved brain structures, without apparent neuronal migration or other structural abnormalities, and with no evidence of degeneration (Figure 2). The patients did not develop ataxia or other neurological symptoms. Routine clinical genetic and metabolic screening showed no abnormalities. Despite careful inquiry, MCSZ patients did not have a higher frequency of common or uncommon infections, offering no clinical evidence of immunodeficiency. Cells from one patient showed sensitivity to irradiation in a standard colony survival assay2,3. However, no patient has developed cancer by age 21 and heterozygous carriers have not developed early onset cancer or any sign of immunodeficiency (see Clinical Information and Summary in Supplementary Note).
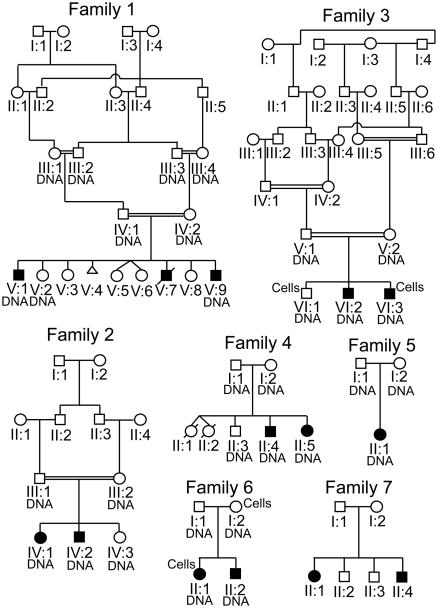
Figure 1. Pedigrees of MCSZ families.
Family 1 represents a consanguineous Palestinian pedigree in Jordan. Family 2 shows another consanguineous Palestinian pedigree reportedly unrelated to Family 1 also in Jordan. Family 3 is also consanguineous and Palestinian but now in the USA. Family 4 is from the Kingdom of Saudi Arabia and the parents were not known to be consanguineous. Family 5 is from Turkey and the parents were not known to be related. Family 6 is of mixed European descent from the USA (German-Irish). Family 7 is also of mixed European (Swedish, Italian, Irish and English) heritage from the USA. The individuals from whom samples were obtained are labeled “DNA”. The individuals from whom we established lymphoid cell lines are labeled “Cells”. Cells and DNA were available for all Family 7 members.
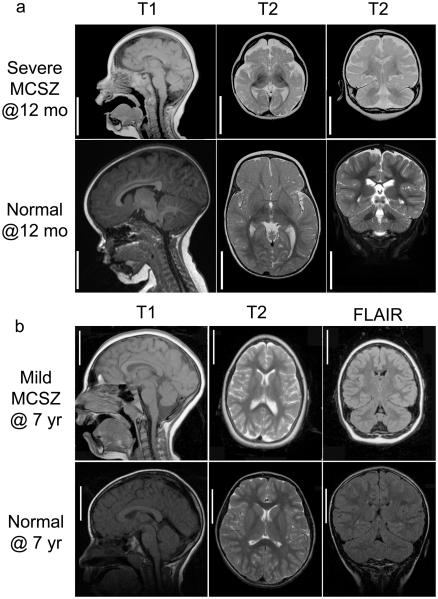
Figure 2. Brain MRIs of MCSZ patients.
Representative MRI images are shown from Families 4 (a, severely affected) and 7 (b, moderately affected) with aged matched controls. MRIs of severely affected patients from other families were similar to the representative images in a. Sagittal images are shown on the left (T1), axial images in the middle (T2) and coronal images on the right (T2 (a) and FLAIR (b)) with the MRI sequence noted above the image. The MRIs illustrate that despite the microencephaly (small brain), the gyral pattern is not clearly abnormal indicating absence of visible neuronal migration abnormality. The cerebellum is proportionately small compared to the cerebrum and the subpallium (basal ganglia or ventral cerebrum) is proportionately with the pallium (dorsal cerebrum). There is no evidence of atrophy or glial scarring. Bar=5 cm for both unaffected and MCSZ images.
Genome-wide linkage screens suggested a single common homozygous region on chromosome 19q in all six MCSZ patients in the consanguineous Families 1-3. This same locus showed homozygosity in the two patients from Family 4 and the single patient from Family 5 whose parents were not known to be related. No other regions of linkage were observed including known primary microcephaly loci. Interestingly, all six patients in the three Palestinian families (Families 1-3) were homozygous for the same 3cM/1.5Mb haplotype (between markers D19S879 and D19S907) at chromosome 19q13.33, suggesting a common ancestor (Supplementary Information Figure 1). Further investigation revealed a common town of origin for these families. Linkage analysis of Families 1-4 and 6 generated a combined maximum two point LOD score of 5.60 at D19S867 with θ =0 (Supplementary Information Table 2) and a maximum multipoint LOD score of 7.12 (Supplementary Information Figure 2).
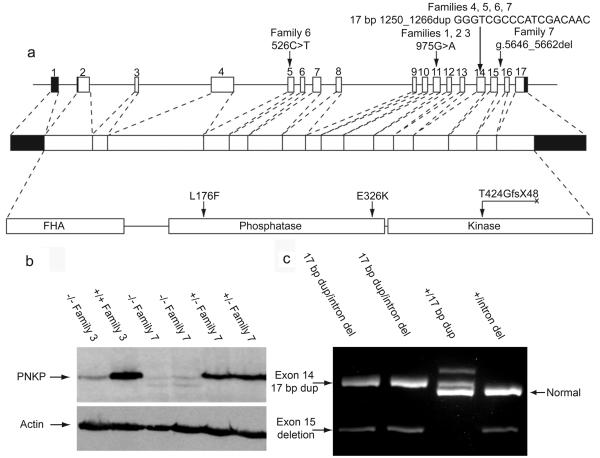
We sequenced 41 genes within the minimal region (see Supplementary Information) and only the PNKP gene contained mutations. Homozygous mutations were present in each of the first five families and compound heterozygous mutations were found in Families 6 and 7 (Figure 3 and Supplementary Information Figure 3 for detailed analyses of all mutations). The three Palestinian Families (Families 1-3) shared a homozygous base pair substitution in exon 11 (975G>A), resulting in the non-conservative amino acid change E326K. The families from the Kingdom of Saudi Arabia (Family 4) and Turkey (Family 5) had the same homozygous 17 bp duplication (17 bp dup) in exon 14 (1250_1266dup), resulting in a frame-shift, T424GfsX48. Family 6 (European) had two heterozygous mutations: the same 17 bp dup in exon 14 as the Saudi and Turkish families and a point mutation in exon 5 (526C>T) resulting in L176F. The moderately affected members of Family 7 (European) displayed compound heterozygosity, carrying the 17 bp dup mutation in exon 14 and a 17 bp deletion in intron 15 that disrupts proper mRNA splicing.
Figure 3. PNKP mutations in MCSZ patients.
(a) Four different mutations diagrammed in human PNKP genomic DNA, mRNA, and protein including domains (forkhead is FHA). The human PNKP gene consists of 17 exons shown as boxes and encodes a peptide of 521 amino acids. Filled boxes represent untranslated regions and open boxes represent coding regions. Lines connecting the exons represent introns. (b) Western blot for PNKP, the first and second lanes show a MCSZ patient (VI:3 with E326K mutation) and unaffected brother (VI:1), respectively, from Family 3. The third and fourth lanes show MCSZ patients (II:1 and 4, both with 17 bp dup and 17 bp intron 15 del) while the fifth and sixth lanes the father (I:1) and brother (II:3) both heterozygous for 17 bp intron 15 del. The band is ~60 kD (57 kD predicted size). Anti-β-actin is a loading control. (c) RT-PCR products of mRNA from members of Family 7 show the expected size from the normal copy of PNKP cDNA, 636 bp, seen in lanes 3 and 4 from non-affected carriers. The 17 bp dup results in a 653 bp long fragment seen in lanes 1, 2 and 3. The 548 bp band in lanes 1, 2, and 4 show samples with the intron 15 deletion lacking exon 15 (determined from sequencing, data not shown). A small amount of normal sized transcript is seen in lanes 1 and 2 with higher exposure (data not shown), indicating that a small amount of normal PNKP mRNA by be produced.
To confirm that these mutations were pathogenic, we analyzed patient-derived, Epstein Bar Virus (EBV)-transformed, lymphocytes from Families 3 (E326K) and 7 (17 bp dup/intron 15 del). The samples from the severely affected patient (Family 3) and the mildly affected patients (Family 7) had greatly diminished PNKP protein compared to unaffected or heterozygous family members (Figure 3b). RT-PCR analysis showed that the intron 15 deletion in Family 7 disrupts mRNA splicing and causes skipping of exon 15; a barely detectable level of properly spliced mRNA remains (Figure 3c). The low levels of PNKP protein and the indistinguishable phenotype among severely affected individuals from Families 1-6 suggest that all of these mutations impair PNKP severely or completely, whereas patients in Family 7 that exhibit a slightly milder phenotype and carry one non-coding mutation may retain some PNKP activity.
Surprisingly, despite their diverse ancestries (Saudi, Turkish, and mixed European), all chromosomes with the 17 bp dup mutation appear to share the same haplotype for 18 SNPs in or near the PNKP locus (Supplementary Figure 4a). The low frequency of this mutant haplotype (Supplementary Figure 4b), suggests that the 17 bp dup in exon 14 is likely of shared origin and very old in all or most of these families. Nonetheless, the mutation shows an extremely low carrier frequency in the general population, as the 17 bp dup mutation was absent in 1080 Middle Eastern or Caucasian control chromosomes, while all other mutations were absent in all of at least 280 control chromosomes screened.
PNKP has been implicated in repair of both double strand breaks (DSBs) and single strand breaks (SSBs) 4,5, since its phosphatase domain removes 3′ phosphates and the kinase domain phosphorylates 5′ hydroxyl groups, required for DNA ligation 6. PNKP's forkhead domain mediates interaction with the non-homologous end joining (NHEJ) complex via XRCC4, or the SSB and base excision repair (BER) pathways via XRCC1 (Supplemental Figure 6 a and b)4-8. PNKP has been further implicated in the repair pathway disrupted in an ataxic neurodegenerative disease, spinocerebellar ataxia with axonal neuropathy, SCAN1 (TDP1) (see Supplementary Information Figure 6 for additional details). The abnormalities in a clinical irradiation sensitivity test already reflect deficiencies in NHEJ required to repair radiation induced DSBs. Because PNKP potentially plays roles in additional repair pathways, we tested cells in response to other DNA damaging agents.
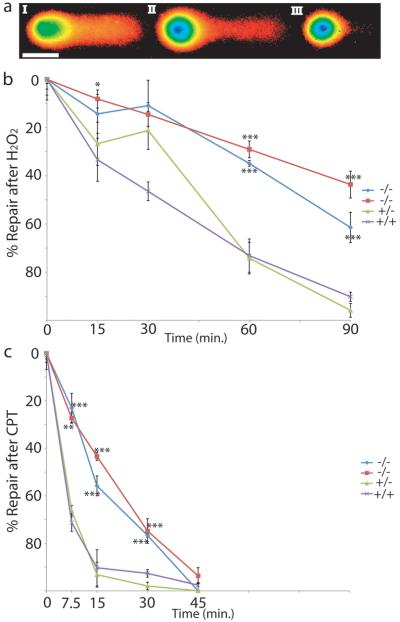
The response to free radical damage from hydrogen peroxide, predominately requiring BER, and camptothecin, requiring TDP1 via its activity on topoisomerase I, was examined via alkaline comet assay which detects both SSBs and DSBs by quantitating the amount of DNA that moves from a nucleus after electrophoresis. Typical nuclei with various amounts of DNA damage are shown in Figure 4a. The relative amount of DNA damage was determined by measuring the % Tail DNA (DNA that has moved out of the nucleus). The relative amount of DNA damage repair was quantitated as % Tail DNA after toxin exposure at various times of recovery (see Supplementary Figure 5 for all data) and normalized to the maximum and minimum damage for each cell line in each experiment. MCSZ derived cell lines were significantly impaired in their ability to repair hydrogen peroxide induced damage (Figure 4b) and were also delayed in repairing camptothecin induced damage (Figure 4c). However, MCSZ derived cells were eventually able to repair the camptothecin induced damage after 45 minutes while they were not able to repair all hydrogen peroxide induced damage even after 90 min. These data suggest that camptothecin induced damage may be easier to repair than free radical damage in the setting of PNKP mutations. Although MCSZ patients do not develop the ataxia that characterizes TDP1 mutations that also have camptothecin sensitivity 9,10, our patients are perhaps too young (oldest 21 years) or maybe there is a compensatory mechanism for the loss of PNKP.
Figure 4. Patient derived lymphocytes show abnormal DNA repair.
(a) Examples of comet assay results are shown. The intensity of the fluorescence is represented in pseudocolor while the electrical field drives damaged, loose DNA from left to right. (I) Shows a cell with 50% Tail DNA with the body of the nucleus (green) on the left and the tail of the comet derived from the damaged DNA extending to the right. (II and III) show progressively less damage, 29% and 11% Tail DNA respectively. (b) After hydrogen peroxide (H2O2) treatment with 0 minutes for recovery, cells show their maximum damage. Cells derived from MCSZ patients (blue, red), show significant impairment in their ability to repair DNA after hydrogen peroxide was removed, while cells derived from unaffected family members were able to repair DNA much more efficiently. (c) After camptothecin (CPT) treatment, there was also statistically significantly slower repair in cells derived from MCSZ patients compared to unaffected family members (green, purple) as well. However, after 45 minutes, the MCSZ derived cells were able to repair all CPT damage, in contrast to H2O2-treated cells. All cells derived from Family 7. Blue diamond and red square were from MCSZ patients with exon 14 17 bp duplication and intron 15 17 bp deletion (II:1 and II:4 respectively), green triangle were from unaffected parent heterozygous for the intron 15 17 bp deletion (I:1) and purple X from unaffected sibling with no mutation (II:2). *(P=0.05), **(P<0.005) and ***(P<0.0005). Scale bar shows 50 μm.
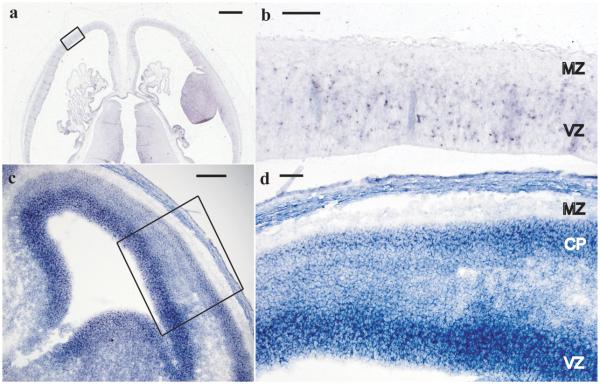
Microcephaly can result from failure to produce enough neurons during development (primary microcephaly) or degeneration after normal development. MCSZ patients have no evidence of brain atrophy or clinical regression, making degeneration unlikely (see Supplemental Note, Clinical Information for further details). In situ hybridization indicated that human and mouse PNKP mRNAs are expressed in both dividing neuronal precursors in the cerebral cortical ventricular zone (Figure 5 a and c VZ) as well as post-mitotic neurons of the cortical plate (Figure 5d CP) consistent with potential roles in both dividing and post-mitotic neurons. In addition, we found that when levels of Pnkp were reduced in dissociated mouse neurons in vitro via RNAi, there was a small but statistically significant increase in apoptosis in both neuronal precursors (Supplemental Information Figure 7c) and differentiated neurons (7d) compared to cells transfected with control plasmids. This indicates that microcephaly could result from apoptosis of precursors, differentiated neurons or both cell types.
Figure 5. PNKP in situ hybridization.
In situ hybridization of Carnegie Stage 22 human embryos (~54 postovulatory days) with anti-sense probe to human PNKP (a). Sense strand (not shown) showed no specific hybridization. Higher magnification image of developing cerebral cortex boxed area in (a) is shown in (b). Ventricular zone (VZ), containing proliferating cells, shows PNKP mRNA expression while the cell-sparse marginal zone (MZ) has no staining. Mouse E14 cerebral cortex (c) with high magnification of boxed region shown in (d) shows a similar staining pattern with high expression within the proliferating VZ and lower but maintained expression within differentiated neurons of the cortical plate (CP). (a) and (b) are in the transverse plane and (c) and (d) are coronal. Scale bars, (a) 1 mm, (b) 100 μm, (c) 150 μm, (d) 75 μm.
Although disruption of genes encoding NHEJ repair proteins leads to microcephaly in human and/or mouse in LIG4 syndrome, Lig411-13, severe combined immunodeficiency with microcephaly, NHEJ1 (cernunnos) 14, Xrcc4 15, Xrcc6 (Ku70)/ Xrcc5 (Ku80)16, and Prkdc (DNA-PKcs) 17, disruption of NHEJ function is not known to cause seizures, which are a prominent clinical feature of MCSZ. In contrast, disruption of the BER/SSB pathways by deletion of Xrcc1 in mice produced normal brain size but the mutant mice developed ataxia, loss of interneurons within the cerebellum, and seizure-like behavior 18. The intriguing similarity of phenotypes between patients with mutations in PNKP and mice with targeted deletion of Xrcc1 does suggest a potential common mechanism demonstrating the requirement for the BER/SSB pathways in the prevention of seizures, potentially via an interneuron specific requirement of the BER/SSB pathway. Therefore, the unique pattern of neurological symptoms of MCSZ may reflect a requirement for PNKP activity in multiple DNA repair pathways.
Online Methods
Genetic screening
Family 1 underwent a genome-wide linkage screen using about 400 microsatellite markers in the ABI linkage mapping set MD v2.5 at 10 cM average density (Applied Biosystems) at the Children's Hospital in Boston (Boston, MA, Genotyping Core facility). Genome-wide screens for Families 3 and 4 were performed at the Center for Inherited Disease Research using microsatellite markers also at 10 cM average spacing. Fine mapping was done using polymorphic microsatellite markers from the ABI linkage mapping set HD v2.5 at 5cM average density (Applied Biosystems) along with additional microsatellite markers identified using the UCSC Human Genome Browser 19 and synthesized primers (Sigma-Genosys). Two point and multipoint LOD scores calculated using Allegro 20 assumed recessive inheritance, full penetrance, and a disease allele frequency of 0.0001. All nucleotide numbers are in reference to cDNA where A (of the ATG start site) is +1 except for the intronic deletion which is in reference to the genomic sequence where A of ATG of the translational start site is +1 (all from UCSC genome browser, NCBI Build 36.1).
PNKP Western blot
For PNKP, EBV transformed lymphocytes were grown in DMEM with 15% FCS plus normocin. 2×105 cells were lysed in sodium dodecyl sulfate (SDS) loading buffer at 90°C and fractionated by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to Hybond-C Extra Nitrocellulose (GE Healthcare), stained with Ponceau S solution (Sigma), and washed in TBST before immunoblotting with anti- PNK (SK3195) 21 at a dilution of 1 in 1000 in 5% milk. Blots were washed and probed with goat anti-rabbit IgG HRP secondary antibody (DakoCytomation) and detected with ECL detection reagents (GE Healthcare). The blot washed in TBST and re-probed with anti-β-actin monoclonal antibody (Sigma) using rabbit anti-mouse IgG HRP (DakoCytomation) as a secondary antibody.
PNKP RT-PCR
EBV transformed lymphocytes were grown as above. RNA was isolated via RNeasy Mini Kit (Qiagen). 5 μg of total RNA was used for first strand synthesis via oligo dT primers via SuperScript III First-Strand Synthesis SuperMix (Invitrogen Corporation). 1 μl of the product of the RT reaction was used in PCR using primers from Exon 10 to Exon 17.
Comet Assays
Alkaline comet assays were performed via the protocol from CometAssayTM ES unit (Trevigen, Inc.) Briefly, patient derived EBV transformed lymphocytes were grown as described. Cells were obtained prior to treatment or exposed to 100 μM hydrogen peroxide (Sigma) or 10 μM camptothecin (Sigma) for 30 and 60 minutes respectively at 37°C. After exposure, cells were immediately collected for 0 min recovery time point or washed once, resuspended in growth media and incubated at 37°C. Cells were subsequently collected at times described. Cells were embedded in low melt agarose, plated upon microscope slides, lysed, treated with alkaline solution, and slides electrophoresed in alkaline solution at 1 Volt/cm (21 V) with ~300 mAmps for 30 minutes. Slides were washed, dried, and DNA stained with SYBR green. Images of nuclei and tails were taken with a Nikon TE2000-E fluorescent microscope with CCD camera and % Tail DNA determined with CometScore 1.5 software (TriTek, Corp.).
In situ hybridization
Human in situ hybridizations were performed at the Human Developmental Biology Resource (Institute of Human Genetics, International Centre for Life, Newcastle upon Tyne, NE1 3BZ) with probes to anti-sense human PNKP cDNA (+488-1500). Mouse in situs were done as previously published 22, with anti-sense probe from mouse Pnkp cDNA (+386-1566).
Statistics were done within Microsoft Excel software unless otherwise described. The % DNA Repair in Figure 4 was determined by measuring mean of % Tail DNA in 3 separate wells (raw data in Supplementary Information Figure 5) and using combined % Tail DNA at 0 minute recovery (maximum damage) and minimum damage measured as full recovery for each individual cell line. P-values were determined by comparing the baseline adjusted DNA repair level from 3 separate wells using T-Test comparing each MCSZ deficient line to 2 normal control cell lines with two-tailed distribution and homoscedastic test.
PNKP mutation analysis
PNKP protein alignments were performed with MegAlign (Lasergene). Three dimensional structure of mouse Pnkp was examined in MacPyMOL from previously published information 23. The Branch Point analysis was done with Human Splicing Finder version 2.3 (http://www.umd.be/HSF/).
Exon 14 17 bp dup haplotype determination
Sequenom SNP genotypes were determined in patient and controls samples at the Molecular Genetics Core Facility at Children's Hospital Boston (Boston, MA, USA). Haplotypes were determined by Mendelian inheritance patterns for parents and offspring and Phase haplotype determining software24-26 for control samples.
Pnkp RNAi studies
Silencer 1.0 GFP had RNAi oligos targeted to mouse Pnkp mRNA identified via the Broad Institute RNAi Consortium shRNA Library, ligated into EcoRI and ApaI sites (see Supplemental Information (Invitrogen)). Cerebral cortices from E13.5 day old Swiss-Webster mice were dissected and dissociated with Papain Dissociation System (Worthington Biochemical Corp.), cells were washed twice in HBSS buffer and transfected via the Amaxa Nucleofactor 96 well shuttle system (Amaxa Biosystems) with either pSilencer mouse Pnkp-RNAi GFP, pSilencer GFP, with or without pSport-human PNKP (Open Biosystems, Entrez clone ID BC057659). Cells were grown on poly-L-ornithine (Sigma-Aldrich) treated Lab-Tek chamber slides with Permox (Nalge Nunc International) in DMEM with 10% FBS for the Pax6 studies and for the NeuN studies cultures were changed to Neurobasal complete media after 4 hours. Cultures were fixed with 4% paraformaldehyde 24 hours post-transfection. Immunostaining was performed in PBS with 0.2% Triton X-100 and 10% goat serum with chick anti-GFP (Abcam), rabbit anti-cleaved caspase 3 (Asp175) (Cell Signaling Technology, Inc.), mouse anti-Pax6 (The Developmental Studies Hybridoma Bank) or mouse anti-NeuN (Chemicon). Primary antibodies were detected with anti-chicken IgG Alexa 488 (Invitrogen), and donkey anti-mouse IgG Cy3 or donkey anti-rabbit IgG Cy5 (Jackson Immunoresearch Inc.). Images were obtained by looking for GFP+ cells with a Nikon TE2000-E fluorescent microscope and Metamorph imaging software. Cell counts were made from digital images with the counter blinded as to whether the cultures were derived from Pnkp-RNAi or control transfections. P-value in Supplementary Figure 7 were calculations were calculated using G-test of goodness-of fit with 5 degrees of freedom, two-tailed P-value and no Williams correction27.
Mouse Pnkp-RNAi testing
Mo-Pnkp RNAi construct was tested against Mo-Pnkp-DsRed or Hu-PNKP-DsRed both expressed by pCAG-DsRed vector 28. RNAi control vector (empty vector)/mouse (Mo)-Pnkp-DsRed fusion protein, MoPnkp-RNAi/MoPnkp-DsRed, RNAi-control/human (Hu)-PNKP-DsRed fusion, and MoPnkp-RNAi/HuPNKP-DsRed were transfected into NIH-3T3 cells with Lipofectamine 2000 (Invitrogen) per manufacturer protocol. Cells were counted after 24 hours in culture. Random fields were chosen by expression of GFP, the number of high GFP expressing cells were counted followed by the number of Ds-Red fusion protein expressing cells. DsRed expression was nuclear as expected from PNKP localization (data not shown). For Western Blot analysis, NIH-3T3 cells were transfected as above. After 24 hours of culture, cells were lysed via the protocol described in the methods of the main body of the manuscript. After protein quantitation via BCA assay, 25 μg of protein per lane was separated via PAGE. Protein was transferred to membrane, blocked with Odyssey blocking buffer (Li-Cor Biosciences) and probed with Mo anti-Actin (AC-15), Rb anti-GFP (Molecular Probes) and Rb anti-DsRed (Clontech). Primary antibody was detected with anti-mouse labeled with IRDye 700 or anti-rabbit IRDye 800 (Li-Cor Biosciences) and detected with Li-Cor imaging system. In the bar graph protein amounts are normalized with RNAi Control conditions.
Supplementary Material
Acknowledgements
JS was supported by the Victoria and Stuart Quan Fellowship. Research was supported by grants from the National Institute of Neurological Disorders and Stroke to ECG (5K08NS059673-02) and to CAW (NSR01-35129), the Fogarty International Center (R21 NS061772), by the Manton Center for Orphan Disease Research, the Simons Foundation, and by the Dubai Harvard Foundation for Medical Research. ECG was also supported by a K12 Child Health Research Center awarded to Children's Hospital Boston (1 K12 HD052896-01A1) and a Research in Training Award from the Child Neurology Foundation. JR and KWC are supported by Medical Research Council (G0600776 & G0400959). CAW is an Investigator of the Howard Hughes Medical Institute.
We thank Sue Lindsay, and Steve Lisgo from the Medical Research Council-Wellcome Trust Human Developmental Biology Resource (HDBR), Institute of Human Genetics, International Centre for Life, Newcastle upon Tyne, UK for performing human in situ hybridizations.
We are grateful for genotyping services provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C and NIH N01-HG-65403. Genotyping at Children's Hospital Boston is supported by the IDDRC (CHB MRDDRC, P30 HD18655).
Footnotes
The genetic study was approved by Beth Israel Deaconess Medical Center and Children's Hospital Boston Institutional Review Boards. Appropriate informed consent was obtained from all involved human subjects. All animal work was approved by Harvard Medical School, Beth Israel Deaconess Medical Center, and Children's Hospital Boston Institutional Animal Care and Use Committees.
ELECTRONIC-DATABASE INFORMATION
UCSC Human Genome Browser: http://genome.ucsc.edu/
References
- 1.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–12. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huo YK, et al. Radiosensitivity of ataxia-telangiectasia, X-linked agammaglobulinemia, and related syndromes using a modified colony survival assay. Cancer Res. 1994;54:2544–7. [PubMed] [Google Scholar]
- 3.Sun X, et al. Early diagnosis of ataxia-telangiectasia using radiosensitivity testing. J Pediatr. 2002;140:724–31. doi: 10.1067/mpd.2002.123879. [DOI] [PubMed] [Google Scholar]
- 4.Jilani A, et al. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J Biol Chem. 1999;274:24176–86. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 5.Karimi-Busheri F, et al. Molecular characterization of a human DNA kinase. J Biol Chem. 1999;274:24187–94. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 6.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J. 2002;21:2827–32. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitehouse CJ, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–17. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 8.Ali AA, Jukes RM, Pearl LH, Oliver AW. Specific recognition of a multiply phosphorylated motif in the DNA repair scaffold XRCC1 by the FHA domain of human PNK. Nucleic Acids Res. 2009;37:1701–12. doi: 10.1093/nar/gkn1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I–dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 10.El-Khamisy SF, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–13. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 11.Frank KM, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–7. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 12.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–8. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 13.O'Driscoll M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–85. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 14.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–99. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, et al. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc Natl Acad Sci USA. 2000;97:2668–73. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vemuri MC, Schiller E, Naegele JR. Elevated DNA double strand breaks and apoptosis in the CNS of scid mutant mice. Cell Death Differ. 2001;8:245–55. doi: 10.1038/sj.cdd.4400806. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–80. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–3. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 21.Breslin C, Caldecott KW. DNA 3′-phosphatase activity is critical for rapid global rates of single-strand break repair following oxidative stress. Mol Cell Biol. 2009;29:4653–62. doi: 10.1128/MCB.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger UV, Hediger MA. Differential distribution of the glutamate transporters GLT-1 and GLAST in tanycytes of the third ventricle. J Comp Neurol. 2001;433:101–14. doi: 10.1002/cne.1128. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein NK, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–70. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–62. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald JH. Handbook of biological statistics. Vol. 293. Sparky House Publishing; Baltimore: 2008. [Google Scholar]
- 28.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.