Abstract
Objectives
A new method of determining protein turnover by labeling protein with 15N amino acids was used in conjunction with serum-free cell culture to profile secreted proteins that are released by MIA PaCa-2 pancreatic cancer cells in culture.
Methods
MIA PaCa-2 cells were first cultured in Dulbecco modified Eagle medium (Gibco by Invitrogen, Carlsbad, Calif) with 10% fetal bovine serum, then in serum-free modified Eagle medium with or without 50% 15N algal amino acid mixture. The effect of oxythiamine chloride on secreteome was studied. Secreteome from cell culture media was analyzed by 2-dimensional (2D) gel electrophoresis. Differentially expressed proteins were detected and identified. Protein turnover rates were calculated according to the newly established method. Western blot and enzyme-linked immunosorbent assay were used to validate identified proteins.
Results
Among the 14 differentially expressed proteins after oxythiamine treatment, tissue inhibitor of metalloproteases-1 and cytokeratin-10 were identified as 2 newly synthesized secreted proteins caused by substantial 15N incorporation. The inhibition of tissue inhibitor of metalloproteases-1 expression in MIA PaCa-2 cells by oxythiamine treatment was first demonstrated by 2D gel electrophoresis and further validated by Western blotting and enzyme-linked immunosorbent assay analyses.
Conclusions
Our method of labeling protein with 15N amino acids in conjunction with serum-free cell culture allows the identification of actively secreted proteins from pancreatic cancer cells and is a useful method for serum biomarker discovery.
Keywords: quantitative proteomics, pancreatic cancer, 15N stable isotope, oxythiamine, secreted proteome
Secreted proteins and their multidirectional interactions are essential to the growth and invasiveness of tumor cells. Therefore, secreted proteins are potential serum biomarkers in clinical diagnosis of cancer in patients and in monitoring patients during anticancer drug treatment.1 Although rapid identification of proteins using proteomics is a powerful tool for biomarker discovery, few effective cancer biomarkers have been discovered using serum or plasma from cancer patients because of the complexity of serum or plasma. The presence of highly abundant proteins in plasma or serum makes it difficult to discover the subtle changes of secreted protein biomarkers with low abundance. To eliminate the problem of high plasma protein background, cultured cancer cells are commonly used as a platform to identify novel candidate tumor biomarkers. Medium from cancer cell culture is one of the sources to search for potential biomarkers of serum or plasma of cancer patients.2
To further reduce the interference of high abundant proteins in cell-cultured medium with proteomic analysis, the use of serum-free tissue culture was introduced. Secreted proteins can be detected in the concentrate of the medium and identified using mass spectrometry–based proteomic techniques. Stable isotope labeling of proteins in cells (SILAC) has been used in conjunction with serum-free medium to further distinguish serum protein contamination in the medium from secreted proteins by the presence of isotope peaks.3
As reported in our previous study, in contrast with SILAC, the labeling of proteins with low enrichment of 15N amino acid mixture (mSILAC) provides a characteristic isotope envelop of the peptide spectrum, which can be used for the determination of newly synthesized fraction.3 The importance of the secreted protein can be ranked by its synthesis rate. In this paper, we hope to identify secreted proteins from cultured MIA PaCa-2 pancreatic cancer cells and their response to oxythiamine (OT) treatment using 15N amino acids and serum-free media. Using 2D gel electrophoresis (2DE), we detected 14 differentially expressed proteins in media of control (untreated) and OT-treated cells. Among these proteins identified, tissue inhibitor of metalloproteases-1 (TIMP-1) and cytokeratin-10, 2 known pancreatic cancer biomarkers, were 2 proteins heavily labeled with 15N, and their expressions were inhibited by OT treatment. The results from enzyme-linked immunosorbent assays (ELISAs) with human pancreatic cancer sera showed that TIMP-1, although with a signal weaker than CA19-9, has the characteristics of a biomarker of pancreatic cancer. Thus, the labeling of proteins with 15N amino acids and serum-free medium is a potentially useful approach in serum biomarkers discovery using cell culture.
MATERIALS AND METHODS
Cell Culture and Isotopic Labeling
MIA PaCa-2 cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco by Invitrogen, Carlsbad, Calif) supplemented with 10% fetal bovine serum (FBS; Irvine Scientific, Santa Ana, Calif), 1% antibiotic antimycotic (Gibco by Invitrogen), 5% CO2 at 37°C until 85%–95% confluence was attained. Cells were then washed with phosphate-buffered saline (PBS) 3 times to remove all FBS and cultured in 10 mL of serum-free DMEM.4,5 In pilot experiments, we found that the MIA PaCa-2 cells’ proliferation rate was maintained for at least 2 days without significant change of expressions of proteins when cultured without FBS (data not shown). 15N algal amino acid mixture (Cambridge Isotope, Inc, Andover, Mass) was added to the medium at the beginning of the experiment as in the following: group A, cells were cultured in DMEM containing 1 mg/mL of natural amino acid as unlabeled control; group B, cells with 50% enrichment of 15N algal amino acid mixture (98% 15N); and group C, cells with 50% enrichment of 15N algal amino acid mixture (98% 15N) plus 10 μmol/L oxythiamine chloride. Each group was cultured for 48 hours, and each treatment was repeated 3 times.
Processing of Cell Culture Supernatants
After 48 hours of incubation, cell culture supernatants were collected from 4 dishes in each sample group and centrifuged at 12,000g and 4°C for 10 minutes. The supernatants were concentrated by Microcon YM-3 centrifuge tube (Millipore, Billerica, Mass). The concentrated supernatants were then desalted by using desalting columns (Pierce, Rockford, Ill). The desalted supernatants were vacuum dried in Vacufuge plus vacuum concentrator (Eppendorf, Westbury, NY) and suspended in 2D lysis buffer (7 mol/L urea (Amresco, Solon, Ohio), 2 mol/L thiourea (Acros, Morris Plains, NJ), 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (J.T. Baker, Inc, Phillipsburg, NJ) 100 mmol/L dithiothreitol (DTT) (Fisher, Fair Lawn, NJ), 5% glycerol (Sigma, St. Louis, Mo), protease inhibitor cocktail III (EMD Chemicals, Inc, Gibbstown, NJ), and phoshatase inhibitor cocktail set II [EMD Chemicals, Inc, Gibbstown, NJ]). The protein concentration was measured by using Bradford measurement.6
2D Gel Electrophoresis
A 250-μg sample was diluted with rehydration buffer (7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 50 mmol/L DTT, and 0.2% Biolyte [Bio-Rad, Hercules, Calif] with pH 3–10) and then loaded to an 11-cm immobilized pH gradient (IPG) strip (3/10 non-linear; Bio-Rad) followed by rehydration in passive mode for 17 hours. Pre-isoelectric focusing (IEF) was performed at 250 V for 3 hours, then followed by further IEF on a protein IEF system (linear increase of 250–1000 V for 1 hour, 1000 V held for 1 hour, linear increase of 1000–8000 V for 1.5 hours, and 8000 V held until 60,000 kVh was attained; Bio-Rad). IPG strips were then equilibrated with equilibration buffers 1 and 2 (37.5 mmol/LTris-Cl, pH 8.8, 20% glycerol, 2% sodium dodecyl sulfate (SDS [Sigma, St. Louis, Mo]), and 6 mol/L urea, with 2% DTT in buffer 1 and 2.5% iodoacetamide (Bio-Rad) in buffer 2, respectively) 2 times (2 × 15 minutes) each. Protein separation in the second dimension was then performed by using 8–16% SDS-polyacrylamide gel electrophoresis were used to perform the second dimension electrophoresis in a Protean Plus Dodeca cell (Bio-Rad). The gels were then fixed with 20% methanol/7% acetic acid and stained with SYPRO-Ruby (Bio-Rad) overnight in the dark and imaged with a Pharox FX molecular imager (Bio-Rad).
Protein Identification
Proteins spots were selected by using PDQuest software (version 8.0.1; Bio-Rad) and excised by an EXQuest cutter (Bio-Rad). Gel slices were washed in 100 mmol/L NH4HCO3/50% acetonitrile and digested with 20 μg/mL trypsin gold (Promega, Madison, Wis). The digested peptides were extracted once with distilled water and twice with 50% acetonitrile/5% trifluoride acetic acid. The extracted peptides were concentrated and purified by using ZipTip pipette tips with μ-C18 Resin (Millipore) and analyzed with Ultraflex III matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF)/TOF mass spectrometry (MS; Bruker, Bremen, Germany) for peptide fingerprint and sequence analysis using α-cyano-4-hydroxycinamic acid (Sigma-Aldrich, St Louis, Mo) as the matrix. The tandem mass spectrometry (MS/MS) spectra were used against Homo sapiens in SWISS-PROT database to search for protein identity using Mascot search engine (www.matrixscience.com).
Spectral Data Analysis
A well-established algorithm was used based on our newly published method3 to separate natural and 15N-labeled peptide spectra with the inverse concatenation function. The newly synthesized proteins were represented by the fraction of labeled isotopomers. The 15N isotope enrichment was 50%. The isotopomers of the 15N-labeled peptides were constructed using concatenation operation because the natural isotopomers and the 15N isotopomers were known. Thus, the resultant distributions were the distributions of the new peptides and could be used to determine newly synthesized fractions.3
Western Blot Analysis
A 50-μg sample of each group was resolved by 10% SDS-polyacrylamide gel electrophoresis. Proteins were transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore) and stained with Ponceau S (Sigma-Aldrich) to show that all samples were loaded equally.7-9 Ponceau S–stained blots were destained and blocked using Tween-20 (0.1%) PBS blocking buffer (Pierce). Primary monoclonal antibodies TIMP-1 (Yn5) or cytokeratin 10 (clone VIK-10 (Santa Cruz Biotechnology, Inc, Santa Cruz, Calif) were used at 1:500 dilution overnight at 4°C. The secondary antibodies rabbit polyclonal IgG (Millipore) were used at 1:10,000 dilution for 2 hours at room temperature. Western blots were then incubated with SuperSignal West Dura Substrate (Pierce) and detected by using FluorChem Imager (Alpha Innotech, San Leandro, Calif).10
ELISA Analysis
Blood was drawn from 12 healthy control subjects or patients into EDTA tubes and then centrifuged at 1000g for 10 minutes. The level of TIMP-1 in plasma was quantified by human TIMP-1 Immunoassay kit (R&D Systems, Inc, Minneapolis, Minn) following the manufacturer’s instruction. TIMP-1 levels in plasma were expressed in nanogram per milliliter.11,12 Absorbance was measured on an ELISA reader (Bio-Rad) at 450 nm. Serum CA19-9 (Alpha Diagnostics, San Antonio, Tex) was also detected by following the manufacturer’s protocol. All the experiments were performed in triplicates.
RESULTS
Dynamics of Secreted Proteome in Pancreatic Cancer Cells in Response to Oxythiamine Treatment
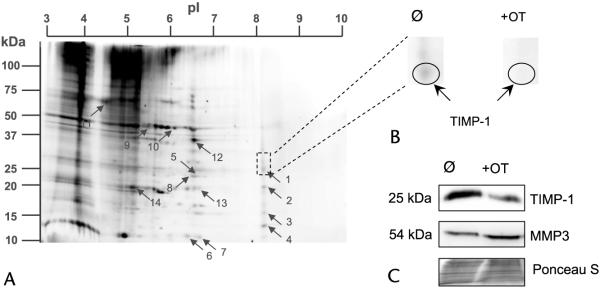
Cancer cells extensively use glucose for providing energy and substrates for proliferation. Transketolase (TK) is a key enzyme in nonoxidative pentose cycle (PC) pathway and plays a crucial role in cancer cell proliferation. It was reported that OT as an inhibitor of TK can specifically inhibit the activity of TK, leading to about 85% loss of PC pathway activity. The MIA PaCa-2 cell line is a commonly used cell model of pancreatic cancer and was used in this study. The secreted proteins were extracted from the cell-conditioned media after 48 hours of culture and separated by 2D electrophoresis. Based on 2D gel images, a total of 14 protein spots were significantly suppressed in OT-treated cells (Fig. 1). Stable isotope labeling does not affect normal protein metabolism given that we did not detect a significant difference in the protein expression between groups A and B (data not shown). The differentially expressed 14 spots were then digested with trypsin and analyzed with MALDI-TOF/TOF MS in the reflector mode. Three typical fragments from each protein spot were further identified in the lift mode. The MALDI-TOF/TOF spectra were further analyzed for their mass isotopomer distribution.3 Results are summarized in Table 1. Four of these proteins (1, 6, 9, and 11) are functional proteins of surface receptors or matrix proteins. Proteins 7 and 14 are cytokeratins. Only 2 of these differentially expressed proteins, TIMP-1 and cytokeratin-10, had high isotope incorporation with a newly synthesized fraction greater than 90%.
FIGURE 1.
Two-dimensional electrophoresis of secreted proteins from MIA PaCa-2 cells. MIA PaCa-2 cells were metabolically labeled by supplementing cell culture media with 15N-labeled amino acids. A, The 14 proteins differentially expressed in OT-treated cells were separated by 2DE and identified by MALDI-TOF/TOF (left). B, An example of selection of one of differentially expressed proteins, TIMP-1, in pancreatic cancer cells in response to OT treatment (top right). C, Functional validation of TIMP-1 and its target protein MMP3 in untreated and OT-treated cell media as analyzed by Western blot assays (lower right). Ponceau S staining is shown to demonstrate equal protein loading between conditioned media samples. Please see the Materials and Methods section for details. All the experiments were repeated 3 times.
TABLE 1.
Identification of the Secreted proteins (>2-Fold Decrease ↓ by OT Treatment in MIA PaCa-2 cells) by MALDI-TOF/TOF Mass Spectrometry
| Sample Spot* |
Accession Number |
Protein Name |
15N Labeling |
Score† | Pep‡ | Molecular Weight/ Isoelectric Point |
Possible Role |
|---|---|---|---|---|---|---|---|
| 1 | P01033 | ↓Tissue inhibitor of metalloproteinases-1 (TIMP-1) |
Y | 82 | 3 | 23,156/8.46 | Caner enhancer |
| 2 | P00760 | ↓Cationic trypsin | N | 147 | 2 | 25,769/8.40 | Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa |
| 3 | P62937 | ↓Peptidyl-prolyl cis-trans isomerase A |
N | 91 | 7 | 18,001/7.68 | Related protein folding |
| 4 | P07737 | ↓Profilin-1 | N | 73 | 6 | 15,045/8.44 | Binds to actin and affects the structure of the cytoskeleton |
| 5 | P60174 | ↓Triosephosphate isomerase | N | 207 | 16 | 26,653/6.45 | Catalytic activity |
| 6 | Q9NT22 | ↓EMILIN-3 | N | 49 | 8 | 82,596/7.84 | Extracellular matrix |
| 7 | P13645 | ↓Cytokeratin-10 | Y | 89 | 12 | 59,475/5.13 | Predictive marker for poor prognosis in liver cancer |
| 8 | P60174 | ↓Triosephosphate isomerase | N | 222 | 16 | 26,653/6.45 | Catalytic activity |
| 9 | Q8CHL0 | ↓Frizzled-5 | N | 82 | 7 | 64,069/8.51 | Receptor for Wnt proteins |
| 10 | P00760 | ↓Cationic trypsin | N | 168 | 2 | 25,769/8.40 | Preferential cleavage: Arg+Xaa, Lys+Xaa |
| 11 | P16070 | ↓CD44 antigen | N | 41 | 1 | 81,503/5.13 | Receptor for hyaluronic acid |
| 12 | P04075 | ↓Fructose-bisphosphate aldolase A |
N | 110 | 10 | 39,395/8.30 | Catalytic activity |
| 13 | P00760 | ↓Cationic trypsin | N | 88 | 2 | 25,769/8.40 | Preferential cleavage: Arg+Xaa, Lys+Xaa |
| 14 | P04264 | Cytokeratin-1 | N | 75 | 15 | 65,978/8.16 | May regulate the activity of kinases such as protein kinase C and v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) |
Protein numbers on the 2D gel (Fig. 1).
MOWSE (for MOlecular Weight SEarch) scores obtained by the combined search (peptide mass fingerprinting and “LIFT” data) using Mascot search engine.
Number of peptides identified by ultraflex MALDI-TOF/TOF in lift mode.
Measuring Protein Synthesis in Pancreatic Cancer Cell Supernatants
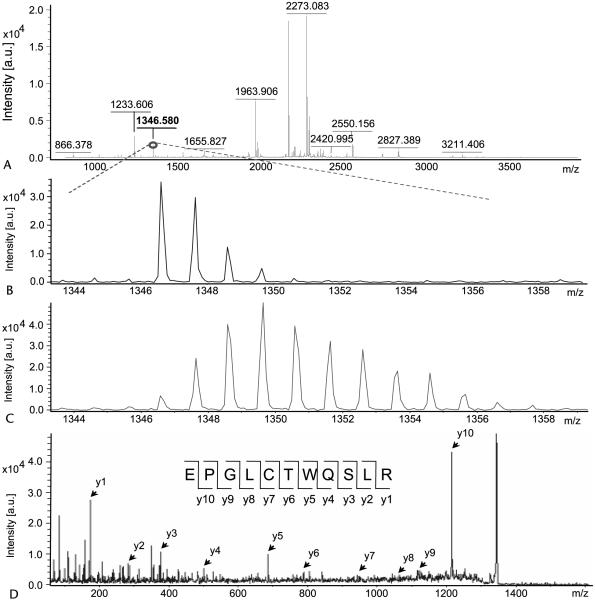
The 14 proteins identified in the culture supernatants of MIA PaCa-2 pancreatic cells can be grouped into 2 categories, secreted proteins and medium proteins, by the presence or absence of mass shift on their mass spectra due to 15N incorporation. No differences were observed between labeled and unlabeled cells in terms of rates of cell division, viability, or cell morphology (data not shown). We previously showed that cells cultured in the presence of 15N amino acids (~50% enriched) incorporate 15N into proteins during protein synthesis. Isotopomer distribution of the resultant peptides shows a characteristic isotope envelop, which is the hallmark of a secreted protein and can be used to calculate fraction of new synthesis of the corresponding protein.3,13 The spectra of peptide (m/z 1346.580) from TIMP-1 with and without 15N incorporation are shown in Figure 2. Among the secreted proteins, TIMP-1 and cytokeratin-10, 2 known pancreatic cancer biomarkers, were identified as actively secreted proteins because of the mass shift in their respective peptide spectrum with a rate of protein synthesis greater than 99% by multiple linear regression analysis.
FIGURE 2.
Calculation of protein synthesis rate and identification of TIMP-1 by mass spectrometry. Total mass spectra (A) of the 1346 m/z fragment from spot 1 (TIMP-1) of cell media in the presence of natural amino acids (B) and 50% enriched 15N algal amino acid mixtures (C). Panel B shows the distribution of the unlabeled fragment from media group A. Panel C shows obvious spectrum shift in mass from the same peptide obtained from media group B with 50% 15N amino acids (group C not shown). Panel D shows the MS/MS spectrum of this fragment in lift mode. Fragmentation spectrum with y ions labeled and the peptide bond cleavages noted in the peptide sequence. Please see the Materials and Methods section for details. All the experiments were repeated 3 times.
Up-Regulation of TIMP-1 in Pancreatic Cancer
The differential expression of TIMP-1 in MIA PaCa-2 pancreatic cell supernatants, when treated with OT, was first seen on 2D gels (Fig. 1B) and later verified by western blotting (Fig. 1C). OT suppresses up to 85% of cell proliferation via the TK pathway.14,15 The suppression of TIMP-1 with the presence of OT suggests that TIMP-1 is highly expressed in pancreatic cancer. Given that TIMP-1 is actively synthesized and secreted by MIA PaCa-2 pancreatic cells, TIMP-1 could be used as a biomarker in the clinical diagnosis of pancreatic cancer or in the management of pancreatic cancer. The gene TIMP-1 belongs to the TIMP family, which encodes natural inhibitors of active matrix metalloproteinases (MMPs), a group of peptidases required for degradation of the extracellular matrix.16-18 In addition to its suppression against most of the MMPs, TIMP-1 is suggested to be antiapoptosis because it is known to promote cell proliferation in a wide range of cell types.19 A previous research suggests that MMP3, a member of MMPs, is inhibited by TIMP-1 by analysis of crystal structure of an MMP-TIMP complex formed between the catalytic domain of human MMP3 and human TIMP-1.20 We found that MMP3 had an opposite expression level as TIMP-1 between control (untreated) and OT-treated MIA PaCa-2 pancreatic cancer cells (Fig. 1C).
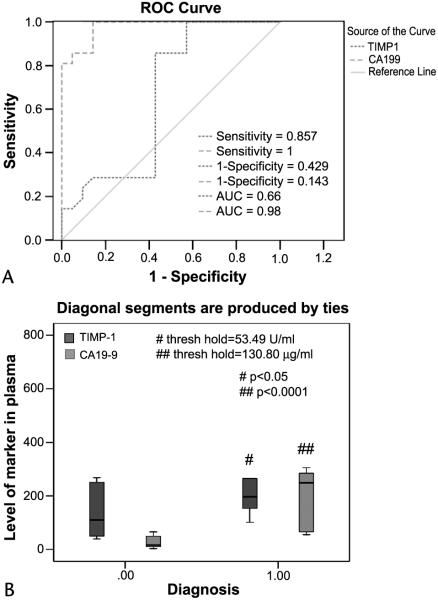
To determine the relevance to human pancreatic cancer, ELISAs of TIMP-1 and CA19-9, a marker that is currently in clinical trial as a pancreatic cancer marker,21 were carried out in 12 plasma samples from patients with pancreatic adenocarcinoma and an equal number of plasma samples from control subjects. Receiver operating characteristic (ROC) curve analyses were used to investigate the sensitivity and specificity of TIMP-1 and CA19-9 as separate diagnostic tests for pancreatic adenocarcinoma. Figure 3 depicts the TIMP-1 and CA19-9 ROC curves for the discrimination of patients with pancreatic cancer and of non–pancreatic cancer control subjects (true-negative cases). An ROC analysis estimates a curve, which describes the inherent tradeoff between the sensitivity and specificity of a diagnostic test. Each point on the ROC curve is associated with a specific diagnostic criterion. The area under the ROC curve (AUC) may be regarded as a mean of the sensitivity of all possible specificities. The diagnostic measure with the higher AUC is typically regarded as better. Thus, TIMP-1 (AUC = 0.66) had a good accuracy of all possible cutoffs. The cutoff (using 130.30 μg/mL TIMP-1 in serum as shown in Fig. 3B) yielded a sensitivity of 85.57 % and a specificity of 57.5% for TIMP-1 when considering pancreatic cancer as true-positive cases and all non–pancreatic cancer subjects as true-negative cases (Fig. 3B). Compared with CA19-9, the specificity and sensitivity of TIMP-1 are lower, suggesting that CA19-9 could be an option for the clinical diagnosis of pancreatic cancer.
FIGURE 3.
Functional validation of TIMP-1 using ELISA. A, Receiver operator characteristic curves for diagnosis of pancreatic adenocarcinoma versus noncancerous cases. Curves demonstrate the relative accuracy for the individual serum levels of both TIMP-1 and CA-19-9 (as reference) to discriminate between pancreatic cancer and control cases. Serum levels of healthy control subjects (n = 12) were considered true-negative cases, whereas serum levels of patients with cytologically confirmed pancreatic adenocarcinoma (n = 12) were considered true-positive cases. The AUC was 0.66 (95% confidence interval [CI]: 0.342–0.964) for TIMP-1, 0.98 (95% CI: 0.917–1.00) for CA19-9. At a cutoff of 130.80 ng/mL plasma (B), the sensitivity was 85.7% (CI: 81.0%–99.3%) and the specificity was 81.5% (CI: 70.4%–81.5%) for TIMP-1, whereas the sensitivity and specificity for CA19-9 at a cutoff of 53.49 U/mL plasma (B) were 86.7% and 100%, respectively. B, Box plot of relative serum concentrations of TIMP-1 and CA19-9 for control subjects and for patients with pancreatic cancer for an independent sample set. The lines inside the boxes denote the medians. The boxes represent the interval between the 25th and 75th percentiles, and the whiskers indicate the interval between maximum and minimum. Please see the Materials and Methods section for details. All the experiments were repeated 3 times.
DISCUSSION
High throughput technologies such as proteomics have been proposed to be the new ways for biomarker discoveries.22 The coupling of proteomics and in vitro labeling of proteins in cells (SILAC) further improves the efficacy in biomarker discovery. However, previous studies using SILAC are proved to be time consuming and costly because of the expensive fully labeled 15N amino acids and the special amino acid–depleted media required.23,24 The use of the newly established technique by stable isotope labeling with low enrichment 15N amino acids in cells (mSILAC)3,13 has been shown to be an effective approach for quantitative proteomics without the associated costs of expensive isotopes and medium. The combination of serum-depleted medium and mSILAC was applied in this study of MIA PaCa-2 pancreatic cancer cell supernatants. We were able to identify secreted proteins by their isotope signature and by the rate of synthesis. Using this approach, we identified TIMP-1 and cytokeratin-10 as 2 actively synthesized and secreted proteins. The secretion of these proteins was highly sensitive to OT, which is known to suppress MIA PaCa-2 cell proliferation. TIMP-1 is a member of the TIMP gene family, which encodes inhibitors of the matrix metalloproteinases (MMPs), a group of proteolytic enzymes involved in the degradation of the extracellular matrix (ECM).16,17 The degradation of adjacent ECM of malignant epithelial cells is an essential step in the processes of invasion and metastasis.25 Bramhall et al26 reported the imbalance of expression of MMPs and TIMPs in human pancreatic carcinoma. Most of the pancreatic cancer cells with overexpression of MMPs were localized to adenocarcinoma nests and vascular lumens, indicating that these tumor cells have a strong potential to local invasion and distant metastasis.27
Secreted proteome of cancer cells in culture is often considered to be the source of serum biomarkers in cancer patients. The plasma steady-state concentration of such a protein depends on the balance between its production and removal. If the concentration is dependent on the protein’s production rate, the concentration of the biomarker in plasma reflects the rate of synthesis. On the other hand, if the removal rate is the limiting factor, concentration of the protein in plasma is then a less sensitive biomarker of the presence of the tumor. However, when the protein synthesis can be simultaneously determined, the rate of synthesis (or turnover) can be used reliably as a tumor biomarker. Such a consideration may be relevant to our observation of TIMP-1 in the sera from patients with pancreatic cancer (Fig. 3) that TIMP-1 was a less sensitive biomarker than CA19-9.
In conclusion, labeling protein with 15N amino acids in conjunction with depleted serum allows the identification of actively secreted proteins from pancreatic cancer cells, and the rate of production of a secreted protein may be used as an independent biomarker of the presence of tumor.
Acknowledgments
This study was jointly supported by grants awarded to Dr Gary Guishan Xiao from the Bone Biology Program of the Cancer and Smoking Related Disease Research Program and the Nebraska Tobacco Settlement Biomedical Research Program (LB692, LB595, and LB506) and partially supported by a grant awarded to WNPL from the UCLA Center of Excellence in Pancreatic Disease (P01 AT003960-01), Harbor-UCLA GCRC Mass Spectrometry Core (M01 RR00425) and the Hirshberg Foundation for Pancreatic Cancer Research.
REFERENCES
- 1.Bonin-Debs AL, Boche I, Gille H, et al. Development of secreted proteins as biotherapeutic agents. Expert Opin Biol Ther. 2004;4:551–558. doi: 10.1517/14712598.4.4.551. [DOI] [PubMed] [Google Scholar]
- 2.Kulasingam V, Diamandis EP. Tissue culture–based breast cancer biomarker discovery platform. Int J Cancer. 2008;123:2007–2012. doi: 10.1002/ijc.23844. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y, Lee WN, Xiao GG, et al. Quantitative proteomics: measuring protein synthesis using 15N amino acid labeling in pancreatic cancer cells. Anal Chem. 2009;81:764–771. doi: 10.1021/ac801905g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flora JW, Edmiston J, McKinney W, et al. Identification of in vitro differential cell secretions due to cigarette smoke condensate exposure using nanoflow capillary liquid chromatography and highresolution mass spectrometry. Anal Bioanal Chem. 2008;391:2845–2856. doi: 10.1007/s00216-008-2197-3. [DOI] [PubMed] [Google Scholar]
- 5.Haqqani AS, Kelly J, Stanimirovic DB, et al. Protein markers of ischemic insult in brain endothelial cells identified using 2D gel electrophoresis and ICAT-based quantitative proteomics. J Proteome Res. 2007;6:226–239. doi: 10.1021/pr0603811. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook J, Russell D. Molecular cloning: a laboratory manual. ed 3 Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 7.Senechal KR, Thaller C, Noebels JL. ADPEAF mutations reduce levels of secreted LGI1, a putative tumor suppressor protein linked to epilepsy. Hum Mol Genet. 2005;14:1613–1620. doi: 10.1093/hmg/ddi169. [DOI] [PubMed] [Google Scholar]
- 8.Toussaint-Smith E, Donner DB, Roman A. Expression of human papillomavirus type 16 E6 and E7 oncoproteins in primary foreskin keratinocytes is sufficient to alter the expression of angiogenic factors. Oncogene. 2004;23:2988–2995. doi: 10.1038/sj.onc.1207442. [DOI] [PubMed] [Google Scholar]
- 9.Moore MK, Viselli SM. Staining and quantification of proteins transferred to polyvinylidene fluoride membranes. Anal Biochem. 2000;279:241–242. doi: 10.1006/abio.2000.4482. [DOI] [PubMed] [Google Scholar]
- 10.Rockenstein E, Mante M, Masliah E, et al. High beta-secretase activity elicits neurodegeneration in transgenic mice despite reductions in amyloid-beta levels: implications for the treatment of Alzheimer disease. J Biol Chem. 2005;280:32957–32967. doi: 10.1074/jbc.M507016200. [DOI] [PubMed] [Google Scholar]
- 11.Sundaram R, Lynch MP, Kaumaya PT, et al. De novo design of peptide immunogens that mimic the coiled coil region of human T-cell leukemia virus type-1 glycoprotein 21 transmembrane subunit for induction of native protein reactive neutralizing antibodies. J Biol Chem. 2004;279:24141–24151. doi: 10.1074/jbc.M313210200. [DOI] [PubMed] [Google Scholar]
- 12.Hui EP, Sung FL, Chan AT, et al. Plasma osteopontin, hypoxia, and response to radiotherapy in nasopharyngeal cancer. Clin Cancer Res. 2008;14:7080–7087. doi: 10.1158/1078-0432.CCR-08-0364. [DOI] [PubMed] [Google Scholar]
- 13.Xiao GG, Garg M, Lee WN, et al. Determination of protein synthesis in vivo using labeling from deuterated water and analysis of MALDITOF spectrum. J Appl Physiol. 2008;104:828–836. doi: 10.1152/japplphysiol.00976.2007. [DOI] [PubMed] [Google Scholar]
- 14.Boros LG, Puigjaner J, Schirmer WJ, et al. Oxythiamine and dehydroepiandrosterone inhibit the nonoxidative synthesis of ribose and tumor cell proliferation. Cancer Res. 1997;57:4242–4248. [PubMed] [Google Scholar]
- 15.Rais B, Comin B, Cascante M, et al. Oxythiamine and dehydroepiandrosterone induce a G1 phase cycle arrest in Ehrlich’s tumor cells through inhibition of the pentose cycle. FEBS Lett. 1999;456:113–118. doi: 10.1016/s0014-5793(99)00924-2. [DOI] [PubMed] [Google Scholar]
- 16.Cottam DW, Rees RC. Regulation of matrix metalloproteinases: their role in tumor invasion and metastasis. Int J Oncol. 1993;2:861–872. doi: 10.3892/ijo.2.6.861. [DOI] [PubMed] [Google Scholar]
- 17.Sato H, Takino T, Seiki M, et al. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 18.Stetler-Stevenson WG. Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities. Sci Signal. 2008;1:re6. doi: 10.1126/scisignal.127re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singla DK, McDonald DE. Factors released from embryonic stem cells inhibit apoptosis of H9c2 cells. Am J Physiol Heart Circ Physiol. 2007;293:H1590–1595. doi: 10.1152/ajpheart.00431.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomis-Ruth FX, Maskos K, Bode W. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature. 1997;389:77–81. doi: 10.1038/37995. [DOI] [PubMed] [Google Scholar]
- 21.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266–270. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Kageyama S, Isono T, Yoshiki T, et al. Identification by proteomic analysis of calreticulin as a marker for bladder cancer and evaluation of the diagnostic accuracy of its detection in urine. Clin Chem. 2004;50:857–866. doi: 10.1373/clinchem.2003.027425. [DOI] [PubMed] [Google Scholar]
- 23.Ong SE, Blagoev B, Mann M, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Gruhler S, Kratchmarova I. Stable isotope labeling by amino acids in cell culture (SILAC) Methods Mol Biol. 2008;424:101–111. doi: 10.1007/978-1-60327-064-9_9. [DOI] [PubMed] [Google Scholar]
- 25.Liotta LA, Stetler-Stevenson WG. Tumor invasion and metastasis: an imbalance of positive and negative regulation. Cancer Res. 1991;51:5054s–5059s. [PubMed] [Google Scholar]
- 26.Bramhall SR, Neoptolemos JP, Lemoine NR, et al. Imbalance of expression of matrix metalloproteinases (MMPs) and tissue inhibitors of the matrix metalloproteinases (TIMPs) in human pancreatic carcinoma. J Pathol. 1997;182:347–355. doi: 10.1002/(SICI)1096-9896(199707)182:3<347::AID-PATH848>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 27.Gong YL, Xu GM, Chen LB, et al. Expression of matrix metalloproteinases and the tissue inhibitors of metalloproteinases and their local invasiveness and metastasis in Chinese human pancreatic cancer. J Surg Oncol. 2000;73:95–99. doi: 10.1002/(sici)1096-9098(200002)73:2<95::aid-jso7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]