Abstract
The mesolimbic dopamine system and cAMP-dependent/protein kinase A (PKA) pathways are strongly implicated in addictive behaviors. Here we determine the role of dopamine D2 receptors (D2) in PKA signaling responses to δ-opioid (DOR) and cannabinoid (CB1) receptors. We find in NG108-15/D2 cells and in cultured primary neurons that a brief exposure to saturating concentrations of DOR and CB1 agonists increases cAMP, promotes PKA Cα translocation and increases cAMP-dependent gene expression. Activation of PKA signaling is mediated by Gi-βγ dimers. Importantly, subthreshold concentrations of DOR or CB1 agonists with D2 agonists, which are without effect when added separately, together activate cAMP/PKA signaling synergistically. There is also synergy between DOR or CB1 with ethanol, another addicting agent. In all instances, synergy requires adenosine activation of adenosine A2 receptors and is mediated by βγ dimers. Synergy by this molecular mechanism appears to confer hypersensitivity to opioids and cannabinoids while simultaneously increasing the sensitivity of D2 signaling when receptors are expressed on the same cells. This mechanism may account, in part, for drug-induced activation of medium spiny neurons in the nucleus accumbens.
Although addicting drugs produce different clinical responses, they share the common characteristic of causing addiction. This suggests that a molecular mechanism shared by addicting drugs could contribute to the development of addiction. All addicting substances increase extracellular dopamine in the nucleus accumbens (NAc) (1, 2), a striatal component of reward and addiction (3). cAMP/protein kinase A (PKA) signaling is also involved in addiction (2).
Dopamine D2 (D2), δ-opioid (DOR), and cannabinoid (CB1) receptors inhibit adenylyl cyclase (AC) activity (2) by activating inhibitory GTP-binding proteins, Gi/o. Gi/o consists of αi/o and βγ subunits; αi/o inhibits AC. βγ dimers have several effects (4), including stimulation of AC isozymes II and IV (5-7). D2 activates AC II and IV, apparently via βγ dimers (8). βγ dimers are required to maintain voluntary alcohol consumption in rats (9).
NG108-15/D2 (NG) cells express functional DOR (10) and CB1 (11), as do rat primary hippocampal neurons (PHN). We report here that DOR and CB1 increase cAMP and stimulate PKA Cα translocation in these cells at 10 min followed by increased cAMP-dependent gene transcription 5 h later. We also find synergy for PKA signaling and gene expression between subthreshold concentrations of DOR or CB1 agonists with ethanol or D2. In all instances, synergy requires adenosine and is mediated by βγ dimers. Synergy appears to confer hypersensitivity simultaneously to DOR, CB1, and D2 when expressed on the same neurons with adenosine A2 (A2) receptors.
Methods
Materials. Reagents were from Sigma except where indicated. Ham's F-12 medium was from GIBCO; R(-)-2,10,11-trihydroxy-N-propylnorapomorphine hydrobromide (NPA), [d-Ala2, d-Leu5]enkephalin (DADLE), methanandamide (Met), UK 14304 (UK), carbachol (Carb), spiperone (Spip), naltrindole (Nal), AM 281 (AM), 1,3-dipropyl-8-cyclopentylxanthine (DPCPX), 3,7-dimethyl-1-(2-propynyl)xanthine (DMPX), and pertussis toxin (PTX) were from Sigma/RBI (Natick, MA); Rp-cAMPS (Rp) was from BioLog (La Jolla, CA); pCRE-Luciferase (Luc) was from Stratagene; and pCMV β-galactosidase was from Qiagen (Hilden, Germany).
Cell Culture. NG and PHN were prepared and used exactly as described in ref. 9. Drugs were added to NG cells on day 4 and to primary cultures on day 10.
Viral Vectors. Construction and production of recombinant Ad5QEHA, Ad5SKEE, Ad5βARK1, and Ad5LacZ vectors were as described in ref. 9. To make the HSVLacZ/CRE-Luc construct, the LacZ gene was cloned into pHSVPrpUC under control of the HSV1 endogenous IE4/5 promoter. A CRE-Luc expression cassette was then inserted downstream of the LacZ gene in reverse orientation. To produce the virus, 2-2 cells were transfected with the construct by using the Lipofectamine reagent as described by the manufacturer (GIBCO/BRL) and superinfected with the helper virus 5dl1.2. After amplification three times on 2-2 cells, the virus was purified and titrated as described in ref. 12.
CRE-Luciferase Reporter Assay. Luc was assayed in NG cells exactly as described (9). Rat PHN were plated at 2.5 × 104 cells per 24-well plate, grown for 10 days in Neurobasal A medium supplemented with B27 and glutamate, and then cultured in Neurobasal A medium only for 2 days. Cells were then infected overnight with HSVLacZ/CRE-Luc at 1 multiplicity of infection. The cells were then treated with drugs for 10 min, washed, and cultured for 4 h before Luc assay. Luc in PHN was normalized for transfection efficiency as determined by β-galactosidase activity (Stratagene).
All procedures were performed in accordance with protocols approved by the Gallo Center Institutional Animal Care and Use Committee protocols and the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Results
Activation of DOR or CB1 Induces PKA Cα Translocation in NG Cells. NG cells were incubated for 10 min with the DOR agonist, DADLE (1 μM), or the CB1 agonist, Met (2 μM), and translocation of Cα was measured with specific antibodies. DADLE and Met each causes translocation of Cα away from the Golgi to the nucleus and cytoplasm, as does forskolin (Fig. 1A). Nal (10 μM), a DOR antagonist, or AM (10 μM), a CB1 antagonist, blocks PKA Cα translocation induced by DADLE or Met, respectively (Fig. 1 A). By contrast, Gi/o-coupled α2 adrenergic (α2) (10 μM UK) or M4-cholinergic (M4) (10 μM Carb) agonists are without effect (Fig. 1 A). Western blot analysis confirms these findings (Fig. 1B).
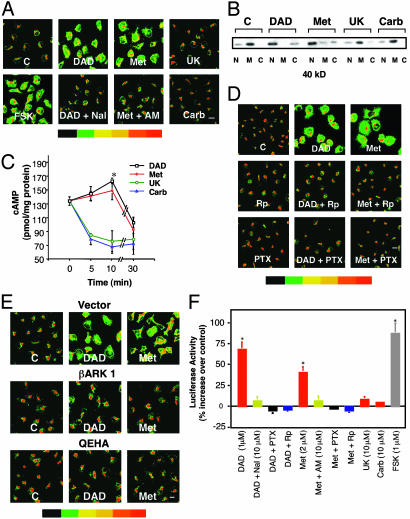
Fig. 1.
Activation of DOR or CB1 promotes PKA Cα translocation and CRE-Luc expression in NG cells. (A) Cα translocation detected by immunostaining. NG cells were incubated with or without 1 μM DADLE (DAD), 2 μM Met, 10 μM UK, or 10 μM Carb or forskolin (FSK) (1 μM) for 10 min. Where indicated, cells were preincubated either with the DOR antagonist Nal (10 μM) or the CB1 antagonist AM (10 μM) for 30 min. Data represent at least three experiments. Staining intensity is indicated by the color bar (red indicates highest concentration) (scale bar, 10 μm; magnification, ×400). (B)Cα translocation detected by Western blots of nuclear (N), membrane (M), and cytosolic (C) fractions from treated cells. (C) Time course of cAMP production. Cells were incubated with or without 1 μM DADLE, 2 μM Met, 10 μM UK, or 10 μM Carb for the indicated times. cAMP was measured by RIA (17). Data are the mean ± SEM of four experiments. *, P < 0.05 compared with time 0 (one-way analysis of variance and Dunnett's test). cAMP levels in the absence of drugs did not change during the experiment. (D) Rp and PTX inhibit PKA Cα translocation. Cells were pretreated with 20 μM Rp for 1.5 h or PTX (50 ng/ml) overnight before incubation with DADLE or Met as in A. (E) βγ dimers are required for PKA Cα translocation. Cells were transfected with the βγ inhibitors Ad5βARK1 or Ad5QEHA or Ad5 vector control and incubated with or without 1 μM DADLE or 2 μM Met. (F) Activation of DOR or CB1 induces CRE-Luc expression. Cells were transiently transfected with a CRE-Luc construct, preincubated with buffer or Nal, AM, Rp, or PTX as above and then treated for 10 min with or without DADLE, Met, or forskolin. Luc was assayed 5 h after drug treatment. Data are the mean ± SEM of at least three experiments. *, P < 0.01 compared with control (one-way analysis of variance and Dunnett's test).
DOR or CB1 Agonists Transiently Increase cAMP. Activation of PKA requires cAMP. DADLE increases cAMP levels at 10 min in NG cells, followed by decreases at 30 min (Fig. 1C). Similar results were found with Met. α2 or M4 agonists, which do not cause Cα translocation (Fig. 1 A and B), only reduce cAMP (Fig. 1C). Minor increases in cAMP can activate PKA (13). Rp (20 μM), which prevents cAMP binding to the regulatory subunit of PKA, blocks DADLE- or Met-induced Cα translocation (Fig. 1D).
βγ Dimers Mediate DOR and CB1-Induced PKA Cα Translocation. We next asked whether βγ released from activated Gi/o mediates DADLE- or Met-induced PKA translocation. PTX (50 ng/ml), which prevents release of βγ from Gi/o, blocked DADLE- and Met-induced PKA Cα translocation (Fig. 1D). Adenoviral vectors expressing the βγ inhibitor peptide βARK1 (7) or QEHA (14), also prevented DOR- and CB1-induced PKA Cα translocation (Fig. 1E). Overexpression of the inactive control peptide SKEE (15) was without effect. These data suggest that βγ dimers mediate DADLE- and Met-induced PKA Cα translocation, presumably by stimulating AC.
DADLE and Met Activate CRE-Mediated Gene Expression. We next asked whether DOR or CB1 induces cAMP-dependent gene expression. DADLE and Met each increases CRE-Luc 5 h after a 10-min exposure, as does 1 μM forskolin (Fig. 1F). α2 or M4 agonists are without effect. DOR and CB1 antagonists block DADLE or Met stimulation of Luc, as does Rp (Fig. 1F). Moreover, PTX (Fig. 1F) or QEHA (data not shown) also prevents this response. These data suggest that DOR and CB1 activate CRE-Luc via PKA Cα translocation to the nucleus.
Synergy Between DOR or CB1 Agonists with a D2 Agonist or Ethanol. PKA Cα translocation. Activation of PKA signaling by DOR and CB1 agonists suggested the possibility of synergy between βγ dimers with ethanol/A2 activation via Gαs (9). Low concentrations of DADLE (0.01 nM), Met (0.02 nM), NPA (0.5 nM), or ethanol (25 mM) alone do not cause PKA Cα translocation (Fig. 2 B and C). However, these subthreshold concentrations caused a synergistic induction of PKA Cα translocation when DADLE or Met were coincubated with NPA or ethanol (Fig. 2 A-C). DADLE with Met did not produce synergy (not shown).
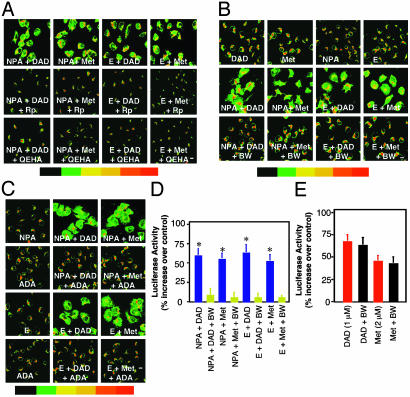
Fig. 2.
Synergy for PKA Cα translocation and CRE-Luc expression between subthreshold concentrations of DADLE or Met with NPA or ethanol. (A) Subthreshold concentrations of NPA (0.5 nM), DADLE (0.01 nM), Met (0.02 nM), or ethanol (E) (25 mM) that did not induce translocation alone were tested for synergy during a 10-min incubation (Top). Cells were also preincubated in Rp or QEHA (Middle and Bottom). (B) Cells were preincubated with or without 10 μM BW before testing for translocation synergy with subthreshold concentrations of NPA, DADLE, Met, or ethanol as in A. (C) Cells were incubated as in A, in the presence or absence of 1 unit/ml adenosine deaminase (ADA). (D) Cells transiently transfected with CRE-Luc were treated for 10 min as in A, and Luc was measured 5 h later. Data are the mean ± SEM of at least three experiments. *, P < 0.01 compared with control (one-way analysis of variance and Dunnett's test). (E) Cells were preincubated with (black bars) or without (red bars) BW (10 μM) for 1 h and then further incubated in 1 μM DADLE or 2 μM Met. Data are the mean ± SEM of at least three experiments.
CRE-mediated gene expression. Subthreshold concentrations of DADLE, Met, or NPA did not stimulate cAMP-dependent gene expression (Table 1). However, coincubation of subthreshold concentrations of DADLE or Met for 10 min with NPA or ethanol increased CRE-Luc 5 h later by 54-67%; forskolin (8) induced an 87% increase (Table 1). Synergy involving DADLE or Met is blocked by Nal (10 μM) and AM (10 μM), respectively (not shown). There is no synergy between DADLE and Met (Table 1).
Table 1. Synergistic increase of CRE-mediated luciferase activity in NG cells.
| Treatment | % Increase over control |
|---|---|
| DADLE (0.01 nM) | 4 ± 3 |
| Met (0.02 nM) | 1 ± 5 |
| NPA (0.5 nM)* | 0 ± 6 |
| EtOH (25 mM)* | 3 ± 5 |
| Forskolin (1 μM)* | 87 ± 12** |
| NPA + DADLE | 60 ± 8** |
| NPA + DADLE + Rp-cAMPS | –3 ± 9 |
| NPA + DADLE + PTX | 4 ± 8 |
| NPA + DADLE + QEHA | 7 ± 8 |
| NPA + Met | 55 ± 3** |
| NPA + Met + Rp-cAMPS | 2 ± 4 |
| NPA + Met + PTX | –3 ± 6 |
| NPA + Met + QEHA | 0 ± 6 |
| EtOH + DADLE | 67 ± 10** |
| EtOH + DADLE + Rp-cAMPS | 3 ± 3 |
| EtOH + DADLE + PTX | 4 ± 5 |
| EtOH + DADLE + QEHA | 8 ± 7 |
| EtOH + Met | 54 ± 6** |
| EtOH + Met + Rp-cAMPS | 1 ± 3 |
| EtOH + Met + PTX | 5 ± 3 |
| EtOH + Met + QEHA | 4 ± 5 |
| DADLE + Met | 6 ± 7 |
Data are the mean ± SEM of at least three experiments.
Previously reported in ref. 9
P<0.01 compared with control (one-way analysis of variance and Dunnett's test)
Synergy for PKA Cα Translocation and CRE-Mediated Gene Expression Is Mediated by βγ-Dependent PKA Activation. Rp inhibits synergy for PKA Cα translocation between DADLE or Met with NPA or ethanol (Fig. 2 A). Synergy is also blocked by QEHA (Fig. 2 A) and by PTX (data not shown). Rp, PTX, or QEHA each inhibits synergistic activation of CRE-Luc (Table 1).
A2 Receptors Are Required for Synergy. We have reported that ethanol increases PKA signaling via A2 receptors (16, 17). A2 synergizes with D2 to increase cAMP (9, 18). We asked whether adenosine also plays a role in synergy between DOR or CB1 with D2. The adenosine receptor antagonist BW A1434U (BW) (10 μM), a gift from Glaxo/Wellcome, does not affect PKA Cα translocation (data not shown) or gene expression (Fig. 2E) induced by saturating concentrations of DADLE (1 μM) or Met (2 μM). In contrast, BW blocks synergy for PKA translocation between subthreshold concentrations of DOR or CB1 agonists with NPA or ethanol (Fig. 2B). Adenosine deaminase (1 unit/ml), which degrades adenosine, prevents synergy (Fig. 2C). Adenosine receptor blockade also prevents synergistic increases in cAMP-dependent gene expression at the same low concentrations of agonists (Fig. 2D). This appears to be caused by A2 because A1 receptors are not expressed in NG cells under these conditions of study (A.S.G. and I.D., unpublished data). Taken together, our data support the model presented in Fig. 4 that synergy for cAMP-dependent gene expression is caused by βγ and A2/Gαs activation and translocation of PKA Cα to the nucleus.
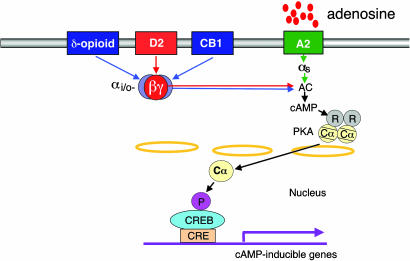
Fig. 4.
Schematic representation of postsynaptic DOR, CB1, and D2 activation-induced PKA Cα translocation and CRE-mediated gene expression via βγ dimers. A central role for βγ subunits released from Gi/o is proposed for DOR, CB1, and D2. This diagram indicates synergy between subthreshold levels of DOR or CB1 (blue arrows) with D2 (red arrows). Synergy for PKA translocation and CRE-mediated gene expression is mediated by βγ dimers from Gi/o. Adenosine A2 activation via Gαs is required for synergy. We propose that colocalization of A2, D2, DOR, and CB1 on the same neurons, like in NAc, confers hypersensitivity to exogenous opioids, cannabinoids, and ethanol because of synergy. In addition, synergy promotes simultaneous hypersensitivity of postsynaptic D2 signaling that is characteristic of addicting drugs.
PHN Exhibit DADLE- and Met-Induced Synergy with NPA or Ethanol. PKA Cα translocation. To validate findings in transformed NG cells, we also studied untransformed PHN. Immature rat PHN express D2 (19), CB1 (20), DOR (21), A2, and A1 (9). Here, saturating levels of DADLE (1 μM) or Met (2 μM) induce PKA Cα translocation to the cytoplasm, neurites, and nuclei in 10 min (Fig. 3 A and B). DOR or CB1 antagonists block DADLE or Met-induced translocation, respectively (Fig. 3A). As in NG cells, Rp, PTX, and QEHA each prevents DADLE- or Metinduced PKA Cα translocation (Fig. 3B). Synergy for translocation occurs at subthreshold concentrations of DADLE (0.01 nM) or Met (0.02 nM) with NPA (0.5 nM) or ethanol (25 mM) (Fig. 3C). Rp, PTX, and QEHA also prevent synergy (not shown).
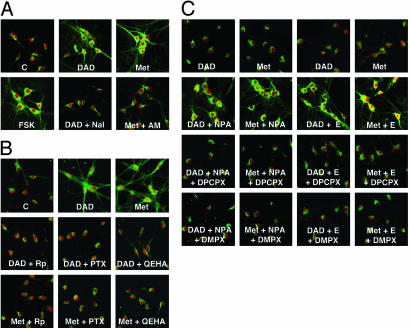
Fig. 3.
DOR or CB1 agonists induce PKA Cα translocation and CRE-Luc expression in rat PHN. (A) PHN were preincubated in the presence or absence of buffer, Nal, or AM, and then incubated in the presence or absence of 1 μM DADLE or 2 μM Met for 10 min. Cα is indicated by green staining, and the neuron-specific marker, NeuN, is indicated by red staining. (B) PHN were preincubated with or without Rp for 1.5 h, or overnight with or without PTX or QEHA before incubation with or without DADLE or Met as in A. (C) PHN were treated with subthreshold concentrations of DADLE, Met, NPA, or ethanol alone or in combination as in Fig. 2 A. Where indicated, the cells were preincubated for 1 h with 1,3-dipropyl-8 cyclopentylxanthine (DPCPX) (100 nM) or 3,7-dimethyl-1-(2-propynyl)xanthine (DMPX) (10 μM). Incubation with subthreshold concentrations of each agent alone was without effect.
CRE-mediated gene expression. Saturating concentrations of DADLE, Met, and NPA each increases Luc in PHN almost as effectively as forskolin (Table 2). DOR-, CB1-, and D2-mediated increases are blocked by their respective antagonists as well as by Rp, PTX, and QEHA (Table 2). Adenosine receptor blockade only prevented ethanol-induced increases in Luc without affecting DOR-, CB1-, or D2-mediated increases (Table 2). M4 and α2 agonists did not induce Luc (Table 2). Subthreshold concentrations of DADLE or Met exhibited synergy with NPA or ethanol (Table 3). Synergy was blocked by Rp, PTX, and QEHA (Table 3), indicating that synergy in untransformed PHN occurs by the same mechanism as in transformed NG cells (Fig. 4).
Table 2. CRE-mediated luciferase activity in PHN.
| Treatment | % increase over control |
|---|---|
| DADLE (1 μM) | 56 ± 10* |
| DADLE + BW A1434U | 56 ± 2* |
| DADLE + Nal (10 μM) | –5 ± 3 |
| DADLE + Rp-cAMPS | 7 ± 5 |
| DADLE + PTX | 0 ± 6 |
| DADLE + QEHA | 4 ± 5 |
| Met (2 μM) | 46 ± 7* |
| Met + BW | 45 ± 9* |
| Met + AM281 (10 μM) | –7 ± 8 |
| Met + Rp-cAMPS | 3 ± 8 |
| Met + PTX | 9 ± 3 |
| Met + QEHA | –4 ± 5 |
| NPA (50 nM) | 51 ± 5* |
| NPA + BW | 48 ± 3* |
| NPA + Spip (10 μM) | 5 ± 3 |
| NPA + Rp-cAMPS | –3 ± 5 |
| NPA + PTX | –1 ± 5 |
| NPA + QEHA | 5 ± 7 |
| EtOH (100 mM) | 41 ± 9* |
| EtOH + BW (10 μM) | –7 ± 2 |
| EtOH + Rp-cAMPS (20 μM) | –5 ± 5 |
| EtOH + PTX (50 ng/ml) | 4 ± 5 |
| EtOH + QEHA | –4 ± 6 |
| Carb (10 μM) | 10 ± 5 |
| UK (10 μM) | 11 ± 8 |
| Forskolin (10 μM) | 78 ± 7* |
P < 0.01 compared with control as in Table 1
Table 3. Synergistic increase of CRE-mediated luciferase activity in PHN.
| Treatment | % increase over control |
|---|---|
| DADLE (0.01 nM) | 8 ± 6 |
| Met (0.02 nM) | 5 ± 8 |
| NPA (0.5 nM) | 3 ± 9 |
| EtOH (25 mM) | 4 ± 7 |
| NPA + DADLE | 46 ± 9* |
| NPA + DADLE + BW | 5 ± 4 |
| NPA + DADLE + Rp-cAMPS | 1 ± 7 |
| NPA + DADLE + PTX | 9 ± 5 |
| NPA + DADLE + QEHA | 3 ± 5 |
| NPA + Met | 42 ± 8* |
| NPA + Met + BW | –7 ± 3 |
| NPA + Met + Rp-cAMPS | 8 ± 4 |
| NPA + Met + PTX | 1 ± 5 |
| NPA + Met + QEHA | –5 ± 4 |
| EtOH + DADLE | 38 ± 8* |
| EtOH + DADLE + BW | 5 ± 2 |
| EtOH + DADLE + Rp-cAMPS | 7 ± 9 |
| EtOH + DADLE + PTX | –4 ± 8 |
| EtOH + DADLE + QEHA | 8 ± 7 |
| EtOH + Met | 43 ± 9* |
| EtOH + Met + BW | 3 ± 4 |
| EtOH + Met + Rp-cAMPS | 9 ± 5 |
| EtOH + Met + PTX | –3 ± 2 |
| EtOH + Met + QEHA | 4 ± 2 |
| DADLE + Met | 8 ± 8 |
P < 0.01 compared with control as in Table 1
Adenosine Receptor Activation Is Required for Synergy. We asked whether adenosine receptors are required for synergy between DADLE or Met with NPA or ethanol in PHN. Three adenosine receptor antagonists were used: the general antagonist BW, the A1 antagonist DPCPX (100 nM), and the A2 antagonist DMPX (10 μM). All three antagonists prevent synergy for PKA Cα translocation (Fig. 3C) (data for BW not shown). These results suggest that both A1 and A2 receptors are required for synergy in immature PHN. In the CNS, A1 are usually presynaptic, whereas A2 are predominantly postsynaptic (38). In our in vitro studies, Gi/o-coupled A1 receptors are probably postsynaptic and may contribute to synergy via βγ dimers. Preincubation with 1 μM tetrodotoxin (TTX) for 2 h was without effect, indicating that presynaptic neurotransmitter release is not required. BW also blocks synergistic increases in CRE-Luc induced by subthreshold concentrations of DADLE or Met with NPA or ethanol (Table 3). Taken together, our data suggest that PHN in culture exhibit DOR-, CB1-, and D2-induced βγ-dependent PKA Cα translocation and increased cAMP-dependent gene expression. Synergy for PKA Cα translocation and gene expression is also observed and appears to require adenosine and βγ dimers.
Discussion
Our major findings are illustrated schematically in Fig. 4. First, activation of Gi/o-coupled DOR and CB1 by saturating concentrations of agonists increases PKA signaling and cAMP-dependent gene transcription via βγ dimers. Second, transient increases in cAMP and PKA Cα translocation to the nucleus observed at 10 min were sufficient to increase cAMP-dependent CRE-mediated gene expression hours later. Third, and most important, there is a remarkable synergy for PKA signaling between DOR or CB1 with D2 agonists or ethanol. Subthreshold concentrations of DADLE or Met, which are without effect alone, when added together with subthreshold concentrations of NPA or ethanol induce a synergistic increase in PKA Cα translocation and cAMP-dependent gene expression. In all instances, synergy appears to require adenosine and is mediated by βγ dimers. Thus, adenosine and βγ dimers appear to confer hypersensitivity simultaneously to DOR, CB1, and D2 agonists when expressed on the same cells.
cAMP/PKA signaling is involved in neural responses to opioids and cannabinoids in cellular (9) and animal (2, 21-23) models of addiction and withdrawal. We find that saturating concentrations of DOR and CB1 agonists increase cAMP levels at 10 min, whereas M4 and α2 agonists do not. Nevertheless, all of these receptors inhibit cAMP production when assayed at 30 min. These findings suggest that short-term activation of Gi/o-coupled receptors involved in addiction appears to stimulate AC and PKA translocation. This leads to cAMP-dependent gene expression hours later. Overexpression of βγ inhibitor peptides blocks DOR- or CB1-induced PKA translocation and gene expression, probably by preventing βγ activation of AC (9, 22). Adenosine is not required. This is in contrast to ethanol, which requires adenosine to activate cAMP production via A2/Gαs (17). Thus, our results suggest that saturating concentrations of DADLE or Met compared with ethanol appear to share a common characteristic; brief exposure to these agents increases PKA signaling by Gi/o βγ dimers or Gαs, respectively.
Our model cell systems allowed us to investigate the consequences of DOR and CB1 activation of AC. We find that cAMP-dependent gene transcription is demonstrable 5 h later, when cAMP levels are no longer increased. This finding is consistent with current concepts that cAMP-dependent gene expression persists long after cAMP produced by receptor activation has been degraded (23). Indeed, prolonged responses involving PKA signaling may help to explain the observation that a single dose of ethanol (which increases cAMP/PKA signaling) potentiates γ-aminobutyric acidergic synaptic function for ≈7 days (24). Potentiation is PKA-dependent and appears to contribute to increased ethanol consumption. Our findings that brief exposures to addicting substances produce significant increases in cAMP-dependent gene expression are consistent with the pathophysiologic importance of PKA in the development of addiction (2).
We also find a striking synergy between subthreshold concentrations of DOR or CB1 agonists with ethanol to potentiate PKA signaling and cAMP-dependent gene expression. Ethanol increases cAMP by promoting A2 activation of Gαs (25). Because Gαs is required for βγ dimer stimulation of AC II and IV (5), we presume that synergy between DOR or CB1 with ethanol is mediated by Gi/o βγ potentiation of ethanol/Gαs-mediated increases in AC activity (Fig. 4).
Addicting drugs use dopaminergic signaling (1). Here we find that subthreshold concentrations of DOR or CB1 agonists hypersensitize D2 signaling by synergy with D2 agonists. Based on these data, we would predict that D2 contributes to opiate-seeking behavior and physical dependence, and that D2 antagonists would have the opposite effect. This has been confirmed in animal studies (29). Dopamine agonists mimic heroin priming (26, 27), and D2 antagonists produce signs of withdrawal in morphine-dependent rodents (28). Mice lacking D2 show reduced morphine self-administration (29) and a lack of opiate rewarding responses (29, 30), but not everyone agrees (31). In a different experimental setting, functional evidence consistent with synergy mediated by βγ dimers has been reported in pain studies. After prior stimulation of Gs-coupled receptors by prostaglandin, opioid activation of a Gi/o-coupled receptor produces a paradoxical cAMP-dependent hyperalgesia in rats that is blocked by a βγ inhibitor (32). There is also evidence that D2 activation increases release of cannabinoids (33) and that cannabinoids potently increase dopaminergic signaling (34). Here our data documenting synergy between CB1 and D2 are supported indirectly by the observation that D2 agonists potentiate cannabinoid-induced sedation at doses that are ineffective alone (35).
The striatum is enriched with postsynaptic A2 (36). Also, A2 and D2 appear to be coexpressed very closely with each other on the same striatal neurons, in contrast to most other brain regions (37-42). Others have shown that synergy between A2 and D2 increases cAMP in PC12 cells (18). Our model (Fig. 4) suggests that the presence of A2 and D2 with DOR and CB1 on the same neurons appears to confer hypersensitivity to exogenous DOR and CB1 agonists for activation of PKA signaling. Because synergy also confers hypersensitivity to D2 agonists, synergy would be expected to hypersensitize drug-induced activation of D2 signaling in the ventral tegmental area (VTA)-NAc pathway.
All instances of synergy appear to require adenosine and A2 receptors. Thus, degradation of adenosine by adenosine deaminase or adenosine receptor blockade prevents synergy. However, it is not clear how synergy develops between DOR or CB1 with D2. The simplest explanation for our data is that synergy of DOR or CB1 with D2 increases extracellular adenosine. However, unlike ethanol (25), NPA does not significantly inhibit adenosine uptake (unpublished observation), and it remains to be determined how synergy promotes A2 activation. Nevertheless, we would predict that A2 agonists potentiate drug-seeking behavior, whereas A2 antagonists would do the opposite. This hypothesis is supported by a recent report in which A2 agonists potentiate morphine self-administration in rats, whereas A2 antagonists block this addictive behavior (43). In addition, opioid agonists appear to decrease adenosine uptake in striatal cells (44), and chronic morphine increases the sensitivity of excitatory postsynaptic currents to adenosine inhibition in the NAc, apparently by decreasing adenosine uptake (45).
Adenosine receptor agonists and morphine produce similar physical dependence and bidirectional cross-withdrawal syndromes in response to receptor antagonists (46). Thus, naloxone, an opioid antagonist, precipitates adenosine withdrawal and an A1/A2 antagonist provokes morphine withdrawal. Also, adenosine receptor antagonists attenuate the development of morphine sensitization in mice (47). In humans, adenosine infusions strikingly reduce the requirement for opiates for postoperative pain (48). These behavioral and clinical observations support our hypothesis that βγ-mediated synergy produced by low concentrations of exogenous opioid agonists in the presence of endogenous adenosine regulates responses to opiates. We propose that adenosine and βγ dimers are components of a postsynaptic molecular mechanism that hypersensitizes dopaminergic signaling in the presence of opioids, cannabinoids, and ethanol and may contribute to their self-administration. This suggests that adenosine receptor antagonists might attenuate the development of addiction.
Acknowledgments
We thank Glaxo/Wellcome for providing BW A1434U, S. Taylor (University of California, San Diego) for the antibodies to PKA Cα, R. J. Lefkowitz (Howard Hughes Medical Institute, Duke University Medical Center, Durham, NC) for the βARK1 minigene, and R. L. Neve (Harvard Medical School, Boston) for the HSVPrpUC vector. We are grateful to A. Bonci, A. Constantinescu, M. Diamond, P. Janak, M. Miles, and R. White for critical reading of the manuscript. We thank Y. Jiang for viral vector construction, L. Wang for preparing primary hippocampal cultures, and S. Karadottir for expert manuscript preparation. This research was supported by National Institutes of Health Grants AA10030 and AA10039 (to A.S.G. and I.D.), funds provided by the State of California for medical research on ethanol and substance abuse through the University of California, San Francisco, and a grant from the Department of the Army, DAMD 17-01-1-0803 (to I.D.).
Abbreviations: Met, methanandamide; Nal, naltrindole; Carb, carbachol; AC, adenylyl cyclase; PKA, protein kinase A; NG, NG108-15/D2; A2, adenosine A2 receptor; D2, dopamine D2 receptor; Luc, luciferase; Rp, Rp-cAMPS; PTX, pertussis toxin; AM, AM 281; UK, UK 14304; BW, BW A1434U; PHN, primary hippocampal neurons; DADLE, [d-Ala2, d-Leu5]enkephalin; DOR, δ-opioid receptor; CB1, cannabinoid receptor; NPA, R(-)-2,10,11-trihydroxy-N-propylnorapomorphine hydrobromide.
References
- 1.Robbins, T. W. & Everitt, B. J. (1999) Nature 398, 567-570. [DOI] [PubMed] [Google Scholar]
- 2.Nestler, E. J. (2001) Nat. Rev. Neurosci. 2, 119-128. [DOI] [PubMed] [Google Scholar]
- 3.Hyman, S. E. & Malenka, R. C. (2001) Nat. Rev. Neurosci. 2, 695-703. [DOI] [PubMed] [Google Scholar]
- 4.Albert, P. R. & Robillard, L. (2002) Cell. Signal. 14, 407-418. [DOI] [PubMed] [Google Scholar]
- 5.Tang, W. J. & Gilman, A. G. (1991) Science 254, 1500-1503. [DOI] [PubMed] [Google Scholar]
- 6.Federman, A. D., Conklin, B. R., Schrader, K. A., Reed, R. R. & Bourne, H. R. (1992) Nature 356, 159-161. [DOI] [PubMed] [Google Scholar]
- 7.Koch, W. J., Hawes, B. E., Inglese, J., Luttrell, L. M. & Lefkowitz, R. J. (1994) J. Biol. Chem. 269, 6193-6197. [PubMed] [Google Scholar]
- 8.Watts, V. J. & Neve, K. A. (1997) Mol. Pharmacol. 52, 181-186. [DOI] [PubMed] [Google Scholar]
- 9.Yao, L., Arolfo, M. P., Dohrman, D. P., Jiang, Z., Fan, P., Fuchs, S., Janak, P. H., Gordon, A. S. & Diamond, I. (2002) Cell 109, 733-743. [DOI] [PubMed] [Google Scholar]
- 10.Sharma, S. K., Klee, W. A. & Nirenberg, M. (1975) Proc. Natl. Acad. Sci. USA 72, 3092-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugiura, T., Kodaka, T., Nakane, S., Miyashita, T., Kondo, S., Suhara, Y., Takayama, H., Waku, K., Seki, C., Baba, N. & Ishima, Y. (1999) J. Biol. Chem. 274, 2794-2801. [DOI] [PubMed] [Google Scholar]
- 12.Lim, F. & Neve, R. L. (2001) in Current Protocols in Neuroscience (Wiley, New York), pp. 4.13.1-4.13.17.
- 13.Houslay, M. D. & Milligan, G. (1997) Trends Biochem. Sci. 22, 217-224. [DOI] [PubMed] [Google Scholar]
- 14.Weng, G., Li, J., Dingus, J., Hildebrandt, J. D., Weinstein, H. & Iyengar, R. (1996) J. Biol. Chem. 271, 26445-26448. [DOI] [PubMed] [Google Scholar]
- 15.Chen, J., DeVivo, M., Dingus, J., Harry, A., Li, J., Sui, J., Carty, D. J., Blank, J. L., Exton, J. H., Stoffel, R. H., et al. (1995) Science 268, 1166-1169. [DOI] [PubMed] [Google Scholar]
- 16.Asher, O., Cunningham, T. D., Yao, L., Gordon, A. S. & Diamond, I. (2002) J. Pharmacol. Exp. Ther. 301, 66-70. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, A. S., Collier, K. & Diamond, I. (1986) Proc. Natl. Acad. Sci. USA 83, 2105-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudlacek, O., Just, H., Korkhov, V. M., Vartian, N., Klinger, M., Pankevych, H., Yang, Q., Nanoff, C., Freissmuth, M. & Boehm, S. (2003) Neuropsychopharmacology 28, 1317-1327. [DOI] [PubMed] [Google Scholar]
- 19.Meador-Woodruff, J. H., Mansour, A., Civelli, O. & Watson, S. J. (1991) Prog. Neuropsychopharmacol. Biol. Psychiatry 15, 885-893. [DOI] [PubMed] [Google Scholar]
- 20.Katona, I., Sperlagh, B., Sik, A., Kafalvi, A., Vizi, E. S., Mackie, K. & Freund, T. F. (1999) J. Neurosci. 19, 4544-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Commons, K. G. & Milner, T. A. (1997) J. Comp. Neurol. 381, 373-387. [PubMed] [Google Scholar]
- 22.Olianas, M. C. & Onali, P. (1999) Biochem. Pharmacol. 57, 649-652. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz, J. H. (2001) Proc. Natl. Acad. Sci. USA 98, 13482-13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melis, M., Camarini, R., Ungless, M. A. & Bonci, A. (2002) J. Neurosci. 22, 2074-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy, L. E., Diamond, I., Casso, D. J., Franklin, C. & Gordon, A. S. (1990) J. Biol. Chem. 265, 1946-1951. [PubMed] [Google Scholar]
- 26.De Vries, T. J. & Shippenberg, T. S. (2002) J. Neurosci. 22, 3321-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wise, R. A., Murray, A. & Bozarth, M. A. (1990) Psychopharmacology 100, 355-360. [DOI] [PubMed] [Google Scholar]
- 28.Funada, M. & Shippenberg, T. S. (1996) Behav. Pharmacol. 7, 448-453. [PubMed] [Google Scholar]
- 29.Elmer, G. I., Pieper, J. O., Rubinstein, M., Low, M. J., Grandy, D. K. & Wise, R. A. (2002) J. Neurosci. 22, RC224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maldonado, R., Saiardi, A., Valverde, O., Samad, T. A., Roques, B. P. & Borrelli, E. (1997) Nature 388, 586-589. [DOI] [PubMed] [Google Scholar]
- 31.Dockstader, C. L., Rubinstein, M., Grandy, D. K., Low, M. J. & van der Kooy, D. (2001) Eur. J. Neurosci. 13, 995-1001. [DOI] [PubMed] [Google Scholar]
- 32.Khasar, S. G., Wang, J. F., Taiwo, Y. O., Heller, P. H., Green, P. G. & Levine, J. D. (1995) Neuroscience 67, 189-195. [DOI] [PubMed] [Google Scholar]
- 33.Giuffrida, A., Parsons, L. H., Kerr, T. M., Rodriguez de Fonseca, F., Navarro, M. & Piomelli, D. (1999) Nat. Neurosci. 2, 303-304. [DOI] [PubMed] [Google Scholar]
- 34.Maldonado, R. & Rodriguez de Fonseca, F. (2002) J. Neurosci. 22, 3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meschler, J. P., Clarkson, F. A., Mathews, P. J., Howlett, A. C. & Madras, B. K. (2000) J. Pharmacol. Exp. Ther. 292, 952-959. [PubMed] [Google Scholar]
- 36.Svenningsson, P., Le Moine, C., Fisone, G. & Fredholm, B. B. (1999) Prog. Neurobiol. 59, 355-396. [DOI] [PubMed] [Google Scholar]
- 37.Fink, J. S., Weaver, D. R., Rivkees, S. A., Peterfreund, R. A., Pollack, A. E., Adler, E. M. & Reppert, S. M. (1992) Brain Res. Mol. Brain Res. 14, 186-195. [DOI] [PubMed] [Google Scholar]
- 38.Le Moine, C., Svenningsson, P., Fredholm, B. B. & Bloch, B. (1997) J. Neurosci. 17, 8038-8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noble, F. & Cox, B. M. (1997) J. Pharmacol. Exp. Ther. 283, 557-565. [PubMed] [Google Scholar]
- 40.Glass, M. & Felder, C. C. (1997) J. Neurosci. 17, 5327-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreau, J.-L. & Huber, G. (1999) Brain Res. Brain Res. Rev. 31, 65-82. [DOI] [PubMed] [Google Scholar]
- 42.Hillion, J., Canals, M., Torvinen, M., Casado, V., Scott, R., Terasmaa, A., Hansson, A., Watson, S., Olah, M. E., Mallol, J., et al. (2002) J. Biol. Chem. 277, 18091-18097. [DOI] [PubMed] [Google Scholar]
- 43.Sahraei, H., Motamedi, F., Khoshbaten, A. & Zarrindast, M.-R. (1999) Eur. J. Pharmacol. 383, 107-113. [DOI] [PubMed] [Google Scholar]
- 44.Halimi, G., Devaux, C., Clot-Faybesse, O., Sampol, J., Legof, L., Rochat, H. & Guieu, R. (2000) Eur. J. Pharmacol. 398, 217-224. [DOI] [PubMed] [Google Scholar]
- 45.Brundege, J. M. & Williams, J. T. (2002) J. Neurophysiol. 87, 1369-1375. [DOI] [PubMed] [Google Scholar]
- 46.Coupar, I. M. & Tran, B. L. T. (2001) Life Sci. 69, 779-790. [DOI] [PubMed] [Google Scholar]
- 47.Weisberg, S. P. & Kaplan, G. B. (1999) Neurosci. Lett. 264, 89-92. [DOI] [PubMed] [Google Scholar]
- 48.Fukunaga, A. F., Alexander, G. E. & Stark, C. W. (2003) Pain 101, 129-138. [DOI] [PubMed] [Google Scholar]