Abstract
Oxythiamine (OT), a transketolase inhibitor, is known to inhibit pancreatic cancer cell proliferation. In this study, we investigated the effect of inhibition of the transketolase pathway on signaling pathways in MIA PaCa cancer cells using in-house proteomic techniques. We hypothesized that OT alter protein phosphorylation thus affecting cell cycle arrest and cell proliferation. MIA PaCa-2 cells were cultured in media containing an algal 15N amino acid mixture at 50% enrichment, with and without OT, to determine protein expression and synthesis. Analysis of cell lysates using two-dimensional gel electrophoresis matrix assisted laser desorption and ionization time-of-flight and time-of-flight mass spectrometry (2-DE-MALDI-TOF/TOF MS) identified 12 phosphor proteins that were significantly suppressed by OT treatment. Many of these proteins are involved in regulation of cycle activities and apoptosis. Among the proteins identified, expression of the phosphor heat shock protein 27 (Hsp27) was dramatically inhibited by OT treatment while the level of its total protein remained unchanged. Hsp27 expression and phoshporylation is known to be associated with drug resistance and cancer cell survival. The changes in phosphorylation of key proteins of cancer proliferation and survival suggest that protein phosphorylation is the confluence of the effects of OT on metabolic and signaling pathways.
Keywords: Quantitative proteomics, Pancreas cancer, 15N stable isotope, phosphorylation, turnover rate, oxythiamine
Introduction
The pentose phosphate cycle is the source of pentose for synthesis of important coenzymes ATP, CoA, NAD(P)+, FAD, and genetic material, RNA and DNA. It consists in reactions in the oxidative and non-oxidative branches. The oxidative pathway is catalyzed by glucose-6-phosphate 1-dehydrogenase, and converts a 6-carbon lactone intermediate to a 5-carbon ribose phosphate and reducing equivalents (NADPH). The non-oxidative pathway is catalyzed by transketolase (TK) and transaldolase (TA) in the pentose phosphate pathway. The transketolase reaction involves the TK cofactor thiamine diphosphate-mediated transfer of a 2-carbon fragment from D-xylulose-5-P to the aldose erythrose-4-phosphate. The near equilibrium reactions of TK and TA allow redistribution of glucose carbons among glucose-6-phosphate, fructose-6-phosphate, pentose phosphate and glyceraldehyde-3-phosphate to meet the substrate need of intersecting pathways of glycolysis/gluconeogenesis. In proliferating tumor cells, the non-oxidative pathways play an important role in regulating glucose carbon recruitment toward de novo nucleic acid ribose synthesis and cell proliferation (1, 2) to sustain cancer growth. High expression levels of transketolase in invasive tumors have been observed, suggesting it can be used as a target for treatment of cancer (1-4). Furthermore, inhibition of TK with oxythiamine (OT) has been shown to reduce tumor cell proliferation in vitro and in vivo leading to cell cycle arrest and apoptosis (3).
Oxythiamine (OT) has been demonstrated in many studies to inhibit cancer cell growth through suppression of the cell cycle. However, the molecular mechanisms that mediate the inhibitory effect of oxythiamine treatment are not known. In this study, we investigated the molecular mechanisms of the anti-proliferative action of OT at both translational and post-translational levels in MIA PaCa cancer cells using proteomic techniques to determine differential expression. In addition, the synthesis rates of these proteins in MIA PaCa cells were also determined using a new method for metabolic labeling of proteins. This study, using the newly developed quantitative proteomic methodology (5, 6), was designed to investigate whether OT treatment can alter the protein expression profile, especially the phosphoprotein profile, in hopes of advancing our understanding of the molecular mechanism of pancreatic cancer, and finding new drug targets for this devastating disease. Abnormal protein phosphorylation has been shown to correlate positively with the development of the cancer phenotype (7). We found that OT treatment caused cell cycle arrest associated with the deactivation of mitogen-activated protein kinase (MAPK) pathways, and, further, found a significant suppression of phosphorylation of heat shock protein 27 at serine 78 in MIA cancer cells.
Materials and Methods
Cell culture
MIA PaCa cells were grown in MEM supplemented with 10% fetal bovine serum, 1% antibiotic antimycotic, 5% CO at 37° 2C until 85-95% confluence when the experiment started. Experiments were set up in three groups: group 1 cells were cultured in MEM medium containing 1 mg/ml of natural amino acids; group 2 cells with 50% of 15N algal amino acid mixture (98% 15N); and group 3 cells with 50% of 15N algal amino acid mixture (98% 15N) plus 10 μM OT according to a previous report (1). Each treatment was repeated four times and had 4 flasks with 10 ml/flask. After incubation for 72 hours, the cell pellets were collected and stored at −80°C.
Assessment of cell cycle
The cell cycle distribution was analyzed by using flow cytometry based on cellular intake on propidium iodide. Briefly, 106 cells were washed with PBS, incubated with 0.1% sodium citrate dihydrate, 0.1% Triton X-100, 200 μg/mL RNase A, 50 mg/mL propidium iodide (Roche Molecular Biochemicals, NM), and finally analyzed by using a flow cytometer (Becton Dickinson, Carsbad, CA). The percentage of cells in the various stages of the cell cycle was determined with the ModFitLT software (Becton Dickinson, Carlsbad, CA). Three independent experiments were performed for each assay.
Two-dimensional gel electrophoresis
Cell pellets were solubilized in a lysis buffer consisting of 7 M urea, 2 M thiourea, 65 mM DTT and 4% CHAPS. The protein concentration was evaluated with the Bradford assay. IPG-strips were passively rehydrated using 185 μl samples containing 400 μg protein of each paired preparation (11cm, pH 3-10NL or pH 4-7 NL, Bio-Rad, USA). After 14 h of rehydration, the strips were transferred to an IEF Cell (Bio-Rad, USA). IEF was performed at constant power (50 μA/ IPG-strip) at 250 V for 2 h, linear; linear ramping to 1000V for 1 h, 1000 V hold 1 h, linear ramping to 8000 V for 1 h; and finally 8000 V hold for 5 h. The second dimension was performed on an 8-16% Tris-HCl pre-cast gel (Bio-Rad). The protein spots were first stained with Pro-Q Diamond (8, 9), then stained with SYPRO-Ruby (Molecular Probes, Eugene, OR) or visualized with the Coomassie Brilliant Blue R-250 (Merck, Germany). Each experiment was performed twice to ensure the accuracy of analyses. The images were scanned with a Pharox FX™ molecular imager (Bio-Rad) with a 532 nm laser excitation and a 580 nm band pass emission filter. The differential protein spots were identified using PD-Quest 2-DE analysis software (Bio-Rad, USA). The intensity of each protein spot was normalized to the total intensity of the entire gel image, respectively. Only those spots that changed consistently and significantly (> 1.5 fold) were selected for MALDI-TOF/TOF MS.
In-gel digestion and protein identification
Spots of interest were excised from the gels and transferred to Eppendorf LoBind tubes. Then gel slices were de-stained with 50% acetonitrile (can) in 100 mM NH4HCO3 twice at 37 °C for 45 min each, followed by dehydration with ACN for 5 min and vacuum-dried. Gel slices were fully rehydrated with 10-20 μl of NH4HCO3: ACN (25 mM: 10%) supplemented with trypsin (10 ng/μl) (Promega, Madison, WI, USA) at 4 °C and then incubated at 37°C overnight. The peptides were extracted twice with 50 μl of 5% TFA and 50% ACN with vortexing for 45 min at room temperature. The combined extracts were evaporated to about 5 μl in a SpeedVac.
MALDI samples were prepared according to a thin layer method as described before (10). Mass spectra were recorded on an Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) under the control of FlexControl™ 2.2 software (Bruker Daltonics GmbH, Germany). MALDI-TOF spectra were recorded in the positive ion reflector mode in a mass range of 800-3500 Da and the ion acceleration voltage was 25 kV. After analysis of the peptide mass fingerprinting (PMF) results by FlexAnalysis™ 2.2 (Bruker Daltonics GmbH, Germany), the proteins were subjected to MS/MS analysis in “LIFT” (Second Ion Source in the TOF-TOF instrument) mode. Some strong peaks of each PMF spectra were selected as precursor ions which were accelerated in TOF1 at a voltage of 8 KV and fragmented by lifting the voltage to 19 KV. The MALDI-TOF spectra and the MS/MS spectra were processed by FlexAnalysis™ 2.2 and combined by Biotools 3.1 (in-house version). Each MS/MS spectrum was processed as follows: neither smoothing nor background subtraction was performed since each ‘envelop’ of the raw spectrum that was used for calculation of protein synthesis rate will be taken care by the method developed in our previous papers (5, 6, 11); only peaks of S/N ratio above 6 and a maximum of 200 peaks were selected to build the mass list representative of the MS/MS spectrum. The combined PMF and “lift” MS/MS spectra were searched against Homo sapiens (human) (20405 sequences) in Swiss-Prot database (Release SwissProt 57.1 of April 14, 2009, containing 462764 sequences; 163773385 residues) using the Mascot search program (www.matrixscience.com) (Mascot server version 2.2, released on September 9, 2008) (Matrix Science, London, UK). Search parameters were set as follows: enzyme, trypsin; allowance of up to one missed cleavage peptide; mass tolerance, 50ppm and MS/MS mass tolerance, 0.5 Da; fixed modification parameter, carbamidomethylation (C); variable modification parameters, oxidation (at Met). Protein or peptide score with p < 0.05 was regarded as significant. In case of peptides matching to multiple members of a protein family, the positive identified protein was selected based on both the highest score and the highest number of matching peptides (8).
Phosphorylation site mapping
Alkaline phosphatase treatment was performed directly on the previously analyzed samples after initial MALDI-MS, as described previously (12).
Computation of protein synthesis rate
The synthesis rates of the phosphoproteins identified were calculated according to our in-house algorithms (5, 6)
Western blot analysis
Western blotting analysis was performed as described previously (5, 8, 9). In brief, whole-cell extracts were prepared by lysing cells. Lysates containing 50 μg proteins were subjected to gel electrophoresis. Proteins were then transferred to PVDF membranes (Millipore, CA). The blots were blocked in 5% BSA in TBS–Tween (0.1%) solution for 1 h at room temperature, and then incubated at 4 °C overnight with the primary antibody. Human Hsp90 (1:1000), CDK4 (1:200), cyclin D1 (1:200), p27 (1:200) and phospho Hsp27 (1:500) were purchased from Santa Cruz Biotechnology, Santa Cruz, CA); p38 (ratio1:1000), JNK (ratio 1:1000), α-enolase (1:1000), 60S acidic ribosomal protein P0 (1:1000) and nucleophosmin (1:1000) were purchased from Millipore, USA. Anti-β-actin (1:5000) was obtained from Sigma (Sigma-Aldrich, MA) served as loading control. After incubation with secondary antibodies (1:5000) (GE healthcare, CA) at room temperature for at least 1 h, the blot was visualized with an enhanced chemiluminescence (ECL) detection system (Pierce Biotech Inc., Rockford, IL).
Immunohistochemical staining
A representative surgical specimen of pancreatic cancer was obtained from Department of Pathology at Creighton University Medical Center with the approval of the Institutional Review Board after proper consent. Serial tissue sections (5 μm) from the formalin fixed paraffin embed (FFPE) pancreatic cancer blocks were fixed in acetone for 10 min. Endogenous peroxidase activity was quenched by incubation in 3% hydrogen peroxide for 20 min. Sections were washed in Tris-buffered saline (TBS) and blocked with 4% BSA in TBS for 30 min. Primary phosphor p27 antibodies were diluted (1:50) in TBS containing 1% BSA, and 100 μl was added to each section. Incubation was carried out for 2 h at room temperature in a humidified chamber. The slides were then washed in TBST and were incubated with biotinylated goat anti-mouse secondary antibody (sigma) at 1:100 in TBS containing 1% BSA for 30 min at room temperature. After washes in TBS-Tween, ABC biotinylation complex (1:200) (Vector, Burlingame, CA) was added for 1 h at room temperature. Then, DAB was added to the slide for 1-2 min. The images of the phosphor p27 were taken under a microscope (Leica Microsystems Inc., Bannockburn, IL).
Quantification of Phospho Hsp27 at ser78 by Enzyme-Linked ImmunoSorbent Assay (ELISA)
In addition to the tissue samples, we separately tested a set of serum samples from patients (pancreatic tumor; pT2/pT3) and healthy volunteer subjects with age-matched. The patient serum samples (n=12) were collected by City of Hope National Medical Center and NCI-designated Cancer Center (Duarte, CA) with proper informed consent according to a protocol approved by the Institute Review Board. The healthy volunteer blood samples (n=12) were collected at Creighton University Medical Center (Omaha, NE) under a protocol approved by the Institutioanl Review Board, and with a signed consent form. The blood samples in BD Vacutainer® Blood Collection Tubes (BD Ventures, L.L.C., NJ) were fractionated by centrifuging at 1,000 × g for 10 min. The serum samples were immediately divided into aliquots and frozen at −80 °C.
The mean (±SD) age for the tumor patients was 61.3 (±8.1) years, and for the healthy volunteer group, 60.3(±5.4) years. We measured serum levels of the phosphor Hsp27 (ser78) by use of a PathScan® Phospho-HSP27 (Ser78) Sandwich ELISA Kit (Cell Signaling Technology, Danvers, MA) in duplicate, according to the manufacturer’s instructions. We measured ELISA plates on a microtiter plate reader (MRX II; Dynex Technology) at 450 nm and calculated concentration of the respective protein in serum according to a calibration curve. The levels of the phosphor Hsp27 (ser78) in serum were expressed in mean (OD450/mg protein) ± SEM. P values were calculated by 1-sided t-test; receiver operating characteristic (ROC) curves were constructed for serum phosphor Hsp27 (ser78) concentration by plotting sensitivity vs 1-specificity, and calculated the areas under the ROC curves. All the experiments were performed in triplicates.
Localization Analysis of Hsp27 at ser78 by Confocal Microscope (CFM)
MIA PaCa2 cells were cultured in the same way as mentioned above. Cells (1 × 105 cultured in DMEM containing 10% FBS) were put onto APES-coated slides and fixed in formaldehyde for 30 min, then treated with 0.25% Triton X-100 for 15 min. The slides were then blocked with 5% BSA in TBS for 30 min. Primary antibodies phosphor p27 ser78 (goat Ig G, Santa Cruz, CA) were diluted (1:50) in TBS containing 1% BSA, and 100 μl was added to each section. Incubation was carried out for 2 h at room temperature in a humidified chamber. Following PBS washes, slides were incubated with fluorescein isothiocyanate (FITC)-conjugated mouse anti-goat IgG (working dilution 1:50, Dako Cytomation) in the dark at room temperature for 45 min and stained with 4′-6-diamidino-2-phenylindole (DAPI) in the dark for 5 min. The slides were mounted using fluorescent mounting medium (Dako Cytomation) and observed by CFM (Carl Zeiss, Germany).
Data analysis
The synthesis rate of each protein is the average of two to four fragments. One-way ANOVA with Tukey’s adjustment was used for multiple comparisons in SPSS 16.0 (SPSS Inc., Chicago, IL). The ROC curve was constructed by using SPSS 16.0 (SPSS Inc., Chicago, IL).
Results
Oxythiamine caused cell cycle arrest through the MAPK pathways
The chemotherapeutic effects of oxythiamine (OT) on MIA pancreatic cancer cell proliferation were assessed using flow cytometry and Western blotting analysis (Figure 1). OT was found to increase significantly the percentage of the cancer cells in G1/G0 phase (p<0.02), but dramatically reduce the number of cells in G2/M phase (p<0.008) (Figure 1A). To characterize the effect of OT on cell cycle activity in MIA cells further, we analyzed the expression of both positive (e.g. CDK4, cyclin D1) and negative regulators (e.g. p27) of cell cycles (Figure 1B). Clearly, OT treatment can suppress the expression of both CDK4 and cyclin D1, while the level of p27Kip1 was significantly enhanced in MIA cancer cells (Figure 1B). In addition to the cell cycle inhibition of OT, a dose of 10 μM can also deactivate the intracellular signaling pathways associated with cell growth, proliferation and apoptosis induction (13). This effect of OT treatment is supported by the decreased phosphorylation of p38MAPK (Figure 1C) and the 54-kDa phosphorylated JNK isoform, while the total p38MAPK and JNK proteins remain unchanged (Figure 1C). Interestingly, OT did not interfere with the pro-apoptototic pathways of the ERK1/2 MAPK cascade (data not shown). To find out whether the NF-κB cascade is also involved in OT-stimulated process, the expression levels of key components in this pathway (e.g. p52, p65, and p50) were tested (Figure 1D). We found that the signal levels of these key components were not changed significantly (Figure 1D). Together, these results suggest that MIA cell cycle arrest by OT treatment may be through specific deactivation of the p38MAPK cascades.
Figure 1.
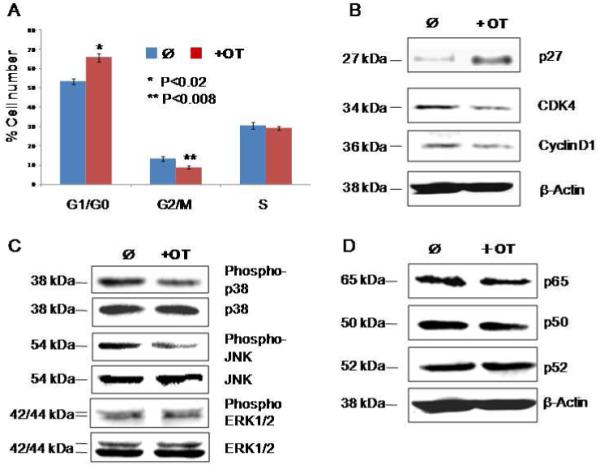
OT causes cell cycle arrest in MIA cells through the MAPK pathway. Panel A shows the alterations of cell cycle distribution by OT treatment as measured by flow cytometry. Panel B shows the altered activity of cell cycle regulators by OT treatment as measured by Western blot. Panel C shows the inhibition of phosphor-p38 and JNK in the MAPK cascades by OT treatment as measured by Western blot. Panel D shows the effect of OT treatment on NF-κB pathway as measured by Western blot. Please see the Materials and Methods section for details. All the experiments were repeated three times.
Oxythiamine suppressed expression of the cellular phosphoproteome
To determine whether the transketolase inhibitor, OT, can also alter cellular phosphoproteome profiles other than those of the MAPK cascade, cell lysates were separated by 2-DE, and the cellular phosphorylated proteins were detected and visualized by Pro-Q Diamond stain and imaged by using an external laser system (upper panel in both Figure 2A and Figure 2B,). In comparison with the phosphoproteome in the OT-treated group (Figure 2B, upper panel) to control (Figure 2A, upper panel), we found that expression levels of the cellular phosphorylated proteins were significantly reduced.
Figure 2.
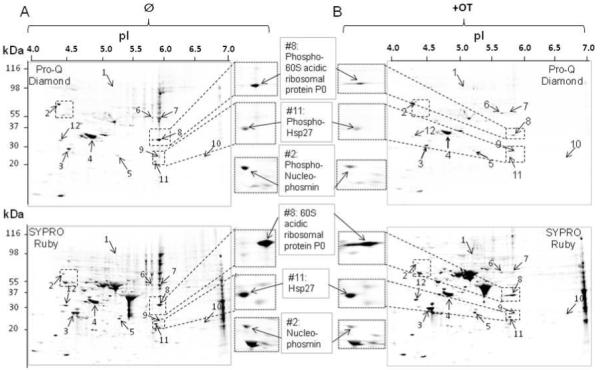
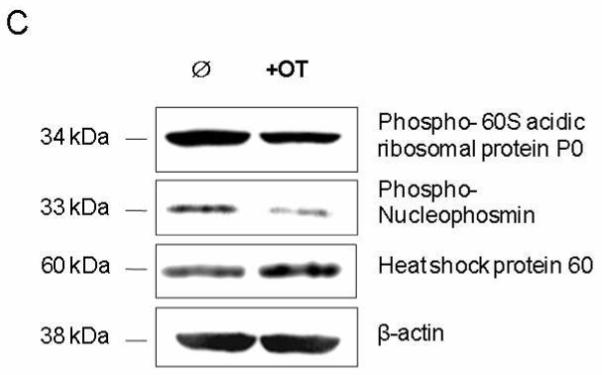
Separation of phosphor proteins by two-dimensional gel electrophoresis (2-DE). Phosphor proteins (A & B, top panel) were separated by 2-DE (4/7) and first stained by using the ProQ-diamond kit, followed by SYPRO Ruby stain for the detection of total proteins on the same gel (A & B, lower panel). The numbered spots with arrows on 2-D gel show proteins suppressed by OT treatment and identified by MS and listed in Table 1. Examples of the suppressed protein spots (p<0.05) by OT treatment (left-hand side of panel B) as compared to control (right-hand side of panel A) were shown. C), Confirmation of proteins suppressed by OT treatment using Western blot analysis. Please see the Materials and Methods section for details. OT experiments were repeated four times and all the 2-DE experiments were performed in triplicate with variation <10%.
Because of the chemical properties of phosphoproteins, we observed that the majority of the phosphoproteome appears in the low pH range of the 2-D gel map (data not shown). To increase the resolution of the phosphoproteins separated by the 2-DE, cell lysates were also separated in the first dimension using the narrow pH range (4-7) of IPG strips (Figure 2A/2B). In the zoom-in separation of the phosphoproteins, 12 cellular phosphoproteins were detected, and significantly suppressed in MIA cancer cells, in response to OT treatment (Figure 2B). These differentially expressed phosphoproteins were further identified by MALDI-TOF/TOF MS. The protein IDs were signed based on the following criteria: a) three tryptic digested peaks must be matched; b) three or more MS/MS de novo sequences from a PMF spectrum must be confirmed if only one or less tryptic digested peak can be matched; c) all the signed protein IDs can be further confirmed by Western blot. All the identified and the confirmed proteins based on the criteria above are listed in Table 1. These include proteins playing a role in the antioxidant (peroxiredoxin-4 and peroxiredoxin-6) involved in cell proliferation, cell cycles, and apoptosis (Hsp90, nuclear autoantigenic sperm protein, and nucleophosmin), two signaling pathway components (14-3-3 protein ε, Hsp27), and one responsible for degradation (proteasome subunit A type 5 (PSMA5)). The suppressive effects of OT on the phosphoproteins were well represented by the higher magnification images of the phosphoproteins such as 60S acidic ribosomal protein P0, nucleophosmin and Hsp27 as shown in both Figure 2A (right-hand side of the upper panel) Figure 2B (left-hand side of the upper panel). Interestingly, the total protein expression of Hsp27 and 60S acidic ribosomal protein P0 remains largely unchanged, while the total protein expression of nucleophosmin showed a trend similar to its phosphor form upon OT treatment (right-hand side of the lower panel in Figure 2A, and left-hand side of the lower panel in Figure 2B).
Table 1.
Identification of differentially expressed phosphoproteins (> two-fold decrease ↓ by OT treatment in MIA PaCa-2 cells) by MALDI-TOF/TOF MS
| Spot #a |
Accession Number |
Protein Name |
Scoreb | m/z | Pep(score)c | Sequence coverage (%) |
Charge | Matched/ Unmatchedd |
Calculated MW/pI |
Possible Role | Turnover Rolee |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phosphor proteins identified | |||||||||||
| 2255.99 |
154HNDDEQYAWESSAGGSFTVR173 (59) |
||||||||||
| 1 | P07900 | ↓Hsp90α | 360 | 2374.179 | 368VFIMDNCEELIPEYLNFIR386 (49) | 42 | mono | 26/66 | 84607/4.94 | Related to tumor growth (24-26, 36) |
0.67 |
| 91348.6 | 328HFSVEGQLEFR338 (20) | ||||||||||
| 2415.210 | 368VFIMDNCEELIPEYLNFIR386 (52) | ||||||||||
| 1342.729 | 536EAQLYAAQAHLK547 (75) | ||||||||||
| 1610.906 | 538EIEELKELLPEIR650 (79) | ||||||||||
| 2 | P49321 | ↓ NASP | 290 | 1988.813 | 77YGETANECGEAFFFYGK93 (61) | 26 | mono | 14/25 | 85186/4.26 | Cellular proliferation (37) |
0.89 |
| 2144.032 | 698KPTDGASSSNCVTDISHLVR717 (89) | ||||||||||
| 1423.788 | 21LFQVEYAIEAIK32 (67) | ||||||||||
| 3 | P28066 | ↓ PSMA5 | 180 | 12084.05 | 67IVEIDAHIGCAMSGLIADAK86 (76) | 39 | mono | 7/39 | 26394/4/74 | H1 hinker histone chaperone that is required for cell proliferation (38) |
0.56 |
| 2162.975 |
150GPQLFHMDPSGTFVQCDAR168 (53) |
||||||||||
| 1819.83 | 278MTDQEAIQDLWQWR291 (60) | ||||||||||
| 4* | P06748 | ↓ NPM | 116 | 2227.22 |
81MSVQPTVSLGGFEITPPVVLR101 (51) |
11 | mono | 3/ | 32555/4.64 | anti-apoptosis (39) |
0.67 |
| 1568.730 | 33VDNDENEHQLSLR45 (40) | ||||||||||
| 5 | P09211 | ↓ GSTP1 | 118 | 1337.55 | 2PPYTVVYFPVR12 (34) | 30 | mono | 4/27 | 23341/5.42 | anti-apoptosis (40) |
0.72 |
| 1883.7 | 56FQDGDLTLYQSNTILR71 (84) | ||||||||||
| 1684.940 | 430AAVEEGIVLGGGCALLR446 (46) | ||||||||||
| 6* | P10809 | ↑ HSP60 | 151 | 2365.335 |
269KPLVIIAEDVDGEALSTLVLNR290 (54) |
11 | mono | 3/ | 61016/5.7 | Regulator in cancer Progression (41) |
0.54 |
| 2560.26 |
97LVQDVANNTNEEAGDGTTTATV LAR121 (67) |
||||||||||
| 1370.689 | 472ELSDFISYLQR482 (80) | ||||||||||
| 7 | P30101 | ↓ PDIA3 | 170 | 91664.75 | 434MDATANDVPSPYEVR448 (36) | 26 | mono | 12/23 | 56747/5.98 | ER stress related (42) |
0.45 |
| 2575.29 |
306TFSHELSDFGLESTAGEIPVVAIR 329 (54) |
||||||||||
| 1896.100 | 248VLALSVETDYTFPLAEK264 (31) | ||||||||||
| 8 | P05388 | ↓ RPLP0 | 90 | 2787.70 |
221NVASVCLQIGYPTVASVPHSIIN GYK246 (23) |
31 | mono | 6/35 | 34252/5.71 | Cancer cell proliferation (43) |
0.72 |
| 1624.790 | 187DYGVYLEDSGHTLR200 (89) | ||||||||||
| 9* | Q13162 | ↓ PRDX4 | 167 | 2443.082 |
46TREEECHFYAGGQVYPGEASR66 (68) |
7 | mono | 2/ | 30521/5.86 | Promote cancer cell growth (44, 45) |
0.67 |
| 1085.600 | 98LPFPIIDDR106 (60) | ||||||||||
| 1395.690 | 42DFTPVCTTELGR53 (87) | ||||||||||
| 10 | P30041 | ↓ PRDX6 | 452 | 1582.72 | 85DINAYNCEEPTEK97 (48) | 32 | mono | 5/30 | 25019/6.0 | Cancer cell proliferation (45) |
0.73 |
| 2031.1 | 25FHDFLGDSWGILFSHPR41 (56) | ||||||||||
| 2098.150 |
2PGGLLLGDVAPNFEANTTVGR22 (111) |
||||||||||
| 960.3980 | 21DWYPHSR27 (43) | ||||||||||
| 11 | P04792 | ↓ HSP27 | 231 | 1163.610 | 28LFDQAFGLPR37 (74) | 58 | mono | 12/57 | 22768/5.9 | Promote tumor growth (45) and drug resistance(46) |
0.66 |
| 1783.920 | 97VSLDVNHFAPDELTVK112 (58) | ||||||||||
| 1905.98 | 172LATQSNEITIPVTFESR188 (57) | ||||||||||
| 1256.65 | 131YLAEFATGNDR141 (42) | ||||||||||
| 12 | P62258 | ↓14-3-3E | 306 | 2088.020 |
197AAFDDAIAELDTLSEESYK215 (96) |
59 | mono | 5/12 | 29155/4.63 | Cell cycle progression and tumor growth (47) |
0.63 |
| 2331.26 |
171LGLALNFSVFYYEILNSPDR190 (60) |
||||||||||
| 1820 | 154AASDIAMTELPPTHPIR170 (75) | ||||||||||
As labeled on the 2-D gel (see Figure. 2).
Molecular Weight Search (MOWSE) scores obtained by the combined search (PMF and LIFT data) that were signed to specific protein identity using Mascot engine.
Peptides identified by Ultraflex MALDI-TOF/TOF in “LIFT” mode, and individual peptide score (LIFT data) in the bracket. The superscript number represents the start and end amino acid.
Number of masses matched and not matched for peptide fingerprint data.
Protein turnover rate was calculated according to a newly developed modified (5, 6).
Only LIFT data is available for sequence coverage and masses matched or unmatched calculation.
To verify the newly induced phosphoproteins identified by MS, western blotting analysis was performed (Figure 2C). Figure 2C shows that phosphor 60S acidic ribosomal protein P0 and phosphor nucleophosmin were significantly suppressed by the introduction of OT into MIA cells. These results confirmed the expression pattern of these two proteins detected by 2-DE analysis (Figure 2B).
OT interrupted protein synthesis rate
Protein turnover in cells is the result of protein synthesis and degradation. The turnover rate of a protein indicates the functional significance of the protein and can explain the incongruence between transcriptome and proteome data. To understand the role the differentially expressed phosphoproteins play in OT-treated MIA cells, we used our recently developed method (5, 6). This method is based on the isotopomer distribution of peptides with the labeled (new) and unlabeled 1 peptides as shown in Figure 3A/B. We measured the synthesis rates of those proteins identified and listed in Table 1. This table shows that the protein synthesis rate in 72 hours was roughly classified into two categories: <60% and >60%. Proteins with a turnover rate <60% include PDIA 3, while proteins with a turnover rate >60% are nuclear autoantigenic sperm protein (89%), 60S acidic ribosomal protein P0 (72%), glutathione S-transferase P (72%), peroxiredoxin-6 (73%) and Hsp 27 (~70%) (Table 1). Because of the low protein concentration recovered from the 2D gel, we were not able to determine accurately the rates of synthesis of these proteins in the OT treated cell extract.
Figure 3.
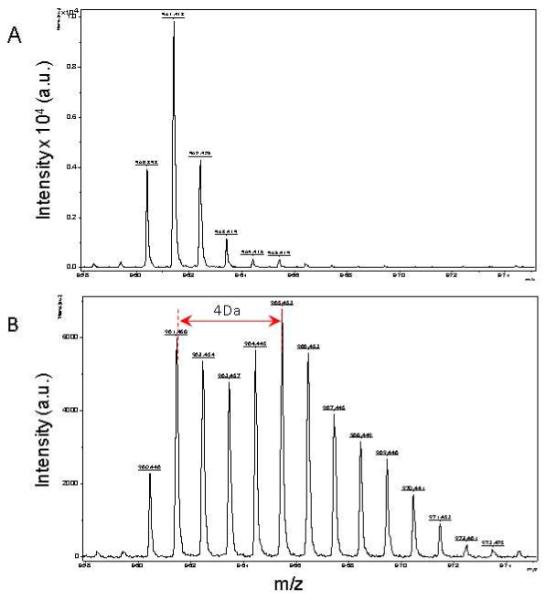
Mass spectra of 960 m/z fragment from Hsp27 of lysates of cells grown in the presence of natural amino acids (Panel A), and 50% enriched 15N algal amino acid mixtures (Panel B). MS spectrum of control in Panel A shows the binomial distribution of isotopomic peaks largely due to natural existence of 13C. Incorporation of 15N resulted in an obvious mass shift in isotopomic distribution in Panel B. The synthesis rate of a protein was based on the isotopomer distribution of these two spectra.
OT selectively inhibits the activity of phosphor Hsp27 isoforms
The specific site of phosphorylation in phosphor Hsp27 was investigated using a method developed by Larsen et al (12), which is based on enzymatic de-phosphorylation and differential mass spectrometric peptide mass to map out the specifically phosphorylated sites in proteins separated by gel electrophoresis (Figure4A). Published studies have reported seven phosphorylation sites in Hsp 27 (14-20). These sites in the protein sequence of Hsp27 are located at serine residues (highlighted in red) shown in Figure 4C. Among the four trypsin-digested peptides identified in this protein (Table 1, Figure 4C underlined sequences), only one peptide showed a 80Da-mass shift after de-phosphorylation of the peptide, indicating that this peptide only carries one phosphorylated site (Figure 4B). The further de novo sequencing analysis of this peptide showed that the phosphorylation site occurs at serine 26 as highlighted in red in Figure 4C (Figure 4B/C). The tryptic digest method does not allow analysis of phosphorylation in the remaining six serine residues. However, three of the residues can be analyzed based on the antibodies commercially available for phosphor-Hsp27 (ser15, ser78, and ser82, respectively). The cell lysates from OT treated and control MIA cells were analyzed by western blot (Figure 4D). Oxythiamine treatment significantly suppressed the activity of the phosphor-Hsp27(ser78) and not that of the other two while the total Hsp27 protein remains unchanged (Figure 4D).
Figure 4.
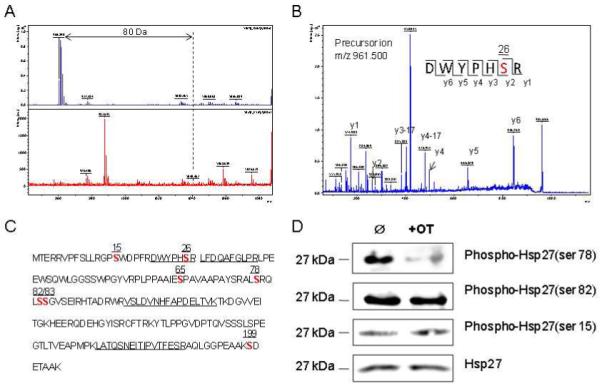
Identification of the phosphorylation site of Hsp27. Panel A shows determination of the phosphorylated site of the peptide by de-phosphorylation with CIAP (Calf Intestinal Alkaline Phosphatase, upper panel in A) and without CIAP (lower panel in A). Panel B shows the MS/MS spectrum of this phosphor fragment in ‘lift” mode. The peptide sequence is easily identified as almost every y ion is observed. Panel C shows the whole sequence of human Hsp27. The number on the top of each bold “S” indicates the possible phosphorylated serine in this protein. Panel D shows overexpression of phosphor Hsp27 at Ser78, Ser82 and Ser15 in MIA cells treated with and without OT by using Western analysis. Please see the Materials and Methods section for details.
Hsp 27 (ser 78) in pancreatic cancer
To validate the role of phosphor Hsp27 (ser78) in pancreatic cancer further, immunohistochemical analysis was performed in a clinical specimen of the pancreatic cancer tissue (n=10) and its adjacent specimen (n=10). We found that positive staining (brown granules) of epithelium cells in pancreatic ductal carcinoma tissue was observed in the cytoplasm and occasionally in the nucleus (Figure 5A). In normal pancreatic tissue, phosphor Hsp27 at Ser-78 was much less stained by its antibody (Figure 5A).
Figure 5.
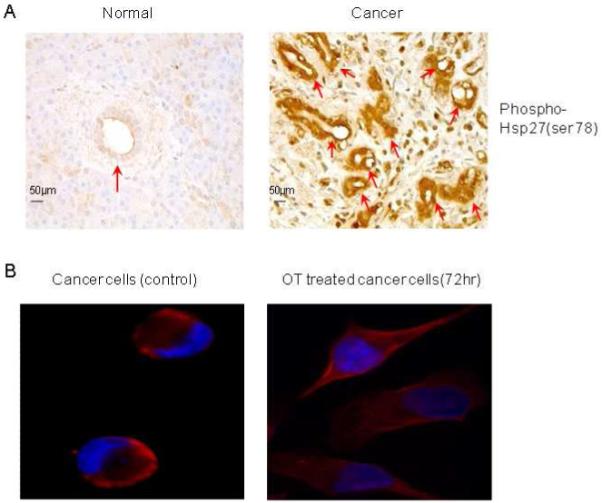
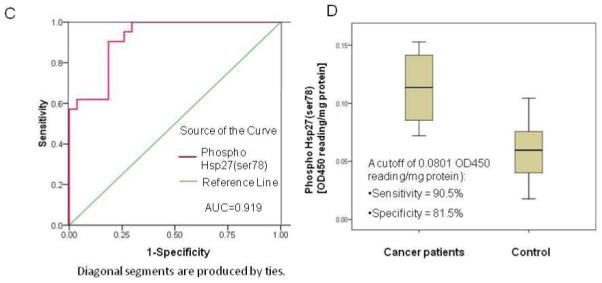
Functional validation of phosphor Hsp27 by analyses of ELISA and immunohistochemistry (IHC). Panel A shows the overexpression of phosphor Hsp27 at Ser78 in human pancreatic cancer tissue and its adjacent tissue (n=10). The arrows indicate overexpression of phosphor Hsp27 at Ser78 in ductal cells. Panel B shows cellular localization of phosphor Hsp27 (ser78) in MIA cancer cells in the presence or absence of OT. Panel C shows receiver operator characteristic curves for the diagnosis of pancreatic adenocarcinoma versus noncancerous cases. Curves demonstrate relative accuracy for individual serum phosphor Hsp27 (ser78) levels to discriminate between pancreatic cancer and control cases. Serum levels in healthy control subjects (n = 12) with age-matched from a new independent sample set were considered true-negative cases, whereas serum levels in patients with cytologically confirmed pancreatic adenocarcinoma (n = 12) from a new independent sample set were considered true-positive cases. The area under the curve (AUC) was 0.919 [95% confidence interval (CI): 0.845 to 0.993]. At a cutoff of 0.0801 OD450 reading/mg protein, the sensitivity was 90.5% (CI 81.0% to 99.3%) and the specificity was 81.5% (CI 70.4% to 81.5%). Panel C shows box plot of relative serum concentrations of phosphor Hsp27 (ser78) for controls and pancreatic cancer patients for an independent sample set. The lines inside the boxes denote the medians. The boxes represent the interval between the 25th and 75th percentiles, and the whiskers indicate the interval between maximum and minimum. The median concentrations for patients and controls were 0.114 OD450 reading/mg protein and 0.06 OD450 reading/mg proteins, respectively. Please see the Materials and Methods section for details. All the experiments were repeated in three times.
To further determine the cellular localization of phosphor Hsp27 (ser78), MIA cells were cultured in the presence and absence of OT for 72 hr, and then stained with phosphor Hsp27 (ser78) and DAPI (Figure 5B). The results in Figure 5B shows that phosphor Hsp27 (ser78) is over-expressed in the cytosol of a cancer cell, indicating cytosolic localization of phosphor Hsp27 (ser78). Intriguingly, in untreated cancer cells, the density of the nucleus is much higher than that in OT treated cancer cells; the cancer cell morphology in untreated cancer cells was different from that in OT treated cancer cells (Figure 5B).
To examine the biological significance of phospho Hsp27 (ser78), ELISA assays of phosphor Hsp27 (ser78) were carried out in a new independent sample set of 12 plasma samples from patients with pancreatic adenocarcinoma and an equal number of plasma samples from age-matched control subjects. The ROC curve analyses were used to investigate the sensitivity and specificity of phosphor Hsp27 (ser78) as separate diagnostic tests for pancreatic adenocarcinoma. Figure 5C depicts phosphor Hsp27 (ser78) ROC curve for the discrimination of pancreatic cancer subjects and non-pancreatic cancer control subjects (true negative cases). ROC analysis estimates a curve, which describes the inherent tradeoff between sensitivity and specificity of a diagnostic test. Each point on the ROC curve is associated with a specific diagnostic criterion. The area under the ROC curve (AUC) may be regarded as an average of the sensitivity over all possible specificities. The diagnostic measure with the higher AUC is typically regarded as better. Thus, Phospho Hsp27 (ser78) (AUC = 0.919) had good accuracy over all possible cutoffs. The cutoff (using a cutoff of 0.0801 OD450/mg protein Hsp27 (ser78) in serum as shown in Figure 5D) yielded a sensitivity of 90.5 % and a specificity of 81.5% for Phospho Hsp27 (ser78) when considering pancreatic cancer as true-positive cases and all non-pancreatic cancer subjects as true-negative cases (Figure 5C/D).
Discussion
Small molecule antimetabolites are among the more effective chemotherapeutic agents in use today. Currently, gemcitabine, a nucleoside analog, is commonly used for the treatment of pancreatic cancer. However, the response rate to gemcitabine, and patient survival, are poor (21, 22); there is an urgent need to discover additional chemotherapeutic targets such as metabolic enzymes that play a crucial role in controlling the growth of cancer cells. Oxythiamine (OT), a transketolase inhibitor, has been demonstrated to inhibit cancer cell growth through suppression of the cell cycle in many in vitro studies. The molecular mechanisms of the anti-proliferative action of these small molecules at both translational and post-translational levels are poorly understood. Transketolase is a key metabolic enzyme in the pentose cycle. Selective inhibition of this enzyme results in redistribution of glucose carbon away from ribose and deoxyribose synthesis required for cell proliferation. In this study, we examined the effect of inhibition of a metabolic pathway (transketolase) by oxythiamine (OT) on key intracellular signaling pathways focusing on the expression of cellular phosphoproteins. We showed by flow cytometry and cell cycle markers that OT treatment specifically induced cell cycle arrest through deactivation of the MAPK pathways, wherein, the key members such as p65, p50, and p52 in NF-κB pathway remained unaffected. The transcription factor NF-κB consists either homo- or heterodimers of different subunits, which are members of a family of structurally related proteins such as Rel/NF-kB proteins. Nowadays, five different Rel/NF-kB proteins have been identified including p50, p52, p65 (also called RelA), RelB, and c-Rel. The p50/p65 heterodimers and the p50 homodimers are the most common dimers found in the NF-κB signaling pathway, which play a crucial role in the regulation of many genes that code for mediators of the immune, acute phase and inflammatory responses. Inhibition of transketolase in the pentose phosphate pathway by OT did not affect the NF-κB signaling pathway, but significantly affected MAPK pathways (Figure 1). This observation was further strengthened by the results from the following phosphor proteomics study. Using a new proteomic technique that allows for the simultaneous determination of protein turnover, we discovered 12 phosphoproteins in the cellular phosphoproteome as being significantly suppressed by OT treatment (Table 1). These proteins can roughly be divided into two groups: those with high turnover rates (>60% in 72 hours), and those with turnover rates of <60%. Proteins with high turnover rates include 60S acidic ribosomal protein P0, glutathione S-transferase P, peroxiredoxin-6, peroxiredoxin-4, Hsp90α, Hsp 27, and sperm protein. Peroxiredoxin-4 (67% turnover) is involved in redox regulation and in the activation of NF-κB in the cytosol by modulating I-κ-B phosphorylation. It is well known that overexpression of NF-κB leads to cancer cell proliferation and blocking NF-κB can cause tumor cell apoptosis (23). Hsp90α (67% turnover rate) is associated with the incidence of non-small cell lung cancer (24) and pancreatic carcinoma (25). Inhibition of Hsp90α appears to induce apoptosis through inhibition of the PI3K/AKT signaling pathway, leading to a predictable clinical outcome (26). A recent study showed that phosphoprotein nuclear autoantigenic sperm protein (spot 2) with a turnover rate of 89%, the highest among those identified in this study, can form a cytoplasmic complex with Hsp90α and H1 linker histones, leading to enhancement of Hsp90 ATPase activity (27).
Hsp27 (66% turnover rate) was further studied since it is considered to be a very active cellular signaling molecule during cancer cell proliferation (28). Hsp27 is a downstream substrate of the activated proteins of PKC β-ERK1/2 and PKC β-p38MAPK in hepatocellular carcinoma (29). It is known to protect cells against apoptosis. An effective chemotherapeutic agent such as TSA (histone deacetylase inhibitor trichostatin A) is known to down-regulate expression of Hsp27 in pancreatic endocrine tumor cell lines (30). Our results showed that down-regulation of the p38MAPK cascade by OT treatment was accompanied by loss of the activity of the phosphor Hsp27 in MIA pancreatic cancer cells. In a recent study of the proteomics of microdissected pancreatic carcinoma tissue, Hsp27 has been identified and validated as a potential serum marker (31). Hsp27 is found to be differentially expressed between pancreatic adenocarcinoma cell lines showing sensitivity and resistance to gemcitabine, validating Hsp27 as a biomarker for resistance of pancreatic cancer cells to gemcitabine (32). There are seven potential serine sites for Hsp27 phosphorylation. Using available antibodies and Western blot analysis, we found that only the phosphor Hsp27 (ser78) was significantly suppressed by OT treatment. The phosphorylation of Hsp27 at serine 26 has not previously been reported. We were not able to evaluate the significance of phosphor Hsp27 at serine 26 because of the lack of available antibodies. Hsp27(ser78) was found to be highly expressed in pancreatic cancer tissue using immunohistochemical staining. Plasma concentrations of this phosphor Hsp27(ser78) were found to be sensitive and specific for the detection of pancreatic cancer in a limited number of subjects, suggesting that phosphor Hsp27(ser78) may be a potential novel drug target for chemotherapy and/or an early biomarker for the diagnosis of pancreatic cancer. Considering the biological significance and clinical importance of phosphor Hsp27, we have chosen phosphor Hsp27 (Ser78) for further characterization in this study.
Interestingly, over-expression of phosphorylated Hsp27 (Ser78) caused the higher density of the nucleus in untreated cancer cells than that in OT treated cancer cells as shown in Figure 5B. As we already demonstrated, OT treatment caused MIA PaCa-2 cancer cell cycle arrest (Figure 1), indicating DNA synthesis in the nucleus was slowing down. This may be the main explanation for why the density of the nucleus in the untreated cancer cells is much higher than that in OT treated cancer cells. In addition, it has been reported that hsp27 phosphorylation was correlated with its entry to the nucleus of cells (33), therefore, nucleus Hsp27 may interact with proteins in nuclei to promote refolding or target damaged proteins for degradation because of the chaperone activity of Hsp27 (33). This may be another explanation for the reason why the density of the nucleus in the untreated cancer cells is much higher than that in OT treated cancer cells. The cell morphological differences observed between untreated and OT treated cancer cells may be because of the interaction of Hsp27 with actin filament in the cell (34) (Figure 5B). It has reported that overexpression of Hsp27 results in cellular actin polymerization (34). Hsp27 phosphorylation modulates the interaction of Hsp27 with microfilaments (34). This may be the explanation for the difference of cell morphology observed in OT-treated and untreated cancer cells (Fig.5B).
Conclusions
Understanding the principles that govern cancer responses to small molecule metabolic inhibitors and to drugs targeting specific signaling pathways remains a major challenge in the development of chemotherapeutic agents. The alteration of intracellular signaling pathways by extrinsic growth factors or by mutation is currently the basic premise on which development of chemopreventive and chemotherapeutic targets is based. The present study revealed that a small molecule metabolic inhibitor such as OT, through its metabolic action can result in specific and non-specific inhibition of translational and post-translational changes in signaling molecules. Our understanding of the cellular metabolic network would suggest that the inhibition of a single metabolic pathway invariably results in metabolic inefficiency of other metabolic reactions in the network (35). Inhibition of the transketolase reaction may cause a global deficiency in high-energy phosphate bonds resulting in non-specific decrease in phosphorylation of proteins. Depending on the enzymatic properties of these kinases, the decrease in phosphorylation may appear to be specific as illustrated by the phosphorylation of Hsp27. We found that the phosphor-proteins that are sensitive to OT inhibition have high turnover rates (>60%), suggesting that the function of these proteins are regulated by both protein synthesis and protein degradation. The intracellular concentration of a protein is dependent on its synthesis and breakdown. Therefore, protein concentration can be controlled either by changes in its synthesis or degradation. It is generally held that the turnover rate of a protein must be high in order for it to change its concentration quickly inside a cell. Our finding of relatively high turnover rates in these signaling proteins is in agreement with this view and suggests that the determination of protein turnover may be useful to identify signaling target in chemotherapy.
ACKNOWLEDGMENT
This work is fully supported by grants awarded to Dr. Gary Guishan Xiao (GGX) from the Bone Biology Program of the Cancer and Smoking Related Disease Research Program and the Nebraska Tobacco Settlement Biomedical Research Program (LB692, LB595, and LB506). It is also partially supported by a grant awarded to WNPL from the UCLA Center of Excellence in Pancreatic disease (P01 AT003960-01), Harbor-UCLA GCRC Mass Spectrometry Core (M01 RR00425) and Hirshberg Foundation for Pancreatic Cancer Research.
ABBREVIATIONS
- MALDI
matrix assisted laser desorption and ionization
- TOF/TOF MS
time-of-flight/time-of-flight mass spectrometry
- OT
oxythiamine
- 2-DE
two dimensional electrophoresis
- TK
transketolase
- TA
transaldolase
- ELISA
Enzyme-Linked ImmunoSorbent Assay
- PMF
Peptide Mass Fingerprinting
- MOWSE
MOlecular Weight Search
- ROC
Receiver Operating Characteristic
- AUC
the Area Under the ROC Curve
Reference List
- (1).Boros LG, Puigjaner J, Cascante M, Lee WN, Brandes JL, Bassilian S, et al. Oxythiamine and dehydroepiandrosterone inhibit the nonoxidative synthesis of ribose and tumor cell proliferation. Cancer Res. 1997;57:4242–8. [PubMed] [Google Scholar]
- (2).Ramos-Montoya A, Lee WN, Bassilian S, Lim S, Trebukhina RV, Kazhyna MV, et al. Pentose phosphate cycle oxidative and nonoxidative balance: A new vulnerable target for overcoming drug resistance in cancer. Int J Cancer. 2006;119:2733–41. doi: 10.1002/ijc.22227. [DOI] [PubMed] [Google Scholar]
- (3).Rais B, Comin B, Puigjaner J, Brandes JL, Creppy E, Saboureau D, et al. Oxythiamine and dehydroepiandrosterone induce a G1 phase cycle arrest in Ehrlich’s tumor cells through inhibition of the pentose cycle. FEBS Lett. 1999;456:113–8. doi: 10.1016/s0014-5793(99)00924-2. [DOI] [PubMed] [Google Scholar]
- (4).Boros Laszlo G. Drug Discovery Today: Therapeutic Strategies. 4. Vol. 1. Elsevier; 2004. NJSMSCW-NPL. Use of metabolic pathway flux information in targeted cancer drug design; pp. 435–443. Ref Type: Generic. [Google Scholar]
- (5).Xiao GG, Garg M, Lim S, Wong D, Go VL, Lee WN. Determination of protein synthesis in vivo using labeling from deuterated water and analysis of MALDI-TOF spectrum. J Appl Physiol. 2008;104:828–36. doi: 10.1152/japplphysiol.00976.2007. [DOI] [PubMed] [Google Scholar]
- (6).Zhao Y, Lee WN, Lim S, Go VL, Xiao J, Cao R, et al. Quantitative Proteomics: Measuring Protein Synthesis Using (15)N Amino Acid Labeling in Pancreatic Cancer Cells. Anal Chem. 2008 doi: 10.1021/ac801905g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- (8).Xiao GG, Wang M, Li N, Loo JA, Nel AE. Use of proteomics to demonstrate a hierarchical oxidative stress response to diesel exhaust particle chemicals in a macrophage cell line. J Biol Chem. 2003;278:50781–90. doi: 10.1074/jbc.M306423200. [DOI] [PubMed] [Google Scholar]
- (9).Xiao GG, Nel AE, Loo JA. Nitrotyrosine-modified proteins and oxidative stress induced by diesel exhaust particles. Electrophoresis. 2005;26:280–92. doi: 10.1002/elps.200406145. [DOI] [PubMed] [Google Scholar]
- (10).Fenyo D, Wang Q, DeGrasse JA, Padovan JC, Cadene M, Chait BT. MALDI sample preparation: the ultra thin layer method. J Vis Exp. 2007:192. doi: 10.3791/192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Xiao J, Lee WN, Zhao Y, Cao R, Go VL, Recker RR, et al. Profiling Pancreatic Cancer-Secreted Proteome Using 15N Amino Acids and Serum-Free Media. Pancreas. 2009 doi: 10.1097/MPA.0b013e3181bc44dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Larsen MR, Sorensen GL, Fey SJ, Larsen PM, Roepstorff P. Phospho-proteomics: evaluation of the use of enzymatic de-phosphorylation and differential mass spectrometric peptide mass mapping for site specific phosphorylation assignment in proteins separated by gel electrophoresis. Proteomics. 2001;1:223–38. doi: 10.1002/1615-9861(200102)1:2<223::AID-PROT223>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- (13).Ichijo H, Nishida E, Irie K, ten DP, Saitoh M, Moriguchi T, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–4. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- (14).Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–48. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- (16).De Souza AI, Wait R, Mitchell AG, Banner NR, Dunn MJ, Rose ML. Heat shock protein 27 is associated with freedom from graft vasculopathy after human cardiac transplantation. Circ Res. 2005;97:192–8. doi: 10.1161/01.RES.0000174815.10996.08. [DOI] [PubMed] [Google Scholar]
- (17).Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Landry J, Lambert H, Zhou M, Lavoie JN, Hickey E, Weber LA, et al. Human HSP27 is phosphorylated at serines 78 and 82 by heat shock and mitogen-activated kinases that recognize the same amino acid motif as S6 kinase II. J Biol Chem. 1992;267:794–803. [PubMed] [Google Scholar]
- (19).Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- (21).Mahalingam D, Giles F. Challenges in developing targeted therapy for pancreatic adenocarcinoma. Expert Opin Ther Targets. 2008;12:1389–401. doi: 10.1517/14728222.12.11.1389. [DOI] [PubMed] [Google Scholar]
- (22).Rivera F, Lopez-Tarruella S, Vega-Villegas MA, Salcedo M. Treatment of advanced pancreatic cancer: From gemcitabine single agent to combinations and targeted therapy. Cancer Treat Rev. 2009 doi: 10.1016/j.ctrv.2008.11.007. [DOI] [PubMed] [Google Scholar]
- (23).Sarkar FH, Li Y. NF-kappaB: a potential target for cancer chemoprevention and therapy. Front Biosci. 2008;13:2950–9. doi: 10.2741/2900. [DOI] [PubMed] [Google Scholar]
- (24).Han JY, Oh SH, Morgillo F, Myers JN, Kim E, Hong WK, et al. Hypoxia-inducible factor 1alpha and antiangiogenic activity of farnesyltransferase inhibitor SCH66336 in human aerodigestive tract cancer. J Natl Cancer Inst. 2005;97:1272–86. doi: 10.1093/jnci/dji251. [DOI] [PubMed] [Google Scholar]
- (25).Ogata M, Naito Z, Tanaka S, Moriyama Y, Asano G. Overexpression and localization of heat shock proteins mRNA in pancreatic carcinoma. J Nippon Med Sch. 2000;67:177–85. doi: 10.1272/jnms.67.177. [DOI] [PubMed] [Google Scholar]
- (26).Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–26. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- (27).Alekseev OM, Widgren EE, Richardson RT, O’Rand MG. Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem. 2005;280:2904–11. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- (28).Matsushima-Nishiwaki R, Takai S, Adachi S, Minamitani C, Yasuda E, Noda T, et al. Phosphorylated heat shock protein 27 represses growth of hepatocellular carcinoma via inhibition of extracellular signal-regulated kinase. J Biol Chem. 2008;283:18852–60. doi: 10.1074/jbc.M801301200. [DOI] [PubMed] [Google Scholar]
- (29).Guo K, Liu Y, Zhou H, Dai Z, Zhang J, Sun R, et al. Involvement of protein kinase C beta-extracellular signal-regulating kinase 1/2/p38 mitogen-activated protein kinase-heat shock protein 27 activation in hepatocellular carcinoma cell motility and invasion. Cancer Sci. 2008;99:486–96. doi: 10.1111/j.1349-7006.2007.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cecconi D, Donadelli M, Rinalducci S, Zolla L, Scupoli MT, Scarpa A, et al. Proteomic analysis of pancreatic endocrine tumor cell lines treated with the histone deacetylase inhibitor trichostatin A. Proteomics. 2007;7:1644–53. doi: 10.1002/pmic.200600811. [DOI] [PubMed] [Google Scholar]
- (31).Melle C, Ernst G, Escher N, Hartmann D, Schimmel B, Bleul A, et al. Protein profiling of microdissected pancreas carcinoma and identification of HSP27 as a potential serum marker. Clin Chem. 2007;53:629–35. doi: 10.1373/clinchem.2006.079194. [DOI] [PubMed] [Google Scholar]
- (32).Mori-Iwamoto S, Kuramitsu Y, Ryozawa S, Mikuria K, Fujimoto M, Maehara S, et al. Proteomics finding heat shock protein 27 as a biomarker for resistance of pancreatic cancer cells to gemcitabine. Int J Oncol. 2007;31:1345–50. [PubMed] [Google Scholar]
- (33).Bryantsev AL, Chechenova MB, Shelden EA. Recruitment of phosphorylated small heat shock protein Hsp27 to nuclear speckles without stress. Exp Cell Res. 2007;313:195–209. doi: 10.1016/j.yexcr.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bitar KN. HSP27 phosphorylation and interaction with actin-myosin in smooth muscle contraction. Am J Physiol Gastrointest Liver Physiol. 2002;282:G894–G903. doi: 10.1152/ajpgi.00141.2001. [DOI] [PubMed] [Google Scholar]
- (35).Lee W. Characterizing phenotype with tracer based metabolomics. Metabolomics. 2006;2:31–9. doi: 10.1007/s11306-006-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Alekseev OM, Widgren EE, Richardson RT, O’Rand MG. Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem. 2005;280:2904–11. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- (37).Nucleophosmin/B23, a nuclear PI(3,4,5)P(3) receptor, mediates the antiapoptotic actions of NGF by inhibiting CAD [2005/05/17] 2005. [DOI] [PubMed] [Google Scholar]
- (38).Richardson RT, Alekseev OM, Grossman G, Widgren EE, Thresher R, Wagner EJ, et al. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem. 2006;281:21526–34. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- (39).Qi W, Shakalya K, Stejskal A, Goldman A, Beeck S, Cooke L, et al. NSC348884, a nucleophosmin inhibitor disrupts oligomer formation and induces apoptosis in human cancer cells. Oncogene. 2008;27:4210–20. doi: 10.1038/onc.2008.54. [DOI] [PubMed] [Google Scholar]
- (40).El-Gibaly I, Meki AM, Abdel-Ghaffar SK. Novel B melatonin-loaded chitosan microcapsules: in vitro characterization and antiapoptosis efficacy for aflatoxin B1-induced apoptosis in rat liver. Int J Pharm. 2003;260:5–22. doi: 10.1016/s0378-5173(03)00149-2. [DOI] [PubMed] [Google Scholar]
- (41).Sanz R, Aragues R, Stresing V, Martin B, Landemaine T, Oliva B, et al. Functional pathways shared by liver and lung metastases: a mitochondrial chaperone machine is up-regulated in soft-tissue breast cancer metastasis. Clin Exp Metastasis. 2007;24:673–83. doi: 10.1007/s10585-007-9124-4. [DOI] [PubMed] [Google Scholar]
- (42).Short DM, Heron ID, Birse-Archbold JL, Kerr LE, Sharkey J, McCulloch J. Apoptosis induced by staurosporine alters chaperone and endoplasmic reticulum proteins: Identification by quantitative proteomics. Proteomics. 2007;7:3085–96. doi: 10.1002/pmic.200600964. [DOI] [PubMed] [Google Scholar]
- (43).Chang TW, Chen CC, Chen KY, Su JH, Chang JH, Chang MC. Ribosomal phosphoprotein P0 interacts with GCIP and overexpression of P0 is associated with cellular proliferation in breast and liver carcinoma cells. Oncogene. 2008;27:332–8. doi: 10.1038/sj.onc.1210651. [DOI] [PubMed] [Google Scholar]
- (44).Kinnula VL, Paakko P, Soini Y. Antioxidant enzymes and redox regulating thiol proteins in malignancies of human lung. FEBS Lett. 2004;569:1–6. doi: 10.1016/j.febslet.2004.05.045. [DOI] [PubMed] [Google Scholar]
- (45).Nordfors K, Haapasalo J, Helen P, Paetau A, Paljarvi L, Kalimo H, et al. Peroxiredoxins and antioxidant enzymes in pilocytic astrocytomas. Clin Neuropathol. 2007;26:210–8. doi: 10.5414/npp26210. [DOI] [PubMed] [Google Scholar]
- (46).Tsuruta M, Nishibori H, Hasegawa H, Ishii Y, Endo T, Kubota T, et al. Heat shock protein 27, a novel regulator of 5-fluorouracil resistance in colon cancer. Oncol Rep. 2008;20:1165–72. [PubMed] [Google Scholar]
- (47).Qi W, Liu X, Qiao D, Martinez JD. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int J Cancer. 2005;113:359–63. doi: 10.1002/ijc.20492. [DOI] [PubMed] [Google Scholar]


