Abstract
Human MOF (MYST1), a member of the MYST (Moz-Ybf2/Sas3-Sas2-Tip60) family of histone acetyltransferases (HATs), is the human ortholog of the Drosophila males absent on the first (MOF) protein. MOF is the catalytic subunit of the male-specific lethal (MSL) HAT complex, which plays a key role in dosage compensation in the fly and is responsible for a large fraction of histone H4 lysine 16 (H4K16) acetylation in vivo. MOF was recently reported to be a component of a second HAT complex, designated the non-specific lethal (NSL) complex (Mendjan, S., Taipale, M., Kind, J., Holz, H., Gebhardt, P., Schelder, M., Vermeulen, M., Buscaino, A., Duncan, K., Mueller, J., Wilm, M., Stunnenberg, H. G., Saumweber, H., and Akhtar, A. (2006) Mol. Cell 21, 811–823). Here we report an analysis of the subunit composition and substrate specificity of the NSL complex. Proteomic analyses of complexes purified through multiple candidate subunits reveal that NSL is composed of nine subunits. Two of its subunits, WD repeat domain 5 (WDR5) and host cell factor 1 (HCF1), are shared with members of the MLL/SET family of histone H3 lysine 4 (H3K4) methyltransferase complexes, and a third subunit, MCRS1, is shared with the human INO80 chromatin-remodeling complex. In addition, we show that assembly of the MOF HAT into MSL or NSL complexes controls its substrate specificity. Although MSL-associated MOF acetylates nucleosomal histone H4 almost exclusively on lysine 16, NSL-associated MOF exhibits a relaxed specificity and also acetylates nucleosomal histone H4 on lysines 5 and 8.
Keywords: Chromatin, Histone Modification; Gene Regulation; Histone Acetylase; Histone Modification; Proteomics
Introduction
In eukaryotic cells, chromosomal DNA is packaged with histones and other proteins into chromatin. Alterations in chromatin structure affect the accessibility of chromosomal DNA to enzymes involved in transcription, replication, and repair. Changes in chromatin structure are regulated in at least three different ways: by ATP-dependent remodeling of nucleosomes, by the incorporation of variants of histones H2A and H3 into nucleosomes, and by post-translational modifications of histones (1–5).
Post-translational modifications of histones include acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP-ribosylation (4–6). Reversible histone acetylation, controlled by histone acetyltransferases (HATs)4 and histone deacetylases, plays an important role in regulation of chromatin structure and function (3, 7). Based on the nature of their catalytic domains, HATs can be grouped into two distinct families: the GCN5-related N-acetyltransferase (GNAT) family, which includes GCN5 and p300/CBP-associating factor (8), and the Moz-Ybf2/Sas3-Sas2-Tip60 (MYST) family, which is characterized by a highly conserved MYST domain composed of an acetyl-CoA binding motif and a zinc finger (9, 10). Some MYST family members also have additional structural features such as chromodomains (MOF, Esa1, and Tip60), plant homeodomain-linked zinc fingers (Moz and MORF), and other domains that bind specifically to modified histones or participate in other protein-protein interactions (10).
Human MOF is an ortholog of the Drosophila MOF HAT. MOF is one of the key components of the dosage compensation or male-specific lethal (MSL) complex. The Drosophila MSL complex is composed of at least five proteins (MSL1, MSL2, MSL3, MLE, and MOF) and two non-coding RNAs (roX1 and roX2). Human cells express an evolutionarily conserved MSL complex that is composed of MOF and at least three additional subunits, including orthologs of MSL1, MSL2, and MSL3. The MOF HAT is believed to be responsible for the majority of histone H4 acetylation at lysine 16 in both Drosophila and human cells (4, 11–13).
Results of recent studies suggest the existence of additional MOF-containing HAT complexes. Roeder and co-workers (14) reported that in addition to the MSL HAT complex, human MOF is stably associated in cells with the mixed lineage in leukemia 1 (MLL1) histone methyltransferase in a multiprotein complex that catalyzes both histone acetylation and methylation and includes the WD repeat protein WDR5 and several other proteins found in mammalian COMPASS-like histone methyltransferases (14). Also present in their MOF- and MLL1-containing preparations were several TATA box-binding protein-associated factor (TAF) proteins, RING2 and other proteins thought to function in the E2F6 repressive complex, and the forkhead domain-containing MCRS1 protein, which we previously identified as a subunit of the human INO80 chromatin-remodeling complex (15).
In an independent line of investigation, Akhtar and co-workers (16) identified a collection of MOF-associated proteins distinct from the subunits of the MSL complex; these proteins included WDR5, the MSL1-like protein NSL1 (KIAA1267, also known as MSLv1 (12, 17)), plant homeodomain-linked finger-containing proteins PHF20 and PHF20L, O-linked N-acetylglucosamine transferase, isoform 1 (OGT1), host cell factor 1 (HCF1), the human INO80 complex subunit MCRS1, and previously uncharacterized proteins FLJ20436 and FLJ10081, which they designated NSL2 and NSL3, respectively. Although they did not determine whether these MOF-associated proteins were present in one or several discrete protein complexes, they proposed that some or all of them were components of a MOF-containing complex they named the nonspecific lethal (NSL) complex. Importantly, Akhtar and co-workers (16) did not detect the MLL1 methyltransferase as a MOF-associated protein in their study.
In an effort to resolve the discrepancy between these previous studies and to explore further the potential contribution of the human INO80 complex subunit MCRS1 to MOF function, we have carried out a systematic proteomic and biochemical analysis of human MOF-containing complexes. As we describe below, our findings are most consistent with those of Akhtar and co-workers (16) and establish that MOF is present in at least two discrete multiprotein complexes, the MSL complex and a second complex, which we refer to as the NSL complex in keeping with their nomenclature. In addition, by comparing the substrate specificities of the MSL and NSL complexes, we obtain evidence that MOF HAT activity is differentially regulated by assembly into the MSL complex, where it acetylates nucleosomal histone H4 on lysine 16, and the NSL complex, where it also acetylates nucleosomal histone H4 on lysines 5 and 8.
EXPERIMENTAL PROCEDURES
Materials
All antibodies were obtained from commercial sources except for anti-MSL1 and anti-MSL3L1 (12), which were a kind gift from E. R. Smith (Stowers Institute).
Immunoaffinity Purification of Protein Complexes from Mammalian Cells
Nuclear extracts were prepared from HEK293/FRT or HeLa S3 cells expressing various FLAG-tagged proteins according to the method of Dignam et al. (18). FLAG-tagged and their associated proteins were purified on anti-FLAG (M2) agarose beads as described (19).
Mass Spectrometry
Proteins were identified by MudPIT mass spectrometry (20–22), and normalized spectral abundance factors (NSAFs) were calculated as described (23, 24).
HAT Assay
Recombinant human histones, histone octamers, and nucleosomes were prepared and HAT assays performed as described (19, 25, 26). Detailed methods for this and other experimental procedures are provided in supplemental information.
RESULTS
Subunit Composition of the NSL Complex
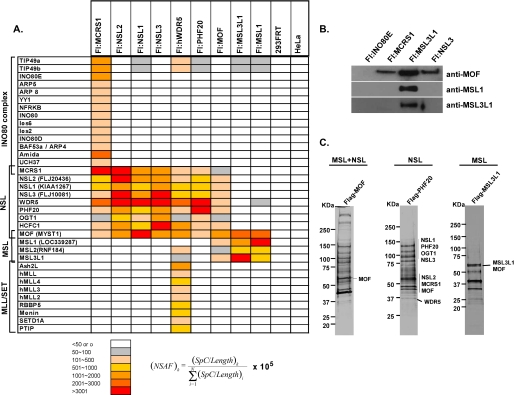
In a previous study, we found that the MCRS1 protein is a subunit of the human INO80 chromatin-remodeling complex (15). In light of evidence that MCRS1 may also be a MOF-associating protein (14, 16), we generated a human cell line stably expressing FLAG-MCRS1, purified FLAG-MCRS1-associating proteins by anti-FLAG agarose immunoaffinity chromatography, and analyzed them by MudPIT mass spectrometry and Western blotting (Fig. 1, A and B). MudPIT analyses of anti-FLAG agarose eluates identified all previously defined subunits of the human INO80 chromatin-remodeling complex (15), as well as a collection of additional proteins not found previously in preparations of the INO80 complex or in control samples. Among these proteins were MOF, the previously established MOF-associating protein NSL1 (16), and several candidate NSL complex subunits (16), including NSL2 (FLJ20436), NSL3 (FLJ10081), WDR5, PHF20, OGT1, and HCF1. Notably, with the exception of MOF, the collection of detectable FLAG-MCRS1-associating proteins did not include subunits of the MSL HAT complex.
FIGURE 1.
Subunit compositions of MOF-containing complexes. A, MudPIT analyses of MOF-containing complexes. Aliquots of anti-FLAG (Fl) agarose eluates prepared from cell lines expressing the indicated proteins or from control HEK293FRT or Hela cells were subjected to MudPIT. The intensity of the coloring corresponds to the NSAF for each protein and is a rough reflection of the relative abundance of the protein in the samples. (NSAF)k = NSAF calculated for an individual protein k; SpC = spectral count; L = protein length in amino acids; and i = all proteins detected in the MudPIT runs. PTIP, Pax transactivation domain-interacting protein; h, human. B, Western blot analysis of MOF-containing complexes. Anti-FLAG agarose eluates from FLAG-INO80E- (as a negative control), FLAG-MCRS1-, FLAG-MSL3L1-, and FLAG-NSL3-expressing HEK293FRT cells were subjected to SDS-PAGE in 4–20% gradient gels, and proteins were identified by Western blotting with the indicated antibodies. C, representative silver-stained gels of proteins associated with MSL and NSL complexes. The positions and relative molecular masses in kDa of protein size standards are indicated at the left of the gels.
To determine (i) which of the FLAG-MCRS1-associating proteins are bona fide subunits of MOF-containing complexes and (ii) whether they reside in a single complex distinct from the MSL complex, we generated human cell lines stably expressing FLAG-MOF, FLAG-NSL1, FLAG-NSL2, FLAG-NSL3, FLAG-WDR5, FLAG-PHF20, or either of the human MSL subunits FLAG-MSL1 or FLAG-MSL3L1. Although the FLAG-tagged proteins are likely overexpressed in these cell lines, it is difficult to estimate the degree of overexpression because we were not able to detect either the endogenous or the epitope-tagged proteins in crude extracts by Western blotting with the available antibodies. Nuclear extracts prepared from each cell line were subjected to anti-FLAG agarose chromatography, and anti-FLAG agarose eluates were analyzed by MudPIT and Western blotting (Fig. 1, A and B).
The results of these analyses argue that MOF is a component of at least two distinct multiprotein HAT complexes and can be summarized as follows. First, consistent with previous studies, anti-FLAG agarose eluates prepared from extracts of cells stably expressing FLAG-MSL1 or FLAG-MSL3L1 contained MSL1, MSL2, MSL3L1, and MOF in a complex corresponding to the well characterized human MSL complex. Second, anti-FLAG agarose eluates prepared from extracts of cells stably expressing FLAG-NSL1, FLAG-NSL2, FLAG-NSL3, or FLAG-PHF20 all contained MOF, NSL1, NSL2, NSL3, MCRS1, PHF20, OGT1, WDR5, and HCF1, arguing that these proteins can all assemble into a single multiprotein MOF-containing complex, which we refer to as the NSL complex. Third, subunits of both the MSL and the NSL complexes copurify with FLAG-MOF, consistent with the presence of MOF in both complexes. Fourth, as expected from the presence of WDR5 in both the MOF-containing NSL complex and the MLL/SET family of histone methyltransferase complexes (27), FLAG-WDR5 copurified with both NSL and MLL/SET subunits. Members of the MLL/SET family of histone methyltransferases were not, however, detected in association with FLAG-MOF or any of the other NSL subunits tested. Finally, we note that the MSL subunits MSL2 and MSL3L1, as well as the TIP49a and TIP49b subunits of the human INO80 complex, were also detected in FLAG-WDR5 immunoprecipitates. We believe that their association with FLAG-WDR5 most likely reflects transient interactions or interactions driven by overexpression of WDR5 because WDR5 either was not detected or was present in relatively low amounts in anti-FLAG agarose eluates prepared from extracts of cells expressing FLAG-MSL1, FLAG-MSL3L1, or INO80 complex subunits other than MCRS1 (15, 28, 29).
NSL- and MSL-associated MOF Exhibits Different Substrate Specificities
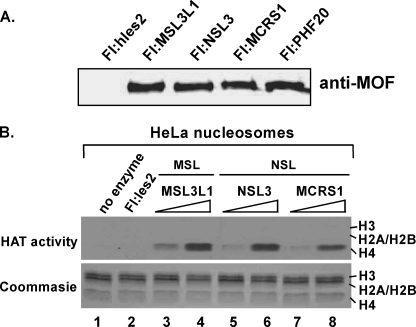
MSL-associated MOF has been shown to acetylate nucleosomal histone H4 specifically on lysine 16 (11–13, 16, 30). To determine whether the NSL complex has the same or a different substrate specificity, we purified NSL-associated MOF from cell lines expressing several FLAG-tagged NSL subunits and MSL-associated MOF from FLAG-MSL3L1-expressing cells. We then assayed nucleosomal HAT activity associated with complexes containing equivalent amounts of the catalytic MOF subunit (Fig. 2A) using [3H]acetyl-CoA. As shown in Fig. 2B, both the NSL and the MSL complexes possessed HAT activity that specifically acetylated nucleosomal histone H4.
FIGURE 2.
Both MSL and NSL complexes acetylate nucleosomal histone H4. A, aliquots of anti-FLAG (Fl) agarose eluates from cells expressing the indicated proteins were subjected to SDS-PAGE in 4–20% gradient gels, and MOF was identified by Western blotting with anti-MOF antibody. h, human. B, HAT activities associated with MOF-containing complexes. Anti-FLAG agarose eluates from equivalent numbers of FLAG-Ies2- (as a negative control), FLAG-MSL3L1-, FLAG-NSL3-, and FLAG-MCRS1-expressing cells were prepared. As determined by Western blotting (panel A), the relative concentrations of the MOF in MSL and NSL complexes were similar in anti-FLAG agarose eluates prepared from the different cell lines and used in HAT assays. HAT assays were performed by mixing HeLa long oligonucleosomes, [3H]acetyl CoA, and 1.0 μl (lanes 2, 3, 5, and 7) or 2.0 μl (lanes 4, 6, and 8) of the indicated anti-FLAG agarose eluates. Proteins were visualized by Coomassie Blue R250 staining (lower gel) and acetylated histones were visualized by autoradiography (top gel).
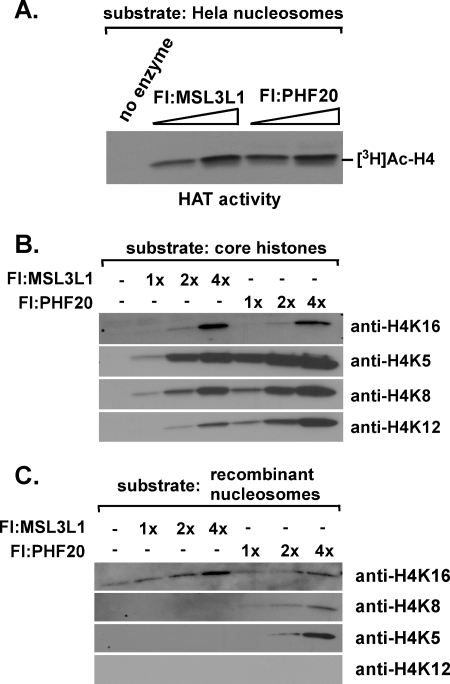
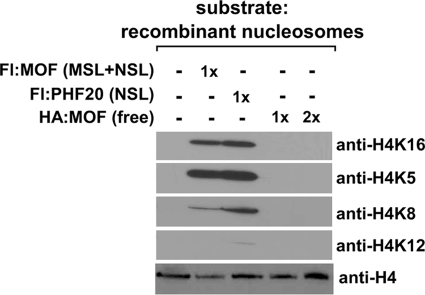
To compare further the substrate specificities of MSL- and NSL-associated MOF, we performed HAT assays with unlabeled acetyl-CoA, using as substrate recombinant core histones or polynucleosomes assembled with recombinant core histones. Histone acetylation was detected using antibodies that specifically recognize histone H4 acetylated at lysines 5, 8, 12, or 16. In these experiments, we used MSL and NSL complexes purified through FLAG-MSL3L1 and FLAG-PHF20, respectively; the HAT activity associated with these complexes catalyzed equivalent amounts of total histone H4 acetylation when assayed in the presence of [3H]acetyl-CoA (Fig. 3A). When presented with recombinant core histones, the HAT activities associated with MSL and NSL complexes catalyzed acetylation of histone H4 on each of the 4 lysines tested (Fig. 3B). As expected from the results of previous studies (11–13, 16, 30), when reactions were performed with nucleosomes reconstituted using the same recombinant core histones, acetylation of histone H4 by the MSL-associated HAT was restricted to lysine 16. In contrast, the NSL-associated HAT activity exhibited a broader, but still restricted, specificity, catalyzing substantial acetylation of histone H4 on lysines 5, 8, and 16 but not on lysine 12 (Fig. 3C). As shown in Fig. 4 and consistent with results of other studies (17, 31), free MOF has little or no HAT activity in our assays, arguing that the broadened substrate specificity of NSL-associated MOF is conferred by its association with one or more of the other NSL subunits.
FIGURE 3.
NSL-associated MOF has a broader substrate specificity than MSL-associated MOF. A, acetylation of Hela long oligonucleosomes by MSL- and NSL-associated MOF. HAT assays were performed with HeLa long oligonucleosomes, [3H]AcCoA, and 1 μl (lane 2 and lane 4) or 2 μl (lane 3 and lane 5) of the indicated anti-FLAG (Fl) agarose eluates, and acetylated hisones were visualized by autoradiography. The MSL and NSL complexes used in these experiments contained approximately equal concentrations of MOF (Fig. 2A). B, acetylation of free recombinant histones by MSL- and NSL-associated MOF. HAT assays were performed with core histones, unlabeled acetyl CoA, and 0.6 μl (lanes 2 and 5), 1.2 μl (lanes 3 and 6), or 2.4 μl (lanes 4 and 7) of the indicated anti-FLAG agarose eluates, and modified residues of histone H4 were detected by Western blotting with acetylation-specific antibodies. C, acetylation of recombinant polynucleosomes by MSL- and NSL-associated MOF. HAT assays were performed with reconstituted polynucleosomes, unlabeled acetyl CoA, and 0.6 μl (lanes 2 and 5), 1.2 μl (lanes 3 and 6), or 2.4 μl (lanes 4 and 7) of the indicated anti-FLAG agarose eluates, and modified histone H4 residues were detected by Western blotting with acetylation-specific antibodies.
FIGURE 4.
HAT activities of free MOF and MSL- and NSL-associated MOF. Anti-FLAG (Fl) agarose eluates from cells stably expressing FLAG-MOF (a mixture of MSL and NSL complexes), FLAG-PHF20 (NSL complex), or free MOF, expressed in and purified from insect cells, were assayed for HAT activity using recombinant polynucleosomes as substrate. Reactions contained the indicated relative molar amounts of free and MSL- and/or NSL-associated MOF. Acetylated histone H4 residues were detected by Western blotting.
DISCUSSION
In this report, we have exploited a MudPIT-based proteomics approach to define the subunit composition of the NSL complex, a MOF-containing HAT complex distinct from the well characterized MSL complex. In addition, we present evidence that the activity and substrate specificity of the MOF HAT is differentially regulated by its assembly into the NSL or MSL complexes. We observe that in contrast to the MSL-associated MOF, which acetylates almost exclusively nucleosomal histone H4 on lysine 16, NSL-associated MOF is capable of catalyzing substantial acetylation of nucleosomal histone H4 on lysines 5, 8, and 16. In this respect, the NSL HAT complex resembles the NuA4 HAT complex, which also specifically acetylates multiple lysines in the H4 N-terminal tail (32, 33).
Prior investigations of the mechanism underlying regulation of MOF HAT activity by subunits of the MSL complex have revealed that association of Drosophila MOF with an MSL1-MSL3 heterodimer leads to strong activation of MOF HAT activity and narrowing of its substrate specificity to just lysine 16 of nucleosomal histone H4 (31). Although it is presently not known how subunits of the NSL complex regulate MOF HAT activity, it was shown previously that the NSL1 protein can bind directly to MOF (16) and, while our manuscript was in preparation, Dou and co-workers (17) reported that binding of NSL1 to MOF enhances MOF acetylation of histone H4 on lysine 16 and of the DNA binding transcription factor p53. Our definition of an apparently complete set of NSL complex subunits should enable a more thorough analysis of the roles of individual NSL complex subunits in allosteric regulation of MOF HAT activity.
It is noteworthy that the NSL complex shares subunits with other chromatin-regulating complexes. The MCRS1 protein is an integral component of both the NSL complex and the INO80 chromatin-remodeling complex. The WDR5 protein is a subunit not only of the NSL complex but also of the MLL/SET1-containing histone H3K4 methyltransferase complexes (34) and of the ATAC (ADA2-containing) HAT complex, which includes as catalytic subunits both the GCN5/KAT2 and the ATAC2/KAT14 HATs (35, 36). The degree to which the presence of these shared subunits argues for functional links between the NSL, INO80, ATAC, and MLL/SET1-containing histone H3K4 methyltransferase complexes remains to be determined. It has been proposed that WDR5 might serve as a physical link between different chromatin regulatory complexes (14, 17); however, others argue that WDR5 most likely functions as a platform on which different chromatin regulatory complexes assemble independently (27). Consistent with this latter possibility, we find no evidence for copurification or stable assembly of the NSL complex with either the INO80 or the MLL/SET1-containing histone H3K4 methyltransferase complexes; however, we cannot rule out the possibility that the NSL complex might interact with these other chromatin regulatory complexes transiently or too weakly to be detected under the stringent washing conditions that we have used in their purifications.
Finally, it is intriguing that the NSL subunit OGT has been shown to be encoded by the Drosophila Polycomb group gene super sex combs (sxc), and evidence suggests that OGT activity helps to regulate Polycomb-mediated repression (37, 38). In the future, it will be of interest to investigate the possibility of cross-talk between the NSL and Polycomb complexes in mammalian cells.
Supplementary Material
Acknowledgments
We thank Edwin R. Smith for a generous gift of valuable antibodies and Edwin R. Smith and Ali Shilatifard for helpful discussions.
This work was supported by Grant R37 GM41628 from the NIGMS, National Institutes of Health Grant (to R. C. C.), and by funds from the Stowers Institute for Medical Research.
- HAT
- histone acetyltransferase
- HCF
- host cell factor
- MLL
- mixed lineage in leukemia
- MOF
- males absent on the first
- MSL
- male-specific lethal
- MYST
- Moz-Ybf2/Sas3-Sas2-Tip60
- NSAF
- normalized spectral abundance factor
- NSL
- nonspecific lethal
- OGT
- O-linked N-acetylglucosamine transferase
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- HA
- hemagglutinin.
REFERENCES
- 1.Clapier C. R., Cairns B. R. (2009) Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 2.Jin J., Cai Y., Li B., Conaway R. C., Workman J. L., Conaway J. W., Kusch T. (2005) Trends Biochem. Sci. 30, 680–687 [DOI] [PubMed] [Google Scholar]
- 3.Lee K. K., Workman J. L. (2007) Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 4.Rea S., Xouri G., Akhtar A. (2007) Oncogene 26, 5385–5394 [DOI] [PubMed] [Google Scholar]
- 5.Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 6.Bhaumik S. R., Smith E., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 8.Dyda F., Klein D. C., Hickman A. B. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillus L. (2008) Curr. Opin. Cell Biol. 20, 326–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avvakumov N., Côté J. (2007) Oncogene 26, 5395–5407 [DOI] [PubMed] [Google Scholar]
- 11.Smith E. R., Pannuti A., Gu W., Steurnagel A., Cook R. G., Allis C. D., Lucchesi J. C. (2000) Mol. Cell. Biol. 20, 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith E. R., Cayrou C., Huang R., Lane W. S., Côté J., Lucchesi J. C. (2005) Mol. Cell. Biol. 25, 9175–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taipale M., Rea S., Richter K., Vilar A., Lichter P., Imhof A., Akhtar A. (2005) Mol. Cell. Biol. 25, 6798–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., Roeder R. G. (2005) Cell 121, 873–885 [DOI] [PubMed] [Google Scholar]
- 15.Jin J., Cai Y., Yao T., Gottschalk A. J., Florens L., Swanson S. K., Gutiérrez J. L., Coleman M. K., Workman J. L., Mushegian A., Washburn M. P., Conaway R. C., Conaway J. W. (2005) J. Biol. Chem. 280, 41207–41212 [DOI] [PubMed] [Google Scholar]
- 16.Mendjan S., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Wilm M., Stunnenberg H. G., Saumweber H., Akhtar A. (2006) Mol. Cell 21, 811–823 [DOI] [PubMed] [Google Scholar]
- 17.Li X., Wu L., Corsa C. A., Kunkel S., Dou Y. (2009) Mol. Cell 36, 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. (1983) Methods Enzymol. 101, 582–598 [DOI] [PubMed] [Google Scholar]
- 19.Cai Y., Jin J., Gottschalk A. J., Yao T., Conaway J. W., Conaway R. C. (2006) Methods 40, 312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 21.Florens L., Washburn M. P. (2006) Methods Mol. Biol. 328, 159–175 [DOI] [PubMed] [Google Scholar]
- 22.Eng J. K., McCormack A. L., Yates J. R., 3rd (1994) J. Am. Soc. Mass. Spec. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 23.Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., Washburn M. P. (2006) J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 24.Paoletti A. C., Parmely T. J., Tomomori-Sato C., Sato S., Zhu D., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen-Hughes T., Utley R. T., Steger D. J., West J. M., John S., Côté J., Havas K. M., Workman J. L. (1999) Methods Mol. Biol. 119, 319–331 [DOI] [PubMed] [Google Scholar]
- 26.Eberharter A., John S., Grant P. A., Utley R. T., Workman J. L. (1998) Methods 15, 315–321 [DOI] [PubMed] [Google Scholar]
- 27.Trievel R. C., Shilatifard A. (2009) Nat. Struct. Mol. Biol. 16, 678–680 [DOI] [PubMed] [Google Scholar]
- 28.Cai Y., Jin J., Yao T., Gottschalk A. J., Swanson S. K., Wu S., Shi Y., Washburn M. P., Florens L., Conaway R. C., Conaway J. W. (2007) Nat. Struct. Mol. Biol. 14, 872–874 [DOI] [PubMed] [Google Scholar]
- 29.Yao T., Song L., Jin J., Cai Y., Takahashi H., Swanson S. K., Washburn M. P., Florens L., Conaway R. C., Cohen R. E., Conaway J. W. (2008) Mol. Cell 31, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akhtar A., Becker P. B. (2000) Mol. Cell 5, 367–375 [DOI] [PubMed] [Google Scholar]
- 31.Morales V., Straub T., Neumann M. F., Mengus G., Akhtar A., Becker P. B. (2004) EMBO J. 23, 2258–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berndsen C. E., Selleck W., McBryant S. J., Hansen J. C., Tan S., Denu J. M. (2007) Biochemistry 46, 2091–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berndsen C. E., Denu J. M. (2008) Curr. Opin. Struct. Biol. 18, 682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. (2007) J. Biol. Chem. 282, 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suganuma T., Gutiérrez J. L., Li B., Florens L., Swanson S. K., Washburn M. P., Abmayr S. M., Workman J. L. (2008) Nat. Struct. Mol. Biol. 15, 364–372 [DOI] [PubMed] [Google Scholar]
- 36.Wang Y. L., Faiola F., Xu M., Pan S., Martinez E. (2008) J. Biol. Chem. 283, 33808–33815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., Honda B. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gambetta M. C., Oktaba K., Müller J. (2009) Science 325, 93–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.