Abstract
Clusterin (CLU) is a ubiquitous protein that has been implicated in tumorigenesis, apoptosis, inflammation, and cell proliferation. We and others have previously shown that CLU is an inhibitor of the NF-κB pathway. However, the exact form of CLU and the region(s) of CLU involved in this effect were unknown. Using newly generated molecular constructs encoding for CLU and various regions of the molecule, we demonstrated that the presecretory form of CLU (psCLU) form bears the NF-κB regulatory activity. Sequence comparison analysis showed sequence motif identity between CLU and β-transducin repeat-containing protein (β-TrCP), a main E3 ubiquitin ligase involved in IκB-α degradation. These homologies were localized in the disulfide constraint region of CLU. We generated a specific molecular construct of this region, named ΔCLU, and showed that it has the same NF-κB regulatory activity as CLU. Neither the α-chain nor the β-chain of CLU had any NF-κB regulatory activity. Furthermore, we showed that following tumor necrosis factor-α stimulation of transfected cells, we could co-immunoprecipitate phospho-IκB-α with ΔCLU. Moreover, we showed that ΔCLU could localize both in the cytoplasm and in the nucleus. These results demonstrate the identification of a new CLU activity site involved in NF-κB pathway regulation.
Keywords: Cytokines, Diseases, Protein/Domains, Protein/Motifs, Signal Transduction, Clusterin
Introduction
Clusterin (CLU)3 is a ubiquitous glycoprotein that shows a very broad pattern of tissue expression in mammals. Several physiological functions have been attributed to CLU, including tissue remodeling, cell-substrate interaction, lipid transport, sperm maturation, and a protective role against tissue damage (1). CLU also plays an important role during tumorigenesis and the progression of human cancers (2, 3). Clu transcription appears complex, generating transcripts of various sizes and producing protein forms targeted to different cellular compartments. These protein forms are also due to the presence of two translation start sites, spaced 100 nucleotides apart within the gene (4). One CLU form is secreted (sCLU), whereas other forms have a cytoplasmic (cCLU) or nuclear (nCLU) localization. Recently, a unified nomenclature has been proposed (5). sCLU is reserved for the main gene transcript identified from the CLU gene locus. It has a predicted molecular mass of 50.1 kDa, which is detected as a 60-kDa glycosylated presecretory form of sCLU and is referred to as psCLU. sCLU is translated from the mRNA including the nine gene exons. Once it forms the mature, secreted heterodimeric 75–80-kDa protein, it should be referred to as sCLU. Thus, mature sCLU is a heterodimeric disulfide-linked glycoprotein (449 amino acids). It is composed of two subunits (α and β) resulting from the cleavage of psCLU (6). sCLU has a cytoprotective and chaperone role (7). sCLU was described as an inhibitor of the cytolytic reaction of terminal complement proteins (8–10).
CLU, probably the psCLU form, also has an essential role in chemoresistance by interacting with activated Bax (11). The Clu gene mRNA may also undergo an alternative splicing of exon 2. These mechanisms are poorly understood. The second CLU gene transcript lacks the endoplasmic reticulum-targeting sequence at exon 2, and its product is detected as a 49-kDa nonglycosylated precursor nCLU protein (pnCLU) in the cytosol and a 55-kDa glycosylated protein (referred to as nCLU) in the nucleus. pnCLU is stored in the cytoplasm; however, upon stress induced by ionizing radiation, pnCLU is activated and becomes the mature form nCLU, which then locates to the nucleus. This “translocation” of nCLU might involve the nuclear localization signal (NLS) sequence identified in exon 3.
Trougakos et al. (12) recently showed that sCLU binds and thereby stabilizes the Ku70-Bax protein complex, thus serving as a cytosol retention factor for Bax. This raises the possibility that elevated sCLU levels may enhance tumorigenesis by interfering with Bax proapoptotic activities. On the other hand, several reports suggest that the nuclear form of CLU (nCLU) is a proapoptotic protein (13–15). Recently, we and others have demonstrated that CLU intracellular forms are involved in NF-κB pathway regulation through IκB-α stabilization (16, 17). We showed that CLU interacts with phospho-IκB-α (pIκB-α) and decreases the translocation of p50/p65 to the nucleus. It has been shown that ectopic CLU expression strongly inhibited NF-κB activity by stabilizing inhibitors of NF-κB (IκB) (16). The presence of clusterin reduced NF-κB activation and expression of NF-κB target genes, such as TNF-α (18). We have also demonstrated a significant intracellular psCLU expression modulation in hyperproliferative synoviocytes of patients with rheumatoid arthritis (17, 19). NF-κB is recognized as a ubiquitous transcription factor that plays a key role in immune response, inflammation, proliferation, and apoptosis and also in tumorigenesis (20). Herein, we generated several molecular constructs coding for various CLU regions. We studied their subcellular localization and tested their role in the NF-κB pathway.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
psCLU, α-chain, and β-chain were obtained by PCR amplification from a pIRES-CLU vector construct (provided by Dr. S. Bettuzzi and M. Scaltriti, Parma, Italy). The pair of primers used for cloning were: psCLU, forward, 5′-TGCGGATCCGACTCCAGAATTGGAGGC- 3′, reverse, 5′-TGCCTCGAGCTCCTCCCGGTGCTT-3′; α-chain, forward, 5′-TGCGGATCCGACTCCAGAATTGGAGGC-3′, reverse, 5′-TGCCTCGAGGCGGACGATGCGGGA-3′; and β-chain, forward, 5′-GCGGCCGCAGCTTGATGCCCTTCTCT-3′, reverse, 5′-TCTAGATCACTCCTCCCGGTGCTT-3′. Amplicons were inserted in pCMV-FLAG (Stratagene) between the BamHI and XhoI restriction sites.
ΔCLU construct (sequence below) was generated by the company Genscript® and cloned in pCMV-FLAG vector. The sizes are as follows: α-chain domain, 1–156 bp; tether sequence (italics), 157–207 bp; β-chain domain, 208–339bp. The sequence was: 5′-GAATCAGAGACAAAGCTGAAGGAGCTCCCAGGAGTGTGCAATGAGACCATGATGGCCCTCTGGGAAGAGTGTAAGCCCTGCCTGAAACAGACCTGCATGAAGTTCTACGCACGCGTCTGCAGAAGTGGCTCAGGCCTGGTTGGCCGCCAGCTTGAGAAGCTTGGGGGATCAGGCGGAGGTGGAGGATCCGGTGGCGGTGGCTCGAGCGAATTCATACGAGAAGGCGACGATGACCGGACTGTGTGCCGGGAGATCCGCCACAACTCCACGGGCTGCCTGCGGATGAAGGACCAGTGTGACAAGTGCCGGGAGATCTTGTCTGTGGACTGTTCCACCAAC-3′. C-terminal FLAG-BAP (Sigma) control protein is a 466-amino acid C-terminal FLAG fusion protein of Escherichia coli bacterial alkaline phosphatase (BAP) with a calculated molecular mass of 49.1 kDa.
Culture Conditions and Transfections
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% heat-inactivated fetal calf serum and 1% penicillin-streptomycin (Invitrogen). Transfections were performed by electroporation using the NucleofectorTM (Amaxa) protocol.
Protein Extracts and Western Blot Analysis
For Western blot analysis, cytoplasmic extracts from HeLa cells were collected after cell lysis in lysis buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, and protease and phosphatase inhibitors (Roche Applied Science)) and spun down at 12,000 rpm for 15 min at 4 °C. The nuclear extracts were prepared using the Nuclear Extraction kit (Active Motif) according to the manufacturer's protocol. Equal amounts of protein (30 μg), as determined by BCATM protein assay kit (Pierce), were resolved on 10–12% polyacrylamide-SDS gels and transferred onto a polyvinylidene difluoride membrane. The membrane was blocked in blocking agent (5% nonfat milk in Tris-buffered saline and 0.1% Tween 20), and immunoblotting was performed using ECL (Amersham Biosciences). Anti-IκB-α Abs (Cell Signaling) and anti-actin Abs (C-11, Santa Cruz Biotechnology) were diluted 1:1000, monoclonal anti-FLAG® M2 Abs (Sigma) were diluted 1:2000, and α-tubulin and lamin B1 Abs (Santa Cruz Biotechnology) were diluted 1:500.
Confocal Microscopy
Twenty-four hours after transfection, cells were fixed with 4% paraformaldehyde for 10 min at room temperature, permeabilized with 0.5% Triton X-100 for 5 min, and incubated with FLAG M2 FITC Abs (Sigma) at 30 μg/ml for 1 h at room temperature. After washing, DAPI solution was added, and cells were mounted with VECTASHIELD HardSet H-1000. Fluorescence localization was detected by confocal microscopy (Leica TCS SP2 AOBS) with a ×40 objective. Images were processed using the ImageJ software.
Reporter Assays
Luciferase reporter assays were performed using the Dual-Luciferase® reporter (DLRTM) assay system (Promega). The NF-κB luciferase reporter plasmid 3E-κB-Cona luc plasmid was generously provided by Dr. Florence Margottin (Cochin Institute, Paris, France). In brief, HeLa cells (2 × 106) were transiently co-transfected with 1 μg of NF-κB-dependent (3E-κB-Cona luc) construct and 2.5 μg of each of the CLU constructs. Twenty-four hours after transfection, luciferase activity was measured and depicted in arbitrary units.
Co-immunoprecipitation
Forty- eight hours after transfection, cells were washed three times in phosphate-buffered saline and left overnight on Dulbecco's modified Eagle's medium supplemented with 1% fetal calf serum. Cells were subsequently stimulated for 10 min with 20 ng/ml recombinant human TNF-α (Thermo Scientific) in the presence of 20 μm MG132 (Calbiochem). Cells were then lysed in lysis buffer. Cell lysates were precleared with protein G-agarose beads (Sigma-Aldrich) for 1 h, and then supernatants were incubated for 2 h at 4 °C with 5 μg/ml mouse monoclonal anti-FLAG® M2 Abs (Sigma) followed by incubation with protein G-agarose beads for 1 h at 4 °C. Immune complexes were eluted with Laemmli buffer, separated by SDS-PAGE gel, and revealed by Western blot using anti-IκB-α (Cell Signaling Technology).
RESULTS
CLU Molecular Constructs
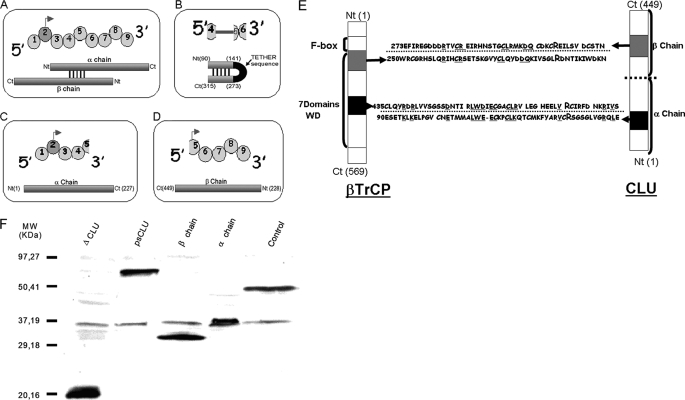
We generated a set of gene constructs corresponding to several regions of clusterin in an expression vector pFLAG to facilitate visualization and homogenize technical parameters for manipulation of the expressed gene. These constructs were psCLU, α-chain, and β-chain (Fig. 1, A–C). Because CLU was shown to be a negative regulator of NF-κB and stabilizer of IκB-α, we performed sequence alignments between clusterin and proteins known to act in the NF-κB pathway. As shown in Fig. 1E, two main homology patterns were found between CLU and β-TrCP. Interestingly, each of these homology regions contains an arginine residue that is involved in either phospho-Ser-32 or His-30 and Arg-29 binding of IκB-α (21) (Fig. 1E). These regions were located in a β-TrCP substrate binding domain, WD40 domain, and in two regions of CLU, one in α-chain and one in β-chain. These regions form, in the CLU mature three-dimensional structure, five disulfide bridges. We then generated a specific construct called ΔCLU in which both homologous regions of CLU and β-TrCP had been linked by a tether junction peptide that facilitated protein folding (Fig. 1D and the nucleotide sequence under “Experimental Procedures”).
FIGURE 1.
CLU constructs and protein expression in HeLa cells following transfection. A–D, schematic representation of psCLU, ΔCLU, α-chain, and β-chain molecular constructs, respectively (top, exonic sequence; bottom, protein structure). Ct, C terminus; Nt, N terminus. E, CLU and β-TrCP homology domains. Bold and capital letter R (for Arg) represents identified IκB-α binding sites (21). Underlined sequence letters represent homologous sequences between CLU and β-TrCP. F, Western blots of cytoplasmic protein extractions after CLU construct transfection were made with anti-FLAG antibody. pFLAG-BAP was used as a control plasmid. MW, molecular weight standards.
Each Region of CLU Has a Different Subcellular Localization
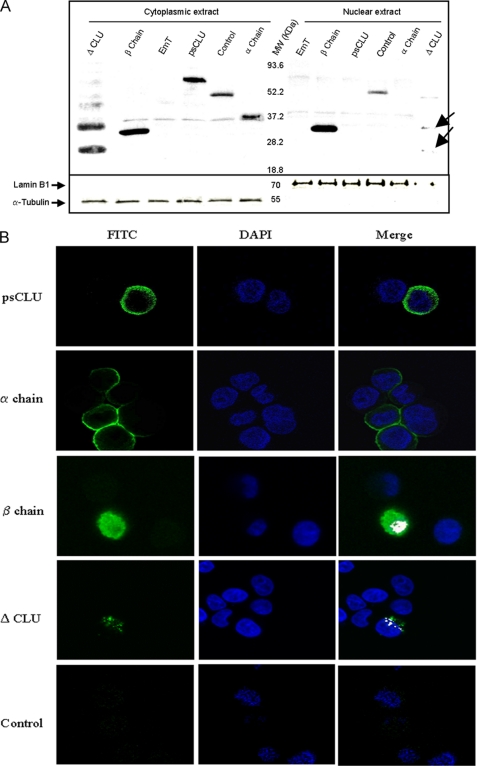
We checked the expression of these CLU molecular constructs (psCLU, α-chain, β-chain, and ΔCLU) following transfection in HeLa cells. Western blot analysis (Fig. 1F) showed that all the constructs were expressed. We observed that the CLU β-chain had both a nuclear and a cytoplasmic localization, whereas psCLU and CLU α-chain were almost exclusively cytoplasmic (Fig. 2A). We also observed that ΔCLU demonstrated both cytoplasmic and nuclear localization, although it has no consensus nuclear localization sequence.
FIGURE 2.
Subcellular localization of CLU constructs in HeLa cells. A, Western blot of nuclear and/or cytoplasmic protein of different CLU constructs: psCLU, ΔCLU, β-chain, and α-chain. Control and EmT correspond to pFLAG-BAP and empty vector transfection, respectively. Lamin B1 and α-tubulin were used as control for nuclear and cytoplasmic protein fractions. MW, molecular weight standards. B, confocal microscopy observation of HeLa cells transfected with psCLU-pFLAG, α-chain pFLAG, β-chain pFLAG, and ΔCLU-pFLAG. Control corresponds to nontransfected cells. The cells were incubated with mouse anti-FLAG antibody-labeled FITC (green) to detect the presence of different constructs and with DAPI (blue) to stain the nucleus. The white color corresponds to the superposition (Merge) of FITC and DAPI fluorescence.
We performed confocal microscopy analysis, and the results depicted in Fig. 2B showed that psCLU and CLU α-chain were cytoplasmic. The β-chain of CLU had a diffuse localization, including the nucleus. The ΔCLU protein had a “patchy” distribution both in the cytoplasm and in the nucleus.
psCLU and ΔCLU Proteins Induced IκB-α Stabilization
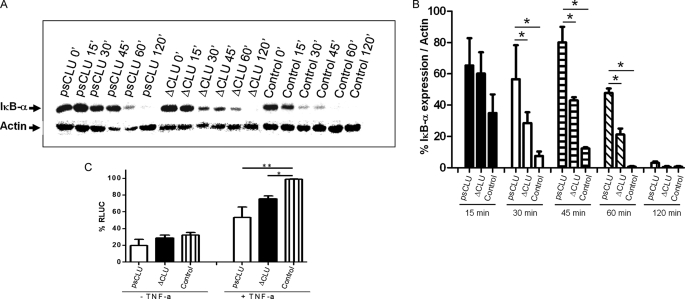
We tested the effect of the expression of the CLU constructs on IκB-α stabilization. Stimulation of cells by TNF-α resulted in a strong and rapid decrease in the expression of IκB-α, which was almost undetectable 30 min following stimulation (Fig. 3A). We observed that the expression of the psCLU form (Fig. 3A), but neither the α-chain nor the β-chain alone (supplemental Fig. S1A), resulted in IκB-α stabilization. IκB-α was still readily detectable at 30 min after stimulation with TNF-α, whereas it was barely detectable in control. Furthermore, the expression of the ΔCLU protein also induced a stabilization of IκB-α (Fig. 3, A and B). Thus, psCLU is the CLU protein isoform that behaves as a negative regulator of NF-κB activation, and this property is conferred by the ΔCLU encoded region.
FIGURE 3.
ΔCLU expression delayed the degradation of IκB-α and modulated NF-κB activation. A, Western blotting of IκB-α expression at 0, 15, 30, 45, 60, and 120 min after TNF-α stimulation (5 ng/ml) in the presence of cycloheximide (100 μg/ml). Control corresponds to empty pFLAG plasmid. B, relative expression levels over time determined by densitometry as mean ± S.E. from two individual experiments. *, p value < 0.05 (Student's t test). C, HeLa cells were co-transfected with NF-κB-dependent (3E-κB-Cona luc) DNA luciferase expression vector (1 μg) and cytomegalovirus vector pFLAG (2.5 μg) expressing either full CLU or ΔCLU. Empty vectors were transfected as control. After transfection, cells were left untreated (−TNF-α) or stimulated with TNF-α (+TNF-α) for 6 h. Experiments were repeated three times. *, p value = 0.0075 (Student's t test), **, p value = 0.0498 (Mann-Whitney test). RLUC, relative luminescent units.
Stabilization of IκB-α by psCLU and ΔCLU proteins prompted us to seek the functional role of these proteins with respect to NF-κB activation. We co-transfected HeLa cells with these constructs along with a 3Enh-κB luciferase plasmid as a reporter system, and the cells were activated by TNF-α (Fig. 3C). In both cases, we observed a reduced level of NF-κB activation after TNF-α stimulation, showing that the stabilization of IκB-α was accompanied by a reduction in NF-κB activation.
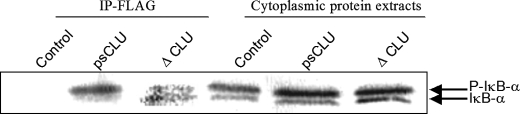
psCLU and ΔCLU Proteins Interact with Phosphorylated IκB-α
We showed that CLU interacts with a phosphorylated form of IκB-α (17). Because ΔCLU is a construct encoding for a homologous region between CLU and β-TrCP, which is also known to interact with pIκB-α, we examined whether ΔCLU could also interact with pIκB-α. We carried out a co-immunoprecipitation following TNF-α stimulation to allow for IκB-α phosphorylation. As shown in Fig. 4, immunoprecipitates precipitated with anti-FLAG (specific for wild-type CLU and ΔCLU) contained the pIκB-α protein. These co-immunoprecipitation data confirmed the presence of an interaction between CLU and pIκB-α and identified psCLU as an interacting form of CLU in this process. Furthermore, these data showed that ΔCLU was also able to interact with pIκB-α.
FIGURE 4.
Identification of the molecular partners of psCLU and ΔCLU. The HeLa cells were transfected with control (empty vector pFLAG) and psCLU- and ΔCLU-encoding plasmids. The immunoprecipitations (IP) were carried out after inhibition of proteasome (MG132 at 20 μm for 4 h) and TNF-α stimulation (20 ng/ml) for 10 min.
DISCUSSION
Clusterin has recently drawn much attention because of its newly discovered functions as a cell death/survival determinant and a regulator of the NF-κB activation pathway (5, 16, 17). The discovery of these new functions of CLU further highlights the importance of this protein, already shown to play a role in cancer and inflammation. Nevertheless, the specific isoform of CLU and the molecular mechanisms involved in these new properties of CLU are poorly documented. We confirmed that CLU is involved in NF-κB regulation and showed that it is a psCLU form of CLU that bears this property. We have found that the CLU α-chain had a cytoplasmic localization, whereas the β-chain had both a cytoplasmic and a nuclear localization, but neither the α-chain nor the β-chain in isolation had a regulatory function on NF-κB or a stabilizing effect on IκB-α. CLU expression could result in IκB-α stabilization (16) (Fig. 3). Following sequence comparison, we found that CLU shared homologous regions with β-TrCP, a major E3 ubiquitin ligase involved in IκB-α degradation (22). We made a molecular construct encoding for this particular CLU region and showed that it was an IκB-α stabilizer and that it interacted with pIκB-α. Interestingly, we observed that ΔCLU localized both in the cytoplasm and in the nucleus, although it has no consensus NLS sequence. This may be explained by the presence of an NLS yet unidentified in CLU, a putative carrier protein that transports ΔCLU to the nucleus by passive transport or by “N-glycosylation” (23), which promotes the translocation. Because we found that psCLU interacted with IκB-α through a homologous domain of β-TrCP, our results also suggest that psCLU could behave as a competitor of β-TrCP for molecular interaction with its substrates and that psCLU could have a putative role in all pathways in which β-TrCP action is proven, such as the Wnt/β-catenin pathway (24).
In summary, our data demonstrate that it is the psCLU form of CLU that has regulatory activity on NF-κB. We identified the site of psCLU that interacts with IκB-α, and we also showed that this region bears the regulatory activity of psCLU on NF-κB.
Supplementary Material
Acknowledgments
We thank S. Mistou and C. Roa-Brith for technical help. We also thank Dr. S. Bettuzzi and M. Scaltriti for the generous gift of FullCLU plasmid. We are grateful to J. P. Girault for helpful discussion on identification of βTrCP substrate binding sites. We are indebted to B. Durel for excellent technical assistance with confocal fluorescence microscopy. We are grateful to E. Walton for critical reading of the manuscript.
This work was supported by a grant from the Société Française de Rhumatologie (SFR) and Fondation Arthritis Courtin.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- CLU
- clusterin
- sCLU
- secreted CLU
- cCLU
- cytoplasmic CLU
- nCLU
- nuclear CLU
- psCLU
- presecretory CLU
- pnCLU
- precursor nCLU
- β-TrCP
- β-transducin repeat-containing protein
- NLS
- nuclear localization signal
- pIκB-α
- phospho-IκB-α
- BAP
- bacterial alkaline phosphatase
- Abs
- antibodies
- FITC
- fluorescein isothiocyanate
- DAPI
- 4′,6-diamidino-2-phenylindole
- luc
- luciferase.
REFERENCES
- 1.Jones S. E., Jomary C. (2002) Int. J. Biochem. Cell Biol. 34, 427–431 [DOI] [PubMed] [Google Scholar]
- 2.Trougakos I. P., Gonos E. S. (2002) Int. J. Biochem. Cell Biol. 34, 1430–1448 [DOI] [PubMed] [Google Scholar]
- 3.Chen X., Halberg R. B., Ehrhardt W. M., Torrealba J., Dove W. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9530–9535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leskov K. S., Klokov D. Y., Li J., Kinsella T. J., Boothman D. A. (2003) J. Biol. Chem. 278, 11590–11600 [DOI] [PubMed] [Google Scholar]
- 5.Trougakos I. P., Djeu J. Y., Gonos E. S., Boothman D. A. (2009) Cancer Res. 69, 403–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapron J. T., Hilliard G. M., Lakins J. N., Tenniswood M. P., West K. A., Carr S. A., Crabb J. W. (1997) Protein Sci. 6, 2120–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French L. E., Wohlwend A., Sappino A. P., Tschopp J., Schifferli J. A. (1994) J. Clin. Invest. 93, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi N. H., Mazda T., Tomita M. (1989) Mol. Immunol. 26, 835–840 [DOI] [PubMed] [Google Scholar]
- 9.Murphy B. F., Saunders J. R., O'Bryan M. K., Kirszbaum L., Walker I. D., d'Apice A. J. (1989) Int. Immunol. 1, 551–554 [DOI] [PubMed] [Google Scholar]
- 10.Choi N. H., Nakano Y., Tobe T., Mazda T., Tomita M. (1990) Int. Immunol. 2, 413–417 [DOI] [PubMed] [Google Scholar]
- 11.Zhang H., Kim J. K., Edwards C. A., Xu Z., Taichman R., Wang C. Y. (2005) Nat. Cell Biol. 7, 909–915 [DOI] [PubMed] [Google Scholar]
- 12.Trougakos I. P., Lourda M., Antonelou M. H., Kletsas D., Gorgoulis V. G., Papassideri I. S., Zou Y., Margaritis L. H., Boothman D. A., Gonos E. S. (2009) Clin. Cancer Res. 15, 48–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaisman-Elbaz T., Sekler I., Fishman D., Karol N., Forberg M., Kahn N., Hershfinkel M., Silverman W. F. (2009) J. Cell Physiol. 220, 222–229 [DOI] [PubMed] [Google Scholar]
- 14.Markopoulou S., Kontargiris E., Batsi C., Tzavaras T., Trougakos I., Boothman D. A., Gonos E. S., Kolettas E. (2009) FEBS. J. 276, 3784–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizzi F., Caccamo A. E., Belloni L., Bettuzzi S. (2009) J. Cell Physiol. 219, 314–323 [DOI] [PubMed] [Google Scholar]
- 16.Santilli G., Aronow B. J., Sala A. (2003) J. Biol. Chem. 278, 38214–38219 [DOI] [PubMed] [Google Scholar]
- 17.Devauchelle V., Essabbani A., De Pinieux G., Germain S., Tourneur L., Mistou S., Margottin-Goguet F., Anract P., Migaud H., Le Nen D., Lequerré T., Saraux A., Dougados M., Breban M., Fournier C., Chiocchia G. (2006) J. Immunol. 177, 6471–6479 [DOI] [PubMed] [Google Scholar]
- 18.Savković V., Gantzer H., Reiser U., Selig L., Gaiser S., Sack U., Klöppel G., Mössner J., Keim V., Horn F., Bödeker H. (2007) Biochem. Biophys. Res. Commun. 356, 431–437 [DOI] [PubMed] [Google Scholar]
- 19.Devauchelle V., Marion S., Cagnard N., Mistou S., Falgarone G., Breban M., Letourneur F., Pitaval A., Alibert O., Lucchesi C., Anract P., Hamadouche M., Ayral X., Dougados M., Gidrol X., Fournier C., Chiocchia G. (2004) Genes Immun. 5, 597–608 [DOI] [PubMed] [Google Scholar]
- 20.Gilmore T. D., Koedood M., Piffat K. A., White D. W. (1996) Oncogene 13, 1367–1378 [PubMed] [Google Scholar]
- 21.Evrard-Todeschi N., Pons J., Gharbi-Benarous J., Bertho G., Benarous R., Girault J. P. (2008) J. Chem. Inf. Model 48, 2350–2361 [DOI] [PubMed] [Google Scholar]
- 22.Margottin F., Bour S. P., Durand H., Selig L., Benichou S., Richard V., Thomas D., Strebel K., Benarous R. (1998) Mol. Cell 1, 565–574 [DOI] [PubMed] [Google Scholar]
- 23.Duverger E., Pellerin-Mendes C., Mayer R., Roche A. C., Monsigny M. (1995) J. Cell Sci. 108, 1325–1332 [DOI] [PubMed] [Google Scholar]
- 24.Liu C., Kato Y., Zhang Z., Do V. M., Yankner B. A., He X. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.