Abstract
Protein-protein interactions represent an important post-translational mechanism for endothelial nitric-oxide synthase (eNOS) regulation. We have previously reported that β-actin is associated with eNOS oxygenase domain and that association of eNOS with β-actin increases eNOS activity and nitric oxide (NO) production. In the present study, we found that β-actin-induced increase in NO production was accompanied by decrease in superoxide formation. A synthetic actin-binding sequence (ABS) peptide 326 with amino acid sequence corresponding to residues 326–333 of human eNOS, one of the putative ABSs, specifically bound to β-actin and prevented eNOS association with β-actin in vitro. Peptide 326 also prevented β-actin-induced decrease in superoxide formation and increase in NO and l-citrulline production. A modified peptide 326 replacing hydrophobic amino acids leucine and tryptophan with neutral alanine was unable to interfere with eNOS-β-actin binding and to prevent β-actin-induced changes in NO and superoxide formation. Site-directed mutagenesis of the actin-binding domain of eNOS replacing leucine and tryptophan with alanine yielded an eNOS mutant that exhibited reduced eNOS-β-actin association, decreased NO production, and increased superoxide formation in COS-7 cells. Disruption of eNOS-β-actin interaction in endothelial cells using ABS peptide 326 resulted in decreased NO production, increased superoxide formation, and decreased endothelial monolayer wound repair, which was prevented by PEG-SOD and NO donor NOC-18. Taken together, this novel finding indicates that β-actin binding to eNOS through residues 326–333 in the eNOS protein results in shifting the enzymatic activity from superoxide formation toward NO production. Modulation of NO and superoxide formation from eNOS by β-actin plays an important role in endothelial function.
Keywords: Cytoskeleton/Actin, Enzymes, Enzymes/Nitric Oxide Synthase, Oxygen/Superoxide, Protein, Protein/Protein-protein interactions, Signal Transduction/Nitric Oxide
Introduction
Nitric oxide (NO) generated by endothelial NO synthase (eNOS)2 plays an important role in a number of physiological and pathophysiological processes including regulation of vascular tone, smooth muscle cell proliferation, and angiogenesis (1–4). The synthesis of NO requires NADPH, tetrahydrobiopterin (BH4), flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and O2 as cofactors and results in NO and the co-product l-citrulline (1). eNOS is tightly regulated by transcriptional, post-transcriptional, and post-translational mechanisms (5, 6). Protein-protein interactions represent an important post-translational mechanism for eNOS regulation (5). We have reported that eNOS is associated with β-actin in endothelial cells and that association of eNOS with β-actin increases eNOS activity (6–8). In endothelial cells, β-actin exists in two forms: filamentous polymerized actin (F-actin) and globular actin (G-actin) (9). In lung endothelial cells, F-actin is in the form of cortical F-actin and actin stress fibers (10). There is a significant amount of eNOS in the insoluble portion of the Triton extraction of endothelial cells (F-actin) (11). We have found that eNOS localized to the plasma membrane is colocalized with cortical F-actin. eNOS that is located in the perinuclear area (probably Golgi) is colocalized with G-actin (5–8). eNOS and actin in endothelial cells can be co-immunoprecipitated, suggesting that eNOS is associated with β-actin protein (8). Studies using a yeast two-hybrid system showed that eNOS has direct interaction with β-actin (7, 12). Incubation of purified eNOS with F-actin and G-actin results in significant increases in eNOS activity (8).
The mechanism for actin association to increase eNOS activity is not clear. The actin-binding site on eNOS has not been identified. eNOS consists of two functional domains: an N-terminal oxygenase domain containing a heme active site and l-arginine- and BH4-binding sites, and a C-terminal reductase domain that contains the FAD-, FMN-, and NADPH-binding sites (13, 14). The heme site is responsible for the dimer formation of eNOS. The reductase domain of eNOS shares a close homology with the cytochrome P450 enzymes, generating electron flow from NADPH through FAD and FMN that is transferred to the oxidase domain of the other monomer where l-arginine oxidation occurs at the heme group in the active site. Using a yeast two-hybrid experiment, we have reported that the eNOS oxygenase domain rather than the reductase domain or the middle part of the eNOS molecule has direct interaction with β-actin, suggesting that the actin-binding site is in the oxygenase domain of the eNOS protein (7). Interestingly, three putative ABSs exist in the eNOS oxygenase domain. In the present study, using synthetic peptides and site-directed mutagenesis we identified amino acid residues 326–333 of eNOS protein as being the β-actin-binding site. Importantly, we found for the first time that β-actin association with eNOS shifts the enzymatic activity from superoxide formation toward NO production. This novel finding indicates that modulation of NO and superoxide generation from eNOS by β-actin may play an important role in endothelial function.
EXPERIMENTAL PROCEDURES
Reagents and Materials
Mouse anti-eNOS and anti-Hsp90 antibodies were obtained from Transduction Laboratory (Lexington, KY). Anti-β-actin monoclonal antibody was obtained from Sigma. Human β-actin was from Cytoskeleton (Denver, CO). BH4 and desferrioxamine was from Calbiochem. Purified Hsp90 was from Stressgen (Ann Arbor, MI) and diethyldithiocarbamate was from Alexis Biochemicals (Lausen, Switzerland). NADPH, calmodulin, and other reagents were purchased from Sigma.
eNOS Purification
Recombinant eNOS protein was purified as described previously by Sud et al. (15) and Rodriguez-Crespo et al. (16) with modifications. Briefly, 4 liters of overnight cell culture of human eNOSpCW were used to inoculate 0.5 liters of TB containing ampicillin (50 μg/ml). The cultures were grown to an A600 of 0.6 at 22 °C (200 rpm) and induced with 0.5 m isopropyl β-d-1-thiogalactopyranoside (IPTG). One hour before IPTG induction, δ-aminolevulinic acid (0.5 mm final) was added, and at the time of induction riboflavin (3 μm final) and ATP (1 mm final) were also added. After induction, cells were kept in the dark at 22 °C and 200 rpm. After 48 h, cell pellets were collected and frozen in −80 °C until purification. When eNOS protein was purified, the cells were resuspended in buffer A (50 mm Tris-HCl, pH 7.8, 1 mm EDTA, 1 mm DTT, 10% glycerol (v/v), 150 mm NaCl, 0.5 mm l-arginine, 4 μm BH4, 2 μm FAD 0.1 mm phenylmethylsulfonyl fluoride, 1 μm leupeptin, and 1 μm pepstatin), lysed by sonication, and then centrifuged. The supernatant was applied to 2′,5′-ADP Sepharose 4B column equilibrated with buffer B (50 mm Tris-HCl, pH 7.8, 0.1 mm EDTA, 0.1 mm DTT, 150 mm NaCl, 10% glycerol, 0.5 mm l-arginine). The column was washed with 20 volumes of buffer B and again with 20 volumes of buffer B containing 300 mm NaCl. Finally, proteins were eluted with buffer B containing 600 mm NaCl and 5 mm 2′-AMP. Repeated dilution/concentration with buffer containing 40 mm Tris buffer, pH 7.6, containing 1 mm l-arginine, 3 mm DTT, 4 μm BH4, 4 μm FAD, 10% glycerol, and 150 mm NaCl were performed to remove 2′-AMP and to achieve a final concentration of 150 mm NaCl. The DTT, BH4, and FAD were removed, and protein-containing fractions were concentrated using Centricon 50 (Millipore, Billerica, MA). The purity of the eNOS protein was verified using SDS-PAGE.
Measurement of NO in Vitro
NO production was determined by measuring NOx (NO2 and NO3). Purified eNOS and β-actin were preincubated at room temperature for 30 min and then added to a 50-μl reaction mix containing 50 mm HEPES buffer, 1 mm NADPH, 100 μm l-arginine, 1 mm CaCl2, 10 μg/ml calmodulin, 4 μm BH4. The mixture was incubated at 37 °C for 30 min. 40 μl of the reaction mix were loaded to the SIEVERS machine for NOx measurement according to standard manufacturer's instructions as previously described (17). For experiments with ABS peptides, β-actin and peptides were preincubated at room temperature for 20 min before eNOS protein was added.
Detection of Superoxide Generation in Vitro
eNOS-derived superoxide generation was measured by electron paramagnetic resonance (EPR) spectroscopy and spin trapping as previously described (18). 50 μl of reaction mix containing 50 mm HEPES buffer, 1 mm NADPH, 100 μm l-arginine, 1 mm CaCl2, 10 μg/ml calmodulin, and 1 μg of purified eNOS were incubated at 37 °C for 60 min. 12.5 μl of spin probe N1-hydroxy-3-methoxy-carbonyl-2,2,5,5-tetramethyl-pyrrolidine (CMH) in EPR buffer were added to the reaction mix. 35 μl of the final reaction mix were loaded into a 50-μl capillary tube and analyzed with a MiniScopeMS200 EPR (Magnetech, Berlin, Germany). A reaction curve was generated by adding 1 unit/ml of xanthine oxidase into 500 μm xanthine solution in buffered PBS (pH 7.4), which contains 5 μm diethyldithiocarbamate and 25 μm desferrioxamine to inhibit any conversion of superoxide into either hydrogen peroxide or hydroxyl radical via Fenton reaction. Reactions were allowed to proceed at 25 °C for up to 40 min. Following incubation, ∼35 μl of each reaction mixture was loaded into a 50-μl capillary tube and analyzed immediately with EPR spectroscopy. EPR spectra were analyzed for amplitude using ANALYSIS software (version 2.02, Magnettech). Given that 1 unit of xanthine oxidase will convert 1 μmol of xanthine per minute at 25 °C, based on this standard curve, we calculated that 1 EPR amplitude units is equivalent of 0.35 pmol of superoxide.
Synthesis of ABS Peptides
To study the function of three putative ABSs of eNOS, peptides corresponding to the amino acid sequences of these three putative ABSs were synthesized by GeneScript Corporation (Piscataway, NJ). A modified version of ABS peptide 326 with hydrophobic leucine and tryptophan substituted for neutrally charged alanine was used as a control peptide for ABS peptide 326. The amino acid sequences of the peptides are NSQLVRYAGYRQQDGSVRGDPANVEITEL for ABS peptide 245, RKKTFKEVANA for ABS peptide 492, LGLRWYAL for ABS peptide 326, AGARAYAA for control peptide for ABS peptide 326, RKKRRQRRRALGLRWYAL for ABS peptide 326 TAT (P326TAT), and RKKRRQRRRAAGARAYAA for control peptide TAT.
Assay of Peptide Binding to β-Actin
The binding capacities of the peptides to β-actin were measured by using an F-actin binding spin-down assay kit from Cytoskeleton, Inc. (Denver, Co). Monomeric human β-actin was polymerized into F-actin in F-actin buffer (5 mm Tris-HCl, pH 7.8, 1 mm ATP, 0.5 mm DTT, 0.2 mm CaCl2, 0.2 mm MgCl2, and 100 mm KCl) for 1 h at 24 °C. ABS peptides at final concentrations of 10 μm were incubated alone or with 23 μm F-actin in F-actin buffer for 30 min in a total volume of 50 μl. The mixtures were centrifuged at 150,000 × g for 2 h at 24 °C in a Beckman TLA-100 rotor. Supernatant and pellet fractions were resuspended in loading buffer and subjected to SDS-PAGE. The gel was stained with Coomassie Blue. The density of the band was measured using Software Image J.
Site-directed Mutagenesis of eNOS and Transfection of COS-7 Cells with Wild Type and eNOS Mutant
Human eNOS gene (GenBankTM M93718.1) was cloned into HindIII and XbaI sites of pcDNA3 vector and regarded as wild-type eNOS. The cDNA of human eNOS was mutated to substitute residues leucine 326, leucine 328, tryptophan 330, and leucine 333 for alanine in the actin-binding site. The site-directed mutagenesis was custom-made by Retrogen (San Diego, CA). The sequences of both strands of the gene in the mutated region were verified by using ABI 3730 automated sequencer. Plasmids containing wild-type eNOS cDNA or eNOS mutant cDNA were transfected into COS-7 cells using Lipofectamine LTX with PLUS reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. 48 h after transfection, cells were used for co-immunoprecipitation and assays for NO and superoxide generation.
Co-immunoprecipitation
Wild-type and eNOS mutant-transfected COS-7 cells were washed in ice-cold PBS and lysed in Tris buffer (50 mm Tris, pH 7.4, 10 mm NaF, 2.5 mm EDTA, 15 mm Na2P2O7, 1% Triton X-100, and 1× protease inhibitor mixture). The lysates or mixtures of eNOS and β-actin were incubated with anti-eNOS antibody at 4 °C overnight. 30 μl of protein A-Sepharose were added, and samples were further incubated for 2 h at 4 °C. Immunoprecipitates were collected by centrifugation and washed three times in buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% Triton X-100. Proteins were eluted from Sepharose beads by boiling the samples in 30 μl of SDS immunoblotting sample buffer. Sepharose beads were pelleted by centrifugation at 10,000 × g, and supernatants were analyzed for eNOS and β-actin by Western blotting.
Determination of NO Production and Superoxide Formation in COS-7 Cells
After wild-type and eNOS mutant transfection, ionomycin (2 μm) was added to the cells. After 30 min of incubation, culture medium was collected and ethanol-precipitated to remove proteins. 50 μl of the reaction mix were loaded to the SIEVERS machine for NOx measurement according to standard manufacturer's instructions (17). In some experiments, NG-nitro-l-arginine methyl ester (l-NAME), a specific eNOS inhibitor, was used to inhibit NO production. Our results indicate that incubation of endothelial cells with l-NAME resulted in a 90% inhibition of NO production, suggesting that this method is reliable to detect NO production. For superoxide assay, after incubation with ABS peptides, the spin trap CMH was added to the cells. Superoxide from cells was trapped for 1 h, and then cells were scraped and subjected to EPR spectroscopy as previously described (18). Protein contents in the cell lysates were determined by Lowry's method.
Determination of Catalytic Activity, NADPH Consumption, and Superoxide Generation from Purified eNOS Mutant
Wild-type and mutant eNOS proteins in COS-7 cells were purified as described in the above section. The catalytic activity of purified eNOS mutant protein was assayed by the measurement of l-[3H]citrulline formation from l-[3H]arginine as reported previously (8). To determine NADPH consumption by eNOS, 0.3 mm NADPH was added to 200 μl of reaction mix containing 1 μg eNOS, 1 μm FAD, 1 μm FMN, 10 μg/ml calmodulin, 100 μm l-arginine, 1 mm CaCl2, 10 μg/ml calmodulin, and 2.5 mm ATP. The reaction was monitored at 340 nm for 10 min. NADPH consumption was calculated by using a molar extinction coefficient 6.22 mm−1·cm−1. The superoxide formation from purified eNOS mutant was measured by spin trapping and EPR spectroscopy as previously described above.
Transfection of Endothelial Cells with ABS Peptides
Pulmonary artery endothelial cells were incubated with ABS peptides at 20 μm final concentration in MEM medium. After 1 h of initial transfection, RPMI medium containing 4% fetal bovine serum (FBS) was added to reach final concentration of 2% FBS. Cells were then incubated for another 2 h before being used for co-immunoprecipitation and eNOS activity assays.
Determination of eNOS-β-Actin Association, NO Production, and Superoxide Formation in Endothelial Cells
eNOS-β-Actin association was evaluated using co-immunoprecipitation. To measure NO production, endothelial cells were incubated with ABS peptides for 3 h. Then ionomycin (2 μm) was added to the cells. After an additional 30 min, culture medium was collected and ethanol-precipitated to remove proteins. 50 μl of the reaction mix were loaded to the SIEVERS machine for NOx measurement as described above. For superoxide assay, after incubation with ABS peptides, the spin trap CMH was added to the cells. Superoxide from cells was trapped for 1 h, and then cells were scraped and subjected to EPR spectroscopy as described above.
Endothelial Monolayer Wound Repair
Pulmonary artery endothelial cells were incubated with ABS peptides for 3 h. Then endothelial monolayer wound repair in the absence and presence of PEG-SOD (100 units/ml) and NOC-18 (10 μm) was measured as previously reported (19). Endothelial monolayer wound repair distance was expressed as the width of the wound before treatment subtracted by that after treatment.
Statistical Analysis
In each experiment, experimental and control cells were matched for cell line, age, seeding density, number of passages, and number of days postconfluence to avoid variation in tissue culture factors that can influence measurements of NO and superoxide production. Results are shown as means ± S.E. for n experiments. Student's paired t test was used to determine the significance of differences between the means of experimental and control cells. A value of p < 0.05 was taken as significant.
RESULTS
An Increase in NO Production Induced by β-Actin Binding to eNOS Is Accompanied by a Decrease in Superoxide Formation
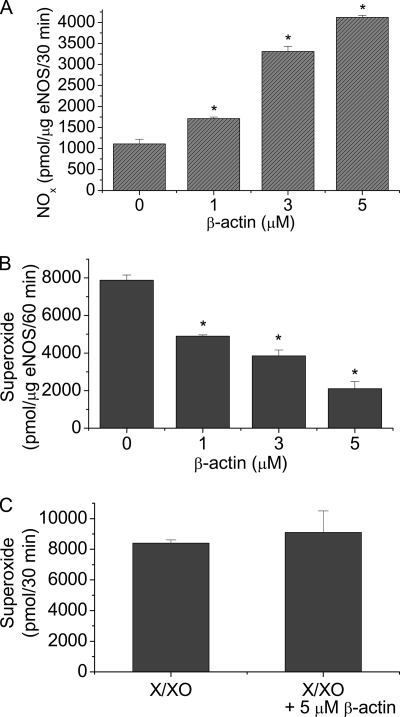
We have reported that the β-actin-binding site on eNOS protein is located at the oxygenase domain (6, 7). The oxygenase domain is involved with superoxide generation when eNOS is uncoupled because of limited BH4 availability (20, 21). To test whether β-actin association with eNOS modulates the formation of superoxide, we measured superoxide and NO production by purified eNOS using EPR spectrometry in the absence and the presence of G-actin. As shown in Fig. 1A, incubation of purified eNOS with β-actin caused an increase in NO production in a dose-dependent manner. However, the level of superoxide produced by eNOS is decreased in the presence of β-actin (1–5 μm) (Fig. 1B). β-Actin did not affect the level of superoxide generated from xanthine oxidase (Fig. 1C), suggesting that β-actin does not scavenge superoxide. These data indicate that β-actin binding to eNOS prevents superoxide generation from eNOS and shifts the enzymatic activity from forming superoxide toward NO production.
FIGURE 1.
β-actin increases NO production and decreases superoxide formation from eNOS. Purified eNOS and β-actin were incubated at room temperature for 30 min before being added to 50 μl reaction mix for NOx measurement using the SIEVERS machine (panel A) and for superoxide analysis using EPR spectroscopy and spin trapping (panel B). Panel C, superoxide was produced from xanthine oxidase (1.0 u) and 500 μm xanthine in 500 μl of PBS (pH 7.4), which contains 5 μm diethyldithiocarbamate and 25 μm desferrioxamine to inhibit any conversion of superoxide into either hydrogen peroxide or hydroxyl radical, in the absence and presence of 5 μm β-actin and then superoxide was quantitated using EPR spectroscopy and spin trapping. Results are expressed as mean ± S.E.; n = 3 experiments. *, p < 0.05 versus control (0).
Sequence Comparison of eNOS and Several Actin-binding Proteins Containing the Calponin Homology (CH-1) Domain
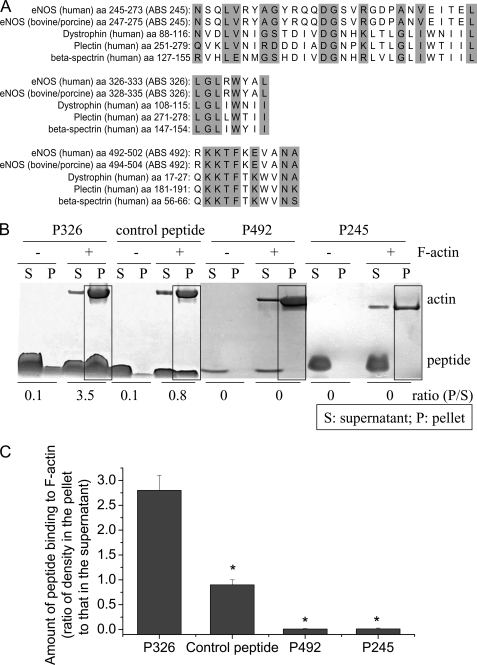
We have reported that the actin-binding region is in the oxygenase domain of eNOS protein based on data from the yeast two-hybrid experiments (7). Using the program VECTOR NTI, we made a sequence alignment analysis of sequences of eNOS protein against the CH-1 domain, an actin-binding region of the calponin homology domain in a number of signaling and actin cross-linking molecules i.e. α-actinin, dystrophin, and utrophin. Three regions in the eNOS oxygenase domain were found to have high consensus with ABSs in these actin-binding proteins (Fig. 2A). These ABSs are actin-binding motifs (22, 23).
FIGURE 2.
Panel A, alignment of the sequences of the three putative ABSs to actin-binding proteins. Three regions in the eNOS oxygenase domain were found to have high consensus with ABS on actin-binding proteins. The highlight indicates same or similar amino acids. Panel B, ABS peptide 326 specifically binds to β-actin. ABS peptides 326 (P326), control peptide, ABS peptides 492 (P492), and ABS peptides 245 (P245) at final concentrations of 10 μm were incubated alone or with 23 μm F-actin in F-actin buffer for 30 min. After high speed centrifugation at 150,000 × g for 2 h, the supernatants and pellets were subjected to SDS-PAGE analysis. The boxes show co-sedimentation of peptides with F-actin in the pellets. The image shown is representative of three experiments. Panel C is a bar graph depicting the changes in the amount of peptide binding to F-actin expressed as the ratio of peptide density in the pellet to that in the supernatant. Results are expressed as mean ± S.E.; n = 3 experiments. *, p < 0.05 versus P326.
Binding of ABS Peptides to β-Actin and Their Effect on eNOS-β-Actin Association
Actin binding capabilities of the synthesized ABS peptides corresponding to putative ABSs in eNOS were evaluated by the F-actin binding spin-down assay. ABS peptides 326, 492, and 245 were incubated with purified F-actin in F-actin buffer containing 2.5 μm ATP and 2.5 μm DTT, and samples were then subjected to high speed centrifugation. The supernatants and pellets were then analyzed by SDS-PAGE. As shown in Fig. 2, B and C, F-actin was pulled down to the pellets by high-speed centrifugation. In the absence of F-actin, ABS peptides remained in the supernatant fraction. However, in the presence of F-actin, a significant portion of peptide 326 was pulled down to the pellets with F-actin. Only a very small amount of the control peptide for peptide 326 was pulled down to the pellets with F-actin. Peptides 492 and 245 were not pulled down to the pellets with F-actin. These results indicate that ABS peptide 326, which is a sequence from the eNOS oxygenase domain, can specifically bind to β-actin.
To further study whether ABS peptide 326 can competitively affect eNOS-β-actin association, purified G-actin was incubated with purified recombinant eNOS protein in the presence of ABS peptide 326 or its control peptide. Then eNOS protein was precipitated using protein G-agarose conjugated with eNOS monoclonal antibody. The amounts of eNOS and β-actin protein in the pellets were measured using Western blot analysis. As shown at Fig. 3, β-actin can be pulled down to the pellets together with eNOS. ABS peptide 326 decreased the amount of β-actin precipitated with eNOS protein in a dose-dependent manner. In contrast, the control peptide for peptide 326, in which residues leucine 326, leucine 328, tryptophan 330, and leucine 333 were replaced by alanine, in the same concentrations did not affect the amount of β-actin precipitated with eNOS protein. Taken together, these data show that ABS peptide 326 specifically binds to β-actin and competitively inhibits eNOS-β-actin association.
FIGURE 3.
ABS peptide 326 inhibits eNOS-β-actin association. Purified eNOS was incubated with β-actin with and without varying concentrations of ABS peptide 326 and its control peptide at room temperature for 30 min, and the eNOS-β-actin mixtures were subjected to immunoprecipitation using anti-eNOS antibody as described under “Experimental Procedures.” eNOS and β-actin in the pellets were measured by Western blot analysis. Panel A is a representative image of a Western blot from three separate experiments. Panel B is a bar graph depicting the changes in the ratio of β-actin to eNOS in the pellets. Results are expressed as mean ± S.E.; n = 3 experiments. *, p < 0.05 versus control peptide.
ABS Peptide 326 Prevents β-Actin-induced Increase in NO and l-Citrulline Production and Decrease in Superoxide Formation in Vitro
To study whether competitive inhibition of eNOS-β-actin association by ABS peptide 326 prevents β-actin-induced increase in NO and l-citrulline production and decrease in superoxide production from eNOS, we analyzed β-actin-induced production of NO, l-citrulline, and superoxide in the absence and presence of ABS peptide 326 and its control peptide. We found that ABS peptide 326 per se did not affect NO and l-citrulline production and superoxide formation (Fig. 4, A–C), suggesting that peptide 326 does not scavenge NO or superoxide. Moreover, ABS peptide 326 prevented β-actin-induced increase in NO and l-citrulline production (Fig. 4, A and B) and decrease in superoxide formation (Fig. 4C). However, control peptide in which residues leucine 326, leucine 328, tryptophan 330, and leucine 333 were replaced by alanine did not affect β-actin-induced changes in the production of NO, l-citrulline, and superoxide from eNOS (Fig. 4, A–C). In addition, incubation of purified eNOS with purified G-actin resulted in a decrease in NADPH consumption in absence of BH4 and ABS peptide 326 prevented β-actin-induced decrease in NADPH consumption (Fig. 4D). Taken together, these data suggest that ABS peptide 326 specifically binds to β-actin and blocks the effects of β-actin on the production of NO, l-citrulline, and superoxide from eNOS.
FIGURE 4.
ABS peptide 326 prevents β-actin-induced increase in NO and l-citrulline production and decrease in superoxide formation and NADPH consumption. Purified eNOS (1.0 μg/500 μl) and β-actin (2.0 μm) were incubated at room temperature in the presence of ABS peptide 326 (P326) or its control peptide (20 μm) for 30 min before being added to 50 μl of master mix for NO measurement using the SIEVERS machine (panel A), l-citrulline assay (panel B), superoxide analysis using spin-trapping EPR spectroscopy (panel C), and NADPH consumption assay with or without BH4 (panel D). Results are expressed as mean ± S.E.; n = 3 experiments. *, p < 0.05 versus WO β-actin (without β-actin).
Mutation of Residues Leucine 326, Leucine 328, Tryptophan 330, and Leucine 333 for Alanine Decreases eNOS-β-Actin Association and NO Production and Increases Superoxide Generation
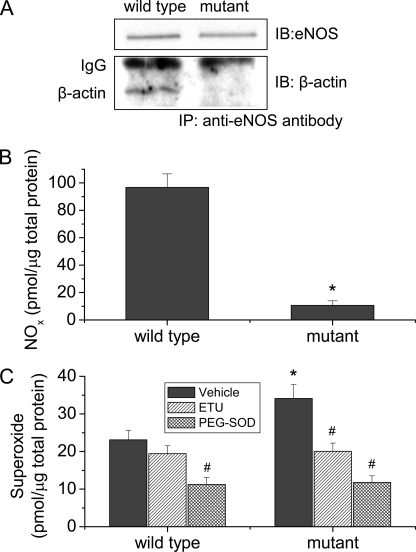
We have demonstrated that the control peptide for peptide 326 in which hydrophobic leucine and tryptophan were replaced by neutrally charged alanine had much lower capacity to bind β-actin (Fig. 2) and did not affect eNOS-β-actin association (Fig. 3) and β-actin-induced changes in NO and superoxide production from eNOS in vitro (Fig. 4), suggesting that hydrophobic leucine and tryptophan in the actin-binding site might be critical for eNOS-β-actin interaction. To further confirm that role of these residues in eNOS-β-actin association, residues leucine 326, leucine 328, tryptophan 330, and leucine 333 were replaced for alanine by site-directed mutagenesis. The plasmids containing wild type and mutant eNOS genes were transfected into COS-7 cells. As shown in Fig. 5A, the amount of β-actin co-precipitated with eNOS mutant was much smaller than that with wild-type eNOS, and the eNOS protein levels were similar. COS-7 cells with mutant eNOS exhibited much lower NO production and higher superoxide generation (Fig. 5, B and C). In the presence of a specific NOS inhibitor 2-ethyl-2-thiopseudourea (ETU, 100 μm) (15, 24) or PEG-SOD, the amounts of superoxide generated from COS-7 cells containing wild type and eNOS mutant were comparable (Fig. 5C), indicating that increased superoxide generation is from expressed eNOS in COS-7 cells.
FIGURE 5.
eNOS mutant exhibits decreased association with β-actin, decreased NO production, and increased superoxide generation in COS-7 cells. COS-7 cells transfected with wild-type and mutant eNOS plasmids, and then eNOS-β-actin association were measured using co-immunoprecipitation (panel A). NOx (panel B) and superoxide production (panel C) were measured using the SIEVERS machine and spin-trapping EPR spectroscopy, respectively. Results are expressed as mean ± S.E.; n = 3 experiments. *, p < 0.05 versus wild-type group. #, p < 0.05 versus vehicle group.
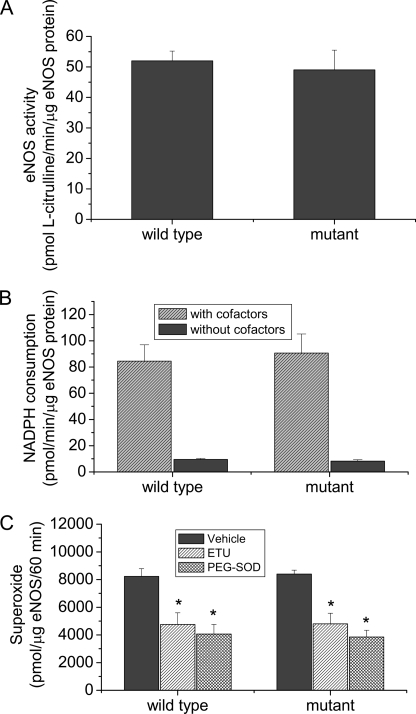
To exclude the possibility that mutation of residues leucine 326, leucine 328, tryptophan 330, and leucine 333 directly causes the alterations in NO and superoxide generation in COS-7 cells, wild-type and mutated eNOS expressed in COS-7 cells were purified. We found that the catalytic activity and rate of NADPH consumption and superoxide generation from purified wild-type and mutated eNOS were comparable (Fig. 6). These data suggest that a decrease in NO production and an increase in superoxide generation in the eNOS mutant are not caused by the direct effect of the mutation.
FIGURE 6.
Purified eNOS mutant protein has the same catalytic activity, the same rate to consume NADPH, and the same rate to generate superoxide as the wild-type eNOS. Wild-type and mutated eNOS expressed in COS-7 cells were purified. The activities (panel A), NADPH consumption (panel B), and superoxide generation (panel C) from these proteins were assayed as described under “Experimental Procedures.” Results are expressed as mean ± S.E.; n = 3 experiments. *, p < 0.05 versus vehicle group.
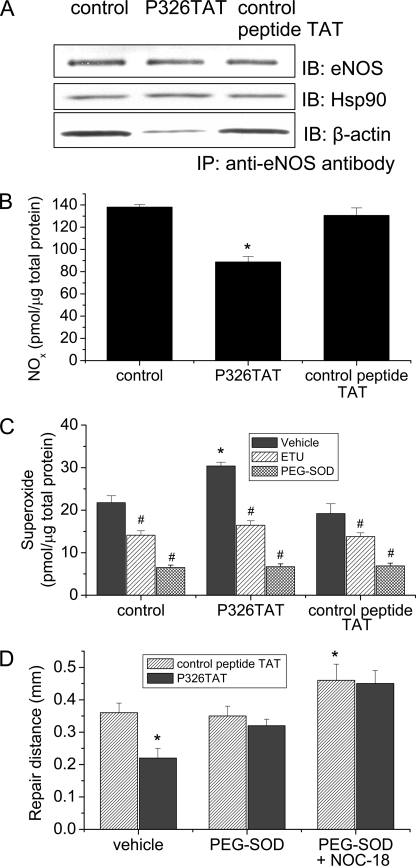
ABS Peptide 326 Decreases eNOS-β-Actin Interaction and NO Production and Increases Superoxide Formation in Intact Endothelial Cells
We have shown that ABS peptide 326 specifically binds to β-actin and competitively inhibits eNOS-β-actin association in vitro. To study whether ABS peptide 326 affects eNOS-β-actin association in intact endothelial cells, pulmonary artery endothelial cells were transfected with ABS peptide 326 linked to an 11-amino acid transduction domain of HIV TAT (P326TAT) as described by Gustafsson et al. (25). This TAT tag is a novel method used to facilitate delivery of biologically active proteins or peptides into cells and tissues through the fusion of a protein transduction domain to the protein or peptide of interest (26). To confirm the efficiency of P326TAT and control peptide TAT to enter endothelial cells, we used FITC-labeled P326TAT and FITC-labeled control peptide TAT. We found that incubation of endothelial cells with FITC-labeled P326TAT and FITC-labeled control peptide TAT (20 μm) for 3 h resulted in marked fluorescence accumulation in endothelial cells (supplemental Fig. S1), suggesting that P326TAT and control peptide TAT can enter endothelial cells efficiently. Endothelial cells were incubated with P326TAT and control peptide TAT for 3 h, and then eNOS-β-actin association was evaluated by co-immunoprecipitation using anti-eNOS antibody. As shown in Fig. 7A, transfection of endothelial cells with P326TAT significantly decreased the amount of β-actin co-immunoprecipitated with eNOS. We then measured NO and superoxide production in P326TAT-transfected endothelial cells. As shown in Fig. 7B, transfection of endothelial cells with P326TAT significantly decreased NO production without any alteration in eNOS protein content (Fig. 7A) and in eNOS protein localization in endothelial cells (supplemental Fig. S2). Meanwhile, superoxide formation was much higher in P326TAT-transfected endothelial cells than control peptide TAT-transfected endothelial cells (Fig. 7C). The specific NOS inhibitor ETU (100 μm) (24) inhibited P326TAT-induced increase in superoxide formation (Fig. 7C), suggesting that the increased superoxide generation was from eNOS rather than mitochondria or xanthine oxidase. Taken together, these results indicate that ABS peptide 326 prevents eNOS-β-actin association, reduces NO production, and increases superoxide formation in intact endothelial cells.
FIGURE 7.
ABS peptide 326 decreases eNOS-β-actin interaction, NO production, and monolayer wound repair, and increases superoxide formation in endothelial cells. Pulmonary artery endothelial cells were incubated with P326TAT (20 μm). Then eNOS-β-actin association was measured by co-immunoprecipitation using eNOS antibody (panel A). The image shown is a representative from four separate experiments. Panel B shows the changes in NO production in endothelial cells incubated with or without P326TAT and control peptide TAT. Panel C shows the changes in superoxide formation in endothelial cells incubated with or without P326TAT and control peptide TAT in the presence and absence of specific NOS inhibitor ETU (100 μm) and superoxide scavenger PEG-SOD (100 units/ml). Panel D shows that endothelial cells incubated with P326TAT exhibit reduction of wound repair and that PEG-SOD and NOC-18 prevent P326TAT peptide-induced decrease in monolayer wound repair. Results are expressed as mean ± S.E.; n = 4 experiments. *, p < 0.05 versus control peptide; #, p < 0.05 versus vehicle.
ABS Peptide 326 Did Not Affect eNOS-Hsp90 Interaction in Endothelial Cells
To rule out the possibility that the effect of P326TAT on eNOS activity in endothelial cells is caused by its effect on eNOS-Hsp90 interaction, we measured eNOS-Hsp90 association by co-immunoprecipitation using eNOS antibody. We found that the amount of Hsp90 co-precipitated with eNOS protein was comparable between P326TAT-transfected cells and control peptide TAT-transfected cells (Fig. 7A), suggesting that P326TAT did not affect eNOS-Hsp90 interaction and that P326TAT-induced inhibition of eNOS activity was not caused by alteration in eNOS-Hsp90 interaction.
ABS Peptide 326 Decreases Endothelial Wound Repair
To investigate whether P326TAT-induced alterations in eNOS-β-actin interaction and in NO and superoxide generation from eNOS result in functional changes in endothelial cells, endothelial monolayer wound repair was evaluated in endothelial cells incubated with P326TAT and its control peptide. As shown in Fig. 7D, endothelial monolayer incubated with P326TAT (20 μm) exhibited lower capacity of wound repair and PEG-SOD and NOC-18 prevented P326TAT-induced decrease in monolayer wound repair.
DISCUSSION
β-Actin exists in non-muscle cells and is traditionally considered to be a structural protein that organizes and maintains the shape of cells. More recent experimental data indicate that β-actin is also a signaling molecule. We and others (5–8, 11, 27) have demonstrated that β-actin associates with and regulates eNOS in vascular endothelial cells and platelets and have provided evidence supporting the concept that β-actin is an activator of eNOS. Using a yeast two-hybrid experiment, we have reported that the eNOS oxygenase domain rather than the reductase domain or the middle part of the eNOS molecule has direct interaction with β-actin, suggesting that the actin-binding site is in the oxygenase domain of the eNOS protein (7). Interestingly, three putative ABSs exist in the eNOS oxygenase domain. Three regions in the eNOS oxygenase domain were found to have high consensus with ABSs in actin-binding proteins. The first region is at residues 245–273 for which no specific function has been found so far. ABS peptide 245 with an amino acid sequence corresponding to this region was found not to bind to β-actin. The second region is at residues 492–502, which is part of the calmodulin-binding site. However, peptide 492 with an amino acid sequence corresponding to this region has very low actin binding capability (Fig. 2, B and C). Therefore, the actin-binding site in eNOS is unlikely to be at residues 245–273 or residues 492–524. The third region is at residues 326–333. We found that synthetic ABS peptide 326, which has an amino acid sequence corresponding to residues 326–333 of the eNOS oxygenase domain, specifically binds to β-actin and inhibits eNOS-β-actin interaction. Moreover, ABS peptide 326 prevents β-actin-induced increase in NO and l-citrulline production and β-actin-induced decrease in superoxide formation and NADPH consumption. These results indicate that the actin-binding site in eNOS might be at residues 326–333 of the eNOS oxygenase domain.
Several studies suggest that hydrophobic residues inside the actin-binding region of actin cross-linking proteins are essential for actin binding. For example, hydrophobic pockets in gelsolin facilitate its binding to the sides of actin filaments (28). ABD-120 binds to actin cross-linking protein via hydrophobic interaction (29). Replacing hydrophobic amino acids leucine 326, leucine 328, tryptophan 330, and leucine 333 with neutral alanine in ABS peptide 326 results in a dramatic reduction of its ability to bind β-actin (Fig. 2, B and C). The modified peptide is unable to interfere with eNOS-β-actin binding and to prevent β-actin-induced increase in NO production and β-actin-induced decrease in superoxide formation. Furthermore, mutation of hydrophobic residues leucine 326, leucine 328, tryptophan 330, and leucine 333 for neutral alanine results in decreases in eNOS-β-actin association and NO production and increases in superoxide generation. These data indicate that hydrophobic amino acids leucine and tryptophan in residues 326–333 of eNOS oxygenase domain are essential for actin binding.
eNOS is capable of generating both NO and superoxide (20, 21, 30). Oxygenase domain is involved with superoxide generation when eNOS is uncoupled due to limited BH4 availability (20, 21). We found that an increase in NO production induced by β-actin binding to eNOS is accompanied by a decrease in superoxide production, suggesting that β-actin binding to eNOS shifts the enzymatic activity from superoxide formation toward NO production. Specific block of eNOS-β-actin association using ABS peptide 326 decreases β-actin-induced increase in NO production and decrease in superoxide formation and NADPH consumption in vitro. Moreover, inhibition of eNOS-β-actin association by ABS peptide 326 decreases NO production and endothelial monolayer wound repair and increases superoxide formation from eNOS in intact endothelial cells. Thus, eNOS-β-actin association might mediate the balance of nitric oxide and superoxide generation from eNOS. Further investigations are needed to determine whether β-actin binding to eNOS may cause conformational change in the eNOS protein which subsequently alters the electron transfer in the heme.
The regulatory mechanisms of eNOS-β-actin interaction are not completely understood. What is the signaling mechanism that initiates an increase or decrease in eNOS-β-actin association in endothelial cells? Alterations in eNOS-β-actin interaction could be caused by changes in the availability of β-actin, in the physical state of actin (e.g. F- or G-actin), or in the affinity between eNOS and β-actin protein. Manipulating G-actin contents using pharmacological agents causes changes in eNOS-β-actin interaction and eNOS activity in platelets and endothelial cells (7, 8, 27). Treatment with cytochalasin D increases cellular G-actin content and eNOS-β-actin interaction and eNOS activity (7). Treatment with phalloidin, an F-actin stabilizer, decreases cellular G-actin content and eNOS-β-actin interaction and eNOS activity (8). The affinity between eNOS and β-actin protein would surely affect eNOS-β-actin interaction. Transfection of endothelial cells with P326TAT prevents eNOS-β-actin association and reduces eNOS activity and endothelial monolayer wound repair. Besides these pharmacological agents, a number of physiological and pathological conditions have been reported to alter the availability of β-actin and/or the affinity between eNOS and β-actin protein. For example, hypoxia and corticotropin have been shown to decrease β-actin gene expression (8, 31, 32). β-actin interaction with eNOS in platelets and endothelial cells is decreased during platelet aggregation (27) and hypoxia (8). Hypoxia reduces NO release and increases superoxide formation in endothelial cells (33, 34). Thus, decreased eNOS-β-actin association may contribute to decreased NO release and increased superoxide generation from eNOS during hypoxia. Therefore, identifying the mechanism for β-actin-induced activation of eNOS has significant implications for vascular biology and disorders such as systemic and pulmonary hypertension, cor pulmonale, atherosclerosis, and thrombotic diseases.
In conclusion, we have identified a novel actin-binding site in the eNOS protein. Hydrophobic amino acid residues leucine 326, leucine 328, tryptophan 330, and leucine 333 in fragment 326–333 of eNOS oxygenase domain are essential for actin binding. β-Actin binding to eNOS shifts the enzymatic activity from superoxide formation toward NO production. Modulation of NO and superoxide formation from eNOS by β-actin plays an important role in endothelial function.
Supplementary Material
Acknowledgment
We thank Dr. Sanjiv Kumar for technical assistance in the superoxide assay.
This work was supported, in whole or in part, by National Institutes of Health Grant R01HL088261. This work was also supported by Flight Attendants Medical Research Institute Grants 032040 and 072104 and American Heart Association Greater Southeast Affiliate Grants 0555322B and 0855338E.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- eNOS
- endothelial nitric-oxide synthase
- ABS
- actin-binding sequence
- BH4
- tetrahydrobiopterin
- Hsp90
- heat shock protein 90
- l-NAME
- NG-nitro-l-arginine methyl ester
- EPR
- electron paramagnetic resonance
- ETU
- 2-ethyl-2-thiopseudourea
- PEG-SOD
- polyethylene glycol-superoxide dismutase
- DTT
- dithiothreitol
- CH-1
- calponin homology
- F-actin
- filamentous polymerized actin
- G-actin
- globular actin
- CMH
- N1-hydroxy-3-methoxy-carbonyl-2,2,5,5-tetramethyl-pyrrolidine
- PBS
- phosphate-buffered saline
- FITC
- fluorescein isothiocyanate.
REFERENCES
- 1.Moncada S., Higgs A. (1993) N. Engl. J. Med. 329, 2002–2012 [DOI] [PubMed] [Google Scholar]
- 2.Michel T., Feron O. (1997) J. Clin. Invest. 100, 2146–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papapetropoulos A., Rudic R. D., Sessa W. C. (1999) Cardiovasc. Res. 43, 509–520 [DOI] [PubMed] [Google Scholar]
- 4.Ignarro L. J., Buga G. M., Wood K. S., Byrns R. E., Chaudhuri G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 9265–9269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su Y., Kondrikov D., Block E. R. (2005) Cell Biochem. Biophys. 43, 439–449 [DOI] [PubMed] [Google Scholar]
- 6.Su Y., Kondrikov D., Block E. R. (2007) Sci. STKE 2007, e52–1-e52– 3 [DOI] [PubMed] [Google Scholar]
- 7.Kondrikov D., Han H. R., Block E. R., Su Y. (2006) Am. J. Physiol. Lung Cell Mol. Physiol. 290, L41–L50 [DOI] [PubMed] [Google Scholar]
- 8.Su Y., Edwards-Bennett S., Bubb M. R., Block E. R. (2003) Am. J. Physiol. Cell Physiol. 284, C1542–C1549 [DOI] [PubMed] [Google Scholar]
- 9.dos Remedios C. G., Chhabra D., Kekic M., Dedova I. V., Tsubakihara M., Berry D. A., Nosworthy N. J. (2003) Physiol. Rev. 83, 433–473 [DOI] [PubMed] [Google Scholar]
- 10.Dudek S. M., Garcia J. G. (2001) J. Appl. Physiol. 91, 1487–1500 [DOI] [PubMed] [Google Scholar]
- 11.Venema V. J., Marrero M. B., Venema R. C. (1996) Biochem. Biophys. Res. Commun. 226, 703–710 [DOI] [PubMed] [Google Scholar]
- 12.Kondrikov D., Su Y., Han H.-R., Block E. R. (2004) FASEB J. 15, 1026 [Google Scholar]
- 13.Chen P. F., Wu K. K. (2003) J. Biol. Chem. 278, 52392–52400 [DOI] [PubMed] [Google Scholar]
- 14.Abu-Soud H. M., Ichimori K., Presta A., Stuehr D. J. (2000) J. Biol. Chem. 275, 17349–17357 [DOI] [PubMed] [Google Scholar]
- 15.Sud N., Sharma S., Wiseman D. A., Harmon C., Kumar S., Venema R. C., Fineman J. R., Black S. M. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 293, L1444–L1453 [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez-Crespo I., Gerber N. C., Ortiz de Montellano P. R. (1996) J. Biol. Chem. 271, 11462–11467 [DOI] [PubMed] [Google Scholar]
- 17.Church J. E., Fulton D. (2006) J. Biol. Chem. 281, 1477–1488 [DOI] [PubMed] [Google Scholar]
- 18.Sud N., Wells S. M., Sharma S., Wiseman D. A., Wilham J., Black S. M. (2008) Am. J. Physiol. Cell Physiol. 294, C1407–C1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su Y., Cui Z., Li Z., Block E. R. (2006) FASEB J. 20, 1443–1451 [DOI] [PubMed] [Google Scholar]
- 20.Vásquez-Vivar J., Kalyanaraman B., Martásek P., Hogg N., Masters B. S., Karoui H., Tordo P., Pritchard K. A., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9220–9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia Y., Tsai A. L., Berka V., Zweier J. L. (1998) J. Biol. Chem. 273, 25804–25808 [DOI] [PubMed] [Google Scholar]
- 22.Keep N. H. (2000) Neurol. Sci. 21, S929–S937 [DOI] [PubMed] [Google Scholar]
- 23.Garciá-Alvarez B., Bobkov A., Sonnenberg A., de Pereda J. M. (2003) Structure 11, 615–625 [DOI] [PubMed] [Google Scholar]
- 24.Lakshminrusimha S., Wiseman D., Black S. M., Russell J. A., Gugino S. F., Oishi P., Steinhorn R. H., Fineman J. R. (2007) Am. J. Physiol. Heart Circ. Physiol 293, H1491–H1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustafsson A. B., Gottlieb R. A., Granville D. J. (2005) Methods Mol. Med. 112, 81–90 [PubMed] [Google Scholar]
- 26.Gustafsson A. B., Sayen M. R., Williams S. D., Crow M. T., Gottlieb R. A. (2002) Circulation 106, 735–739 [DOI] [PubMed] [Google Scholar]
- 27.Ji Y., Ferracci G., Warley A., Ward M., Leung K. Y., Samsuddin S., Lévêque C., Queen L., Reebye V., Pal P., Gkaliagkousi E., Seager M., Ferro A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8839–8844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwiatkowski D. J., Janmey P. A., Yin H. L. (1989) J. Cell Biol. 108, 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bresnick A. R., Warren V., Condeelis J. (1990) J. Biol. Chem. 265, 9236–9240 [PubMed] [Google Scholar]
- 30.Pritchard K. A., Jr., Ackerman A. W., Gross E. R., Stepp D. W., Shi Y., Fontana J. T., Baker J. E., Sessa W. C. (2001) J. Biol. Chem. 276, 17621–17624 [DOI] [PubMed] [Google Scholar]
- 31.Cheitlin R., Ramachandran J. (1981) J. Biol. Chem. 256, 3156–3158 [PubMed] [Google Scholar]
- 32.Ostergaard L., Stankevicius E., Andersen M. R., Eskildsen-Helmond Y., Ledet T., Mulvany M. J., Simonsen U. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H2894–H2903 [DOI] [PubMed] [Google Scholar]
- 33.Le Cras T. D., McMurtry I. F. (2001) Am. J. Physiol. Lung Cell Mol. Physiol. 280, L575–L582 [DOI] [PubMed] [Google Scholar]
- 34.Maulik N., Das D. K. (2002) Free Radic. Biol. Med. 33, 1047–1060 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.