Abstract
Hypoxia, a driving force in neovascularization, promotes alterations in gene expression mediated by hypoxia-inducible factor (HIF)-1α. Connective tissue growth factor (CTGF, CCN2) is a modulator of endothelial cell growth and migration, but its regulation by hypoxia is poorly understood. Therefore, we analyzed signaling pathways involved in the regulation of CTGF by hypoxia in endothelial cells. Exposure to low oxygen tension or treatment with the hypoxia-mimetic dimethyloxalyl glycine (DMOG) stabilized HIF-1α and up-regulated CTGF in human umbilical vein endothelial cells and in a murine microvascular endothelial cell line. Induction of CTGF correlated with a HIF-dependent increase in protein and mRNA levels, and nuclear accumulation of the transcription factor FoxO3a. By contrast, gene expression and cellular localization of FoxO1 were not significantly altered by hypoxia. Expression of CTGF was strongly reduced by siRNA silencing of FoxO1 or FoxO3a. Furthermore, nuclear exclusion of FoxO1/3a transcription factors by inhibition of serine/threonine protein phosphatases by okadaic acid inhibited CTGF expression, providing evidence for both FoxO proteins as regulators of CTGF expression. The DMOG-stimulated induction of CTGF was further increased when endothelial cells were co-incubated with transforming growth factor-β, an activator of Smad signaling. Activation of RhoA-Rho kinase signaling by the microtubule-disrupting drug combretastatin A4 also enhanced the DMOG-induced CTGF expression, thus placing CTGF induction by hypoxia in a network of interacting signaling pathways. Our findings provide evidence that FoxO1, hypoxia-stimulated expression of FoxO3a and its nuclear accumulation are required for the induction of CTGF by hypoxia in endothelial cells.
Keywords: G Proteins/Low Molecular Weight, Growth Factors, Oxygen/Hypoxia, Phosphorylation/Phosphatases/Serine-Threonine, Signal Transduction, Cells/Endothelial
Introduction
Connective tissue growth factor (CTGF, CCN2)3 belongs to a group of secreted proteins termed matricellular proteins (1). These proteins are secreted and sequestered in the extracellular matrix, where they interact with cell surface receptors such as integrins, and with growth factors, proteases, cytokines, or extracellular matrix proteins. In endothelial cells, CTGF has been characterized as a modulator of adhesion, migration, and growth (2, 3). The functional outcome of CTGF activation or inhibition seems to depend largely on the microenvironment, i.e. the presence of other growth factors. Gene expression of CTGF is modulated by the activation of multiple signaling pathways, some of which are independent of the cell type, such as transforming growth factor (TGF)-β-Smad signaling, or are shared by several stimuli such as activation of RhoA Rho-kinase signaling (3, 4). Other signaling pathways seem to activate CTGF expression in specific cell types. An example is Rac-1 activation, which has only been related to CTGF expression in gingival fibroblasts and chondrocytes (5, 6).
In recent studies we have shown that the expression of CTGF is modulated by proteins of the forkhead family of transcription factors FoxO (forkhead family of transcription factors group O) in endothelial cells but not in epithelial cells (7, 8). FoxO1, FoxO3a, and FoxO4 have been implicated in the regulation of endothelial cell biology. All three FoxO proteins regulate gene expression in endothelial cells, showing overlapping as well as isoform-specific actions (9). Activation of phosphatidylinositol 3-kinase (PI-3K)/AKT signaling leads to phosphorylation of FoxO proteins and thus their exclusion from the nucleus. In endothelial cells, inhibition of PI-3K/AKT signaling led to up-regulation of CTGF expression (8). Moreover, knockdown of FoxO1 and FoxO3a by siRNA almost completely inhibited the induction of CTGF, providing evidence for a role of FoxOs in CTGF regulation (7). In accordance with our results, microarray data showed that CTGF was down-regulated upon overexpression of constitutively active AKT, whereas active FoxO1 up-regulated CTGF in HUVEC (10). Furthermore, we observed a role for FoxO transcription factors in CTGF expression induced by TGF-β, alterations of the cytoskeleton, or inhibition of histone deacetylases (7, 8). These data suggested a central role for FoxO transcription factors in endothelial CTGF regulation.
Hypoxia is a driving force in neovascularization in physiology and disease. Accumulation of hypoxia-inducible factor (HIF) leads to the induction of multiple proteins that modulate different aspects of angiogenesis, among them vascular endothelial growth factor (for a review see Ref. 11). Vascular endothelial growth factor has been reported to promote survival and growth of endothelial cells via inhibition of FoxO family members through the PI-3K/AKT signaling system (12). Thus far, interaction between FoxO3a and HIF has been analyzed in nonendothelial cells. A direct protein-protein interaction was described between HIF-1α, p300, and FoxO3a in mouse embryonic fibroblasts, leading to a reduction of HIF-1α transcriptional activity (13). Functional interference of FoxO3a with HIF-1α-induced apoptosis was detected in fibroblasts and breast cancer cells (14). In these cells, HIF-1α-dependent up-regulation of FoxO3a led to transcription of CITED2, a functional inhibitor of HIF-1α. The function of FoxO proteins has been shown to be highly context-specific and may differ even in endothelial cells obtained from different vascular beds (15). Therefore, the interaction between HIF-1α and FoxO proteins in endothelial cells needs further investigation.
The signaling pathways by which hypoxic conditions regulate CTGF expression are far from being understood. Hypoxic conditions were related to up-regulation of CTGF in a variety of tumor cells, among them human breast cancer cells (16, 17) or chondrosarcoma cells (18). These results were in contrast to other reports that showed hypoxia-mediated down-regulation of CTGF in human renal tubular cells (19, 20) or a lack of change in the human breast cancer cell line MCF7 (21). In mouse renal epithelial cells, HIF-1α was described as transcription factor directly activating CTGF transcription (22), whereas an increase of CTGF mRNA stability by factors binding to the 3′-untranslated region was observed in a chondrosarcoma cell line (18). Hypoxic regulation of CTGF in endothelial cells has not yet been investigated.
Therefore, this study was set up to analyze the regulation of CTGF by hypoxia in endothelial cells. The cells were exposed to low oxygen tension or the hypoxia mimetic dimethyloxalyl glycine (DMOG), which leads to the accumulation of active HIF-1α by inhibition of 2-oxoglutarate-dependent dioxygenases including HIF prolyl hydroxylases and the asparagyl hydroxylase FIH-1 (factor inhibiting HIF-1) (23). Given the links between HIF-1α and FoxO transcription factors, we hypothesized that FoxOs might be involved in the hypoxic regulation of CTGF in endothelial cells.
EXPERIMENTAL PROCEDURES
Materials
Cell culture materials were purchased from PAA Laboratories (Pasching, Austria); fetal calf serum was from PAN Biotech (Aidenbach, Germany). DMOG was obtained from Cayman Chemical (Ann Arbor, MI), LY294002 was from Biomol (Hamburg, Germany), Y27632 and UO126 were from Calbiochem (Merck, Darmstadt, Germany), and TGF-β was from Tebu (Frankfurt, Germany). Combretastatin A4 phosphate (CA-4P) was kindly provided by OxiGENE (Waltham, MA).
Cell Culture
The murine glomerular microvascular endothelial cell line (glEND.2) was kindly provided by R. Hallmann (Muenster, Germany). The cells were characterized by positive staining for endothelial cell markers MECA-32 and CD-31 and the lack of staining for mesangial cell markers such as α-smooth muscle actin and α8-integrin, as well as epithelial cell markers such as WT-1 and cytokeratin (24). The cells were cultured as described (25). HUVECs were isolated from freshly delivered umbilical cords and grown on 0.1% gelatin-coated dishes as described (25). In all of the experiments, HUVEC passages 2–4 were used.
Western Blot Analysis
The cells were lysed in buffer containing 50 mm HEPES (pH 7.4), 150 mm NaCl, 1% Triton X-100, 100 mm EDTA, and 10% glycerol for the detection of CTGF or FoxO proteins. To detect HIF-1α, the cells were lysed using 6.65 m urea, 10% glycerol, 10% SDS, 1 m Tris-HCl (pH 6.7), 5 mm dithiothreitol, and protease inhibitors (Complete EDTA-free; Roche Applied Science).
For preparation of nuclear extracts, the cells were washed twice with cold PBS and resuspended in 10 mm HEPES (pH 7.9), 0.1 mm EDTA, 10 mm KCl, 1 mm dithiothreitol, 0.7% Nonidet P-40, and protease inhibitor mixture. After incubation on ice for 10 min, the nuclei were pelleted by centrifugation (8000 rpm, 30 s), washed, and finally resuspended in 50 mm HEPES (pH 7.9), 10% glycerol, 0.3 m NaCl, 50 mm KCl, 0.1 mm EDTA, 1 mm dithiothreitol, and protease inhibitors. After incubation on ice for 20 min, the nuclear extracts were centrifuged at 13,000 rpm for 10 min, and the supernatant was used for Western blot.
The proteins were separated by 10% or 12% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (CTGF, Macherey-Nagel, Dueren, Germany; and HIF-1α, Bio-Rad) or nitrocellulose membranes (FoxO, Protran nitrocellulose membrane; Whatman, Maidstone, UK).
The following antibodies were used: rabbit polyclonal anti-HIF-1α (NB100–449; Novus Biologicals, Littleton, CO), rabbit polyclonal anti-FoxO1 (antibody 9462), rabbit monoclonal anti-FoxO1 (antibody 2880), rabbit polyclonal anti-FoxO3a (antibody 2497), rabbit polyclonal anti-pFoxO1(Thr24)/pFoxO3a(Thr32) (antibody 9464), mouse polyclonal anti-phospho-AKT (antibody 9271), and rabbit polyclonal anti-lamin A/C (antibody 2032) from Cell Signaling (Danvers, MA). Rabbit polyclonal anti-AKT (SC-8312), goat polyclonal anti-CTGF (SC-14939), rabbit polyclonal anti-vinculin (SC-5573), and donkey anti-goat IgG (SC-2020) conjugated to horseradish-peroxidase were from Santa Cruz Biotechnology (Heidelberg, Germany). Mouse monoclonal anti-tubulin antibody (T0198; Sigma), peroxidase-conjugated sheep anti-mouse IgG, and donkey anti-rabbit IgG secondary antibodies (Amersham Biosciences).
Immunoreactive proteins were visualized by the enhanced chemiluminescence detection system (ECL Plus; Amersham Biosciences). Immunoreactive bands were quantified using the luminescent image analyzer LAS-1000 (Fujifilm, Berlin, Germany) and AIDA 4.15 image analyzer software (Raytest, Berlin, Germany). To correct for equal loading and blotting, all of the blots were redetected with antibodies directed against tubulin, vinculin, or lamin A/C. For quantification purposes, the ratio of the specific protein band and a control protein was calculated.
RNA Isolation and Real Time Reverse Transcription-PCR
Total RNA was prepared using TriFastTM reagent from Peqlab (Erlangen, Germany). 200 ng of RNA was reverse transcribed and amplified with TaqMan reverse transcription reagents according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). The following primers were used: murine CTGF forward 5′-GCC CTA GCT GCC TAC CGA CT, reverse 5′-CAT AGT TGG GTC TGG GCC AA; 18 S rRNA forward 5′-TTG ATT AAG TCC CTG CCC TTT GT, reverse 5′-CGA TCC GAG GGC CTC ACT A; murine FoxO1 forward 5′-TAC TTC AAG GAT AAG GGC GAC AGC, reverse 5′-TTC ATT CTG CAC TCG AAT AAA CTT GC; and murine FoxO3a forward 5′-CAA AGC AGA CCC TCA AAC TGA CG, reverse 5′-CAA AGG TGT CAA GCT GTA AAC GG. Quantification of mRNA expression was carried out with respect to 18 S rRNA as a reference as described by PerkinElmer Life Sciences.
siRNA Transfection
To down-regulate HIF-1α, FoxO1, or FoxO3a expression, endothelial cells (glEND.2) were transfected with HIF-1α siRNA (sense 5′-GCC ACU UCG AAG UAG UGC U), FoxO1 siRNA (sense 5′-GCG GGC UGG AAG AAU UCA A), FoxO3a siRNA (sense 5′-GCU CUU GGU GGA UCA UCA A) or luciferase siRNA (Eurogentec, Seraing, Belgium) 3 h after seeding, using HiPerFect (Qiagen) according to the manufacturer's instructions. The experiments were done 24 h after transfection.
Flow System
HUVEC were seeded in flow-through cell culture slides (Ibidi®, Munich, Germany) and grown until confluence. Using a programmed peristaltic pump (Ismatec), the cell monolayer inside the slide channel was exposed to steady laminar shear stress for 18 h at 10 dyne/cm2 (26). Subsequently, the cells were treated with DMOG for 6 h at flow. Protein expression was detected by indirect immunocytochemistry as described below.
Immunocytochemistry
HUVEC were fixed with 3.5% paraformaldehyde in Dulbecco's PBS (140 mm NaCl, 2.68 mm KCl, 1.47 mm KH2PO4, and 8.1 mm Na2HPO4) for 10 min and afterward permeabilized by Triton X-100 in Dulbecco's PBS for 10 min. The cells were incubated with primary antibodies against CTGF (1:100 in PBS) for 1 h and with secondary antibody anti-goat IgG coupled to Alexa Fluor 488 (1:500; Molecular Probes) for 45 min. The images were obtained using an inverted fluorescence microscope combined with a digital camera and Meta-Morph software. For quantification, MetaVue software was used as described in detail in Ref. 26.
Statistics
The data are presented as the means ± S.D. To compare multiple measurements, analysis of variance with Tukey Kramer multiple comparison test or Dunnett post hoc test was used. The paired Student t test was used to compare two conditions. A p value < 0.05 was considered significant.
RESULTS
Up-regulation of CTGF by Hypoxia in Endothelial Cells
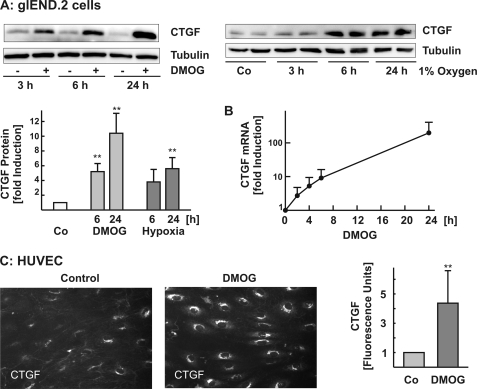
In a first set of experiments, we studied the regulation of CTGF by hypoxia in mouse and human endothelial cells. The cells were exposed to low oxygen tension (1% oxygen) or treated with DMOG (1 mm). Both incubation of glEND.2 cells with DMOG and culture of the cells under hypoxic conditions induced CTGF expression (Fig. 1A). Up-regulation of CTGF was detectable 3 h after incubation with DMOG and was consistently observed after 6 and 24 h of exposure to DMOG or hypoxia (graph in Fig. 1A). Up-regulation of CTGF was confirmed at the mRNA level (Fig. 1B).
FIGURE 1.
Up-regulation of CTGF in response to hypoxia. A, glEND.2 cells were treated with DMOG (1 mm) or exposed to hypoxia (1% oxygen) for the times indicated. The protein levels of CTGF were detected in cellular homogenates by Western blotting. To compare different experiments, CTGF expression in control cells was set to 1. The data presented in the graph are the means ± S.D. of three to six experiments. **, p < 0.01 compared with control cells. B, after incubation of glEND.2 cells with DMOG (1 mm) for the indicated times, mRNA levels of CTGF were analyzed by real time reverse transcription-PCR. The graph summarizes the data of two experiments. C, HUVEC were seeded in flow-through cell culture slides and exposed to steady laminar shear stress at 10 dyne/cm2 for 24 h. For the last 6 h of flow, the cells were treated with DMOG (1 mm). Protein expression was determined by immunofluorescence. The photos are representative of four independent experiments performed in duplicate. The graph summarizes the data of four experiments. Data quantification: six or seven images at objective magnification 20× were taken for every experiment and analyzed using MetaVue software. The thresholded protein expression levels were expressed as arbitrary fluorescence units. **, p < 0.01, t test versus untreated controls.
In contrast to glEND.2, CTGF is highly expressed in HUVEC cultured under static conditions (26). Therefore, to reduce base-line CTGF levels, HUVEC were exposed to laminar flow conditions for 18 h in flow-through slides and then stimulated with DMOG for 6 h under continuous flow. Detection of CTGF by immunocytochemistry showed very low staining in control flow-exposed HUVEC, whereas treatment with DMOG strongly up-regulated CTGF expression (Fig. 1C). Having established the differences in the basic conditions to up-regulate CTGF by hypoxia in human and mouse endothelial cells, all further analyses were performed with the mouse microvascular cells, which allowed analysis of CTGF expression under static conditions.
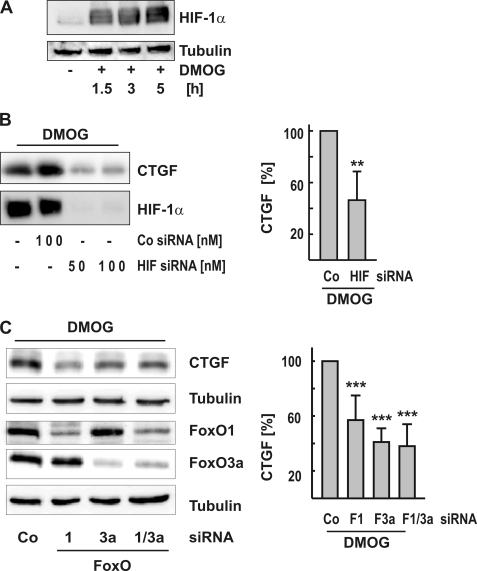
HIF-1α rapidly accumulated in DMOG-treated glEND.2 cells (Fig. 2A). DMOG-mediated up-regulation of HIF-1α was reduced by more than 90%, when glEND.2 cells were transiently transfected with HIF-1α siRNA (Fig. 2B). Concomitantly, DMOG-mediated expression of CTGF was significantly decreased, indicative of a role for HIF-1α in DMOG-stimulated induction of CTGF in endothelial cells (Fig. 2B).
FIGURE 2.
HIF-1α- and FoxO1/3a-dependent up-regulation of CTGF. A, glEND.2 cells were treated with DMOG (1 mm) for the times indicated. HIF-1α was detected by Western blot analysis in cellular homogenates. B, glEND.2 cells were preincubated with siRNA directed against HIF-1α (HIF-1α siRNA; 50 or 100 nm) or siRNA against luciferase (Co siRNA; 100 nm) overnight and then stimulated with DMOG for 6 h. The graph summarizes the results of five experiments. CTGF expression in cells treated with siRNA against luciferase was set to 100%. **, p < 0.01, paired Student t test using the original data. C, glEND.2 cells were pretreated with FoxO siRNA (50 nm FoxO1 and/or 50 nm FoxO3a siRNA) or siRNA against luciferase (Co) overnight. Then the cells were treated with DMOG (1 mm) for 6 h. The graph summarizes the data of eleven (FoxO1/3a) and four (FoxO1 or FoxO3a) experiments. CTGF expression in cells treated with siRNA against luciferase was set to 100%. ***, p < 0.001 compared with control siRNA-treated cells.
In recent studies, we provided evidence that CTGF is regulated by FoxO proteins in endothelial cells (8, 7, 27). Therefore, to investigate whether FoxOs were also relevant for the hypoxic induction of CTGF, FoxO1 and FoxO3a were silenced by small interfering RNA as described (7). DMOG-induced expression of CTGF was significantly reduced upon silencing of FoxO1 and FoxO3a by 40 and 60%, respectively (Fig. 2C). This suggested that both FoxO1 and FoxO3a played a role in CTGF expression in the hypoxic response in endothelial cells.
Regulation of FoxO Expression and Intracellular Localization by Hypoxia
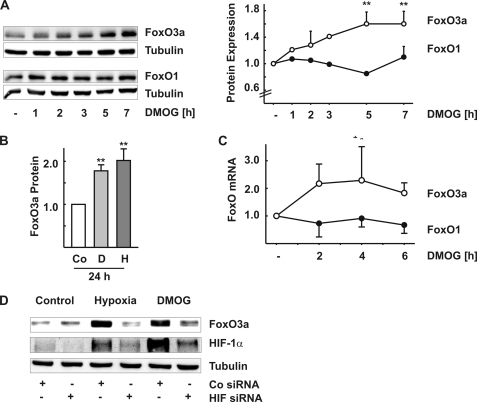
Incubation of glEND.2 cells with DMOG led to a time-dependent increase of the expression of FoxO3a protein, whereas FoxO1 levels remained essentially unaltered (Fig. 3A). After 24 h, up-regulation of FoxO3a was ∼2-fold when the cells were exposed to DMOG (D) or to low oxygen tension (Fig. 3B, column H, 1% oxygen), whereas FoxO1 expression was not increased (data not shown). The increased levels of FoxO3a mRNA in DMOG-treated cells indicated that the regulation occurred at the transcriptional level or by mRNA stabilization (Fig. 3C). Moreover, up-regulation of FoxO3a was dependent on HIF-1α stabilization, because interference with HIF-1α, using siRNA, using inhibited hypoxia as well as DMOG mediated FoxO3a induction (Fig. 3D).
FIGURE 3.
Regulation of FoxO1 and FoxO3a expression by DMOG and hypoxia. A, glEND.2 were stimulated with DMOG (1 mm) for the times indicated. FoxO protein was detected in cellular homogenates by Western blotting. The data are the means ± S.D. of three independent experiments. **, p < 0.01 compared with control cells. B, FoxO3a protein was analyzed by Western blotting in glEND.2 cells stimulated with DMOG (column D, 1 mm) or exposed to hypoxia (column H, 1% oxygen) for 24 h. **, p < 0.01 compared with control (Co) cells. C, FoxO1 and FoxO3a mRNA were analyzed by real time reverse transcription-PCR in cells treated with DMOG (1 mm) for the times indicated. The graph summarizes the data of at least four independent experiments. *, p < 0.05 compared with control cells. D, glEND.2 cells were preincubated with siRNA directed against HIF-1α (HIF siRNA, 100 nm) or directed against luciferase (Co siRNA; 100 nm) overnight and then treated with DMOG or exposed to hypoxia for 24 h. FoxO3a and HIF-1α were detected by Western blot analysis. The blot is representative of two experiments.
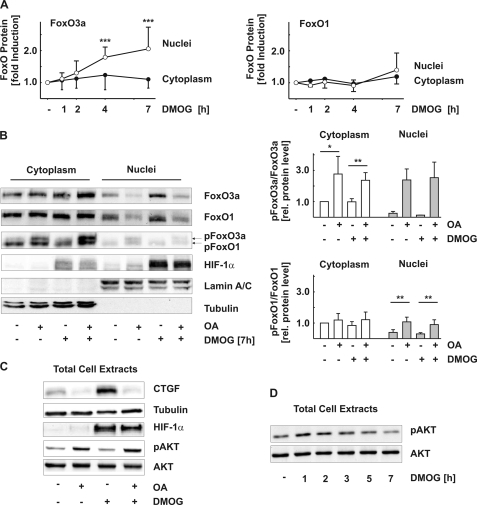
To be active as transcription factor, FoxO proteins need to be localized to the nucleus. In resting cells, FoxO3a was located primarily in the cytosol with only ∼10% of the protein detectable in the nuclear fraction. Upon treatment with DMOG, FoxO3a was significantly increased in the nuclear compartment with hardly any change detectable in the cytosolic fraction (Fig. 4, A and B). By contrast, the distribution of FoxO1 between cytosol and nucleus was not significantly changed during the first 7 h (Fig. 4, A and B) and remained essentially unaltered even after 24 h of exposure to DMOG (data not shown).
FIGURE 4.
Intracellular localization of FoxO1 and FoxO3a under hypoxic conditions. A, glEND.2 cells were incubated with DMOG (1 mm) for the times indicated. FoxO1 and FoxO3a were detected in nuclei and cytoplasm by Western blot analysis. The graphs summarize the data of six experiments (means ± S.D.). ***, p < 0.001 compared with control cells. B, glEND.2 cells were treated with okadaic acid (OA, 50 nm) and/or DMOG (1 mm) for 7 h. 10% of cytosolic protein and 100% of nuclear protein were loaded on the gel. FoxO1, FoxO3a, pFoxO1 (Thr24), pFoxO3a (Thr32), and HIF-1α were detected in cytosolic and nuclear extracts. Lamin A/C and tubulin were used as controls. The blot is representative of four experiments. The graphs summarize the data of four (pFoxO1/FoxO1), three (cytosolic pFoxO3a/FoxO3a), and two (nuclear pFoxO3a/FoxO3a) independent experiments. *, p < 0.05, **, p < 0.01, paired Student t test using the original data. C, glEND.2 cells were treated as in B. CTGF, HIF-1α, pAKT, and AKT were detected in total cellular extracts, and tubulin was used as a control. D, glEND.2 cells were incubated with DMOG (1 mm) for the times indicated. pAKT and AKT were detected in total cell extracts by Western blot analysis. The blot is representative of two experiments.
Nuclear localization of FoxO proteins is regulated by different protein modifications including acetylation and phosphorylation. Inhibition of the calcium- and calmodulin-dependent phosphatase PP2B by cyclosporine A or inhibition of tyrosine phosphatases by vanadate did not affect FoxO3a expression or localization (data not shown). In contrast, the concentration of okadaic acid (50 nm), which primarily inhibits serine/threonine phosphatases PP2A and PP5 caused a significant increase of FoxO3a phosphorylation (Fig. 4B). Phosphorylated FoxO3a was not detectable or was barely detectable in the nuclear fraction confirming a critical role of phosphorylation in the regulation of FoxO3a intracellular localization. In the absence of okadaic acid, the levels of nuclear phospho-FoxO3a were so low that quantification was not possible in all of the experiments. Phosphorylated FoxO3a was excluded from the nucleus as shown by a significant reduction of nuclear FoxO3a protein upon treatment with okadaic acid. DMOG-mediated nuclear localization of FoxO3a was reduced by 64 ± 15% (n = 8; p < 0.001). FoxO1 protein was detectable in the nucleus under normoxic and hypoxic conditions. Treatment with okadaic acid also excluded FoxO1 from the nucleus, reducing nuclear FoxO1 by 60 ± 13% (n = 5; p < 0.001; Fig. 4B), whereas stability and nuclear localization of HIF-1α were not affected (Fig. 4, B and C). There was a correlation between FoxO nuclear accumulation and CTGF expression. Nuclear exclusion of FoxO proteins by okadaic acid reduced basal CTGF expression and prevented the increase in CTGF protein levels under hypoxic conditions (Fig. 4C).
Inhibition of PI-3K/AKT signaling, which leads to the dephosphorylation of FoxO proteins, is a strong stimulus for nuclear localization of FoxO proteins. Furthermore, AKT is a target of okadaic acid-sensitive phosphatases, and the phosphorylation of AKT was increased in the presence of okadaic acid (Fig. 4C). Therefore, we analyzed whether the nuclear localization of FoxO3a upon treatment with DMOG was due to an inhibition of AKT by DMOG. During the first 2 h of incubation with DMOG, there was a small increase in AKT phosphorylation, which was no longer observed at later time points (Fig. 4D). Therefore, modulation of AKT activity by DMOG did not contribute to the retention of FoxO3a in the nucleus.
Modulation of DMOG-induced CTGF Expression by Kinase Pathways
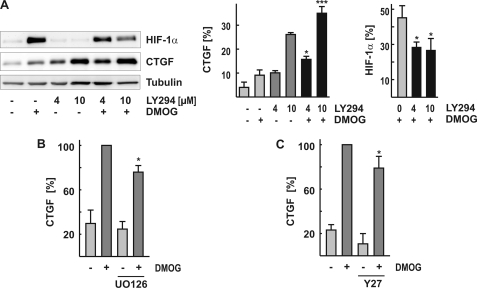
To get an insight into signaling pathways involved in DMOG-mediated CTGF up-regulation, several kinases implicated in CTGF induction (4) were inhibited pharmacologically. We previously observed FoxO-dependent up-regulation of CTGF expression when PI-3K/AKT signaling was inhibited (27). In the present study, co-incubation of glEND.2 cells with LY294002 and DMOG further increased CTGF expression, as compared with the induction obtained by the individual treatments (Fig. 5A). At the same time, inhibition of PI-3K/AKT signaling reduced the accumulation of HIF-1α (Fig. 5A, right panel) in line with data reported by Jiang et al. (28) in carcinoma cells. The remaining levels of HIF-1α correlated with the cooperative induction of CTGF by DMOG and the PI-3K inhibitor LY294002.
FIGURE 5.
Involvement of PI-3K/AKT, MAPK, and ROCK signaling in CTGF expression. A, glEND.2 cells were co-incubated with the PI-3K inhibitor LY294002 (4 or 10 μm) and DMOG (1 mm) for 3 h. HIF-1α and CTGF were analyzed in cellular homogenates by Western blot analysis. The graph summarizes the results of three experiments. *, p < 0.05; ***, p < 0.001 CTGF expression in cells stimulated by LY294002 and DMOG compared with either stimulus alone. B and C, glEND.2 cells were pretreated with UO126 (1 μm, 30 min) and Y27632 (10 μm, 30 min), respectively, and then incubated in the presence or absence of DMOG for 6 h. CTGF expression was detected by Western blot analysis. The graphs summarize the data of three experiments. CTGF expression in cells treated with DMOG alone was set to 100%. *, p < 0.05 inhibitor-treated cells to cells treated with DMOG.
To assess the contribution of mitogen-activated protein kinases (MAPKs) to hypoxia-mediated CTGF expression, we used SB203580 and UO126 to inhibit the activity of p38 and the activation of p42/44 MAPK, respectively. Inhibition of p38 did not alter DMOG-mediated CTGF expression (data not shown), whereas inhibition of p42/44 MAPK activation reduced CTGF expression by ∼20% (Fig. 5B) without affecting either protein levels of HIF-1α and FoxO3a or their nuclear localization (data not shown).
Endothelial CTGF expression is also modulated by alterations of the cytoskeleton. However, pretreatment of glEND.2 cells with the inhibitor of Rho-associated kinases (ROCK) Y27632 (Fig. 5C) only slightly reduced DMOG-induced CTGF expression (20 ± 11%, n = 4, p < 0.05).
Cooperative Induction of CTGF by DMOG, RhoA-ROCK, and TGF-β Signaling
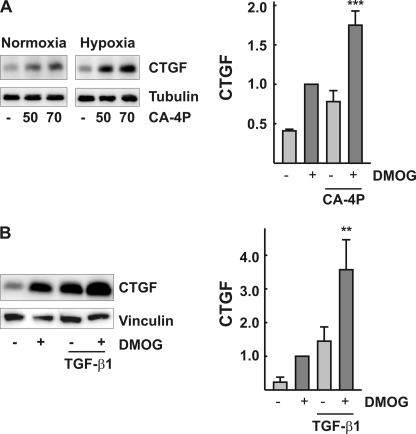
Activation of RhoA-ROCK-serum response factor signaling by the microtubule-disrupting agent combretastatin A4 phosphate led to up-regulation of CTGF in endothelial cells (Fig. 6A). When the cells treated with combretastatin A4 were pre-exposed to hypoxia or co-incubated with DMOG, induction of CTGF was more than additive (Fig. 6A). These data are in line with previous findings showing increased expression of CTGF when the cells are stimulated with combretastatin A4 in the presence of PI-3K inhibitors to activate FoxO-dependent CTGF induction (27).
FIGURE 6.
Cooperative induction of CTGF by hypoxia and RhoA-ROCK, or hypoxia and TGF-β signaling. A, glEND.2 cells were cultured overnight under hypoxic (1% oxygen) or normoxic conditions. The cells were subsequently incubated with combtretastatin A4 (CA-4P; 50 and 70 nm) for 5 h. CTGF was detected by Western blot analysis. The graph summarizes the data of three experiments. glEND.2 cells were stimulated with combretastatin A4 (50 nm) for 5 h in the presence or absence of DMOG (1 mm) as indicated. CTGF expression was detected by Western blot analysis. ***, p < 0.001, compared with cells stimulated with each compound alone. B, glEND.2 cells were incubated with TGF-β1 (5 ng/ml) for 6 h in the presence or absence of DMOG as indicated. CTGF expression was detected by Western blot analysis. The graph summarizes data of three experiments. **, p < 0.01, compared with cells stimulated with either compound alone.
It has been shown in keratinocytes that Smad proteins interact with FoxO transcription factors and may contribute to retention of FoxO proteins in the nucleus (29, 30). Therefore, we analyzed the Smad 2/3 localization in glEND.2 cells. As expected, treatment of the cells with TGF-β led to a rapid translocation of Smad 2/3 into the nucleus. In contrast, there was no alteration of Smad 2/3 localization upon treatment with DMOG alone for up to 7 h (data not shown). Confirming an interaction between Smads and FoxO proteins, we detected a more than additive induction of CTGF in glEND.2 cells treated with DMOG plus TGF-β (Fig. 6B).
DISCUSSION
In this study, we describe molecular mechanisms of hypoxia-mediated induction of CTGF in endothelial cells. Transcription factor HIF-1α and members of FoxO family FoxO1 and FoxO3a were identified to play a central role in hypoxia-inducible CTGF expression. Furthermore, we provide evidence for the differential regulation of FoxO proteins under hypoxic stress, expression and localization of FoxO3a being modulated by HIF-1α.
In human and mouse endothelial cells CTGF was up-regulated, when the cells were exposed to low oxygen tension or treated with the hypoxia mimetic DMOG. Similar to hypoxia, DMOG leads to the stabilization and activation of HIF-1α subunits through the inhibition of the 2-oxoglutarate-dependent dioxygenases. The results of our present study show that DMOG induces CTGF in mouse microvascular endothelial cells in a HIF-1α- and FoxO-dependent manner. Hypoxic stress led to activation of HIF-1α and increased FoxO3a at mRNA and protein levels in glEND.2. Using HIF-1α siRNA, we showed that the observed up-regulation of FoxO3a was indeed mediated by HIF-1α. These data are in line with an earlier report identifying FoxO3a as a direct target of HIF-1α in fibroblasts (14), whereas FoxO1 protein expression was not affected, as also observed in our study.
Activation of HIF-1α not only increased FoxO3a gene expression but also affected its intracellular localization. Both FoxO3a and FoxO1 were localized predominantly in the cytoplasm in resting cells, but only FoxO3a accumulated in the nucleus in DMOG-treated cells. Nuclear-cytoplasmic shuttling of FoxO proteins is a dynamic process, controlled via multiple levels of post-translational modifications. In the nucleus, FoxO proteins control transcriptional programs, whereas in the cytoplasm FoxO proteins become transcriptionally inactive and undergo proteasomal degradation. Phosphorylation of FoxO proteins by kinases such as AKT and serum and glucocorticoid-regulated kinase is generally assumed to be an essential step to keep FoxO proteins in the cytosol by supporting transport to the cytosol and preventing relocation and DNA binding (31). The repression of PI-3K/AKT signaling by LY294002 increased accumulation of FoxO1 and FoxO3a in the nucleus in human and mouse endothelial cells (8, 27). Because serum and glucocorticoid-regulated kinase was barely detectable in glEND.2, we assumed that AKT might be involved in regulation of FoxO3a nuclear accumulation under hypoxic conditions. However, AKT activity was slightly increased rather than decreased by hypoxia, suggesting that other regulatory mechanisms must be responsible for the preferential nuclear localization of FoxO3a.
Serine/threonine-specific protein phosphatases have been reported to be activated by hypoxia and may directly or indirectly modulate FoxO3a activity. As reported by Barreyro et al. (32), FoxO3a was dephosphorylated and thus activated by PP2A in hepatocytes, incubated with free fatty acids. To determine whether phosphatases were involved in hypoxia-mediated FoxO3a nuclear accumulation, we used okadaic acid, a specific inhibitor of PP1, PP2A, and PP5 serine/threonine phosphatases (33–35). Inhibition of protein phosphatases by okadaic acid prevented FoxO3a dephosphorylation and reduced DMOG-mediated FoxO3a nuclear accumulation. The concentration of okadaic acid used in these experiments (50 nm) was effective for PP2A and PP5, although a partial inhibition of PP1 could not be excluded (36). So far, little is known about the regulation of okadaic acid-sensitive phosphatases by hypoxia. PP5 has been shown to be a direct target of HIF-1α in human A549 lung carcinoma cells and Hep3B hepatoma cells (37), whereas PP2A protein activity was reduced rather than increased in neuronal cells under hypoxic conditions (38). Furthermore, the protein phosphatase-1 nuclear targeting subunit has been described as hypoxia-responsive gene (39). In our study, the expression of PP2A and PP5 protein was not altered by hypoxia (data not shown). Given the fact that phosphorylation of AKT, which is also a substrate of okadaic acid-sensitive phosphatases, was not decreased but transiently increased by hypoxia, more subtle changes in phosphatase activities have to be envisaged in terms of hypoxia-mediated regulation of FoxO3a.
It was interesting to note that okadaic acid also reduced FoxO1 nuclear localization under normoxic and hypoxic conditions, underlining the importance of FoxO phosphorylation for localization and thus activity of FoxO transcription factors. The stability and nuclear localization of HIF-1α were not affected by okadaic acid. Nuclear localization of HIF-1α was thus not sufficient to sustain CTGF expression.
Regulation of CTGF by hypoxia seems to be cell type-dependent (summarized in Ref. 3). Although up-regulation of CTGF has been observed, e.g. in some tumor cell lines (40) or trophoblasts (41), our recent studies in renal tubular epithelial cells provided evidence for a down-regulation of CTGF in a HIF-1α-dependent manner (20). These data argue against CTGF as a direct target gene of HIF-1α. The results of the present study argue in favor of an indirect mechanism that involves basal FoxO1, well detectable in nuclear fractions, and hypoxia-regulated FoxO3a proteins. First, the induction of CTGF correlated with hypoxia-induced nuclear accumulation of FoxO3a. Second, it was inhibited when FoxO proteins were excluded from the nucleus upon treatment with okadaic acid, and, third, siRNA-targeted knockdown of FoxO1 and/or FoxO3a suppressed hypoxic induction of CTGF. The CTGF gene contains forkhead-responsive element-binding sequences. Gomis et al. (29) defined several putative FoxO-binding sites in the promoter of CTGF in human keratinocytes, and Paik (42) reported direct binding of FoxO to the promoter of the CTGF gene in human and mouse liver endothelial cells. Whether FoxO-dependent expression of CTGF under hypoxic conditions is mediated via direct FoxO binding to its FoxO-responsive elements or is regulated indirectly via interaction with other transcription factors remains to be investigated. Of interest, FoxO transcription factors are regulated in a highly cell type-specific manner, and their involvement in CTGF regulation by hypoxia may contribute to the cell type-specific regulation of CTGF under hypoxic conditions.
We have shown earlier that FoxO proteins are involved in the regulation of CTGF by histone deacetylases inhibitors or TGF-β (8) and also increase the up-regulation of CTGF by substances that activate RhoA-Rho kinase signaling (7). In those studies, activation of FoxOs was achieved by pharmacological inhibition of PI-3K/AKT signaling, which leads to a strong activation of both FoxO1 and FoxO3a. Therefore, it was of interest to note that activation of RhoA-Rho kinase signaling by combretastatin A4 or activation of Smad signaling by TGF-β increased the endothelial response to hypoxia. These data indicate the existence of functional interactions between signaling pathways that may be relevant in the pathophysiological situation. Combretastatin A4 is currently tested for its ability to reduce tumor angiogenesis, which is often associated with hypoxic conditions in the tumor environment. It may be envisaged that the interaction between FoxO proteins and RhoA-Rho-kinase signaling is not restricted to CTGF but may affect other proteins involved in the anti-angiogenic effect of combretastatin A4, which seems to be rather specific for tumor vessels in vivo (43). Another interaction between Smad and FoxOs has been analyzed in detail in keratinocytes and, based on our data, also seems to be operative in endothelial cells. Further studies are necessary to define the set of proteins regulated by FoxO proteins and Smads in the context of hypoxia.
Taken together, our data provide evidence for an essential role for FoxO proteins in hypoxia-induced up-regulation of CTGF in endothelial cells, which may contribute to the cell type specificity of hypoxic CTGF regulation. Furthermore, hypoxia-induced CTGF expression is enhanced when RhoA or Smad signaling pathways are activated by mechanical stress or growth factors. These interactions may contribute to the increased endothelial CTGF expression observed under pathophysiological conditions.
Acknowledgment
The expert technical assistance of M. Rehm is highly acknowledged.
This work was supported by Project D5 from the Interdisciplinary Center for Clinical Research at the University Hospital of the University of Erlangen-Nuremberg.
- CTGF
- connective tissue growth factor (CCN2)
- DMOG
- dimethyloxalyl glycine
- HIF
- hypoxia-inducible factor
- HUVEC
- human umbilical vein endothelial cells
- PI-3K
- phosphatidylinositol 3-kinase
- ROCK
- Rho-associated kinase(s)
- TGF
- transforming growth factor
- siRNA
- small interfering RNA
- PBS
- phosphate-buffered saline
- PP
- protein phosphatase
- MAPK
- mitogen-activated protein kinase.
REFERENCES
- 1.Bornstein P., Sage E. H. (2002) Curr. Opin. Cell Biol. 14, 608–616 [DOI] [PubMed] [Google Scholar]
- 2.Brigstock D. R. (2002) Angiogenesis 5, 153–165 [DOI] [PubMed] [Google Scholar]
- 3.Cicha I., Goppelt-Struebe M. (2009) BioFactors 35, 200–208 [DOI] [PubMed] [Google Scholar]
- 4.Chaqour B., Goppelt-Struebe M. (2006) FEBS J. 273, 3639–3649 [DOI] [PubMed] [Google Scholar]
- 5.Black S. A., Jr., Trackman P. C. (2008) J. Biol. Chem. 283, 10835–10847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods A., Pala D., Kennedy L., McLean S., Rockel J. S., Wang G., Leask A., Beier F. (2009) Osteoarthritis Cartilage 17, 406–413 [DOI] [PubMed] [Google Scholar]
- 7.Samarin J., Rehm M., Krueger B., Waschke J., Goppelt-Struebe M. (2009) Mol Cancer Res. 7, 180–188 [DOI] [PubMed] [Google Scholar]
- 8.Komorowsky C., Ocker M., Goppelt-Struebe M. (2009) J. Cell Mol. Med. 13, 2353–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papanicolaou K. N., Izumiya Y., Walsh K. (2008) Circ. Res. 102, 16–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly C., Wong V., Burova E., Wei Y., Zabski S., Griffiths J., Lai K. M., Lin H. C., Ioffe E., Yancopoulos G. D., Rudge J. S. (2004) Genes Dev. 18, 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza G. L. (2007) Sci. STKE. 2007, cm8. [DOI] [PubMed] [Google Scholar]
- 12.Abid M. R., Guo S., Minami T., Spokes K. C., Ueki K., Skurk C., Walsh K., Aird W. C. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 294–300 [DOI] [PubMed] [Google Scholar]
- 13.Emerling B. M., Weinberg F., Liu J. L., Mak T. W., Chandel N. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2622–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakker W. J., Harris I. S., Mak T. W. (2007) Mol. Cell 28, 941–953 [DOI] [PubMed] [Google Scholar]
- 15.Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., Jiang S., Gilliland D. G., Chin L., Wong W. H., Castrillon D. H., DePinho R. A. (2007) Cell 128, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimo T., Kubota S., Kondo S., Nakanishi T., Sasaki A., Mese H., Matsumura T., Takigawa M. (2001) Cancer Lett. 174, 57–64 [DOI] [PubMed] [Google Scholar]
- 17.Kondo S., Kubota S., Shimo T., Nishida T., Yosimichi G., Eguchi T., Sugahara T., Takigawa M. (2002) Carcinogenesis 23, 769–776 [DOI] [PubMed] [Google Scholar]
- 18.Kondo S., Kubota S., Mukudai Y., Moritani N., Nishida T., Matsushita H., Matsumoto S., Sugahara T., Takigawa M. (2006) Oncogene 25, 1099–1110 [DOI] [PubMed] [Google Scholar]
- 19.Chi J. T., Wang Z., Nuyten D. S., Rodriguez E. H., Schaner M. E., Salim A., Wang Y., Kristensen G. B., Helland A., Børresen-Dale A. L., Giaccia A., Longaker M. T., Hastie T., Yang G. P., van de Vijver M. J., Brown P. O. (2006) PLoS. Med. 3, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroening S., Neubauer E., Wessel J., Wiesener M., Goppelt-Struebe M. (2009) Nephrol. Dial. Transplant 24, 3319–3325 [DOI] [PubMed] [Google Scholar]
- 21.Elvidge G. P., Glenny L., Appelhoff R. J., Ratcliffe P. J., Ragoussis J., Gleadle J. M. (2006) J. Biol. Chem. 281, 15215–15226 [DOI] [PubMed] [Google Scholar]
- 22.Higgins D. F., Biju M. P., Akai Y., Wutz A., Johnson R. S., Haase V. H. (2004) Am. J. Physiol. Renal Physiol. 287, F1223–F1232 [DOI] [PubMed] [Google Scholar]
- 23.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 24.Li Z. D., Bork J. P., Krueger B., Patsenker E., Schulze-Krebs A., Hahn E. G., Schuppan D. (2005) Biochem. Biophys. Res. Commun. 334, 1049–1060 [DOI] [PubMed] [Google Scholar]
- 25.Muehlich S., Cicha I., Garlichs C. D., Krueger B., Posern G., Goppelt-Struebe M. (2007) Am. J. Physiol. Cell Physiol. 292, C1732–C1738 [DOI] [PubMed] [Google Scholar]
- 26.Cicha I., Goppelt-Struebe M., Muehlich S., Yilmaz A., Raaz D., Daniel W. G., Garlichs C. D. (2008) Atherosclerosis 196, 136–145 [DOI] [PubMed] [Google Scholar]
- 27.Samarin J., Cicha I., Goppelt-Struebe M. (2009) J. Cell Commun. Signal. 3, 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang B. H., Jiang G., Zheng J. Z., Lu Z., Hunter T., Vogt P. K. (2001) Cell Growth & Differ. 12, 363–369 [PubMed] [Google Scholar]
- 29.Gomis R. R., Alarcón C., He W., Wang Q., Seoane J., Lash A., Massagué J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seoane J., Le H. V., Shen L., Anderson S. A., Massagué J. (2004) Cell 117, 211–223 [DOI] [PubMed] [Google Scholar]
- 31.Van Der Heide L. P., Hoekman M. F., Smidt M. P. (2004) Biochem. J. 380, 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barreyro F. J., Kobayashi S., Bronk S. F., Werneburg N. W., Malhi H., Gores G. J. (2007) J. Biol. Chem. 282, 27141–27154 [DOI] [PubMed] [Google Scholar]
- 33.Bialojan C., Takai A. (1988) Biochem. J. 256, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen M. X., McPartlin A. E., Brown L., Chen Y. H., Barker H. M., Cohen P. T. (1994) EMBO J. 13, 4278–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawson J. F., Holmes C. F. (1999) Front. Biosci. 4, D646–D658 [DOI] [PubMed] [Google Scholar]
- 36.Li D. W., Liu J. P., Schmid P. C., Schlosser R., Feng H., Liu W. B., Yan Q., Gong L., Sun S. M., Deng M., Liu Y. (2006) Oncogene 25, 3006–3022 [DOI] [PubMed] [Google Scholar]
- 37.Zhou G., Golden T., Aragon I. V., Honkanen R. E. (2004) J. Biol. Chem. 279, 46595–46605 [DOI] [PubMed] [Google Scholar]
- 38.Truttmann A. C., Ashraf Q., Mishra O. P., Delivoria-Papadopoulos M. (2004) Neuroscience 127, 355–363 [DOI] [PubMed] [Google Scholar]
- 39.Lee S. J., Lim C. J., Min J. K., Lee J. K., Kim Y. M., Lee J. Y., Won M. H., Kwon Y. G. (2007) Cell Death Differ. 14, 1106–1116 [DOI] [PubMed] [Google Scholar]
- 40.Chintalapudi M. R., Markiewicz M., Kose N., Dammai V., Champion K. J., Hoda R. S., Trojanowska M., Hsu T. (2008) Carcinogenesis 29, 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rimon E., Chen B., Shanks A. L., Nelson D. M., Sadovsky Y. (2008) Endocrinology 149, 2952–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paik J. H. (2006) Biochem. Soc. Trans. 34, 731–734 [DOI] [PubMed] [Google Scholar]
- 43.Dark G. G., Hill S. A., Prise V. E., Tozer G. M., Pettit G. R., Chaplin D. J. (1997) Cancer Res. 57, 1829–1834 [PubMed] [Google Scholar]