Abstract
Strongly dominant negative mutant actins, identified by An and Mogami (An, H. S., and Mogami, K. (1996) J. Mol. Biol. 260, 492–505), in the indirect flight muscle of Drosophila impaired its flight, even when three copies of the wild-type gene were present. Understanding how these strongly dominant negative mutant actins disrupt the function of wild-type actin would provide useful information about the molecular mechanism by which actin functions in vivo. Here, we expressed and purified six of these strongly dominant negative mutant actins in Dictyostelium and classified them into three groups based on their biochemical phenotypes. The first group, G156D, G156S, and G268D actins, showed impaired polymerization and a tendency to aggregate under conditions favoring polymerization. G63D actin of the second group was also unable to polymerize but, unlike those in the first group, remained soluble under polymerizing conditions. Kinetic analyses using G63D actin or G63D actin·gelsolin complexes suggested that the pointed end surface is defective, which would alter the polymerization kinetics of wild-type actin when mixed and could affect formation of thin filament structures in indirect flight muscle. The third group, R95C and E226K actins, was normal in terms of polymerization, but their motility on heavy meromyosin surfaces in the presence of tropomyosin-troponin indicated altered sensitivity to Ca2+. Cofilaments in which R95C or E226K actins were copolymerized with a 3-fold excess of wild-type actin also showed altered Ca2+ sensitivity in the presence of tropomyosin-troponin.
Keywords: Actin, ATPases, Calcium-binding Proteins, Cytoskeleton, Mutant, Skeletal Muscle, Protein Expression
Introduction
Actin filaments are major components of the cytoskeletons of all eukaryotic cells and play key roles in a variety of cellular functions, including cell migration, organelle transport, and muscle contraction. These diverse processes depend on the dynamic nature of actin filaments and their interactions with various actin-binding proteins (1), which are thought to involve conformational changes in the subunits that make up actin filaments. For instance, the stability of actin filaments is affected by the conformational difference between actin subunits carrying ATP or ADP and those that do not (2, 3), whereas cofilin, a major actin filament severing protein, alters the twist of actin filaments (4). In addition, modification of actin filaments through chemical cross-linking causes motility defects with myosin, suggesting a possible link between actin-myosin interaction and the conformational dynamics of actin (5, 6). Moreover, the myosin-induced conformational changes appear to be cooperative (7–9), e.g. Ca2+-triggered conformational changes in the skeletal muscle actin filaments are enhanced by cooperative conformational changes induced by myosin (10). In many cases, however, details of the molecular relationship between conformational changes in actin and its physiological function remain unclear.
Actin is a highly conserved protein, so that Dictyostelium actin 15 shares 91% amino acid identity with rabbit skeletal muscle actin. This implies that the majority of amino acid residues in actin are essential for its function. A mutational approach is a powerful tool to investigate the structural basis for the functions of actin. In particular, dominant negative actin mutants may be useful for revealing unknown molecular mechanisms that underlie the function of actin, as they have been with other proteins. The yeast Saccharomyces cerevisiae and the fly Drosophila melanogaster are well known sources of dominant negative actin mutants. A haploid S. cerevisiae cell has one actin gene, act1 (11), which is essential for cell viability (12); and certain mutations to act1 were judged to be dominant lethal, because cells carrying both the mutant and a wild-type (WT)4 copy of the gene were still unable to grow (13, 14). In Drosophila, mutation of the act88F gene, which is expressed only in the indirect flight muscle (IFM), impairs flight without impairing viability, which has enabled identification of numerous dominant negative mutant actin alleles (15, 16).
An and Mogami (16) introduced random mutations into act88F and isolated 10 strongly dominant negative alleles (G63D, R95C, G156D, G156S, E226K, G268D, G301D, G302D, Q353@, and W356@, respectively, where @ denotes a stop codon) that impair the ability of Drosophila to fly, even in the presence of a 3-fold excess of WT actin genes (antimorphic). These investigators speculated that the dominant negative mutant actins are incorporated into actin filaments in the IFM, where they disrupt the interactions between the filaments and myosin, tropomyosin (Tm), and/or adjacent actin molecules within the filaments (16). Detailed biochemical characterization of these mutant actins has not yet been achieved because, up to now, they could not be obtained in sufficient quantities from flies (17) or in vitro translation systems (18). Recently, however, we developed a novel expression system in which a fusion protein of actin and His-tagged thymosin β is expressed in Dictyostelium cells, and this system has proven particularly useful for expressing dominant negative actin mutants. Indeed, it enabled us to purify sufficient amounts of protein to characterize dominant negative mutant actins that were originally identified in yeast but could not be purified (19). In this study, we introduced the strongly dominant negative mutations reported by An and Mogami (16) into the Dictyostelium act15 gene and expressed the proteins using our novel expression system. This approach enabled us to purify six of the aforementioned mutant actins (G63D, R95C, G156D, G156S, E226K, and G268D; Fig. 1) and to characterize their biochemical properties. Our results show that these mutations impair polymerization of actin subunits and confer a tendency to aggregate; they also affect filament ends or Ca2+ regulation via tropomyosin-troponin (Tm·Tn), suggesting that these perturbations of actin function cause the observed dominant negative properties.
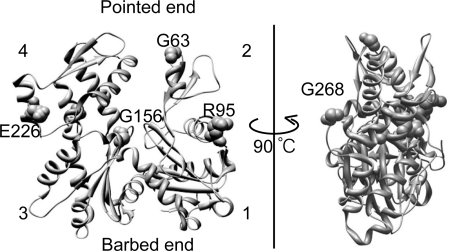
FIGURE 1.
Mutated residues in the actins characterized in this study. The Dictyostelium actin structure (58) (Protein Data Bank code 1c0f) is depicted using Chimera software. Mutated residues are highlighted by space-filling representation. The numbers indicate the subdomains.
EXPERIMENTAL PROCEDURES
Plasmid Construction
pTIKL ART, the plasmid used for our novel expression system (19), contains a G418 resistance gene for selection and the ART gene, which is the Dictyostelium act15 gene modified to carry four unique restriction sites (AR), followed by a Gly-based linker, a synthetic human thymosin β gene, and a His tag. Fragments of the actin sequence, separated by unique restriction sites, were cloned into a cloning vector. Mutations were introduced using a PCR-based method with one of the vectors serving as a template, after which they were subcloned into pTIKL ART using unique restriction sites. GFP-fused mutant actin genes were generated by subcloning the mutated actin gene fragment into pTIKL GFP-AR (19).
Cell Culture
pTIKL-based plasmids were electroporated into Dictyostelium discoideum Ax2 cells (20). Cells carrying pTIKL-based plasmids were selected on plates at 21–22 °C in HL5 medium containing 60 μg/ml each of penicillin and streptomycin and 40 μg/ml G418. For biochemical purification of actin, cells were grown on plates in medium containing 100 μg/ml G418. Large scale cultures were grown for 2–3 days to 0.5–1.0 × 107 cells/ml in 1.5 liters of HL5 medium without G418 in 5-liter conical flasks on a rotary shaker operating at 110–120 rpm.
Observation of GFP Mutant Actin in Live Dictyostelium Cells
Ax2 cells expressing GFP-actin were observed using a confocal laser-scanning microscope (19).
Western Blotting and RT-PCR Analyses
RT-PCR analysis of the RNA expression of mutant ART was performed as described previously (21) using ART-specific primers (5′-GGTACCACTATGTTCCCAGGTATTG-3′ and 5′-ACGCGTTAATGATGGTGATGATGGTGATGATGTGATTCACCT-3′). To assess the protein expression of mutant GFP-AR and ART, cells were lysed in SDS sample buffer and subjected to Western blotting analysis using anti-GFP (22) or anti-actin antibodies (clone C4, Chemicon, Temecula, CA).
Purification of Actin
Recombinant WT and mutant actins were purified as described previously (19) with some modifications. Briefly, cells expressing ART were harvested, washed, resuspended in 2 volumes/g binding buffer (10 mm HEPES (pH 7.4), 40 mm imidazole (pH 7.4), 2 mm MgCl2, 500 mm NaCl, 1 mm ATP, and 7 mm β-mercaptoethanol), and then lysed by adding 2 volumes of binding buffer containing 1.25% Triton X-100 and protease inhibitors. The resultant cell lysate was centrifuged for 30 min at 36,000 × g, after which the supernatant was incubated with nickel-Sepharose 6 Fast Flow (GE Healthcare) for 1 h at 4 °C before washing the resin with washing buffer (10 mm HEPES (pH 7.4), 40 mm imidazole (pH 7.4), 800 mm NaCl, 0.5 mm MgCl2, 0.1 mm ATP and 7 mm β-mercaptoethanol). The crude ART fraction was eluted from the resin in elution buffer (10 mm HEPES (pH 7.4), 500 mm imidazole (pH 7.4), 100 mm NaCl, 0.5 mm MgCl2, 0.1 mm ATP and 7 mm β-mercaptoethanol) and dialyzed against G-buffer (2 mm Tris-HCl (pH 7.4), 0.2 mm CaCl2, 0.2 mm ATP, 0.5 mm DTT, 0.01% NaN3). This was then digested with 1-chloro-3-tosylamido-7-amino-2-heptanone-treated chymotrypsin (Sigma) for 10 min at 25 °C, and the reaction was stopped by the addition of 0.4 mm phenylmethylsulfonyl fluoride. Actin is relatively resistant to chymotrypsin, so this treatment primarily cleaves ART immediately after final residue Phe-375 of native actin (19). However, some mutant actins were more easily cleaved internally, and therefore the ratio of chymotrypsin to total protein and the reaction time were optimized for each mutant actin. The solution of digested protein was applied to an Econo-High Q Cartridge (Bio Rad) pre-equilibrated with G-buffer, and bound proteins were eluted with a linear 0–0.5 m NaCl gradient in G-buffer. Fractions containing the released intact actin were pooled and ultracentrifuged (300,000 × g for 15 min at 25 °C) after being supplemented with 1 mm ATP and 4 mm MgCl2. In the case of polymerization-competent actin (WT, R95C, and E226K), the pellets were dissolved and dialyzed against G-buffer, and the supernatants after ultracentrifugation (300,000 × g for 15 min at 4 °C) were used as the purified actin fraction. In the case of actin with poor polymerization competence (G63D and G268D), the supernatants from the first ultracentrifugation were applied to a nickel-Sepharose column, and the flow-through was dialyzed against G-buffer and then used as the purified actin solution. In the case of the G156D and G156S mutants, which had low but significant polymerization competence, actin fractions from the Econo-High Q column were pooled and directly applied to a nickel-Sepharose column, and the flow-through was collected. After dialysis against G-buffer, the dialysate was ultracentrifuged (300,000 × g for 15 min at 4 °C), and the supernatant was used as the purified actin solution. Before use, Ca2+-actin in G-buffer was converted to Mg2+-actin by adding 1 mm EGTA and 50 μm MgCl2 to the actin solution, which was then incubated for at least 10 min on ice.
Other Protein Preparation
Rabbit skeletal heavy meromyosin (HMM), Tm, Tm·Tn complex, and actin were prepared as described previously (23–25). Gelsolin was purified from bovine plasma (26).
Sedimentation Assay
WT and mutant actins were allowed to polymerize in F-buffer (10 mm HEPES (pH 7.4), 4 mm MgCl2, 100 mm KCl, 1 mm ATP, 0.5 mm DTT), alone or with a 2-fold molar excess of phalloidin (Sigma) or an equal molar concentration of WT, for 1 h at the desired temperature. They were then ultracentrifuged for 10 min at 300,000 × g at the same temperature, and the resultant pellet and supernatant fractions were subjected to SDS-PAGE.
Electron Microscopy
Mutant actins (G156D, G156S, and G268D) were polymerized, negatively stained by 1% uranyl acetate, and observed as described previously (19).
Complexes of Actin and Gelsolin
To form actin·gelsolin complexes, 0.4 μm WT or G63D G-actin was added to 0.5 mg/ml BSA and 20 nm gelsolin in G-buffer, after which the buffer was incubated for 30 min at room temperature. To confirm complex formation, the solutions were mixed with an equal volume of 2 μm rhodamine-phalloidin (RhPh)-labeled skeletal actin filaments in buffer containing 10 mm HEPES (pH 7.4), 4 mm MgCl2, 100 mm KCl, 0.2 mm CaCl2, 0.5 mg/ml BSA, and 0.5 mm DTT. After incubation for 15 min at room temperature and dilution, the mixtures were introduced into HMM-coated flow cells (see below) for microscopic observation of fragmentation.
Pyrene Fluorescence Measurement
Labeling of actin with N-(1-pyrene)iodoacetamide (Molecular Probes, Eugene, OR) was carried out as described previously (27). Actin polymerization was then monitored based on the increase in pyrene fluorescence following the addition of 100 mm KCl and 2 mm MgCl2 at 23 °C. The final buffer contained 5 mm HEPES (pH 7.4), 100 mm KCl, 2 mm MgCl2, 0.4 mm EGTA, 0.5 mg/ml BSA, 0.5 mm DTT, and 1 mm ATP.
Elongation rates of WT actin from gelsolin·actin complexes were measured as follows. The actin·gelsolin complex was formed by mixing 2.5 μm G-actin (WT or G63D) and 250 nm gelsolin in G-buffer and then incubating the mixture for 30 min at room temperature. Twenty microliters of the actin/gelsolin solution were added to 1 μm WT actin (15% of which was pyrene-labeled), and polymerization was immediately initiated by adding 100 mm KCl and 2 mm MgCl2 (total volume of 500 μl). The final buffer contained 5 mm HEPES (pH 7.4), 100 mm KCl, 2 mm MgCl2, 0.4 mm EGTA, 0.5 mg/ml BSA, 0.5 mm DTT, and 0.6 mm CaCl2.
ATPase Activity of Mutant Actin Filaments
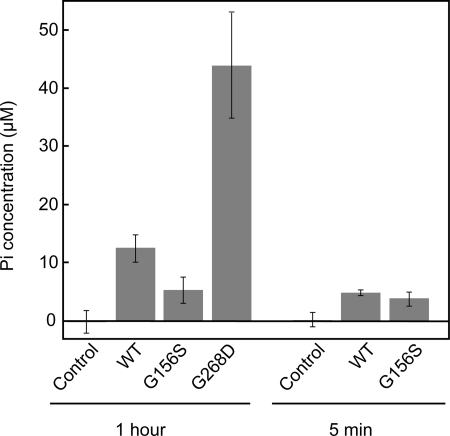
ATPase activities of actin during polymerization were measured by using malachite green (28). G-Buffer (control) and G-Buffer containing 5 μm WT, 5 μm G156S, or G268D actin were incubated at 37 °C for 5 min or 1 h in 10 mm HEPES (pH 7.4), 2 mm MgCl2, 100 mm KCl, 1 mm EGTA, 0.25 mm DTT, and 0.5 mm ATP. After quenching with 0.3 m perchloric acid, Pi concentrations were measured.
In Vitro Motility Assay
Motility assays using nitrocellulose-coated coverslips were performed as described previously (29).
When measuring the Ca2+ sensitivity of the Tm·Tn regulation system, it was critically important to minimize the stalling of actin filaments due to denatured HMM heads, so that we could be sure that stalling was due to the Ca2+ regulation. For this purpose, after washing out unbound HMM, AB/BSA (10 mm HEPES (pH 7.4), 25 mm KCl, 4 mm MgCl2, 0.5 mm DTT, and 0.5 mg/ml BSA) containing phalloidin-stabilized actin filaments (either WT or mutant, to be assayed) was introduced and incubated for 1 min to block inactive HMM heads, followed by incubation with AB/BSA/ATP (AB/BSA plus 1 mm ATP) for 2 min. Thereafter, the flow cell was washed with AB/BSA and incubated with a solution of RhPh-labeled actin filaments for 1 min. This was followed by the addition of AB/BSA containing 1 μg/ml Tm·Tn complex, which was incubated for 3 min. Finally, ATP/Tm·Tn solution (10 mm HEPES (pH 7.4), 4 mm MgCl2, 25 mm KCl, 2 mm EGTA, 1 μg/ml Tm·Tn complex, 0.5% methylcellulose, 0.5 mg/ml BSA, 1 mm ATP, 10 mm DTT, 200 μg/ml glucose oxidase, 30 μg/ml catalase, 3 mg/ml glucose, and various concentrations of CaCl2) was introduced, and movements were observed as above. The free Ca2+ concentration in the ATP/Tm·Tn solution was controlled using a Ca2+-EGTA buffer system containing 2 mm EGTA and 44 μm to 2 mm CaCl2. Individual filaments were classified into the following three groups based on their motility, according to the criteria of Homsher et al. (30): moving at a uniform speed, moving erratically, and not moving. Filaments were judged not to be moving if the measured velocity was below a cutoff value, 0.5 μm/s, which is the mean + 2σ of the measured “velocities” of control filaments immobilized to the HMM-coated surface in the absence of ATP. The mean filament gliding speeds at different pCa values were fitted to the following Hill equation: V = Vmax/(1 + 10n(pCa50−pCa)).
Measurement of Actin-activated S1 ATPase Activity
Actin-activated S1 ATPase activity was measured using an EnzChek phosphate assay kit (Invitrogen). Reactions were started by the addition of 1 unit/ml purine nucleoside phosphorylase, 0.2 mm 2-amino-6-mercapto-7-methylpurine riboside, and then 0.87 μm S1 to a solution of phalloidin-stabilized 4 μm WT or R95C filaments in ATPase buffer (10 mm imidazole (pH 7.0), 50 mm KCl, 2 mm EGTA, 3 mm MgCl2, 97 μg/ml Tm·Tn, various concentrations of CaCl2 and 1 mm ATP) at 25 °C.
RESULTS
In Vivo Polymerization of GFP-actin and Expression of ART
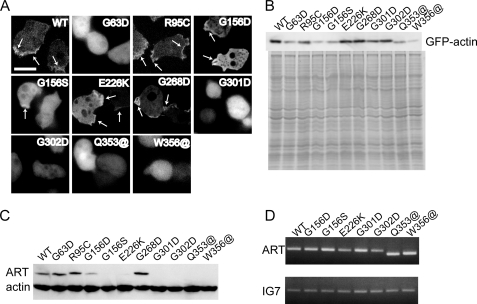
To assess their polymerization competence in vivo, fusion proteins in which GFP was fused to the N terminus of WT or mutant actin via a flexible linker were expressed in Dictyostelium and observed in live cells under a fluorescence microscope (Fig. 2A). GFP-WT actin localized in the cortical layer of cells, indicating it is able to copolymerize with endogenous actin in vivo. GFP-R95C, -G156D, -G156S, -G268D, and -E226K mutant actins showed localization similar to GFP-WT actin. By contrast, the remaining mutant GFP actins (G63D, G301D, G302D, Q353@, and W356@) were diffusely distributed in the cytoplasm, indicating that polymerization of these mutant actins was severely disrupted in vivo. Western blot analysis of whole cell lysates using anti-GFP antibodies showed that the expression levels of GFP-G156D, -G156S, -Q353@, and -W356@ actins were much lower than that of GFP-WT actin (Fig. 2B).
FIGURE 2.
Expression of GFP-mutant actins and mutant ARTs in Dictyostelium cells. A, fluorescence from GFP-actins in Dictyostelium cells observed using a confocal laser-scanning microscope. For GFP-G156D, -G156S, -Q353@, and -W356@ mutant actins, the majority of cells were too dark for observation, and the cells shown here have atypically high expression. Regions of cortical GFP fluorescence are indicated by arrows. Scale bar, 10 μm. B, Western blot (top) and Coomassie Brilliant Blue staining (bottom) following SDS-PAGE of total cell lysates from Dictyostelium cells expressing GFP-mutant actins. Polyclonal anti-GFP antibodies were used for Western blotting. C and D, levels of ART protein and mRNA were examined by Western blot analysis using a monoclonal anti-actin antibody (C) and RT-PCR using ART-specific primers (D).
We next used RT-PCR with specific primers and Western blot analysis with a monoclonal anti-actin antibody to determine the expression levels of mutant ARTs (Fig. 2, C and D). The protein expression levels of some mutant ARTs (i.e. G156D, G156S, and E226K) were significantly lower than that of WT, whereas G301D, G302D, Q353@, and W356@ ARTs were not detected at all. In fact, purification of the latter four mutant ARTs was unsuccessful. Nonetheless, the expression levels of the mutant ART mRNAs were not significantly lower than that of WT mRNA. This suggests that these mutant ARTs are probably rapidly degraded after translation due to inappropriate or unstable folding or because they assume a conformation that is easily attacked by proteases. This is probably why expression levels of the corresponding mutant GFP-actins were also low.
Polymerization Competence of Purified Mutant Actins in Vitro
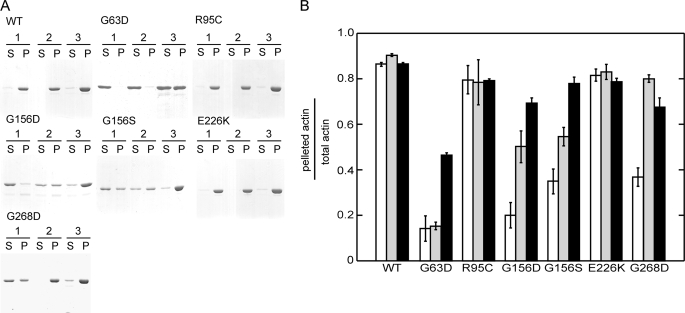
We purified six dominant negative mutant actins in amounts sufficient for biochemical analyses. The six mutants were allowed to polymerize under three conditions as follows: alone, in the presence of excess phalloidin, and in the presence of an equal amount of WT actin. They were then ultracentrifuged and analyzed by SDS-PAGE (Fig. 3). R95C and E226K actins sedimented like WT actin under all conditions. Conversely, G63D actin did not sediment under any conditions, including in the presence of the Tm·Tn complex (not shown). In the presence of WT actin, approximately half of a total actin mixture containing equal amounts of WT and G63D actins pelleted, but this pellet almost certainly consisted mainly of WT actin, as the addition of WT protein did not noticeably reduce the amount of actin in the supernatant. G156D, G156S, and G268D actins did not completely sediment alone, but the addition of phalloidin significantly enhanced their sedimentation. Moreover, supernatants of all three mutants decreased when WT actin was added, suggesting that they copolymerized with WT actin, which is consistent with the behavior of the GFP-fused mutant actins in vivo.
FIGURE 3.
Sedimentation assay of mutant actins. Each purified mutant actin was allowed to polymerize under three conditions and then ultracentrifuged. Condition 1, polymerization of actin alone. Condition 2, polymerization with phalloidin. Condition 3, polymerization with additional equimolar concentration of WT actin. The resulting supernatant (S) and pellet (P) fractions were analyzed using 10% PAGE and stained with Coomassie Brilliant Blue (A). The band intensities were then quantified using ImageJ software (B). White, gray, and black bars represent the data obtained under conditions 1, 2 and 3, respectively. Error bars indicate S.D. (n = 3).
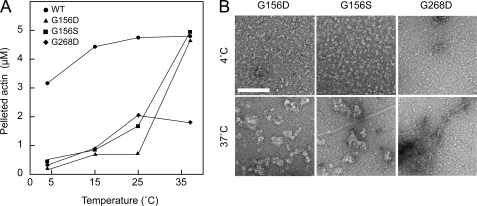
Mutant actins showing defective polymerization (G63D, G156D, G156S, and G268D) were incubated in F-buffer at four different temperatures ranging from 4 to 37 °C and then ultracentrifuged. The sizes of the G156D, G156S, and G268D actin pellets increased with increasing temperature (Fig. 4A), and the G156D and G156S actins precipitated almost completely at 37 °C. Electron microscopic observation revealed that G156D and G156S actins formed small structures, presumably oligomers, in F-buffer at 4 °C (Fig. 4B). At 37 °C, large, amorphous aggregates were formed, suggesting the pellets obtained after ultracentrifugation contained improperly polymerized filaments. G268D actin formed filaments after incubation at 37 °C, but aggregates presumably consisting of denatured G268D actin molecules were also seen around the normal filaments. G63D actin did not precipitate at any of the temperatures tested (data not shown).
FIGURE 4.
Polymerization of mutant actins at different temperatures. WT, G156S, G156D, and G268D actins were incubated in F-buffer at 4, 15, 25, and 37 °C and ultracentrifuged. The resulting supernatant and pellet fractions were subjected to SDS-PAGE and quantified (A). B, electron micrographs of G156D, G156S, and G268D actins incubated in F-buffer at 4 or 37 °C. Scale bar, 100 nm.
ATP Hydrolysis by G156S and G268D Actins
To know whether aggregations of G156 and G268D actins occurred upon transfer to the polymerization condition or after ATP hydrolysis, we determined the amount of Pi released from G156S and G268D actins during polymerization. Molar concentrations of Pi released from G156S and WT actins during the initial 5 min after transfer to the polymerization condition were both similar to that of actin (Fig. 5), suggesting that both actins hydrolyzed ATP once in 5 min. However, G156S actin did not release further Pi during the following 55 min of incubation, whereas WT actin released 2.5-fold more Pi during this period, presumably representing treadmilling or cycling between G- and F-actin. In contrast, G268D actin released 3.5-fold more Pi than the WT in 1 h of incubation under the polymerization condition (Fig. 5).
FIGURE 5.
Actins (5 μm) or G-buffer (control) were incubated in solution including 10 mm HEPES (pH 7.4), 2 mm MgCl2, 100 mm KCl, 1 mm EGTA, 0.25 mm DTT, and 0.5 mm ATP at 37 °C for 5 min or 1 h. Following acid quenching, the amounts of total Pi were determined by the malachite green method. Values shown are after subtracting Pi concentration at 0 min. Error bars indicate S.D. (n = 4–7).
Pyrene Fluorescence Assay
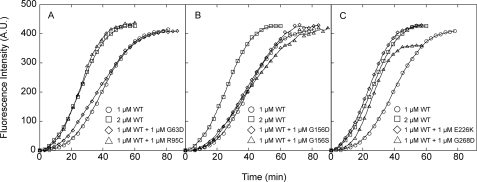
We examined the effects of the mutant actins on polymerization of WT actin by monitoring the increase in the fluorescent signal from pyrene attached to Cys-374 of WT actin polymerized in the presence of mutant actin (Fig. 6).
FIGURE 6.
Effects of mutant actin on the polymerization kinetics of WT actin. 1 μm WT actin (15% pyrene-labeled) was mixed with 1 μm WT or mutant actin, after which polymerization was initiated by adding 100 mm KCl and 2 mm MgCl2 at 23 °C. Polymerization was followed for 60–90 min by monitoring the pyrene fluorescence intensity.
R95C and E226K actins accelerated polymerization of WT actin to the same extent as WT actin (Fig. 6, A and C), suggesting these mutants can interact normally with WT actin, which is consistent with the results of the pelleting assay. In contrast, G156D and G156S actins did not significantly affect the overall polymerization kinetics of WT (Fig. 6B). Numerous aggregates observed by electron microscopy suggest that the majority of these mutant actin molecules aggregated without properly interacting with the WT actin.
Addition of G268D actin accelerated polymerization, but the final intensity of pyrene fluorescence was lower than was seen with WT actin alone (Fig. 6C). Based on this finding and the results of electron microscopic observations, we suggest that under our polymerization conditions, a fraction of the G268D actin molecules polymerized properly, but the remainder did not, some of which formed aggregates. In this scenario, the normal fraction of the mutant molecules interacts with the WT actin properly, copolymerizing and accelerating the process; however, the aggregating fraction draws in WT molecules, which then do not enhance pyrene fluorescence, resulting in the observed reduction in the final fluorescence intensity.
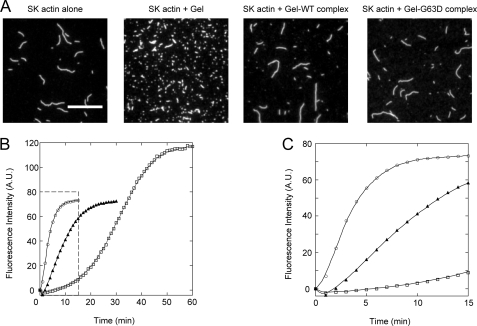
Addition of 1 μm G63D actin accelerated polymerization of WT actin slightly during the early phase of polymerization (Fig. 6A), and this accelerating effect was much more pronounced with 5 μm G63D actin (data not shown), indicating this mutant can interact with WT actin during the nucleation stage of polymerization. Gly-63 is on the surface of the actin molecule and is at the pointed end when incorporated into filaments. Gelsolin is a potent actin filament severing and nucleating protein that caps the barbed end of actin filaments (31, 32), so that elongation of the filaments from the actin·gelsolin complex occurs only from the pointed end surface of the complex. We therefore examined the rates of elongation of WT actin from the gelsolin·G63D actin complex to assess the interaction between the pointed end surface of G63D and the barbed end surface of WT actin. To do this, we first tested whether G63D actin binds to gelsolin. Gelsolin and WT or G63D actin were incubated in the presence of Ca2+ to allow formation of the complex, after which the solutions containing the complexes were added to a solution of RhPh-labeled rabbit skeletal actin filaments (Fig. 7A). Addition of gelsolin-G63D actin did not lead to significant fragmentation of the filaments nor did the addition of gelsolin-WT actin. On the other hand, addition of gelsolin alone caused significant fragmentation. This inhibitory effect of G63D actin on the ability of gelsolin to sever actin filaments confirms that G63D actin is able to bind to gelsolin normally and that the barbed end surface of G63D actin is properly masked within the complex. The gelsolin·actin complex reportedly accelerates polymerization in the presence of Ca2+ but reduces the final extent of polymerization because the elongation occurs only at the pointed end of the filaments (33, 34). The rate of elongation from gelsolin-G63D actin was significantly slower than the rate from gelsolin-WT actin (Fig. 7, B and C), indicating that the pointed end surface of G63D actin is able to interact with the barbed end surface of WT actin, although this interaction is weaker than between WT molecules.
FIGURE 7.
Interaction between G63D actin and gelsolin and the effect of the gelsolin·G63D complex on polymerization of WT actin. A, gelsolin·WT actin complex (Gel·WT complex), gelsolin·G63D actin complex (Gel·G63D complex), or gelsolin alone was added to 1 μm RhPh-labeled rabbit skeletal muscle actin filaments (SK actin). The mixture was then diluted and visualized on HMM-coated coverslips using a fluorescence microscope. Scale bar, 10 μm. B, rates of WT actin elongation in the presence of Gel·G63D or Gel·WT complex were measured to examine the interaction between the barbed ends of WT actin and the pointed ends of G63D actin. 1 μm WT actin (15% pyrene-labeled) was allowed to polymerize in the presence of the following additives: gelsolin·G63D complex (filled triangles), gelsolin·WT actin complex (open circles), and G-buffer (open squares). Polymerization was followed by monitoring the pyrene fluorescence intensity for 60 min. The boxed area is enlarged in C.
In Vitro Motility and Actin-activated S1 ATPase Activity
We tested the ability of filaments of each mutant actin to slide over immobilized rabbit skeletal muscle HMM in an in vitro motility assay. G63D actin filaments were not observed after labeling with RhPh. Few RhPh-labeled G156D and G156S filaments were observed, and these rare filaments were very short (data not shown). Therefore, these three mutant actins were not subjected to the motility assay. E226K filaments had a tendency to form bundles, and we chose single filaments for the velocity measurements. The velocities of E226K and G268D filaments were slightly slower than the WT filaments, whereas R95C filaments moved at speeds indistinguishable from the WT (Table 1). However, higher concentrations of KCl were required to release R95C filaments from the HMM surface in the presence of ATP than were required to release WT filaments (60 versus 45 mm KCl; data not shown), indicating that the affinity of R95C filaments for myosin in the presence of ATP is higher than that of WT filaments.
TABLE 1.
Gliding velocities of mutant actin filaments
Motility assays were carried out in AB (10 mm HEPES (pH 7.4), 4 mm MgCl2, 25 mm KCl, 0.5 mg/ml BSA, 10 mm DTT, and the oxygen scavenger system) at 25 °C.
| WT | R95C | E226K | G268D | |
|---|---|---|---|---|
| Velocity (μm/s) | 4.2 ± 0.41 | 4.3 ± 0.43 | 3.4 ± 0.42 | 3.9 ± 0.47 |
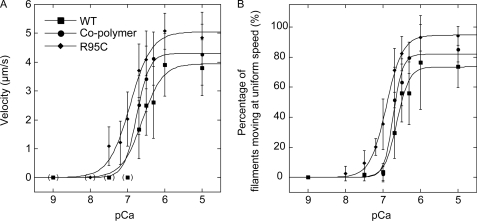
Because the R95C (35–37) and E226K mutation (38, 39) sites overlap the proposed interaction site for Tm, we next examined the Ca2+ sensitivity of movements of R95C and E226K filaments in the presence of Tm or the Tm·Tn complex (Figs. 8 and 9). We used rabbit skeletal Tm and Tm·Tn complex for these experiments because we previously found that Dictyostelium actin is efficiently regulated by Ca2+ in the presence of rabbit Tm·Tn (38). The Hill equation parameters obtained by fitting the velocities and percentages of filaments moving at uniform speeds are shown in Tables 2 and 3. The calculated maximum velocity (Vmax) and percentage of filaments moving at uniform speeds (Fmax) of R95C filaments in the presence of Tm·Tn (Tm·Tn R95C filaments) was 5.1 μm/s and 95%, respectively. These were somewhat higher than 3.9 μm/s and 74% obtained with WT filaments (Tm·Tn WT filaments). The concentration of Ca2+ required for activation of the movement of Tm·Tn R95C filaments was lower than that required for movement of Tm·Tn WT filaments, and the Ca2+ concentrations required for one-half Vmax and Fmax (pCa50) were both shifted by 0.3 (from 6.6 to 6.9). We also tested the effects of Tm·Tn on filaments in which R95C and WT actins were copolymerized at a molar ratio of 1:3 (Tm·Tn R95C/WT cofilaments). This ratio was selected because it is the ratio of the copy numbers of R95C and WT actin genes in Drosophila IFM showing impaired flight (16). The pCa50 of Tm·Tn R95C/WT cofilaments in velocity and percentages of filaments moving at uniform speeds also shifted upward both by 0.2 to 6.8.
FIGURE 8.
Effects of R95C mutation on Tm·Tn function. Velocities of moving filaments (A) and percentage of filaments moving at uniform speeds (B) of RhPh-labeled WT, R95C filaments, and cofilaments over HMM surfaces were measured in the presence of Tm·Tn and various Ca2+ concentrations (pCa) at 25 °C. The Hill coefficient, pCa50, maximum velocity (Vmax (μm/s)), and maximum percentages of filaments moving at uniform speeds (Fmax (%)) were obtained by fitting each data set to the Hill equation (the parameters used are given in Table 2). Error bars indicate S.D. The differences of velocities and percentages of filaments moving at uniform speeds at pCa 6.7 between each pair of WT filaments and R95C filaments or cofilaments were significant (p < 0.05, t test). A significant fraction of filaments was not moving at intermediate pCa, and to avoid very large S.D., those stalled filaments were excluded from velocity analysis (30). However, this procedure resulted in excessive high velocity at high pCa if a very small number of filaments moved at velocities above the cutoff, even though the vast majority of the filaments had stalled. Therefore, those data points were excluded from fitting to the Hill equation, and zero values were used instead at those data points, which are shown in parentheses. Shown here is a representative of three independent experiments.
FIGURE 9.
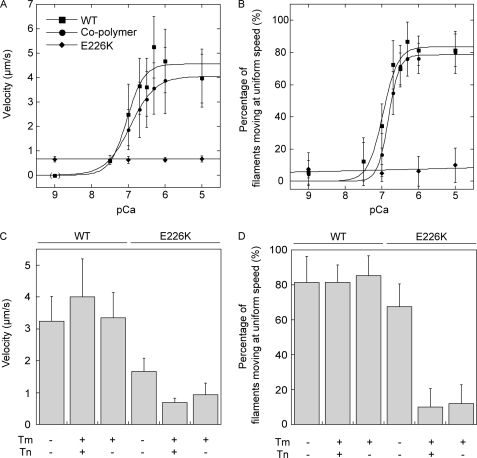
Effects of E226K mutation on Tm·Tn function. Velocities of moving filaments (A) and percentage of filaments moving at uniform speeds (B) of RhPh-labeled WT, E226K filaments, and cofilaments over HMM surfaces were measured in the presence of Tm·Tn and analyzed as in Fig. 8. The differences of the velocities and percentages of filaments moving at uniform speeds between cofilaments and WT filaments at pCa 6.7 were significant (p < 0.05, t test). The velocities (C) and percentages of filaments moving at uniform speeds (D) in the presence or absence of regulatory proteins shown in the bar graphs are those at pCa 5, except that the data in the presence of Tm alone was obtained in the absence of Ca2+.
TABLE 2.
Hill parameters for WT and R95C actin filaments
Each parameter ± S.E. was determined by fitting the data presented in Fig. 8 to the Hill equation. Shown here is a representative of three independent measurements.
| Velocity |
Filaments moving at uniform speeds |
|||||
|---|---|---|---|---|---|---|
| WT | Mutant | Cofilament | WT | Mutant | Cofilament | |
| Hill coefficient | 1.6 ± 0.70 | 1.5 ± 0.28 | 2.5 ± 0.54 | 2.7 ± 0.63 | 2.0 ± 0.14 | 3.5 ± 0.94 |
| pCa50 | 6.6 ± 0.09 | 6.9 ± 0.06 | 6.8 ± 0.03 | 6.6 ± 0.04 | 6.9 ± 0.02 | 6.7 ± 0.04 |
| Vmax (μm/s) | 3.9 ± 0.38 | 5.1 ± 0.27 | 4.3 ± 0.14 | |||
| Fmax (%) | 74 ± 4.1 | 95 ± 1.8 | 82 ± 4.7 | |||
TABLE 3.
Hill parameters for WT and E226K actin filaments
Each parameter ± S.E. was determined by fitting the data presented in Fig. 9 to the Hill equation. The WT data shown here is different from the one in Table 2, and this presumably reflects a minor difference in the experimental conditions (e.g. the Ca-EGTA buffer and the HMM batch).
| Velocity |
Filaments moving at uniform speeds |
|||
|---|---|---|---|---|
| WT | Cofilament | WT | Cofilament | |
| Hill coefficient | 1.9 ± 0.80 | 1.3 ± 0.07 | 2.1 ± 0.58 | 2.9 ± 0.68 |
| pCa50 | 7.0 ± 0.11 | 6.9 ± 0.02 | 7.0 ± 0.06 | 6.8 ± 0.04 |
| Vmax (μm/s) | 4.6 ± 0.40 | 4.0 ± 0.05 | ||
| Fmax (%) | 84 ± 4.4 | 79 ± 3.6 | ||
We next investigated the effects of R95C mutation on actin-activated skeletal muscle S1 ATPase activity in the presence of Tm·Tn and found that the steady state ATPase activity stimulated by R95C filaments was higher than that by WT filaments at all pCa points measured in this study (Table 4). This enhanced activation by R95C mutation in the presence of Tm·Tn parallels the results of the motility assay. However, we were unable to detect increased sensitivity to Ca2+, i.e. larger pCa50, in the present ATPase measurements.
TABLE 4.
Actin-activated S1 ATPase activity
Data are average and S.D. of three independent measurements.
| pCa | WT filaments | R95C filaments |
|---|---|---|
| s−1 | s−1 | |
| 4.0 | 1.41 ± 0.09 | 2.83 ± 0.02 |
| 6.4 | 1.13 ± 0.24 | 3.31 ± 0.24 |
| 6.7 | 1.10 ± 0.13 | 2.70 ± 0.11 |
| 7.0 | 0.6 ± 0.05 | 1.17 ± 0.20 |
E226K mutation also significantly impaired regulation by the Tm·Tn system, but the effect was opposite to that of the R95C mutation. Even at pCa 5, very few Tm·Tn E226K filaments were moving, and those that were moving did so very slowly in the presence of the Tm·Tn complex. Furthermore, inhibition of motility was also seen in the presence of tropomyosin only (Fig. 9), indicating that E226K mutation impairs properly regulated interaction with tropomyosin. With the cofilaments of WT and E226K mutant actins, the pCa50 of the percentage of filaments moving at uniform speeds was reduced slightly (6.8 ± 0.04 for Tm·Tn E226K/WT cofilaments and 7.0 ± 0.06 for Tm·Tn WT filaments), but the pCa50 of velocity did not change significantly (6.9 ± 0.02 for Tm·Tn E226K/WT cofilaments and 7.0 ± 0.11 for Tm·Tn WT filaments).
DISCUSSION
Using our novel system, which enables expression of toxic actin mutants, we successfully purified six of the strongly dominant negative mutant actins identified by An and Mogami (16). Subsequent biochemical analyses enabled their classification into three functional groups as follows: poor polymerization with a tendency to aggregate; no polymerization presumably due to disrupted filament ends; and perturbed regulation by Tm·Tn.
Mutant Actins Showing Reduced Polymerization and a Tendency to Aggregate
G156D and G156S actins formed large aggregates at higher incubation temperatures during polymerization. Moreover, sedimentation assays indicated a strong interaction between the mutant and WT actins, whereas pyrene fluorescence assays did not show significant copolymerization between the mutant and WT actins. IFM fibers of heterozygous Drosophila in which G156D or G156S actin was expressed were normal (16); therefore, we have no specific idea how G156D and G156S actins affect WT actin in IFM. Interestingly, G156S mutant actin appeared to aggregate after ATP hydrolysis under the polymerization condition. Gly-156 is located within the ATP binding loop, which is thought to sense the nucleotide state and to be involved in the conformational change accompanying the release of Pi (40). Thus, aggregation of G156D or G156S actin may correlate with structural changes coupled to ATP hydrolysis or Pi release during polymerization.
In sedimentation assays, G268D actin showed a weak polymerization defect. Gly-268 is in the “hydrophobic loop,” which interacts with the “hydrophobic pocket” formed by two subunits in the opposing strand within filaments. Introduction of a hydrophilic residue into this region could destabilize filaments by disrupting the hydrophobic interaction, as has been suggested for yeast mutant actins (41). This instability may accelerate “treadmilling,” which may be why G268D actin released larger amounts of Pi than WT during extended incubation under the polymerization condition. Release of Pi from V266G/L267G yeast actin, which only forms oligomers even under the polymerization condition, continues in the steady state. This result was suggested to represent unstable cycling of oligomers driven by the hydrolysis of ATP (42).
Polymerization of G268D actin was cold-sensitive. Chen et al. (41) reported that the L266D mutant of yeast actin is cold-sensitive, presumably due to the cold sensitivity of hydrophobic interactions. This same argument may be applicable to the cold sensitivity of G268D actin. Furthermore, the conformational dynamics of the hydrophobic loop accompanying polymerization is also important. The hydrophobic loop detaches from the main body of actin and is inserted into the hydrophobic pocket, stabilizing the filaments (43, 44). Thus, cross-linked actin, in which extension of the hydrophobic loop was inhibited, was unable to polymerize (45). Because Gly residues are generally important to the conformational dynamics of proteins, and because Gly-268 is completely conserved among actin sequences, Gly-268 may contribute to the conformational dynamics of the hydrophobic loop. Conformational changes of the hydrophobic loop are unnecessary for ATPase activity accelerated under the polymerization condition (46). This is consistent with the high ATPase activity of G268D actin.
Electron microscopic observation and pyrene assays suggested that a fraction of WT actin molecules are taken into the aggregates when they are allowed to copolymerize with G268D actin, which may cause reduction of functional actin in IFM. It is also possible that G268D, as well as G156D and G156S mutant actins, take in actin-binding proteins into aggregates, which may perturb the actin turnover in vivo.
Mutation That Affects Filament Ends
Sedimentation assays showed that G63D actin only minimally copolymerized with WT actin in the steady state, whereas pyrene assays indicated that G63D actin accelerated nucleation and polymerization of WT actin. G63D actin bound to gelsolin, but polymerization of WT actin from G63D-gelsolin was slow. Because Gly-63 is located on the pointed end surface of the actin molecule, where it participates in the formation of the hydrophobic pocket within filaments, we suggest that the polymerization defect of G63D actin derives from the reduced hydrophobicity on the pointed end surface of the molecules, whereas the barbed end surface remains normal. Polymerization of β-actin with R62D mutation is similarly defective, reflecting a weak interaction with WT actin (47).
In the early copolymerization phase of WT and G63D actins, a fraction of the G63D molecules may have been incorporated into the filaments from the growing pointed ends. In the steady state, however, G63D molecules were not found in the filaments. This is presumably because G63D molecules with defective pointed end surfaces would not be able to polymerize if, as is generally assumed, polymerization occurs slowly only at the barbed ends during treadmilling. Moreover, those G63D molecules that had been incorporated into filaments would eventually be expelled from the filaments by treadmilling. In this scenario, G63D actin disrupts the initial phase of the formation of thin filament structures in IFM. In fact, An and Mogami (16) reported that IFM fibers of heterozygous Drosophila fly containing the G63D mutation were wavy. That said, we are unable to rule out the possibility that G63D actin sequesters some actin monomer-binding protein, thereby indirectly disturbing the turnover of actin in IFM cells. However, this model predicts that most, if not all, polymerization-incompetent actin alleles would be dominant negative, but we have no evidence to that effect.
Mutations at Tropomyosin-binding Sites
R95C and E226K mutations did not affect polymerization of the mutant actin alone or in the presence of WT actin. This is reasonable because these mutation sites are located away from the surface facing adjacent actin subunits within the filament. However, RhPh-labeled E226K filaments tended to form bundles in buffers for motility assays. Because actin is highly acidic, the charge reversal resulting from E226K mutation either weakens repulsion between filaments or attracts clusters of negative charges in neighbor filaments. The bundling of mutant actin filaments was also associated with mutations of the N-terminal acidic residues (48, 49). However, we do not think that this bundling of E226K filaments was the cause of the dominant negative phenotype, as E226K filaments did not bundle in physiological salt concentrations or when copolymerized with WT actin (data not shown).
The acto-S1 docking model (50) suggests that Arg-95 of actin is involved in the interaction with myosin, and previous mutational studies suggest that negative charges in this region are important for myosin binding (48, 51–53). Consistent with this, R95C filaments, in which the net negative charge is increased in the region, showed greater affinity for myosin in in vitro motility assays. The difference was subtle, however, and was unlikely to cause the dominant negative effect seen in vivo. We therefore examined Tm·Tn regulation of R95C and E226K filaments, as these mutation sites are near tropomyosin-binding sites. Tropomyosin equilibrates in three states within thin filaments, blocked, closed, and open, and equilibrium among these states is affected by Ca2+ binding to troponin and/or myosin binding to thin filaments (54). In the blocked state, the myosin-binding sites on actin filaments are sterically blocked by Tm; in the closed state they are partially exposed, allowing weak binding of myosin to actin. Finally, the open state is induced from the closed state by transition of myosin to the strongly bound state, leading to force generation. Arg-95 and Glu-226 are near the proposed position occupied by Tm in the blocked (35–37) and open states (38, 39), respectively.
Motility of Tm·Tn R95C filaments exhibited a higher sensitivity to Ca2+ than Tm·Tn WT filaments. This may be caused by disruption of equilibrium among the Tm states due to the loss of a positive charge and/or the higher affinity for myosin. With respect to the first mechanism, it is known that the equilibrium among tropomyosin states is shifted to the blocked state on E93K mutant filaments even in the presence of Ca2+, suggesting that the introduced positive charge stabilizes the blocked state (55). In the R95C mutant, a positive charge is removed, so that Tm may tend to shift away from the blocked state. With respect to the second mechanism, it is known that the number of cross-bridges per unit length of the thin filament alters Ca2+ sensitivity (30). Because R95C filaments have a higher affinity for myosin than WT filaments, a larger number of HMM molecules may interact with R95C filaments, so that the equilibrium of the Tm state may shift to the open state at a lower Ca2+ concentration. At present we have no data with which to estimate the relative contributions of these two mechanisms.
Interestingly, enhanced sensitivity to Ca2+ was not detected in the ATPase measurements in solution. In motility assays, filaments were stretched by HMM molecules. It is thus tempting to speculate that stretching affected sensitivity of the filaments to Ca2+ and that this is related to stretch activation of IFM (see below).
Motility of Tm·Tn E226K filaments were severely inhibited even in the presence of high Ca2+ concentrations. Furthermore, a similar motility defect was observed with Tm only, whereas those filaments moved normally in the absence of Tm. Thus, these motility defects derive from a disruption of Tm function. Presumably the equilibrium among tropomyosin states on E226K filaments is biased to the closed or blocked state.
Beating IFM undergoes a high frequency oscillation that is dependent on stretch and is not synchronized to the release of Ca2+ (stretch activation) (56). Nonetheless, a certain level of Ca2+ is maintained during beating (57), which suggests that a certain level of Tm·Tn activation is required for stretch activation. Both R95C and E226K filaments showed altered sensitivity to Ca2+ in the presence of Tm·Tn in in vitro motility assays but appeared more or less normal in the absence of Tm·Tn. Thus, a relatively minor perturbation of the Tm·Tn function may affect the intricate stretch activation system, as the pCa50 of Tm·Tn cofilaments with either R95C or E226K and WT actins were shifted by only 0.1–0.2. Additionally, the two types of cofilaments moved 10% faster and 13% slower than Tm·Tn WT filaments, respectively, and these differences in sliding velocities might enhance the effects of increased and decreased sensitivities of the cofilaments to Ca2+ in terms of stretch activation.
Finally, if R95C and E226K mutant actin molecules are randomly distributed within the thin filaments, there would be segments of higher concentration of the mutant actin, resulting in nonhomogeneous activation at intermediate pCa. This could interfere with the muscle function, including the stretch activation mechanism, more profoundly than can be suggested from in vitro motility assays reporting averaged motile functions of thousands of actin subunits.
Fibers in IFM expressing E226K were normal, but those expressing R95C actin were disrupted (16), and this may be an indirect consequence of the disturbed stretch activation mechanism.
Mutant Actins That We Failed to Purify
The four mutant actins that we were unable to purify were expressed at very low but detectable levels when fused with GFP. They were presumably expressed in the IFM cells as well, because they exhibited strongly dominant negative effects there. Different approaches will be needed to express these mutant actins and assess their effect on muscle function.
Acknowledgments
We thank Dr. K. Mogami for help in the initial phase of this work, Dr. A. Nagasaki for technical assistance with the RT-PCR, and Dr. S. Iwai for comments on the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Science, Culture, Sports, and Technology (to T. U. and T. N.).
- WT
- wild type
- GFP
- green fluorescent protein
- DTT
- dithiothreitol
- BSA
- bovine serum albumin
- IFM
- indirect flight muscle
- HMM
- heavy meromyosin
- RT
- reverse transcription
- Tm
- tropomyosin
- Tn
- troponin
- RhPh
- rhodamine-phalloidin.
REFERENCES
- 1.Pollard T. D., Blanchoin L., Mullins R. D. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 [DOI] [PubMed] [Google Scholar]
- 2.Carlier M. F. (1992) Philos. Trans. R. Soc. Lond. B Biol. Sci. 336, 93–97 [DOI] [PubMed] [Google Scholar]
- 3.Schüler H. (2001) Biochim. Biophys. Acta 1549, 137–147 [DOI] [PubMed] [Google Scholar]
- 4.McGough A., Pope B., Chiu W., Weeds A. (1997) J. Cell Biol. 138, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prochniewicz E., Yanagida T. (1990) J. Mol. Biol. 216, 761–772 [DOI] [PubMed] [Google Scholar]
- 6.Kim E., Bobkova E., Hegyi G., Muhlrad A., Reisler E. (2002) Biochemistry 41, 86–93 [DOI] [PubMed] [Google Scholar]
- 7.Oosawa F., Fujime S., Ishiwata S., Mihashi K. (1972) Cold Spring Harbor Symp. Quant. Biol. 37, 277–285 [Google Scholar]
- 8.Hegyi G., Belágyi J. (2006) FEBS J. 273, 1896–1905 [DOI] [PubMed] [Google Scholar]
- 9.Miki M., Wahl P., Auchet J. C. (1982) Biochemistry 21, 3661–3665 [DOI] [PubMed] [Google Scholar]
- 10.Gordon A. M., Homsher E., Regnier M. (2000) Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 11.Ng R., Abelson J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 3912–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shortle D., Haber J. E., Botstein D. (1982) Science 217, 371–373 [DOI] [PubMed] [Google Scholar]
- 13.Wertman K. F., Drubin D. G., Botstein D. (1992) Genetics 132, 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannes F. J., Gallwitz D. (1991) EMBO J. 10, 3951–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiromi Y., Okamoto H., Gehring W. J., Hotta Y. (1986) Cell 44, 293–301 [DOI] [PubMed] [Google Scholar]
- 16.An H. S., Mogami K. (1996) J. Mol. Biol. 260, 492–505 [DOI] [PubMed] [Google Scholar]
- 17.Anson M., Drummond D. R., Geeves M. A., Hennessey E. S., Ritchie M. D., Sparrow J. C. (1995) Biophys. J. 68, 1991–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon T. L., Solomon L. R., Gay L. S., Rubenstein P. A. (1988) J. Biol. Chem. 263, 19662–19669 [PubMed] [Google Scholar]
- 19.Noguchi T. Q., Kanzaki N., Ueno H., Hirose K., Uyeda T. Q. (2007) J. Biol. Chem. 282, 27721–27727 [DOI] [PubMed] [Google Scholar]
- 20.Egelhoff T. T., Titus M. A., Manstein D. J., Ruppel K. M., Spudich J. A. (1991) Methods Enzymol. 196, 319–334 [DOI] [PubMed] [Google Scholar]
- 21.Nagasaki A., Uyeda T. Q. (2008) Exp. Cell Res. 314, 1136–1146 [DOI] [PubMed] [Google Scholar]
- 22.Nagasaki A., Uyeda T. Q. (2004) Mol. Biol. Cell 15, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margossian S. S., Lowey S. (1982) Methods Enzymol. 85, 55–71 [DOI] [PubMed] [Google Scholar]
- 24.Murray J. M. (1982) Methods Enzymol. 85, 15–17 [DOI] [PubMed] [Google Scholar]
- 25.Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 26.Kurokawa H., Fujii W., Ohmi K., Sakurai T., Nonomura Y. (1990) Biochem. Biophys. Res. Commun. 168, 451–457 [DOI] [PubMed] [Google Scholar]
- 27.Kouyama T., Mihashi K. (1981) Eur. J. Biochem. 114, 33–38 [PubMed] [Google Scholar]
- 28.Kodama T., Fukui K., Kometani K. (1986) J. Biochem. 99, 1465–1472 [DOI] [PubMed] [Google Scholar]
- 29.Uyeda T. Q., Kron S. J., Spudich J. A. (1990) J. Mol. Biol. 214, 699–710 [DOI] [PubMed] [Google Scholar]
- 30.Homsher E., Kim B., Bobkova A., Tobacman L. S. (1996) Biophys. J. 70, 1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurth M. C., Wang L. L., Dingus J., Bryan J. (1983) J. Biol. Chem. 258, 10895–10903 [PubMed] [Google Scholar]
- 32.Wang L. L., Bryan J. (1981) Cell 25, 637–649 [DOI] [PubMed] [Google Scholar]
- 33.Harris D. A., Schwartz J. H. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 6798–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doi Y., Frieden C. (1984) J. Biol. Chem. 259, 11868–11875 [PubMed] [Google Scholar]
- 35.Lehman W., Vibert P., Uman P., Craig R. (1995) J. Mol. Biol. 251, 191–196 [DOI] [PubMed] [Google Scholar]
- 36.Murakami K., Yumoto F., Ohki S. Y., Yasunaga T., Tanokura M., Wakabayashi T. (2005) J. Mol. Biol. 352, 178–201 [DOI] [PubMed] [Google Scholar]
- 37.Johnson P., Blazyk J. M. (1978) Biochem. Biophys. Res. Commun. 82, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 38.Saeki K., Sutoh K., Wakabayashi T. (1996) Biochemistry 35, 14465–14472 [DOI] [PubMed] [Google Scholar]
- 39.Vibert P., Craig R., Lehman W. (1997) J. Mol. Biol. 266, 8–14 [DOI] [PubMed] [Google Scholar]
- 40.Belmont L. D., Orlova A., Drubin D. G., Egelman E. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., Cook R. K., Rubenstein P. A. (1993) J. Cell Biol. 123, 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao X., Rubenstein P. A. (2001) J. Biol. Chem. 276, 25598–25604 [DOI] [PubMed] [Google Scholar]
- 43.Holmes K. C., Popp D., Gebhard W., Kabsch W. (1990) Nature 347, 44–49 [DOI] [PubMed] [Google Scholar]
- 44.Feng L., Kim E., Lee W. L., Miller C. J., Kuang B., Reisler E., Rubenstein P. A. (1997) J. Biol. Chem. 272, 16829–16837 [DOI] [PubMed] [Google Scholar]
- 45.Shvetsov A., Musib R., Phillips M., Rubenstein P. A., Reisler E. (2002) Biochemistry 41, 10787–10793 [DOI] [PubMed] [Google Scholar]
- 46.Shvetsov A., Galkin V. E., Orlova A., Phillips M., Bergeron S. E., Rubenstein P. A., Egelman E. H., Reisler E. (2008) J. Mol. Biol. 375, 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Posern G., Sotiropoulos A., Treisman R. (2002) Mol. Biol. Cell 13, 4167–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johara M., Toyoshima Y. Y., Ishijima A., Kojima H., Yanagida T., Sutoh K. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2127–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cook R. K., Blake W. T., Rubenstein P. A. (1992) J. Biol. Chem. 267, 9430–9436 [PubMed] [Google Scholar]
- 50.Rayment I., Holden H. M., Whittaker M., Yohn C. B., Lorenz M., Holmes K. C., Milligan R. A. (1993) Science 261, 58–65 [DOI] [PubMed] [Google Scholar]
- 51.Miller C. J., Reisler E. (1995) Biochemistry 34, 2694–2700 [DOI] [PubMed] [Google Scholar]
- 52.Miller C. J., Wong W. W., Bobkova E., Rubenstein P. A., Reisler E. (1996) Biochemistry 35, 16557–16565 [DOI] [PubMed] [Google Scholar]
- 53.Bookwalter C. S., Trybus K. M. (2006) J. Biol. Chem. 281, 16777–16784 [DOI] [PubMed] [Google Scholar]
- 54.McKillop D. F., Geeves M. A. (1993) Biophys. J. 65, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cammarato A., Craig R., Sparrow J. C., Lehman W. (2005) J. Mol. Biol. 347, 889–894 [DOI] [PubMed] [Google Scholar]
- 56.Pringle J. W. (1981) J. Exp. Biol. 94, 1–14 [Google Scholar]
- 57.Gordon S., Dickinson M. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4311–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuura Y., Stewart M., Kawamoto M., Kamiya N., Saeki K., Yasunaga T., Wakabayashi T. (2000) J. Mol. Biol. 296, 579–595 [DOI] [PubMed] [Google Scholar]