Abstract
The newly described F-BAR (Fer/CIP4 and Bin, amphiphysin, Rvs) family of proteins includes Cdc42-interacting protein-4 (CIP4), formin-binding protein-17 (FBP-17) and transactivator of cytoskeletal assembly-1 (Toca-1), and drives membrane deformation and invagination. Membrane remodeling affects endocytosis, vesicle budding, and cargo selection. The F-BAR family presents a novel family of proteins, which little is known about their in vivo function. We investigated the physiological role of CIP4, by creating Cip4-null mice through homologous recombination. Compared with their wild-type littermates, the Cip4-null mice displayed lower early post-prandial glucose levels. Adipocytes isolated from Cip4-null mice exhibited increased [14C]2-deoxyglucose uptake compared with cells from wild-type mice. The enhanced insulin sensitivity was not due to higher levels of insulin or phospho-Akt, a critical player in insulin signaling. However, higher glucose transporter 4 (GLUT4) levels were detected in muscle membrane fractions in Cip4-null mice under insulin stimulation. Mouse embryonic fibroblasts from Cip4-null mice demonstrated decreased transferrin uptake, fluorescein isothiocyanate-dextran, and horseradish peroxidase uptake, indicating that CIP4 affects multiple modes of endocytosis. These studies demonstrate a physiological role for CIP4 in endocytosis leading to a whole animal phenotype.
Keywords: Cytoskeleton, Cytoskeleton/Actin, GENE/Knockout, Membrane, Membrane/Function, Membrane/Trafficking, Membrane/Turnover
Introduction
CIP4 is a member of the newly recognized group of proteins related to the Bin/amphiphysin/Rvs (BAR)3 superfamily of proteins. Other members of the F-BAR subfamily closely related to CIP4 are transactivator of cytoskeletal assembly-1 (Toca-1) and formin-binding protein-17 (FBP-17) (1–3). These proteins contain the previously identified N-terminal Fes/Fer/CIP4 homology domain and an extended region comprising the F-BAR domain (4, 5). Structural analysis of the F-BAR domain reveals a positively charged, banana-shaped α-helical dimer. The concave surface of the dimer interacts with the negatively charged phospholipids to either promote or sense membrane curvature (6). Recently, cryoelectron microscopy suggests that F-BAR proteins are recruited to membranes and through tail-tail interactions form helical scaffolds (7). These helical scaffolds deform membranes into tubules potentially aiding in endocytotic processes. Membrane deformation and scission are facilitated through recruitment of cofactors through the C-terminal SH3 interactions of CIP4. Its SH3 domain interacts with proline-rich protein Wiskott Aldrich syndrome protein (WASp) and the GTPase dynamin (8, 9). WASp recruitment to plasma membrane generates filamentous actin or cortical actin through Arp2/3 activation, which is crucial in receptor-mediated endocytosis (10). Dynamin mediates scission of emerging vesicles from the plasma membrane (8). Thus, the novel family of CIP4 proteins belongs to a unique class of proteins that regulate endocytosis (8, 11).
CIP4 was first identified using the activated form of Cdc42 or the non-catalytic domains of the Src kinase Lyn as bait in yeast two-hybrid screens (12, 13). An alternate splice form (CIP4h or CIP4/2) was discovered in a PCR screen (14) and in a yeast two-hybrid of 3T3L1 cells with the RhoGTPase Tc10 as bait (15). CIP4 was reported to colocalize with Tc10 and GLUT4, the insulin-responsive glucose transporter in muscle and fat tissue of mammals. Glucose homeostasis requires dynamic control of the number of GLUT4 transporters in the plasma membrane, which the cell achieves by controlling GLUT4 membrane expression through a complex cascade of signaling pathways that regulate exo- and endocytosis (16). Under resting conditions, prior to a glucose load, GLUT4 resides primarily in intracellular storage vesicles. Upon insulin stimulation, the insulin receptor phosphorylates the insulin receptor substrate-1, which facilitates the activation of phosphatidylinositol 3-kinase and its downstream target Akt/protein kinase B. These signaling events lead to immediate trafficking of GLUT4 to the cell surface. Although the signaling events underlying the translocation of GLUT4 to the plasma membrane are well established, the molecular events that lead to endocytosis of GLUT4 are less well understood (17). Like exocytosis, endocytosis requires dynamic remodeling of the cytoskeleton of the cell through overlapping sets of proteins. Key components of actin-associated cytoskeletal assembly are Src protein-tyrosine kinases, the RhoGTPases, the Arp2/3 activator (N-)WASp, and scaffolding proteins such as cortactin.
Based on that observation and the growing recognition that F-BAR domains play a role in regulating biological processes that require membrane curvature deformation, we hypothesized that CIP4 may regulate GLUT4 endocytosis. We generated Cip4-null mice by homologous recombination and found that deletion of Cip4 resulted in enhanced early glucose uptake. These physiological observations were substantiated at the cellular level by demonstrating that surface expression of GLUT4 on muscle cells under insulin stimulation is altered in the absence of CIP4. We found Cip4−/− adipocytes took up more glucose and Cip4−/− murine embryonic fibroblast (MEF) cells showed slower endocytosis of transferrin. We concluded that greater glucose tolerance of the Cip4-null mice was due to decreased and delayed GLUT4 endocytosis. This occurred despite redundant tissue expression of Toca-1 and FBP-17. These results suggest that specific F-BAR proteins play a non-redundant physiological role in vesicle trafficking from the membrane.
MATERIALS AND METHODS
Generation of Cip4-null Mice
A Cip4 gene targeting vector was constructed by subcloning a 1.1-kb segment (short arm) immediately upstream of the ATG site of the murine Cip4 gene into the BamHI and XhoI sites of the pKI-lacZ vector (14). Subsequently the long arm, a 5-kb fragment containing part of intron 3 through exon 9a, was subcloned into the SalI and EcoRI sites. The short and long arms were generated by PCR using a BAC clone containing the murine Cip4 gene as a template. Mutant Cip4 embryonic stem cells were generated by the MD Anderson Cancer Center Genetically Engineered Mouse Facility by electroporating a total of 25 μg of the pKI-lacZ Cip4 targeting vector linearized with NotI into R1 embryonic stem cells derived from 129 mice. Cells were selected in the presence of G418 and ganciclovir, and two clones positive for the mutant Cip4 were identified by PCR screening. Homologous recombination was confirmed by PCR with primers specific for the mutant allele. Each clone was expanded, aggregated with ICR morulas, and implanted in ICR females. One of the litters had a germline competent 85% chimeric male that was bred to Institute for Cancer Research (ICR) and 129 females. ICR heterozygotes were identified by PCR. Experiments were performed on 129 mice and approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas MD Anderson Cancer Center.
Southern Blot
To perform Southern blotting, 10 μg of DNA was digested by NcoI overnight, resolved on 0.8% agarose gel, and transferred to a nitrocellulose membrane using the standard method. A 1.1-kb sequence on the long arm was amplified by PCR and labeled with [32P]dATP using Prime It® II Random Primer Labeling Kit (Stratagene). After hybridization and washing, the membrane was exposed to x-ray film at −80 °C for 3 days with intensifying screen.
Genotyping
Genomic DNA was isolated from mouse tail snips by Qiagen DNeasy tissue kit. For PCR analysis, the primers were 5′-CCTGAAGTCAAGTAAGAAT-3′ and 5′- CACACACAGTTTTTTCCAATCCTCG-3′ for the wild-type reaction. For the knock-out reaction the primers were 5′- GAGTCCTCAGATCTGGAAGAG-3′ and 5′-CTTGTTGGTCAAAGTAAACGAC-3′. Reaction conditions were: 94 °C for 1 min for 1 cycle, 94 °C for 1 min, 61 °C for 1 min, 72 °C for 1 min for 30 cycles, and 72 °C for 5 min for 1 cycle. PCR products were resolved on a 2% agarose gel.
mRNA Isolation and Quantitative Real Time RT-PCR
25–50 mg of tissues from wild-type and knock-out mice were homogenized using a mechanical homogenizer. Total RNAs were isolated using Qiagen RNeasy Mini Kit following the manufacturer's protocol. 2 μg of total RNA was processed with DNase I (Invitrogen) for 10 min to remove the genomic DNA. The digestion reaction was stopped by adding EDTA and heating at 65 °C for 10 min. First-strand cDNA was synthesized using Superscript II Reverse Transcriptase (Invitrogen) and used as the template to perform the qPCR. iQ SYBR Green Supermix (Bio-Rad) was used for the qPCR. Both cDNA synthesis and qPCR followed the manufacturer's manuals. We used these primers for qPCR: CIP1340F, 5′-TCC TCA GTA ACC GAG GGG ACA GC; CIP1470R, 5′-TCT TCT GAG CTC TCC TTA TTG TCC; RT-TOCAF, 5′-CGA GAA AGC CCT GAG GGA AGT; RT-TOCAR, 5′-CGA AGG CAG GGG ATC ATC ATC TTC G; RT-FBPF, 5′-GCC TTG GTC ACG TTG ATG TCA G; RT-FBPR, 5′-CGC TCA GTC AAG TAA GAG GAG GTT TGA GCG; ACTIN138F, 5′-CAA TAG TGA TGA CCT GGC CGT; ACTIN138R, 5′-AGA GGG AAA TCG TGC GTG AC.
Antibodies, Constructs, and Western Blot Analysis
Various mouse tissues were harvested and placed directly into 1% Nonidet P-40 lysis buffer supplemented with protease and phosphatase inhibitors and homogenized. Protein concentrations were determined by Bradford assay (Bio-Rad) and equivalent quantities of protein were mixed with 2× Laemmli buffer (Bio-Rad) and boiled for 5 min. Lysates were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA), blocked in 5% milk, TBS-T, and probed with specific antibodies available from commercial sources. Monoclonal antibodies CIP4 (BD Biosciences, San Jose, CA) and actin (Santa Cruz Biotechnology, Santa Cruz, CA) were purchased from commercial sources. Immunoblotting for components of the Akt signaling pathway was performed with antibodies from Cell Signaling Technology (Danvers, MA): pAkt S473, Akt. Membranes were washed in phosphate-buffered saline with 0.1% Tween, incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology), and visualized by enhanced chemiluminescence (Western Lightning®, PerkinElmer Life Sciences). Membranes were stripped by incubating in Western blot stripping buffer (Restore®, Pierce) at 37 °C, and re-blocked in 5% milk for further probing.
Metabolic Evaluation
Blood glucose concentration was determined by directly measuring the tail blood using a One Touch Ultra glucose meter (LifeScan Inc.). For the glucose tolerance test, 5–8 age- and sex-matched mice were fasted for 12 h with free access to water prior to the glucose challenge. 1.5 mg of glucose/1 g of body weight was injected intraperitoneally, and blood glucose levels were measured at 0, 20, 50, and 120 min. 100 μl of blood was collected from the mouse tail and centrifuged to obtain serum, and serum insulin levels were measured by an enzyme-linked immunosorbent assay using a commercial kit (Ultra Sensitive Rat Insulin ELISA Kit, Crystal Chem Inc., Chicago, IL). For the insulin tolerance test, 3-month-old male mice with a n = 3 were fasted for 4 h with free access to water and challenged with 0.25 units/kg of novolin insulin injected intraperitoneally. A basal glucose level was recorded and measured at 15, 30, 60, 90, and 120 min. For metabolic work-up, serum samples were collected from the tail of randomly fed mice, and serum triglyceride and cholesterol levels were measured by the Section of Veterinary Laboratory Medicine at University of Texas MD Anderson Cancer Center.
Creation of Immortalized Cell Lines from Cip4+/+ and Cip4−/− Mice
Primary mouse brown fat cells were isolated from a newborn mouse that was 1–2 days old. The brown fat pad was removed from the sacrificed mouse, minced, and digested with 1.5 mg/ml of collagenase (Roche) in 37 °C for 40 min. The digested tissues were filtered and centrifuged to harvest the primary brown fat cells. The brown fat cells are suspended with DMEM, high glucose, 20% fetal bovine serum, without sodium pyruvate. Two days later the cells were immortalized in a solution containing 50% pBABE-puro SV40 large T virus stock (obtained from Ronald Kahn, Harvard Medical School), 50% culture medium, and 4 μg/ml of Polybrene. The immortalized cells were cultured and selected by DMEM with high glucose, 20% fetal bovine serum, and 2 μg/ml of puromycin. The immortalized brown fat cells are grown in DMEM with high glucose and 10% fetal bovine serum. MEF were isolated from day 13.5 mouse embryo. Embryos were minced and digested for 15 min at 37 °C with trypsin (Invitrogen). The suspension cells were collected and cultured in MEF medium (DMEM with 10% fetal bovine serum, 100 μm non-essential amino acids with 2 mm l-glutamine). MEFs were immortalized by pBABE-puro SV40 large T virus using puromycin selection.
Glucose Transport in Immortalized Adipocytes
In 6-well plates, fully differentiated wild-type or Cip4-null adipocytes were incubated at 37 °C for 2 h with DMEM containing 1% bovine serum albumin, and then washed with Krebs-Ringer buffer (130 mm NaCl, 5 mm KCl, 1.3 mm CaCl2, 1.3 mm MgSO4, 25 mm HEPES, pH 7.4). The cultures were further incubated without glucose in Krebs-Ringer buffer containing 1% bovine serum albumin for 2 h. Subsequently, cultures were stimulated with or without insulin (100 nm) for 15 min. Glucose uptake was initiated by addition of 200 μCi/ml of [14C]2-deoxy-d-glucose (GE Radiochemicals, Piscataway, NY) to a final assay concentration of 0.6 μCi/ml for a further 5 min. Transport was terminated by transferring cultures to ice followed by three washes with cold (4 °C) PBS. The cells were solubilized with 0.05% SDS, 0.1% Triton X-100 and incorporated 14C was determined by scintillation counting. Nonspecific uptake and trapping in the extracellular space was determined by measuring uptake in the presence of 10 μm cytochalasin B (Biomol, Plymouth Meeting, PA). Specific uptake was normalized to total protein content as described (18).
Subcellular Fractionation
Subcellular fractionation was performed as described elsewhere (19, 20). Briefly, quadriceps muscles from wild-type and knock-out mice were isolated, minced, and homogenized 3 times for 10 s in ice-cold buffer (20 mm HEPES, 250 mm sucrose, 1 mm EDTA, 5 mm protease inhibitor mixture, 1 mm phenylmethylsulfonyl fluoride, pH 7.4). Tissue homogenates were spun at 2000 × g for 10 min at 4 °C to pellet any remaining large debris. Supernatant was recovered and protein concentration was determined by a Bradford assay. Equivalent amounts of protein were spun at 9,000 × g for 20 min at 4 °C. The resulting pellet (pellet 1) was resuspended in 100 μl of PBS with 5 mm protease inhibitor mixture, EDTA, and 1 mm phenylmethylsulfonyl fluoride. Supernatant was spun at 60,000 × g in an SW60 rotor (Beckman Coulter) for 90 min at 4 °C. The supernatant was recovered (cytosol), and the pellet was resuspended in PBS with 5 mm protease inhibitor mixture, 1 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride and loaded on a 10–30% sucrose gradient and spun at 48,000 × g for 55 min at 4 °C in an SW60 rotor. Nine equal volume fractions were isolated from the gradient beginning at the top. The pellet (Pellet 2) was resuspended in PBS with 5 mm protease inhibitor mixture, 1 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride. Fractions were subsequently immunoblotted for GLUT4 (rabbit polyclonal, Cell Signaling).
Transferrin Endocytosis
Bovine holo-transferrin (Invitrogen) was labeled with 125I (GE Radiochemicals), using IODO-BEADS (Pierce Biotechnology) according to the manufacturer's protocol. Mouse embryonic fibroblasts were depleted of transferrin by washing with serum-free growth medium for 1–2 h at 37 °C. After starvation, cultures were exposed to a saturating concentration (5 μg/ml) of 125I-labeled transferrin for the indicated times. After the indicated time points, plates were removed, placed on ice, and washed six times with cold, neutral pH buffer (150 mm NaCl, 5 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 20 mm HEPES). Monolayers were then incubated with pH 2.0 buffer (500 mm NaCl, 0.2 n acetic acid) for 5 min at 4 °C. pH 2.0 buffer was then discarded and wells were washed with neutral pH buffer 3 more times. Monolayers were then lysed in 0.05% SDS, 0.1% Triton X-100, scraped, and incorporated 125I-transferrin measured by γ-counter and normalized to total protein content. Nonspecific, cell-associated 125I-transferrin was assessed in parallel by exposing cells to 5 μg/ml of 125I-labeled transferrin in the presence of a 200-fold excess of unlabeled holo-transferrin.
FITC-Dextran Internalization Assays
The uptake of FITC-dextran was performed as described (18, 28). FITC-dextran was diluted in pre-warmed medium and added to Cip4+/+ or Cip4−/− MEF and incubated for 15 min. After internalization experiments, cells were washed 3 times and gently scraped in PBS containing 4% bovine serum albumin. FITC-dextran was then determined by flow cytometry. Flow cytometry was performed using a 4-color FACSCalibur (BD Biosciences). Side scatter, forward scatter, and FITC outputs, minimally 10,000 events, were collected in list mode form and analyzed with FloJo (Treestar, Inc., Ashland, OR). All values are corrected for cultures exposed to FITC-dextran at 4 °C, as a negative (i.e. no endocytosis) control.
HRP Uptake Assay
The extent of horseradish peroxidase (HRP) internalization in Cip4+/+ and Cip4−/− MEF was assessed using protocols described previously (21, 22). Cells were cultured in 6-well dishes followed by overnight starvation in 0.4% serum. Cells were incubated with 4 mg/ml of HRP in DMEM with 20 mm HEPES, 0.2% bovine serum albumin, pH 7.4, for 7 min at 37 or 4 °C. The uptake was stopped by quick aspiration of the medium and five PBS washes at 4 °C. The cells were then treated with 0.1% Pronase in PBS, to detach cells and remove residual HRP nonspecifically bound to the plasma membrane. Cells were then spun at 4 °C through a sucrose cushion (0.5 m in PBS, pH 7.4) for 5 min at 1,000 × g followed by 5 min at 14,000 × g. The cells were lysed in PBS, 0.5% Triton X-100, and aliquots were assayed for enzyme activity (A490 nm) by using o-phenylenediamine as a substrate and normalized to protein concentration.
Statistical Analysis
Statistical analysis of the difference between wild-type and knock-out mice was performed with a Student's t test for individual glucose tolerance, serum insulin, the area under the curve, and endocytosis assays. All tests were carried out at the 95% confidence interval using Microsoft Excel.
RESULTS
Generation of CIP4 Knock-out Mice
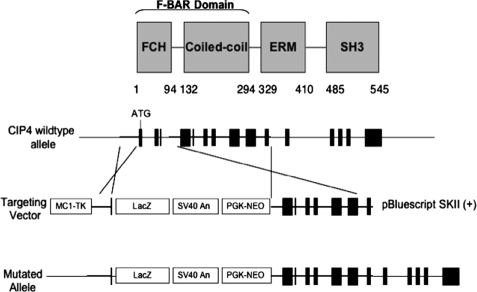
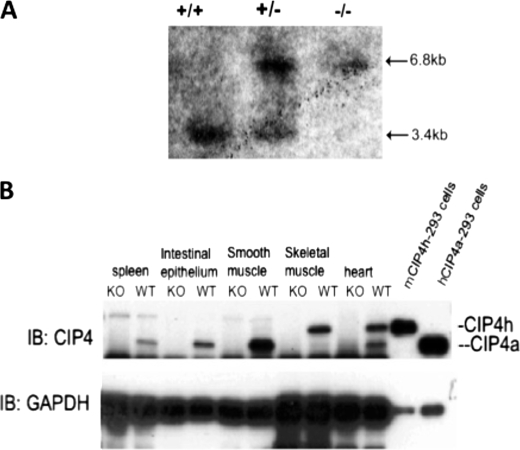
The murine Cip4 gene consists of 15 exons, encoding a protein of 546 amino acids. There is high degree of conservation in both nucleotide (84%) and amino acid (87%) sequences between murine and human CIP4 (14). All four CIP4 isoforms result from alternative splicing at the 3′ region of the Cip4 gene. To abrogate all CIP4 isoforms, we constructed a targeting vector that replaced the first three exons of the murine Cip4 gene with a neomycin-resistant pKI-lacZ vector (Fig. 1). Chimeric mice were mated with ICR females to generate heterozygous mice, which were subsequently bred onto the 129 genetic background. Genotype was determined Southern blotting with DNA isolated from tail snips (Fig. 2A). After NcoI digestion of genomic DNA, the wild-type genomic DNA produces a 3.4-kb fragment that can be detected by probe from a sequence on the long arm, and the targeted (knock-out) genomic DNA produces a 6.8-kb fragment. Gene disruption resulted in loss of transcript as detected by RT-PCR (data not shown) and protein expression as detected by Western blotting (Fig. 2B). The two most prevalent isoforms are CIP4a and CIP4h (also known as CIP4/2). CIP4a, consisting of 545 amino acids, is ubiquitously expressed, but CIP4h, consisting of 601 amino acids, is found in highest levels in skeletal muscle, and cardiac muscle. As shown in Fig. 2B, both isoforms were absent in the Cip4−/− mice. Genotypes of heterozygous litters followed the predicted Mendelian frequency. Cip4-null mice showed no obvious defects in structural or behavioral development and were fertile. Postnatal growth and weight patterns were comparable among wild-type and Cip4-null mice (data not shown).
FIGURE 1.
Strategy for targeted disruption of the Cip4 gene. Upper panel, outlines the protein domains in CIP4, whereas lower panels, the genomic organization of the mouse Cip4 gene with the detailed strategy for gene disruption.
FIGURE 2.
Demonstration of loss in expression of Cip4. A, genomic DNA was harvested from tail-snips and digested with NotI and BamHI. Digests were hybridized to the cDNA probe corresponding to a 1.1-kb fragment of the CIP4 gene. B, whole cell lysates were prepared from progeny of the backcross, resolved by SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, and immunoblotted for CIP4. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IB, immunoblot; WT, wild-type; KO, knock-out.
Enhanced Glucose Tolerance of CIP4-null Mice
CIP4 has been reported to co-localize with GLUT4 in 3T3 adipocytes and has been implicated in GLUT4 trafficking in response to insulin (15). To investigate the function of CIP4 in the null mouse, we studied insulin-sensitive tissues muscle and fat.
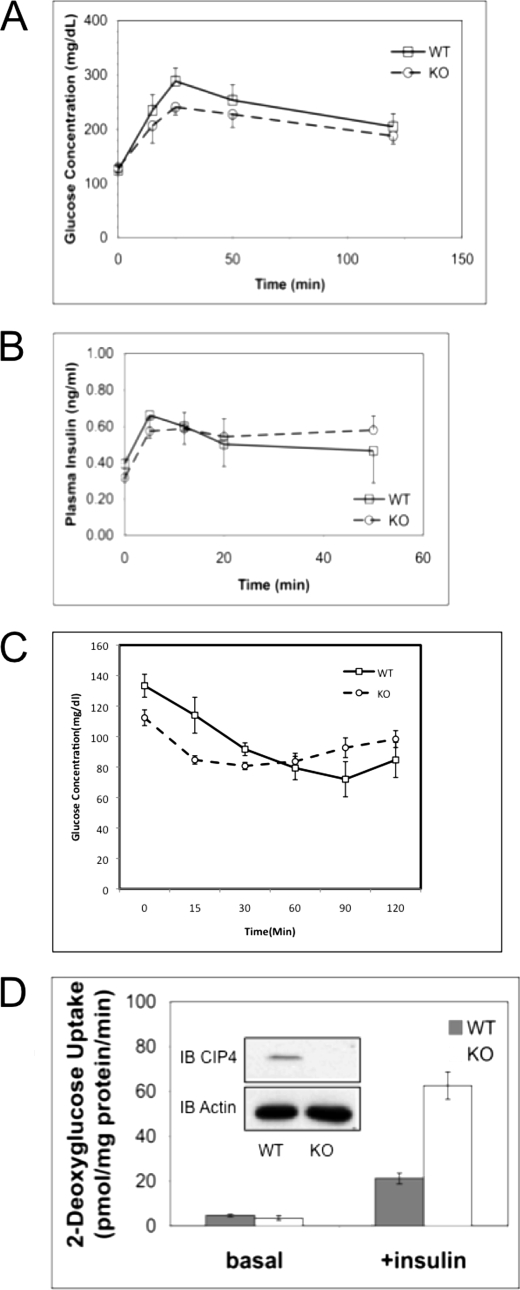
We performed a glucose tolerance test using 5–8 age- and sex-matched mice. After a 12-h fast, mice were given a glucose challenge. 20 min after the challenge, the blood glucose level in Cip4−/− mice was 22% lower than that of the wild-type mice (p < 0.05). A time course showed that post-peak serum glucose levels were comparable (Fig. 3A), suggesting that absence of CIP4 promoted rapid glucose uptake but did not affect counter-regulatory processes. Measurement of serum insulin levels showed no significant difference between Cip4−/− and wild-type mice (Fig. 3B) during the glucose challenge. To monitor glucose levels following insulin challenge, 3-month-old male mice were fasted for 4 h. Following fasting Cip4−/− mice showed a lower initial blood glucose. The mice were then challenged with 0.25 units/kg of insulin and the blood glucose levels measured. Cip4−/− mice displayed a rapid clearance of blood glucose in comparison to wild-type mice. This data suggest increased glucose uptake following insulin challenge.
FIGURE 3.
The absence of CIP4 promotes glucose uptake. A, cohorts of age and sex-matched mice (Cip4+/+ and Cip4−/−) were starved overnight and then challenged with 2.0 mg/kg of body weight glucose. Serum glucose was measured by glucometer. B, cohorts of age and sex-matched mice were randomly fed, starved overnight, or glucose challenged for 25 min followed by analysis of serum insulin concentrations. Values are expressed as mean ± S.E. (n = 5, p < 0.05). C, cohorts of 3-month-old male mice were intraperitoneally injected with 0.25 units/kg of insulin and blood glucose monitored over 0–120 min post-injection. Values represent mean ± S.E. (n = 3, p < 0.05). D, the insulin-mediated uptake of 2-deoxyglucose was measured by serum and glucose starvation of fully differentiated Cip4−/− or Cip4+/+ adipocytes followed by stimulation with 100 nm for 15 min. Subsequent to the 15-min stimulus, [14C]2-deoxy-d-glucose was added at a final concentration of 0.6 μCi/ml for a further 5 min. Cells are then lysed for protein normalization and collected for scintillation counting. Basal indicates cultures without insulin stimulation. Values represent mean ± S.E. (n = 3, p < 0.05). IB, immunoblot; WT, wild-type; KO, knock-out.
Enhanced glucose uptake at the cellular level confirmed the effect seen in the whole organism, as determined by 2-deoxyglucose uptake in adipocytes derived from Cip4−/− and Cip4+/+ mice (Fig. 3C). However, there were no significant differences in mouse mass or fat content (data not shown). Triglyceride and serum cholesterol levels were similar between Cip4−/− and wild-type mice (data not shown). The metabolic studies indicate that the absence of CIP4 results in enhanced early uptake of glucose in insulin responsive cells.
CIP4 Affects Surface Expression of GLUT4
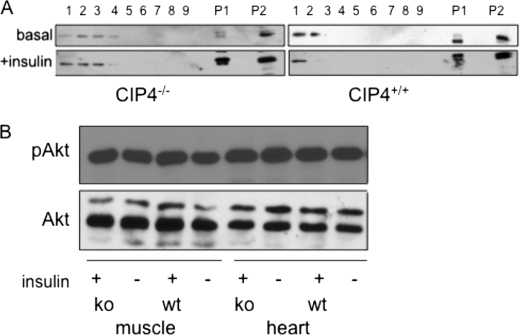
Following a glucose load, skeletal muscle provides the major vehicle for glucose clearance through insulin-stimulated trafficking of GLUT4. We hypothesized that decreased levels of post-prandial glucose in Cip4-null mice might be due to increased levels of GLUT4 on the cell surface, facilitating a greater acute clearance. Subcellular fractionation was performed on skeletal muscle from wild-type and Cip4-null mice. As shown in Fig. 4A, there were greater GLUT4 levels in plasma membrane fractions (P1, P2, and the early fractions) in the Cip4−/− mice at both basal and 25 min post-insulin stimulation. Immunofluorescence microscopy of adipocytes derived from either wild-type or Cip4-null mice suggested greater surface expression of GLUT4 in the absence of CIP4 (supplemental Fig. S1). Using a quantitative flow cytometric approach with L6 myoblasts that express myc-tagged GLUT4, we showed that RNA interference-mediated knockdown of CIP4 increased insulin-stimulated glucose uptake, which was associated with greater surface expression of GLUT4 (23).
FIGURE 4.
CIP4 negatively regulates surface GLUT4 expression through Akt-independent signaling. A, subcellular fractions were isolated from whole tissue lysate of mouse skeletal muscle (quadriceps femoralis) collected from the top of the gradient (fraction 1) to the bottom of the gradient (fraction 9), with the most buoyant fractions (fractions 1, 2, and 3) representing plasma membrane. P1 and P2 contain almost all of the plasma membrane and T tubule marker proteins (20). P1 contains most of the nuclei, mitochondria, and other heavy organelles. P1 also contains surface membrane. GLUT4 also resides within the intracellular membrane fractions. Protein concentration was normalized for each fractionation. B, wild-type or homozygous Cip4-null mice were fasted overnight followed by intraperitoneal injection of insulin. 20 min after injection, muscle and heart tissues were harvested and lysates prepared. The lysates were used for Western blotting for pAkt and Akt. wt, wild-type; ko, knock-out.
Increased surface expression of GLUT4 could either be due to increased insulin signaling, or decreased endocytosis. We first determined whether enhanced insulin signaling accounted for increased GLUT4 exocytosis. We measured the activity of phosphatidylinositol 3-kinase, a critical mediator for insulin signaling, by Western blotting for the phosphorylated form of Akt, phospho-Akt levels were not different between wild-type and Cip4-null cardiac or skeletal muscle tissue (Fig. 4B).
CIP4 Delays Endocytosis
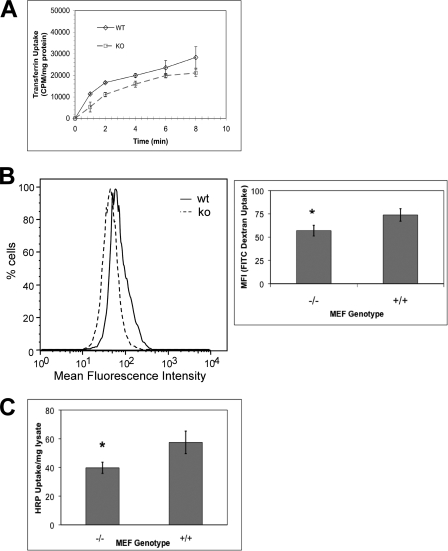
To further verify the role of CIP4 in multiple modes of endocytosis, we isolated and immortalized MEF from both Cip4−/− and Cip4+/+ mice. Using CIP4 wild-type and Cip4-null MEF cells, we measured 125I-transferrin uptake to establish a role of CIP4 in endocytosis. As shown in Fig. 5A, Cip4−/− MEF displayed delayed and decreased transferrin receptor endocytosis. At the earliest time point (1 min), endocytosis was reduced by 55%. As time increases, the inhibition of endocytosis via Cip4 knock-out lessens. Upon calculating the Km, we determined the rate of transferrin receptor endocytosis is significantly decreased in comparison to wild-type. Together the data suggests both delayed and decreased endocytosis in Cip4−/− MEFs.
FIGURE 5.
Clathrin-mediated and fluid phase endocytosis is decreased in the absence of CIP4. A, transferrin endocytosis in SV40-transformed Cip4−/− and Cip4+/+ MEF was measured following stimulation with a saturating concentration of 125I-transferrin for the indicated times ± S.E. (n = 3, p < 0.05). Asterisks indicate means that differ significantly from the scrambled/vector control. B, Cip4 wild-type (WT) or knock-out (KO) MEF were treated for 30 min with 0.25 mg/ml of FITC-dextran and the amount internalized determined by flow cytometry ± S.E. (n = 5, p < 0.05). C, fluid phase uptake in Cip4+/+ and Cip4−/− MEF was assessed by serum starvation and treatment with 4 mg/ml of HRP for 7 min. Cells were collected, nonspecifically bound HRP removed, and internalized HRP determined via enzymatic assay ± S.E. (n = 4, p < 0.05).
We also analyzed the role of CIP4 in fluid phase endocytosis by two methods: HRP uptake (21, 22) and accumulation of FITC-dextran (Fig. 5, B and C). In both cases, Cip4 deletion led to an ∼30% decrease in fluid phase macropinocytosis, establishing the role of CIP4 in multiple modes of endocytosis.
DISCUSSION
Endocytosis involves multiple molecules that work together in a highly coordinated spatiotemporal process to drive the deformation of the plasma membrane. A newly recognized component in membrane reorganization is the F-BAR family of proteins. Structural and biochemical analysis of the F-BAR domain provides support that these proteins function to sense and affect membrane curvature (6, 24). The F-BAR domain dimerizes, and through positively charged amino acid residues on the concave surface interact with membrane phospholipids (7). In addition, through its C-terminal SH3 domain, CIP4 interacts with N-WASp and dynamin and acts as a multifunctional adapter (5, 9, 23, 25). Through these interactions CIP4 has the potential to act in a diverse range of internalization processes.
As shown in primary cells, CIP4 promotes internalization through both macropinocytosis and endocytosis. We observed defects in both processes even though there is ubiquitous expression of CIP4, Toca-1, and FBP-17 by RT-PCR and Western blotting (data not shown). This is potentially through the ability of CIP4 to promote both membrane deformation and endocytosis through the coordinatation of membrane deformation and SH3-mediated association with N-WASp and dynamin. During endocytosis, CIP4 could potentially be involved in two potentially important steps: vesicle formation and scission. At formation, CIP4 could be recruited and enhance membrane curvature. During scission, CIP4 could accumulate around the neck along with dynamin to constrict the neck. Disruption of these processes could have severe consequences on vesicle formation including endocytosis of cell surface proteins and macropinocytosis.
Endocytosis regulates several biological processes through regulation of cell surface receptors. Following insulin stimulation, GLUT4 is rapidly redistributed from intracellular storage vesicles to the plasma membrane. This facilitates glucose uptake in skeletal and adipose tissues. The insulin receptor triggers this process through phosphatidylinositol 3-kinase dependent and independent mechanisms (26). GLUT4 remains at the cell surface until glucose levels fall, whereupon it is internalized by endocytosis. Little is known about the precise mechanisms that regulate GLUT4 endocytosis. Because the number of patients with insulin-resistant diabetes is increasing, a greater understanding of the molecular interactions that control glucose levels is important for development of effective therapies.
We have also demonstrated a unique role of CIP4 in macropinocytosis. Macropinocytosis is involved in fluid phase endocytosis of substrates including antigens, low density lipoproteins, and cell surface adhesion proteins (27–29). Macropinocytosis does not involve the identical set of effectors and regulators as receptor-mediated endocytosis. One major difference is the size of the membrane deformation. Potentially CIP4 could be contributing similar functions in divergent internalization processes. Our data also suggest that CIP4 possesses a unique and non-redundant ability to deform membranes and recruit binding partners contributing to different internalization processes. This is potentially through contributions to both vesicle formation and scission by recruiting N-WASp and dynamin. The contribution of distinctive CIP4 proteins in membrane internalization requires further systemic analysis.
Supplementary Material
Acknowledgments
We thank Christopher Thompson for model illustration, Laura Nelson for assistance in managing the breeding colony, Kevin Voelker for advice on preparing muscle sections, Amira Klip for discussions on GLUT4, and Ronald Kahn for pBABE large T SV40 viral stocks.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1HL080052 (to S. J. C.). This work was also supported by a grant-in-aid from the American Heart Association, Charlotte Geyer Foundation, and grants from the University of Texas M.D. Anderson Cancer Center Odyssey Fellowship and the Tennessee Law Foundation (to S. M. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- BAR
- Bin/amphiphysin/Rvs
- Toca-1
- transactivator of cytoskeletal assembly-1
- FBP
- formin-binding protein
- SH3
- Src homology domain 3
- MEF
- murine embryonic fibroblast
- RT
- reverse transcriptase
- qPCR
- quantitative PCR
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- HRP
- horseradish peroxidase
- ICR
- Institute for Cancer Research
- GLUT4
- glucose transporter 4
- WASp
- Wiskott Aldrich syndrome protein.
REFERENCES
- 1.Chitu V., Stanley E. R. (2007) Trends Cell Biol. 17, 145–156 [DOI] [PubMed] [Google Scholar]
- 2.Dawson J. C., Legg J. A., Machesky L. M. (2006) Trends Cell Biol. 16, 493–498 [DOI] [PubMed] [Google Scholar]
- 3.Ho H. Y., Rohatgi R., Lebensohn A. M., Le, Ma, Li J., Gygi S. P., Kirschner M. W. (2004) Cell 118, 203–216 [DOI] [PubMed] [Google Scholar]
- 4.Itoh T., De Camilli P. (2006) Biochim. Biophys. Acta 1761, 897–912 [DOI] [PubMed] [Google Scholar]
- 5.Tsujita K., Suetsugu S., Sasaki N., Furutani M., Oikawa T., Takenawa T. (2006) J. Cell Biol. 172, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimada A., Niwa H., Tsujita K., Suetsugu S., Nitta K., Hanawa-Suetsugu K., Akasaka R., Nishino Y., Toyama M., Chen L., Liu Z. J., Wang B. C., Yamamoto M., Terada T., Miyazawa A., Tanaka A., Sugano S., Shirouzu M., Nagayama K., Takenawa T., Yokoyama S. (2007) Cell 129, 761–772 [DOI] [PubMed] [Google Scholar]
- 7.Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E. H., De Camilli P., Unger V. M. (2008) Cell 132, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamioka Y., Fukuhara S., Sawa H., Nagashima K., Masuda M., Matsuda M., Mochizuki N. (2004) J. Biol. Chem. 279, 40091–40099 [DOI] [PubMed] [Google Scholar]
- 9.Tian L., Nelson D. L., Stewart D. M. (2000) J. Biol. Chem. 275, 7854–7861 [DOI] [PubMed] [Google Scholar]
- 10.Lamaze C., Fujimoto L. M., Yin H. L., Schmid S. L. (1997) J. Biol. Chem. 272, 20332–20335 [DOI] [PubMed] [Google Scholar]
- 11.Bu W., Chou A. M., Lim K. B., Sudhaharan T., Ahmed S. (2009) J. Biol. Chem. 284, 11622–11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dombrosky-Ferlan P., Grishin A., Botelho R. J., Sampson M., Wang L., Rudert W. A., Grinstein S., Corey S. J. (2003) Blood 101, 2804–2809 [DOI] [PubMed] [Google Scholar]
- 13.Aspenström P. (1997) Curr. Biol. 7, 479–487 [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Rudert W. A., Grishin A., Dombrosky-Ferlan P., Sullivan K., Deng X., Whitcomb D., Corey S. (2002) Biochem. Biophys. Res. Commun. 293, 1426–1430 [DOI] [PubMed] [Google Scholar]
- 15.Chang L., Adams R. D., Saltiel A. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12835–12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryant N. J., Govers R., James D. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 267–277 [DOI] [PubMed] [Google Scholar]
- 17.Hou J. C., Pessin J. E. (2007) Curr. Opin. Cell Biol. 19, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moyers J. S., Bilan P. J., Reynet C., Kahn C. R. (1996) J. Biol. Chem. 271, 23111–23116 [DOI] [PubMed] [Google Scholar]
- 19.Yang C., Coker K. J., Kim J. K., Mora S., Thurmond D. C., Davis A. C., Yang B., Williamson R. A., Shulman G. I., Pessin J. E. (2001) J. Clin. Invest. 107, 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou M., Sevilla L., Vallega G., Chen P., Palacin M., Zorzano A., Pilch P. F., Kandror K. V. (1998) Am. J. Physiol. Endocrinol. Metab. 38, E187–E196 [DOI] [PubMed] [Google Scholar]
- 21.Schlunck G., Damke H., Kiosses W. B., Rusk N., Symons M. H., Waterman-Storer C. M., Schmid S. L., Schwartz M. A. (2004) Mol. Biol. Cell 15, 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarar D., Waterman-Storer C. M., Schmid S. L. (2007) Dev. Cell 13, 43–56 [DOI] [PubMed] [Google Scholar]
- 23.Hartig S. M., Ishikura S., Hicklen R. S., Feng Y., Blanchard E. G., Voelker K. A., Pichot C. S., Grange R. W., Raphael R. M., Klip A., Corey S. J. (2009) J. Cell Sci. 122, 2283–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henne W. M., Kent H. M., Ford M. G., Hegde B. G., Daumke O., Butler P. J., Mittal R., Langen R., Evans P. R., McMahon H. T. (2007) Structure 15, 839–852 [DOI] [PubMed] [Google Scholar]
- 25.Itoh T., Erdmann K. S., Roux A., Habermann B., Werner H., De Camilli P. (2005) Dev. Cell 9, 791–804 [DOI] [PubMed] [Google Scholar]
- 26.Tengholm A., Meyer T. (2002) Curr. Biol. 12, 1871–1876 [DOI] [PubMed] [Google Scholar]
- 27.Bryant D. M., Kerr M. C., Hammond L. A., Joseph S. R., Mostov K. E., Teasdale R. D., Stow J. L. (2007) J. Cell Sci. 120, 1818–1828 [DOI] [PubMed] [Google Scholar]
- 28.Kruth H. S., Jones N. L., Huang W., Zhao B., Ishii I., Chang J., Combs C. A., Malide D., Zhang W. Y. (2005) J. Biol. Chem. 280, 2352–2360 [DOI] [PubMed] [Google Scholar]
- 29.Sallusto F., Cella M., Danieli C., Lanzavecchia A. (1995) J. Exp. Med. 182, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.