Abstract
Human antigen R (HuR) is an RNA-binding protein with protective activities against cellular stress. This study considers the mechanisms by which HuR transcriptional regulation occurs in renal proximal tubule cells. Under basal conditions, HuR mRNA is expressed in two forms: one that contains a ∼20-base 5′-untranslated region (UTR) sequence and one that contains a ∼150-base, G+C-rich 5′-UTR that is inhibitory to translation. Recovery from cellular stresses such as thapsigargin and ATP depletion induced increased expression of the shorter, more translatable transcript and decreased expression of the longer form. Analysis of HuR upstream regions revealed sequences necessary for regulation of the shorter mRNA. Within the long, G+C-rich 5′-UTR exist multiple copies of the alternate Smad 1/5/8-binding motif GCCGnCGC. Recovery from ATP depletion induced increases in Smad 1/5/8 levels; further, gel shift and chromatin immunoprecipitation analyses demonstrated the ability of these Smads to bind to the relevant motif in the HuR 5′-UTR. Transfection of exogenous Smad 1 increased HuR mRNA expression. Finally, HuR mRNA expression driven by the Smad-binding sites was responsive to BMP-7, a protein with known protective effects against ischemic injury in kidney. These data suggest that transcriptional induction of a readily translatable HuR mRNA may be driven by a mechanism known to protect the kidney from injury and provides a novel pathway through which administration of BMP-7 may attenuate renal damage.
Keywords: Gene/Promoters, Hormones/Growth Factors, Metabolism/Energy, RNA/Turnover, Tissue/Organ Systems/Kidney, Transcription/Promoter, Transcription/Regulation, Transcription/Smad
Introduction
Cellular recovery from the trauma of environmental stress requires multiple levels of regulation. Acute kidney injury is characterized by a decrease in glomerular filtration rate that is associated with high morbidity and mortality rates (1). One common factor of acute kidney injury is acute tubular necrosis, which is induced by hypoxia caused by ischemia/reperfusion. In the kidney, proximal tubules play a central role in the response to kidney insult because they are the most susceptible to injury. These cells are the major site of necrosis associated with renal ischemia, and the recovery of these regions from such an injury is a reliable predictor of clinical outcome (2). Attenuation of acute tubular necrosis is a major factor in the recovery of filtration rate and restoration of nephron function. Following injury, epithelial cells must dedifferentiate and proliferate for restoration of nephron integrity (3). Although this process has been morphologically defined, the underlying molecular mechanisms for this event have yet to be fully described. Following the insult of ischemic injury, the cells of the nephron must up-regulate numerous genes for the purpose of attenuating injury (4).
TGF-β12 and BMP-7 are members of the TGF-β superfamily of regulatory cytokines. Both have been implicated in kidney injury; however, they appear to work in opposing roles, with BMP-7 demonstrating the capacity to reverse deleterious effects of TGF-β1 (5). Members of the TGF-β family of cytokines exert their effects through serine-threonine kinase receptors. Type II receptors bind ligand and recruit type I receptors. This process triggers a unique signaling pathway through the Smad family of proteins. Smads can be divided into three categories, pathway-restricted Smads (R-Smads), common partner Smad, and inhibitory Smads. R-Smads include Smad 1, Smad 5, and Smad 8 stimulated by BMPs and Smad 2 and Smad 3 stimulated by TGF-β. Phosphorylation of the R-Smad by the type I receptor follows ligand binding by the type II receptor (6). The phosphorylated R-Smads form heteromultimers and associate with the common partner Smad, Smad 4. Inhibitory Smads, including Smad 6 and Smad 7, appear to function as competitive inhibitors because they lack the sequences necessary for activation but still bind common partner Smad.
BMP-7 (previously known as osteogenic protein-1) and Smad 1/5/8 signaling are known to play a major role in kidney development. First, kidneys were shown to be the major producers of BMP-7 in both adult animals and embryos (7). It was subsequently found that BMP-7 induces metanephric differentiation (8) and that BMP-7-null mice exhibit a lack of renal mesenchymal differentiation and an absence of nephrons (9, 10). Further, both BMP type I receptors and BMP-responsive Smads are expressed in the nephrogenic zones of developing kidneys (11, 12). More recently, reports have demonstrated that BMP-7 plays an important role in recovery from multiple types of kidney injury, including damage from ischemia and reperfusion (5, 13–16). Taken together, these findings illustrate the critical role of BMP-7/Smad signaling in kidney development and repair.
Recently, we described the behavior of the mRNA-binding protein HuR in an in vitro model of renal ischemia/reperfusion (17, 18) and in native rat kidneys subjected to ischemia/reperfusion (19). HuR is a ubiquitously expressed protein that binds and stabilizes hundreds to thousands of mRNAs bearing uridine-rich or adenine/uridine-rich sequences (20). Normally localized primarily in the nucleus, HuR undergoes translocation to the cytosol when cells undergo various types of stress, including heat shock, UV irradiation, and nutritional stress (21–23). In the cytosol, HuR is able to bind its target transcripts and protect them from degradation; further, it can also play a positive role in the regulation of translation. In doing so, HuR plays a protective role against cellular apoptosis and senescence (24, 25). Indeed, abnormal overexpression of HuR has been correlated with a tumorigenic phenotype in multiple cancer types, and recent studies in a breast tumor model have defined a number of signaling pathways through which HuR can promote its transforming effects, including those involved in cell cycle control, inhibition of apoptosis, and promotion of signaling cascades (26, 27). We recently demonstrated that energy depletion in a renal epithelial cell model induces nucleocytoplasmic translocation of HuR as well as alterations in its expression (17, 18). When subjected to ATP depletion and recovery in vitro, the porcine proximal tubule cell line LLC-PK1 mimics a number of the responses of ischemia/reperfusion-injured proximal tubule cells in native kidney, including disruption of the actin cytoskeleton (28) and activation of heat shock proteins and protein kinases (29, 30). We found that ATP depletion of LLC-PK1 cells induces slow translation-mediated increases in HuR protein, whereas reversion to normal growth medium induces HuR transcription without accompanying increased translation. We hypothesized that this new transcription may act as a preconditioning mechanism, because a second ATP depletion results in rapid new translation of HuR (18). However, the mechanisms by which HuR is transcriptionally regulated following stress have yet to be determined. These data present mechanisms identified for transcriptional regulation of HuR and provide an initial characterization of essential promoter elements required for HuR transcriptional activation during recovery of renal epithelia from cellular stress.
EXPERIMENTAL PROCEDURES
Cell Culture
LLC-PK1 cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium containing penicillin/streptomycin supplemented with fetal bovine serum (10%) at 37 °C in 5% CO2. For depletion of ATP from these cells, the cultures were first grown to confluency; then medium was replenished, and the cultures were incubated overnight. The cells then were rinsed twice with phosphate-buffered saline, and the culture medium was replaced with prewarmed Dulbecco's modified Eagle's medium base supplemented with l-glucose, sodium bicarbonate, and 0.1 μm antimycin A for 0–4 h. Mock treated cells underwent the same procedure, but the culture medium was replaced with normal growth medium. In some experiments, ATP-depleted cells were allowed to recover by returning the cells to normal growth medium. For experiments testing effects of BMP-7 on HuR expression, human recombinant BMP-7 (25 ng/ml) was added for 6 h to LLC-PK1 cells following 16 h of incubation in 0.5% fetal bovine serum-supplemented Dulbecco's modified Eagle's medium. During incubation, the serum content was lowered to 0.2%. BMP-7 was obtained from R & D Systems (Minneapolis, MN). Thapsigargin (an inhibitor of SERCA) was purchased from Sigma and used at a final concentration of 1 μm. LLC-PK1 cells were treated with thapsigargin for 1 h and allowed to recover in normal growth medium for 6 h. After the harvesting of cells, RNA was isolated by TRIzol (Invitrogen) according to the manufacturer's specifications.
Ribonuclease Protection Assay
RNase protection assays (RPAs) were performed using the RPA III kit (Ambion Corp., Austin, TX) according to the manufacturer's specifications. For RPAs of porcine HuR, a cDNA template was generated that spanned from upstream sequences 100 bp into the HuR coding region. This template was subcloned into the vector pCRII-TOPO (Invitrogen) and was transcribed in vitro in the presence of a ribonucleotide mixture containing 40 μCi of [32P]UTP (800 Ci/mmol) and T7 RNA polymerase (Maxiscript; Ambion Corp). Transcription was terminated by the addition of DNase I, and labeled RNA was purified by polyacrylamide gel electrophoresis. The full-length RNA probe was excised and eluted from the gel as specified by the manufacturer. The HuR-specific probe (4 × 104 cpm) was mixed with 20 μg of total LLC-PK1 RNA in the presence of hybridization buffer, then heat denatured, and hybridized for 16 h at 56 °C. As a control, the HuR-specific probe was incubated with 20 μg of yeast tRNA. Following digestion with RNases A and T1, protected double-stranded RNA was precipitated, harvested by centrifugation, and run in a 5% polyacrylamide-urea gel. Protected RNA fragments were visualized by autoradiography and quantified using Image J software (31).
Luciferase Reporter Constructs and Transfected Cell Lines
Promoter constructs were subcloned into the firefly luciferase expression vector pGL4.14 (Promega). The deletions were performed by PCR and confirmed by sequencing. Stable transfections were performed using Lipofectamine and Plus reagent (Invitrogen). Forty-eight hours post-transfection, the cells were treated with 600 μg/ml hygromycin (Invitrogen); following expansion of cells, individual clones were selected and analyzed for basal luciferase activity using Bright-Glo reagent (Promega) and a GloMax 96 microplate luminometer (Turner Biosystems, Sunnyvale, CA) according to the manufacturer's recommendations.
A rat Smad 1 cDNA in expression vector pCMV-SPORT6 was purchased from the American Type Culture Collection. This I.M.A.G.E. clone possesses the GenBankTM accession number BC061757. For an empty vector control, the Smad I cDNA was excised from pCMV-SPORT6 with EcoRV and NotI, 5′ overhangs were filled in with Klenow, and the resulting fragment was religated. Nanogram to microgram quantities of the Smad 1 expression vector or control were transiently transfected into 25 × 104 LLC-PK1 cells using an Amaxa nucleofector and a Cell Line Nucleofector kit L (Lonza Walkersville Inc., Walkersville, MD). RNA was harvested for competitive RT-PCR 24 h post-transfection.
RT-PCR
An internal standard for competitive RT-PCR of porcine HuR was synthesized using a previously described method (18). Total RNA from LLC-PK1 cells was isolated using TRIzol (Invitrogen) following the manufacturer's instructions.
A mixture of internal standard RNA (1 pg) and LLC-PK1 RNA (5 μg) was reverse transcribed using the Super Script first strand synthesis system (Invitrogen). The resulting cDNA was subjected to PCR using Platinum Taq High Fidelity (Invitrogen) with the following primers: 5′-GGTTATGAAGACCACATGGCCG-3′ (sense) and 5′-AAGCCATAGCCCAAGCTGT-3′ (antisense).
For competitive RT-PCR of LLC-PK1 cells stably expressing luciferase expressing vectors, an internal standard was synthesized using the previously described method. PCR was performed using the following primers: 5′-CGTCGTATTCGTGAGCAAGAAAGG-3′ (sense) and 5′-AAGAATAGCTCCTCCTCGAAGCGG-3′ (antisense). The PCR was electrophoresed in a 2% agarose gel and visualized and quantified using a Chemi-Doc image analysis system with Quantity One software. For RT-PCR of ALK2, previously published primers were used (32).
Immunocytochemistry
For immunolocalization of HuR, Smads, or BMP-7, 3 × 104 LLC-PK1 cells were seeded on glass coverslips in 24-well plates and grown to confluence, at which time the cells were given fresh medium and cultured overnight prior to use. Following the necessary treatments, the cells were fixed and permeabilized in 2% formaldehyde in stabilization buffer (33). The cells were probed with primary antibodies and Alexa 488- or Alexa 568-conjugated secondary antibodies from Invitrogen/Molecular Probes. The cells were visualized with a Nikon Eclipse 80i epifluorescent microscope with SPOT software (Diagnostic Instruments, Sterling Heights, MI) or with a Zeiss 510 META laser scanning confocal microscope at the Campus Microscopy and Imaging Facility at The Ohio State University.
Antibodies and Western Blot Procedures
For detection of HuR, a mouse monoclonal antibody (3A2) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Smad (H-465) rabbit polyclonal antibody, anti-Smad 1/5/8 (N-18), and goat polyclonal antibody anti-BMP-7 (L-19) were also purchased from Santa Cruz Biotechnology. The anti-pSmad 1/5 (41D10) rabbit polyclonal antibody was purchased from Cell Signaling Technology (Danvers, MA). Antibodies recognizing ALK2 were purchased from Abcam (Cambridge, MA).
For Western analysis, the proteins were transferred to Hybond P membrane (GE Healthcare) and probed with antibodies at 1:500 (3A2), 1:250 (H-465), 1:250 (N-18), 1:500 (L-19), or 1:250 (41D10) dilutions. Primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies and an ECL Western blotting detection kit (GE Healthcare). Quantification of Westerns was performed using a Chemi-Doc image analysis system with Quantity One software (Bio-Rad).
Gel Mobility Shift Assay
Nuclear extracts were prepared from untreated or ATP-depleted/recovered LLC-PK1 cells as previously described (34). Double-stranded DNA oligonucleotide probes were synthesized that corresponded to −148 to −109 or −77 to −37 of the porcine HuR 5′-UTR (numbered relative to the translational start). Probes were end-labeled with [γ-32P]ATP and T4 polynucleotide kinase using a gel shift assay system (Promega). Fourteen micrograms of nuclear extract were incubated with labeled probe in the provided binding buffer at room temperature for 20 min prior to separation in a nondenaturing 4% polyacrylamide gel. Supershift assays were performed by preincubating 6 μg of anti-Smad 1/5/8 antibody (Santa Cruz Biotechnology) with nuclear extracts for 1 h at 4 °C prior to the addition of radiolabeled probe. Oligonucleotide competition reactions were performed by preincubating the nuclear extracts with unlabeled oligonucleotide for 10 min at room temperature prior to the addition of the labeled probe.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed with the Magna ChIP A kit (Millipore, Billerica, MA) according to the manufacturer's protocol. Control and treated LLC-PK1 cells (10 × 106 each) were fixed for 10 min at room temperature and sonicated according to the manufacturer's protocol. DNA bound to Smad 1/5/8 was precipitated with anti-Smad 1/5/8 antibody N-18 (Santa Cruz Biotechnology). To amplify the Smad-binding region in 5′-UTR of the HuR gene, the precipitated DNA was subjected to PCR using PrimeSTAR® HS DNA polymerase with GC buffer (Takara Bio Inc.) with the following primers under the following conditions: 5′-GCTGAGGAGGAGCCGC-3′ (sense) and 5′-GGCTGCTCCGGGTCG-3′ (antisense); 3 min at 94 °C; and then 30 cycles of 98 °C for 10 s, 66 °C for 5 s, and 72 °C for 30 s; followed by a final extension at 72 °C for 2 min. The PCR was electrophoresed in a 5% polyacrylamide gel and quantified using Image J software. The identities of the resulting bands were confirmed by DNA sequencing.
ELISA
For ELISA, LLC-PK1 cells were subjected to different ATP conditions as indicated. After this incubation, the conditioned medium was centrifuged to remove cellular debris. The concentration of BMP-7 in the supernatant was determined using a Quantikine BMP-7 ELISA kit (R & D Systems).
Statistical Analysis
The graphical data indicate the means ± standard deviations. Pairwise comparisons between data points were performed by Student's t test; p values <0.05 were deemed statistically significant.
RESULTS
Determination of Alternative 5′-Untranslated Regions for HuR mRNAs
To define promoter sequences responsible for transcriptional regulation of HuR, it was necessary to first determine the length of HuR transcripts in our cell model, the porcine proximal tubule line LLC-PK1. Although a murine HuR cDNA containing over 500 bases of 5′-UTR has been detected (accession number AK078333) and we have confirmed its existence by RT-PCR, our previous studies of HuR mRNAs in native rat kidney showed the most abundant HuR transcripts to contain 5′-UTRs of ∼150 and ∼20 bases (19); longer sequences were undetectable by ribonuclease protection assay. We performed similar analysis on LLC-PK1 mRNA. As shown in Fig. 1A (lane 4), the cells under normal growth conditions expressed two HuR mRNAs in roughly equal amounts (arrows). These mRNAs are consistent in size and abundance with those detected in native rat kidney containing 5′-UTRs of ∼150 and ∼20 bases (19).
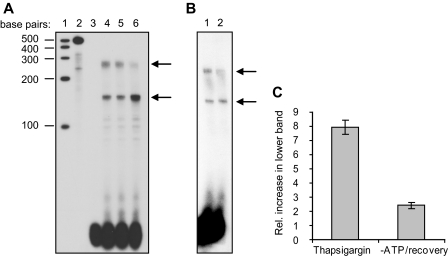
FIGURE 1.
Alternate transcriptional starts of HuR mRNA. A, effects of thapsigargin treatment on HuR mRNA expression were assessed by RPA. Lane 1, molecular size markers; lane 2, probe + yeast tRNA, undigested; lane 3, probe + yeast tRNA, digested; lane 4, mRNA from untreated cells; lane 5, mRNA from cells treated with thapsigargin for 1 h; lane 6, mRNA from cells treated with thapsigargin for 1 h and allowed to recover for 6 h. B, effects of ATP depletion/recovery on HuR mRNA as assessed by RPA. Lane 1, mRNA from untreated cells; lane 2, mRNA from cells ATP depleted for 2 h and allowed to recover for 6 h. C, graphical representation of increases in the smaller HuR transcript relative to the longer mRNA, as demonstrated by RPA analysis.
In the same experiments, we tested whether cellular stress would alter the abundance of these mRNAs. Fig. 1A demonstrates that treatment of LLC-PK1 cells with thapsigargin, an inhibitor of SERCA, for 1 h caused no significant change in HuR mRNA levels (lane 5); however, after 6 h of recovery from thapsigargin treatment, there was a notable increase in the levels of the smaller mRNA accompanied by a decrease in levels of the higher molecular weight form (lane 6). These results indicate that recovery from cellular stress causes a switch in the synthetic rates or stability of the two HuR mRNAs.
Previous studies in nonrenal tissues have suggested that HuR protein expression is under translational control (35, 36). More recently, we found HuR to be under translational control in LLC-PK1 cells (18) and in native rat kidney (19). In vitro transcription/translation of HuR mRNAs from both rats and mice showed the shorter form to be readily translated, whereas the longer form was translated with dramatically less efficiency (19). Therefore, the current RPA experiments demonstrate that recovery of LLC-PK1 cells from stress can decrease steady-state levels of a longer, poorly translatable HuR mRNA while increasing levels of a shorter, readily translatable transcript.
To determine whether recovery from the stress of ATP depletion might cause a shift in expression of HuR mRNAs similar to that following thapsigargin treatment, RPA was performed on LLC-PK1 cells that were either untreated or ATP-depleted for 2 h and allowed to recover for 6 h. As shown in Fig. 1B, a similar switch between transcripts was seen during the recovery following ATP depletion, although the magnitude of the change was more modest than that seen with thapsigargin. No change in HuR mRNA was seen during ATP depletion itself (not shown). Thus, these results indicate that recovery from cellular stresses (either inhibition of SERCA or ATP depletion) induces expression of a readily translated HuR mRNA at the expense of a poorly translated form. The relative increase in expression of the smaller transcript over the longer mRNA during recovery from these stresses is expressed graphically in Fig. 1C.
HuR Promoter Activity during ATP Depletion and Recovery
Our previous studies demonstrated that increased levels of HuR transcription during recovery from ATP depletion permitted a rapid translation of HuR upon a second stress (18). This suggests that the switch in HuR transcripts shown in Fig. 1 is to due to increased activation of a promoter element that drives transcription of the shorter mRNA (rather than changes in transcript stability). To explore this hypothesis and define sequences of the HuR promoter that are necessary for this response, upstream regions of the mouse HuR gene were subcloned into the luciferase reporter plasmid pGL4.14, and the resulting constructs were stably transfected into LLC-PK1 cells. Fig. 2A shows the genomic fragments used, which extended downstream through the entirety of exon I to −16 bp (relative to the translational start site). Upstream boundaries for promoter fragments ranged from −566 bp to −2.69 kb. Four stably transfected LLC-PK1 clones were generated and maintained for each construct and assayed for basal luciferase expression (not shown). Clones expressing similar levels of basal luciferase activity were used for subsequent ATP depletion studies.
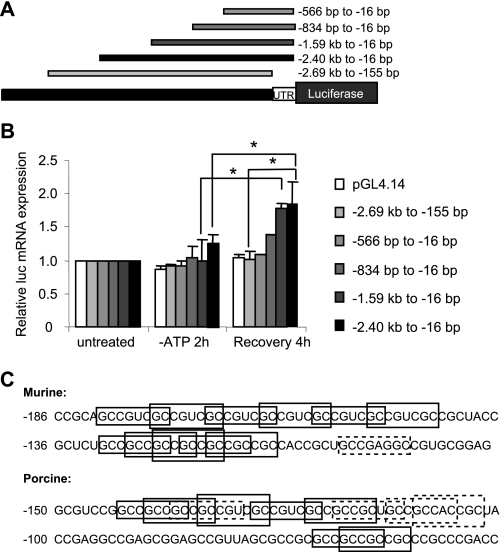
FIGURE 2.
Characterization of the HuR promoter and 5′-UTR. A, transcriptional activation was examined using promoter constructs containing various deletions of upstream sequences as well as deletion of the 5′-UTR. B, stable cell lines were cultured in normal growth medium or ATP depletion medium with or without recovery for 4 h. Total RNA was isolated and subjected to competitive RT-PCR for quantification of luciferase mRNA. The asterisks indicate statistical significance as determined by Student's t test (p < 0.05). C, murine and porcine versions of the HuR 5′-UTR show multiple copies of the Smad 1/5/8-binding site, GCCGnCGC. The solid boxes indicate perfect matches; the dashed boxes indicate motifs with one nucleotide variation.
To assay HuR promoter activity, we quantified levels of luciferase mRNA expression, rather than taking advantage of its luminescent properties. This was due to a couple of features of these experiments that made assays for luciferase protein unreliable. First, as discussed above, we showed that sequences in the HuR 5′-UTR can interfere with translatability (19); second, previous reports demonstrated that luciferase protein is unstable during ATP depletion (37). Therefore, competitive RT-PCR was used to quantify luciferase mRNA levels (Fig. 2B). For any of the constructs tested, ATP depletion alone resulted in no significant changes in reporter mRNA levels. This finding is consistent with our results demonstrating no change in HuR mRNA levels during ATP depletion (this study and Ref. 18). However, increased luciferase expression was seen from some constructs during recovery from ATP depletion. LLC-PK1 cells stably expressing constructs containing at least 1.59 kb of upstream sequence, including the nearly full 5′-UTR (−2.40 kb to −16 bp and −1.59 kb to −16 bp), increased luciferase expression ∼2-fold over base-line levels (Fig. 2B). In contrast, a construct of similar size that lacks sequences corresponding to the HuR 5′-UTR (−2.69 kb to −155 bp) demonstrated no significant increase in mRNA levels. These results suggest the importance of the region between −155 and −16 bp in mediating ATP depletion/recovery-induced HuR expression. Although the region between −1.59 kb and −834 bp also was required for full luciferase expression, these results demonstrate the importance of the 5′-UTR in transcriptional induction during recovery from ATP depletion.
Initial inspection of HuR 5′-UTR sequences within 200 base pairs of the translational start revealed a highly G+C-rich (>80%), highly conserved (77% identity between mouse and pig), ∼120-base region that contains multiple overlapping copies of an alternate Smad 1/5/8 binding motif, GCCGnCGC (reviewed in Ref. 6). Fig. 2C illustrates these sequences, showing perfect matches in solid boxes and motifs that differ by a single base in dashed boxes. This sequence is positioned appropriately to direct transcription of the HuR mRNAs containing only ∼20 bases of 5′-UTR. Based on these findings and on the known importance of bone morphogenetic proteins and Smad 1/5/8 signaling in renal development and recovery from renal ischemia, we further explored the role of these Smads in HuR expression.
Expression and Distribution of Smads 1/5/8 during ATP Depletion and Recovery
Under normal conditions, HuR shows a characteristic nuclear staining (Fig. 3A, top row). Following 2 h of ATP depletion, HuR redistributes to the cytoplasm as previously described and returns to a nuclear distribution following 2 or 4 h in recovery medium (17, 18). To determine whether ATP depletion and recovery cause redistribution of Smads, immunocytochemistry was performed on LLC-PK1 cells subjected to 2 h of ATP depletion followed by 2–4 h of recovery. Probing with an antibody that recognizes all R-Smads (Smads 1, 2, 3, 5, and 8) uncovered a nuclear/cytoplasmic staining in untreated cells (Fig. 3A, middle row). During ATP depletion, many cells showed a loss of nuclear staining. Upon 2 h of recovery, R-Smads redistributed to a nuclear and perinuclear localization that was sustained through 4 h of recovery. An antibody that recognized phosphorylated (activated) Smad 1/5 (pSmad 1/5) revealed that under normal conditions, pSmad 1/5 also was located both in nuclear and perinuclear distributions (Fig. 3A, bottom row). The loss of nuclear staining occurred under ATP depletion conditions. However, after 2 h of recovery, pSmad 1/5 returned to a nuclear location, which became even more intense by 4 h of recovery. These results demonstrate an increased presence of active pSmads 1/5 during recovery from ATP depletion.
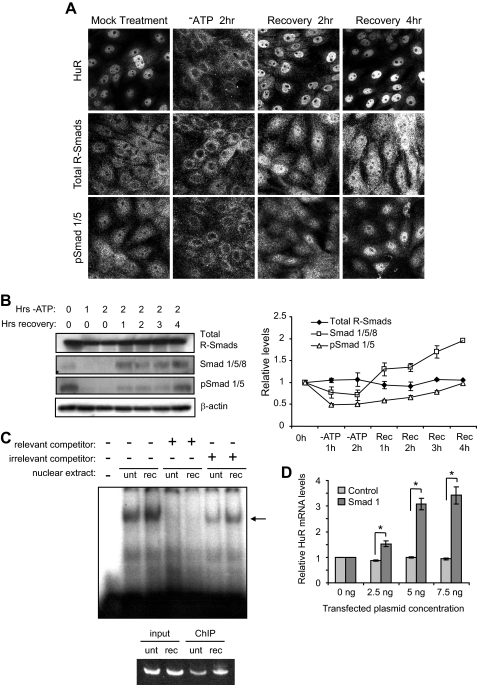
FIGURE 3.
Distribution and expression of Smads 1/5/8 during ATP depletion and recovery. A, LLC-PK1 cells were cultured in normal growth medium or ATP depletion medium for 2 h or allowed to recover for an additional 2 or 4 h. Immunocytochemical localization of HuR, total R-Smads, and pSmad 1/5 was determined. B, total R-Smad, Smad 1/5/8, and pSmad 1/5 were assayed by Western analysis. β-Actin was included as a loading control. Bands from multiple Western blots as shown were quantified and expressed in graphical form. C, gel mobility shift assays and ChIP were performed on nuclear extracts from untreated (unt) or ATP-depleted/recovered (rec) LLC-PK1 cells. In gel mobility shift assays (top), one major band (designated by the arrow) was found to specifically bind Smad 1/5/8 consensus motifs in the porcine HuR 5′-UTR. The intensity of this band increased following ATP depletion/recovery. In ChIP assays (bottom panel), antibody against Smad 1/5/8 precipitated DNA corresponding to the HuR 5′-UTR. D, introduction of exogenous Smad 1 into LLC-PK1 cells increased HuR mRNA expression, as determined by competitive RT-PCR. An empty vector control produced no effect. The asterisks indicate p < 0.005, as determined by Student's t test.
Next, the protein levels of Smads were determined to ascertain whether ATP depletion and recovery caused alterations in their expression (Fig. 3B). The total R-Smad levels remained fairly flat throughout the ATP depletion and recovery periods (see representative Western and graph). In contrast to total R-Smad levels, Smad 1/5/8 and pSmad 1/5 levels decreased during 1–2 h of ATP depletion. Subsequently, during 1–4 h of recovery, the Smad 1/5/8 levels steadily increased to ∼2-fold over base line. This time course correlates with that of HuR mRNA levels during recovery from ATP depletion (18). Although recovery caused pSmad 1/5 levels to only return to base line, the immunocytochemical analysis in Fig. 3A demonstrates that these Smads are more highly concentrated in the nuclear compartment after stress, suggesting an increased availability for transcriptional modulation.
To determine whether the putative Smad 1/5/8-binding sites in the HuR 5′-UTR are indeed capable of binding Smads, gel mobility shift assays were performed on nuclear extracts derived from both untreated LLC-PK1 cells and those that were ATP-depleted and allowed to recover. Fig. 3C shows an example of this method, using a porcine probe corresponding to the sequence −148 to −109 that encompasses five perfect and two imperfect copies of the GCCGnCGC motif (Fig. 2C). As shown, one major band was shifted in the presence of nuclear extracts. This band was routinely more intense in extracts from ATP-depleted/recovered cells than untreated cells (an increase of 87 ± 18%; n = 5). The addition of an excess of unlabeled probe caused loss of this band, whereas addition of an irrelevant unlabeled oligonucleotide did not. Preincubation of extracts with antibody against Smads 1/5/8 resulted in the disappearance of the band, rather than a supershift, indicating potential interference of the antibody with the probe-binding site (data not shown). Therefore, we performed ChIP to confirm the identities of the relevant bands. As shown in the bottom panel of Fig. 3C, immunoprecipitation of chromatin with the Smad 1/5/8 antibody isolated a fragment containing the HuR 5′-UTR sequences. Consistent with the gel mobility shifts, Smads 1/5/8 roughly doubled their association with the HuR 5′-UTR following ATP depletion/recovery (an increase of 86 ± 15%; n = 3). These results demonstrate that the HuR 5′-UTR is indeed capable of binding R-Smads and that binding activity is increased in cells subjected to ATP depletion and recovery.
Finally, we transiently transfected exogenous Smad 1 into LLC-PK1 cells to determine its effect on HuR mRNA expression. Transfection of Smad 1 in increasing amounts produced a dose-dependent increase in mRNA (∼3-fold) as assayed by competitive RT-PCR, whereas transfection of an empty vector control did not. These results are illustrated graphically in Fig. 3D.
Expression of BMP-7 and Effects on HuR Levels
In the homogeneous cell line LLC-PK1, activation of Smad 1/5/8 necessarily would occur through cell autonomous secretion of bone morphogenetic proteins. BMP-7 would be indicated as the prime candidate, because it is the major BMP expressed by kidney and has been shown to play an important role in recovery from renal ischemia/reperfusion injury. Expression of BMP-7 by proximal tubule cells is somewhat controversial, because most of the renal expression of BMP-7 has been shown to come from glomeruli and epithelial cells of medulla (38, 39). However, some studies have indicated low BMP-7 expression in both rodent and human proximal tubule cells (40–42). As a first step toward determining whether LLC-PK1 cells express BMP-7, cells in varying stages of ATP depletion and recovery were immunolabeled with a BMP-7-specific antibody. BMP-7 labeling was present in these cells and revealed a diffuse distribution with some enrichment around the nucleus (Fig. 4A). In contrast, following 2 h of ATP depletion, most BMP-7 was redistributed to the periphery of the cell. Following 2–4 h of recovery in normal growth medium, BMP-7 returned to a more base-line distribution.
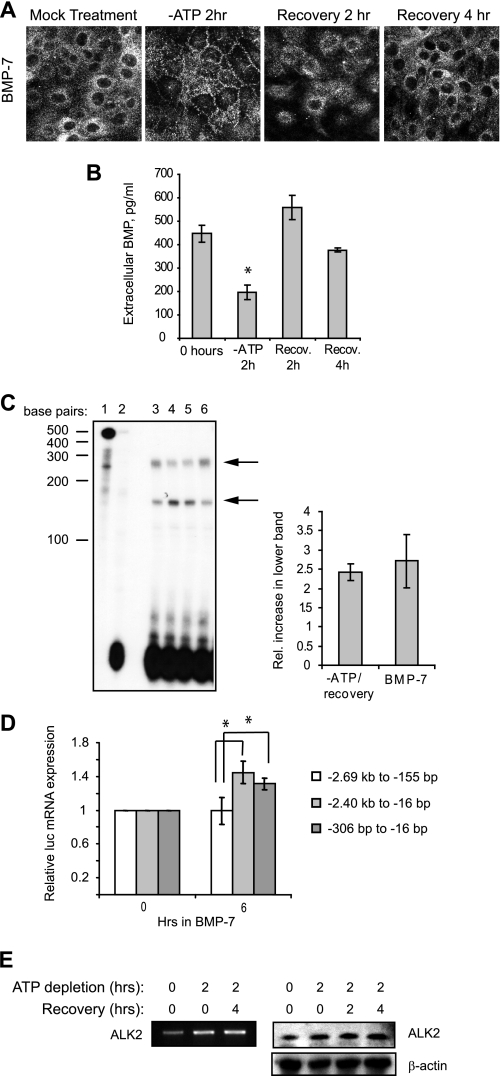
FIGURE 4.
BMP-7 induced Smad 1/5/8 activity during recovery from ATP depletion. A, immunolocalization of BMP-7 in LLC-PK1 cells grown in normal medium or subjected to ATP depletion/recovery. B, ELISA of conditioned medium from normal LLC-PK1 cells or cells subjected to 2 h ATP depletion alone or followed by recovery in normal growth media. The asterisk shows statistically a significant difference from 0 h control (p < 0.05). C, left panel, addition of exogenous BMP-7 to LLC-PK1 cells induced a shift in HuR mRNA expression, as assessed by RPA. Lane 1, probe + yeast tRNA, undigested; lane 2, probe + yeast tRNA, digested; lane 3, mRNA from untreated cells; lane 4, mRNA from cells treated with BMP-7 in 0.2% fetal bovine serum for 6 h; lane 5, mRNA from cells ATP depleted for 2 h and then treated with BMP-7 in 0.2% serum; lane 6, mRNA from cells cultured for 6 h in 0.2% serum alone. Right panel, the relative increase in the smaller HuR transcript relative to the longer mRNA under ATP depletion/recovery or BMP-7 was quantified and graphed. D, treatment of LLC-PK1 cells with exogenous BMP-7 induced luciferase expression in cells stably expressing constructs containing Smad 1/5/8-binding sites. The asterisks show statistically significant differences (p < 0.05). E, ATP depletion and recovery induced expression of ALK2 mRNA (left panel) and protein (right panel). A β-actin immunoblot is included as a loading control.
To determine whether BMP-7 was secreted by LLC-PK1 cells, ELISA analysis of the conditioned medium was employed (Fig. 4B). Conditioned medium from nonstressed cells contained ∼450 pg/ml of BMP-7, similar to the levels normally found in circulation of the blood (14). However, the levels were found to drop to almost 25% of normal during ATP depletion. This is perhaps not surprising given the stresses put on the cell; however, the immunocytochemistry shown in Fig. 4A suggests that redistribution of BMP-7 to the periphery may be an attempt by the cell to secrete more of its BMP-7 content. By 2–4 h of recovery in normal growth medium, extracellular BMP-7 increased to levels not significantly different from those of base-line conditions.
The kinetics of pSmad 1/5 activation and nuclear localization during recovery from ATP depletion are consistent with the timing of increased expression of the shorter HuR transcript. To determine whether the BMP-7 signaling pathway is responsible for this modulation of HuR levels during recovery, LLC-PK1 cells were treated with recombinant BMP-7. As expected, treatment with recombinant BMP-7 induced an increase in the levels of phosphorylated Smad 1/5 (not shown). As shown in Fig. 4C (left panel), exposure of cells to BMP-7 for 6 h caused increased levels of the shorter HuR transcript and loss of the longer transcript (compare lanes 3 and 4). Two hours of ATP depletion prior to the addition of BMP-7 did not change the magnitude of this response (lane 5). Incubation of cells in the low serum levels required for BMP-7 addition did not induce these changes (lane 6), showing the specificity of the response to BMP-7. Multiple such experiments were performed, and the relative increase in the smaller HuR mRNA over the longer transcript was quantified (Fig. 4C, right panel). As shown, BMP-7 treatment induced changes similar to those seen during recovery from ATP depletion, indicating this factor as a stimulator for Smad-mediated changes in ATP-depleted/recovered cells.
In addition, cell lines stably expressing HuR promoter-luciferase constructs were treated with BMP-7 for 6 h. As with the ATP depletion/recovery experiments in Fig. 2, only plasmids containing the HuR 5′-UTR region between −155 and −16 were able to mediate increased luciferase expression. Constructs lacking these sequences were not activated (Fig. 4D).
Finally, to determine whether BMP-7 receptors were present in LLC-PK1 cells, RT-PCR was performed using primers corresponding to ALK2 (also known as Acvr1 and ActRI), a type I receptor that transduces signals from BMP-7 (43) and is expressed in proximal tubule cells (38). Semi-quantitative RT-PCR (Fig. 4E, left panel) showed the existence of this receptor and suggested that ATP depletion and recovery induced higher expression of ALK2 mRNA (an increase of 85 ± 18%; n = 4). To confirm this finding at the protein level, Western analysis was performed on LLC-PK1 cells subjected to various times of ATP depletion/recovery. Fig. 4E (right panel) demonstrates similarly increased levels of ALK2 protein in stressed and recovered cells compared with those under normal growth conditions. This finding provides a mechanism by which ATP-depleted/recovered cells, which express only base-line levels of BMP-7 (Fig. 4B), can amplify their responsiveness to this protein, thus inducing signaling through the Smad 1/5/8 pathway.
DISCUSSION
These studies offer the first description of transcriptional mechanisms regulating stress-mediated expression of HuR, an RNA-binding protein with global effects on cell survival. Although this work has focused specifically on its role in protection against energy depletion in renal epithelia, it may provide additional clues to HuR function in other cells where BMP/Smad signaling is stimulated. For example, a number of recent studies have indicated that BMPs may promote tumor progression in some types of cancers (44–46). Our findings suggest that one mechanism by which this might occur is through transcriptional regulation of HuR, which itself has global effects on tumorigenicity. These studies also suggest a role for HuR in renal development, because BMP-7 is required for metanephric differentiation. Additionally, our finding that transcriptional switches in HuR expression are stimulated by other stresses such as thapsigargin treatment suggests a more global role for Smad 1/5/8 activation in responses to stress.
By stabilizing and promoting translation of specific mRNAs during stress events, HuR is protective against cellular damage. Our recent findings suggest that HuR not only has a role in mRNA stabilization during ATP depletion in renal cells but also appears to be involved in preparation of the cell for a second insult. During recovery from ATP depletion, HuR exhibits an unusual decoupling behavior, during which its protein levels decrease to normal levels while simultaneously increasing expression of a more translatable transcript. This increase in mRNA appears to be necessary for rapid induction of HuR protein during a second stress event (18). These findings are reminiscent of ischemic preconditioning, the phenomenon whereby brief ischemic episodes render cells resistant to subsequent ischemia (47). This study identifies important regulatory sequences, as well as potential transcriptional activators necessary for this regulation.
Examination of sequences driving HuR transcription shows that the HuR promoter is typical of a “TATA-less” gene promoter. Originally thought to be rare, members of this class of promoters are G+C-rich, lack a TATA element, are rich in Sp1-binding sites and are found in ∼46% of human core promoters (48). These promoters routinely contain multiple transcriptional start sites, as demonstrated by the existence of alternate HuR transcripts. These studies indicate that the HuR gene possesses alternate promoters that can be activated or suppressed dependent on the stress experienced by cells. Genome wide analysis has revealed the prevalence of numerous alternative promoters, in fact suggesting that up to half of human genes produce variant transcripts (49). Transcription from alternate promoters has been shown to be induced under multiple circumstances, including developmental activity (50), tissue specificity (51), and stress (52). Further, alternative promoter transcripts previously have been shown effective in the control of translational machinery, as suggested here for HuR (53). However, this study has not completely resolved the significance of the alternate HuR transcripts. Although the HuR mRNA bearing the short 5′-UTR is under control of BMP/Smad signaling and is readily translatable, a function for the longer, poorly translatable transcript is not yet apparent. It is possible that this transcript may possess an internal ribosome entry site, based on the length and G+C content of the 5′-UTR. We are currently investigating the possibility that an internal ribosome entry site is present; if so, this may provide a means to continue HuR translation under stress conditions in which normal cap-dependent ribosome function is impaired (54). This finding would be consistent with our previous studies showing that HuR translation is increased in cells undergoing an almost total depletion of ATP (18).
The data presented here indicate that HuR mRNA expression is responsive to BMP-7 levels. In a recent study, we found that HuR protein levels increase only in the proximal tubules of rat kidneys subjected to ischemia/reperfusion injury. This increase began at 24 h post-ischemia and was maintained for at least 2 weeks (19). Other groups have shown that immediately following ischemic injury to rat kidneys, BMP-7 levels temporarily decreased (14, 39) but recovered and peaked at ∼48 h (41). A similar pattern of expression was seen for Smads and pSmads, which peaked in expression at 24–48 h post-ischemia (41). In both cases, the increased expression was seen in regenerating proximal tubule epithelia. Thus, the Smad/BMP-7 signaling pathway is likely to be responsible for increased HuR expression in proximal tubules of native kidney. However, in our previous study of rats, comparison of kidney cortex from sham-operated animals with 1- or 14-day post-ischemic kidneys did not reveal any differences in ratios between the shorter and longer HuR transcripts; rather, levels of both mRNAs increased (19). It is possible that the time points chosen for this experiment did not represent the time of peak transcription for the shorter mRNA; alternately, it may be that the specific responses of proximal tubule cells were masked by the presence of nonproximal tubules in the cortical preparation. More detailed studies will be required to resolve this discrepancy.
Although HuR has the capacity to bind hundreds to thousands of mRNA targets, it is known to promote mRNA stabilization or translation of a few specific gene products that play important roles in promoting cell proliferation or protecting against apoptosis and senescence (24, 25). Target mRNA ligands for HuR include cell cycle-associated gene products such as c-Fos, p21, p53, and various cyclins, anti-apoptotic proteins including Bcl-2, prothymosin α, and SIRT1, and other stress-related proteins such as Hsp70 and HIF1α (reviewed in Ref. 24; see also Ref. 55). Our own studies have demonstrated the anti-apoptotic nature of HuR in LLC-PK1 cells subjected to ATP depletion and indicate Bcl-2 and Hsp70 as targets (19). Therefore, the expression of HuR in kidney epithelia subjected to ischemia/reperfusion injury may be protective against apoptosis caused by the ischemic insult and may also promote proliferation of newly regenerating proximal tubule epithelia. Although the transcriptionally mediated changes in HuR mRNA expression demonstrated appear modest (only a few-fold, both in LLC-PK1 cells and rat kidney), the effects of HuR on cell survival and proliferation are broad; indeed, overexpression of HuR by only a few-fold can result in malignancy (56). Therefore, its expression necessarily must be tightly controlled.
Modulated expression of HuR during ischemia/reperfusion injury provides a system in which there are multiple levels of regulation working in concert to protect and restore the cell. These findings present the first attempts to understand the complex regulation of HuR mRNA in protecting and preconditioning cells for a subsequent injury. Not only do these findings provide insight into the regulation of HuR during recovery from ATP depletion, they also suggest a mechanism through which BMP-7 exerts pleiotropic effects to protect the kidney from injury. Understanding these pathways may lead to a greater understanding of the pathogenesis of acute kidney injury and aid in finding ways to reduce injury following trauma to the kidney.
Note Added in Proof
Additionally, a functional NF-κB binding site has been identified in the HuR promoter (57). However, this site is positioned outsite of sequences required for the stress response in this model system, so a role for NF-κB in HuR expression in renal cells has yet to be determined.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK052131 and RO1 AR051515 (to B. S. L.).
- TGF
- transforming growth factor
- HuR
- human antigen R
- UTR
- untranslated region
- ChIP
- chromatin immunoprecipitation
- BMP
- bone morphogenetic protein
- R-Smad
- pathway-restricted Smad
- SERCA
- sarco/endoplasmic reticulum Ca2+-ATPase
- RPA
- RNase protection assay
- RT
- reverse transcription
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Schrier R. W., Wang W., Poole B., Mitra A. (2004) J. Clin. Invest. 114, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer B. F. (1997) J. Investig. Med. 45, 346–361 [PubMed] [Google Scholar]
- 3.Bonventre J. V. (2003) J. Am. Soc. Nephrol. 14, (Suppl. 1) S55–S61 [DOI] [PubMed] [Google Scholar]
- 4.Supavekin S., Zhang W., Kucherlapati R., Kaskel F. J., Moore L. C., Devarajan P. (2003) Kidney Int. 63, 1714–1724 [DOI] [PubMed] [Google Scholar]
- 5.Zeisberg M., Hanai J., Sugimoto H., Mammoto T., Charytan D., Strutz F., Kalluri R. (2003) Nat. Med. 9, 964–968 [DOI] [PubMed] [Google Scholar]
- 6.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 7.Ozkaynak E., Schnegelsberg P. N., Oppermann H. (1991) Biochem. Biophys. Res. Commun. 179, 116–123 [DOI] [PubMed] [Google Scholar]
- 8.Vukicevic S., Kopp J. B., Luyten F. P., Sampath T. K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9021–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudley A. T., Lyons K. M., Robertson E. J. (1995) Genes Dev. 9, 2795–2807 [DOI] [PubMed] [Google Scholar]
- 10.Luo G., Hofmann C., Bronckers A. L., Sohocki M., Bradley A., Karsenty G. (1995) Genes Dev. 9, 2808–2820 [DOI] [PubMed] [Google Scholar]
- 11.Martinez G., Loveland K. L., Clark A. T., Dziadek M., Bertram J. F. (2001) Exp. Nephrol. 9, 372–379 [DOI] [PubMed] [Google Scholar]
- 12.Vrljicak P., Myburgh D., Ryan A. K., van Rooijen M. A., Mummery C. L., Gupta I. R. (2004) Am. J. Physiol. Renal Physiol. 286, F625–F633 [DOI] [PubMed] [Google Scholar]
- 13.Hruska K. A., Guo G., Wozniak M., Martin D., Miller S., Liapis H., Loveday K., Klahr S., Sampath T. K., Morrissey J. (2000) Am. J. Physiol. Renal Physiol. 279, F130–F143 [DOI] [PubMed] [Google Scholar]
- 14.Vukicevic S., Basic V., Rogic D., Basic N., Shih M. S., Shepard A., Jin D., Dattatreyamurty B., Jones W., Dorai H., Ryan S., Griffiths D., Maliakal J., Jelic M., Pastorcic M., Stavljenic A., Sampath T. K. (1998) J. Clin. Invest. 102, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Chen Q., Simon T. C., Strebeck F., Chaudhary L., Morrissey J., Liapis H., Klahr S., Hruska K. A. (2003) Kidney Int. 63, 2037–2049 [DOI] [PubMed] [Google Scholar]
- 16.Zeisberg M., Bottiglio C., Kumar N., Maeshima Y., Strutz F., Müller G. A., Kalluri R. (2003) Am. J. Physiol. Renal Physiol. 285, F1060–F1067 [DOI] [PubMed] [Google Scholar]
- 17.Jeyaraj S., Dakhlallah D., Hill S. R., Lee B. S. (2005) J. Biol. Chem. 280, 37957–37964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeyaraj S. C., Dakhlallah D., Hill S. R., Lee B. S. (2006) Am. J. Physiol. Renal Physiol. 291, F1255–F1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayupova D. A., Singh M., Leonard E. C., Basile D. P., Lee B. S. (2009) Am. J. Physiol. Renal Physiol. 297, F95–F105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., Gorospe M. (2000) Mol. Cell. Biol. 20, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallouzi I. E., Brennan C. M., Steitz J. A. (2001) RNA 7, 1348–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaman I., Fernandez J., Sarkar B., Schneider R. J., Snider M. D., Nagy L. E., Hatzoglou M. (2002) J. Biol. Chem. 277, 41539–41546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdelmohsen K., Kuwano Y., Kim H. H., Gorospe M. (2008) Biol. Chem. 389, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelmohsen K., Lal A., Kim H. H., Gorospe M. (2007) Cell Cycle 6, 1288–1292 [DOI] [PubMed] [Google Scholar]
- 26.Mazan-Mamczarz K., Hagner P. R., Corl S., Srikantan S., Wood W. H., Becker K. G., Gorospe M., Keene J. D., Levenson A. S., Gartenhaus R. B. (2008) Oncogene 27, 6151–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazan-Mamczarz K., Hagner P. R., Dai B., Wood W. H., Zhang Y., Becker K. G., Liu Z., Gartenhaus R. B. (2008) Cancer Res. 68, 7730–7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashworth S. L., Southgate E. L., Sandoval R. M., Meberg P. J., Bamburg J. R., Molitoris B. A. (2003) Am. J. Physiol. Renal Physiol. 284, F852–F862 [DOI] [PubMed] [Google Scholar]
- 29.Meldrum K. K., Meldrum D. R., Sezen S. F., Crone J. K., Burnett A. L. (2001) Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R359–R364 [DOI] [PubMed] [Google Scholar]
- 30.Park K. M., Kramers C., Vayssier-Taussat M., Chen A., Bonventre J. V. (2002) J. Biol. Chem. 277, 2040–2049 [DOI] [PubMed] [Google Scholar]
- 31.Rasband W. S. (2008) Image J, National Institutes of Health, Bethesda, MD [Google Scholar]
- 32.Grijelmo C., Rodrigue C., Svrcek M., Bruyneel E., Hendrix A., de Wever O., Gespach C. (2007) Cell. Signal. 19, 1722–1732 [DOI] [PubMed] [Google Scholar]
- 33.Zeng Q., Lagunoff D., Masaracchia R., Goeckeler Z., Côté G., Wysolmerski R. (2000) J. Cell Sci. 113, 471–482 [DOI] [PubMed] [Google Scholar]
- 34.Lee K. A., Bindereif A., Green M. R. (1988) Gene Anal. Tech. 5, 22–31 [DOI] [PubMed] [Google Scholar]
- 35.Nabors L. B., Gillespie G. Y., Harkins L., King P. H. (2001) Cancer Res. 61, 2154–2161 [PubMed] [Google Scholar]
- 36.Okano H. J., Darnell R. B. (1997) J. Neurosci. 17, 3024–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen V. T., Bensaude O. (1994) Eur. J. Biochem. 220, 239–246 [DOI] [PubMed] [Google Scholar]
- 38.Gould S. E., Day M., Jones S. S., Dorai H. (2002) Kidney Int. 61, 51–60 [DOI] [PubMed] [Google Scholar]
- 39.Simon M., Maresh J. G., Harris S. E., Hernandez J. D., Arar M., Olson M. S., Abboud H. E. (1999) Am. J. Physiol. Renal Physiol. 276, F382–F389 [DOI] [PubMed] [Google Scholar]
- 40.Rudnicki M., Eder S., Perco P., Enrich J., Scheiber K., Koppelstätter C., Schratzberger G., Mayer B., Oberbauer R., Meyer T. W., Mayer G. (2007) Kidney Int. 71, 325–335 [DOI] [PubMed] [Google Scholar]
- 41.Villanueva S., Céspedes C., Vio C. P. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R861–R870 [DOI] [PubMed] [Google Scholar]
- 42.Wang S. N., Lapage J., Hirschberg R. (2001) J. Am. Soc. Nephrol. 12, 2392–2399 [DOI] [PubMed] [Google Scholar]
- 43.Macías-Silva M., Hoodless P. A., Tang S. J., Buchwald M., Wrana J. L. (1998) J. Biol. Chem. 273, 25628–25636 [DOI] [PubMed] [Google Scholar]
- 44.Bailey J. M., Singh P. K., Hollingsworth M. A. (2007) J. Cell. Biochem. 102, 829–839 [DOI] [PubMed] [Google Scholar]
- 45.Rothhammer T., Poser I., Soncin F., Bataille F., Moser M., Bosserhoff A. K. (2005) Cancer Res. 65, 448–456 [PubMed] [Google Scholar]
- 46.Yang S., Zhong C., Frenkel B., Reddi A. H., Roy-Burman P. (2005) Cancer Res. 65, 5769–5777 [DOI] [PubMed] [Google Scholar]
- 47.Cochrane J., Williams B. T., Banerjee A., Harken A. H., Burke T. J., Cairns C. B., Shapiro J. I. (1999) Ren. Fail. 21, 135–145 [DOI] [PubMed] [Google Scholar]
- 48.Yang C., Bolotin E., Jiang T., Sladek F. M., Martinez E. (2007) Gene 389, 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuritani K., Irie T., Yamashita R., Sakakibara Y., Wakaguri H., Kanai A., Mizushima-Sugano J., Sugano S., Nakai K., Suzuki Y. (2007) Genome Res 17, 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saleh A., Makrigiannis A. P., Hodge D. L., Anderson S. K. (2002) J. Immunol. 168, 5163–5169 [DOI] [PubMed] [Google Scholar]
- 51.Kamat A., Hinshelwood M. M., Murry B. A., Mendelson C. R. (2002) Trends Endocrinol. Metab. 13, 122–128 [DOI] [PubMed] [Google Scholar]
- 52.Meshorer E., Toiber D., Zurel D., Sahly I., Dori A., Cagnano E., Schreiber L., Grisaru D., Tronche F., Soreq H. (2004) J. Biol. Chem. 279, 29740–29751 [DOI] [PubMed] [Google Scholar]
- 53.Landry J. R., Mager D. L., Wilhelm B. T. (2003) Trends Genet. 19, 640–648 [DOI] [PubMed] [Google Scholar]
- 54.Spriggs K. A., Stoneley M., Bushell M., Willis A. E. (2008) Biol. Cell 100, 27–38 [DOI] [PubMed] [Google Scholar]
- 55.Galbán S., Kuwano Y., Pullmann R., Jr., Martindale J. L., Kim H. H., Lal A., Abdelmohsen K., Yang X., Dang Y., Liu J. O., Lewis S. M., Holcik M., Gorospe M. (2008) Mol. Cell. Biol. 28, 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.López de Silanes I., Lal A., Gorospe M. (2005) RNA Biol. 2, 11–13 [DOI] [PubMed] [Google Scholar]
- 57.Kang M. J., Ryu B. K., Lee M. G., Han J., Lee J. H., Ha T. K., Byun D. S., Chae K. S., Lee B. H., Chun H. S., Lee K. Y., Kim H. J., Chi S. G. (2008) Gastroenterology 135, 2030–2042 [DOI] [PubMed] [Google Scholar]