Abstract
The high mobility group AT-hook 2 (HMGA2), a DNA architectural protein, is highly regulated during development and plays an important role in tumorigenesis. Indeed, HMGA2 was overexpressed in many different kinds of tumors. However, the mechanisms regulating HMGA2 expression remain elusive. Using microarray analysis, we found that HMGA2, along with a dozen of other genes, was co-repressed by ZBRK1, BRCA1, and CtIP. BRCA1 exerts its transcriptional repression activity through interaction with the transcriptional repressor ZBRK1 in the central domain, and with CtIP in the C-terminal BRCT domain. Here, we show that ZBRK1, BRCA1, and CtIP form a repression complex that coordinately regulates HMGA2 expression via a ZBRK1 recognition site in the HMGA2 promoter. Depletion of any of the proteins in this complex via adenoviral RNA interference in MCF10A mammary epithelial cells activates HMGA2 expression, resulting in increased colony formation in soft agar. Similarly, depletion of ZBRK1, or ectopic overexpression of HMGA2, in MCF10A cells induces abnormal acinar size with increased cell number and inhibits normal acinar formation. Consistently, many BRCA1-deficient mouse breast tumors express higher levels of HMGA2 than BRCA1-proficient tumors. These results suggest that activation of HMGA2 gene expression through derepression of the ZBRK1/BRCA1/CtIP complex is a significant step in accelerating breast tumorigenesis.
Keywords: Cancer/Breast, Cell/Epithelial, Transcription/Regulation, Cell differentiation, Chromatin Immunoprecipitation, BRCA1, HMGA2, ZBRK1, Mammary Tumorigenesis, Transcriptional Repression
Introduction
The HMGA family consists of four proteins: HMGA1a, HMGA1b, HMGA1c, and HMGA2.2 HMGA1a, -b, and -c are all encoded by the same gene but vary in length due to alternative splicing (1–3), whereas HMGA2 is encoded by a distinctive gene (4). The common structural motifs in this group include an acidic C terminus and three DNA binding domains called A-T hooks, because they bind short (4 ± 6 bp) AT-rich sequences in the minor groove (5–8). HMGA proteins regulate the expression of many genes through architectural remodeling of the chromatin structure and the formation of multiprotein complexes on promoter/enhancer regions. In accordance with their many roles in transcriptional regulation, aberrant expression of HMGA proteins has been observed in a large number of human cancers (reviewed by Farnet et al. (9)). The HMGA2 architectural protein is critical for a variety of cellular processes, including gene transcription, induction of neoplastic transformation, and promotion of metastatic progression (10, 11). Importantly, HMGA2 overexpression in tumors is associated with poor prognosis and metastasis in breast cancer patients (12). Although it is known that transcriptional repression of HMGA2 may prevent mammary tumorigenesis, the mechanisms governing repression remain elusive.
The potential role of BRCA1 in transcriptional regulation has been revealed by discovering its ability to bind many important transcription factors, including p53, c-Myc, and STAT1 (13–15). Expression of several target proteins, including p21WAF1, cyclin B1, and EGR1, is activated or repressed by the presence of BRCA1. BRCA1 lacks DNA sequence-specific binding activity, suggesting that it may serve as a mediator to regulate a given gene expression in collaboration with transcriptional factors with specific DNA recognition activities. This missing link between BRCA1 and DNA binding was filled by identifying a novel transcriptional repressor, named ZBRK1 (zinc finger and BRCA1-interacting protein with a KRAB domain 1), which physically interacts with BRCA1 and binds to a consensus sequence of GGGxxxCAGxxxTTT (16). Interestingly, sequences closely conforming to this recognition sequence lie within the putative regulatory regions of a subset of BRCA1 target genes (16). Therefore, it appears that BRCA1, together with ZBRK1, may control transcription of at least a subset of its downstream effectors.
BRCA1 is known to bind CtBP-interacting protein (CtIP), a transcriptional co-repressor, via its BRCT domains. This interaction is abolished by tumor-associated mutations that affect these domains, such as A1708E, M1775R, and Y1853Δ. Thus, the in vivo interaction of CtIP and BRCA1 is likely to be important for BRCA1-mediated tumor suppression. Our laboratory previously discovered through microarray analysis that BRCA1 and CtIP depletion in MCF10A cells results in co-activation/overexpression of a common set of genes, including angiopoietin-1 (ANG1) and HMGA2 (17). Furthermore, BRCA1 and CtIP form a repressor complex with ZBRK1 on ANG1 promoter (18). Removal of any one of these proteins derepresses the promoter and activates the transcription of ANG1, which enhances blood vessel size and promotes breast tumor progression (18). However, whether HMGA2 is subject to a similar transcriptional modulation remains to be elucidated.
In this communication, we demonstrate that ZBRK1, BRCA1, and CtIP form a repressor complex at a single recognition site of ZBRK1 in HMGA2 promoter. Inactivation of this repressor complex by depleting any one of these proteins derepresses and activates HMGA2 expression in MECs, leading to increased proliferation, anchorage-independent growth in soft agar, and impairment of mammary acini formation. These results provide a direct functional link between oncogenic activity of HMGA2 and the ZBRK1/BRCA1/CtIP repressor complex.
EXPERIMENTAL PROCEDURES
Cell Culture and Adenoviral RNAi Production
Human mammary epithelial MCF10A cells were cultured as described previously (19). The adenovirus-based RNAi vectors were generated as described (18) via cloning an expression cassette of U6 promoter-BRCA1, -CtIP, or -ZBRK1 short hairpin RNAi (0.4 kb) into pAdTrack plasmid upstream of a cytomegalovirus-green fluorescent protein cassette (1.6 kb). Adenoviruses were produced as described (17). MCF10A cells seeded at 5 × 105 cells/60-mm plate were infected with adenovirus at a designated multiplicity of infection (m.o.i.) for 24 h.
qRT-PCR and RT-PCR
MCF10A cells seeded at 5 × 105 cells/60-mm plate were infected with adenoviral luciferase-, ZBRK1, BRCA1-, or CtIP-RNAi at 10 or 20 m.o.i. for 24 h. Total RNA from RNAi-treated and human breast cancer cells was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The first-strand cDNAs were synthesized using the SuperscriptTM First-Strand Synthesis System for RT-PCR kit (Invitrogen). Triplicate samples were subjected to qRT-PCR using an iCycler (Bio-Rad). α-Tubulin was used as an internal control. Primer sequences used were: HMGA2 qRT-PCR (F), 5′-AAA GCA GCT CAA AAG AAA GCA-3′; HMGA2 qRT-PCR (R), 5′-TGT TGT GGC CAT TTC CTA GGT-3′; α-tubulin RT (F), 5′-TGA CCT GAC AGA ATT CCA GAC CA-3′; and α-tubulin RT (R), 5′-GCA TTG ACA TCT TTG GGA ACC AC-3′. The relative abundance of HMGA2 mRNA was calculated after normalization using α-tubulin mRNA. RT-PCR analysis of HMGA2 expression in control and BRCA1-deficient mouse mammary tumors was performed as described before (18).
Western Blotting
Total cell lysates (20 μg) were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membranes. Each membrane was incubated with specific primary antibody overnight at 4 °C, followed by horseradish peroxidase-conjugated secondary antibody. Membranes were visualized using the ECL Western blotting detection system (Amersham Biosciences). HMGA2 antibody (GTX98058) was purchased from GeneTex (Irvine, CA), whereas the mouse monoclonal antibodies for ZBRK1, BRCA1, CtIP, and p84 were prepared by our laboratory.
Reporter Constructs and Luciferase Reporter Assay
A 5133-bp fragment of the human HMGA2 promoter was obtained from PCR-amplified plasmid DNA isolated from human BAC (bacterial artificial chromosome) clone RP11–677M24 (Invitrogen). Primer sequences used were: HMGA2–5003(S) (5′-GCT CTA GAG ATT GGA CAG AGG AGA GTA CTG G-3′) and HMGA2+114(AS) (5′-ATA TAA GCT TAC AGG CAG AGG ACA GAG TAG TGG-3′). The primers used to amplify −2994/+114 and −5003/−2994 fragments were HMGA2–2994(S) (5′-GCT CTA GAG AGA ATA GCA TGG GAG AAC CAC C-3′), HMGA2+114(AS) (5′-ATA TAA GCT TAC AGG CAG AGG ACA GAG TAG TGG-3′), HMGA2–5003(S) (5′-GCT CTA GAG ATT GGA CAG AGG AGA GTA CTG G-3′), and HMGA2–2994(AS) (5′-ATA TAA GCT TGG TGG TTC TCC CAT GCT ATT CTC-3′), respectively. The amplified DNA fragments were ligated into pGL2-basic plasmid through the NheI and HindIII sites. The primers used for site-directed mutagenesis to inactivate ZBRK1 binding site 1 and 2 were mHMGA2-ZBRK1-1(S) (5′-CTA CAT CAT GTG TCA AGC TTA ATT AAA AAC CTT G-3′), mHMGA2-ZBRK1-1(AS) (5′-CAA GGT TTT TAA TTA AGC TTG ACA CAT GAT GTA G-3′), mHMGA2-ZBRK1–2(S) (5′-GGA GAA AAA AGT TCA ATG AAG AAG TCT AGA CGC TCT GTG TGT GCA CA-3′), and mHMGA2-ZBRK1–2(AS) (5′-TGT GCA CAC ACA GAG CGT CTA GAC TTC TTC ATT GAA CTT TTT TCT CC-3′), respectively. Correct mutations were confirmed by sequencing and restriction digestion.
MCF10A cells seeded at 5 × 105 cells/60-mm plate were transfected with 3 μg of luciferase reporter and 0.5 μg of β-galactosidase plasmids using FuGENE 6 prior to adenoviral RNAi infection. Each of the reporter constructs were cotransfected with an internal control plasmid, pCH110, which carries a β-galactosidase reporter gene under the control of the SV40 promoter. Luciferase activity was measured according to the manufacturer's instructions (Promega, Madison, WI) in a Fluoroscan Ascent FL machine (Thermo Labsystems, Waltham, MA). An aliquot of the same cell lysates was used for the measurement of β-galactosidase activity to normalize luciferase activity.
ChIP Assay
This assay was performed as described (20) using the following HMGA2 and ANG1 promoter primers for PCRs: HMGA2-ZBRK1-1-ChIP(S) (5′-TTT TAC CAC CCA CTA ATG GGC TGA CCT G-3′), HMGA2-ZBRK1-1-ChIP(AS) (5′-GAA GAG GCA GTG AAT GGT TGA GAA ACA TAA AC-3′), ANG1(−2400/−2100)(S) (5′-TCC CTC AGG AAA TTG TGC ATT CCT GC-3′), and ANG1(−2400/−2100)(AS) (5′-CTA TGC ACA GCC ACA AAG ATG AAG TGC-3′). Quantitative PCR was carried out to quantify DNA from ZBRK1-, BRCA1-, CtIP-, or nonspecific IgG-ChIP and input control. Comparisons were normalized to input controls.
HMGA2 Retrovirus
GP2–293 packaging cells were transfected with pWZL-HA-HMGA2 and pVSVG plasmids using Lipofectin, and retrovirus was harvested from the conditioned medium. MCF10A cells were infected with retrovirus using 8 μg/ml Polybrene, and the stable clones (MCF10A/HMGA2) were selected with 50 μg/ml hygromycin (Roche Applied Science). Overexpression of HMGA2 was confirmed by Western analysis.
Silencing HMGA2 Expression by siRNA
MCF10A cells were transiently transfected with the indicated amounts of HMGA2-specific siRNA or scramble siRNA (Dharmacon, Lafayette, CO) using LipofectamineTM RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Cells treated with transfection reagent only were used as a mock control. Inhibition of HMGA2 was determined by Western blotting at 60 h post-transfection. All transfectants were maintained in antibiotic-free complete medium until collection for analysis.
Anchorage-independent Growth Assay
Anchorage-independent cell growth was determined by analyzing colony formation of cells in soft agar. Cells (1 × 104) were resuspended in 1 ml of top agar (MCF10A media containing 0.35% Noble agar (USB Corp., Cleveland, OH) warmed to 40 °C. The cell suspension was layered onto 1 ml of set bottom agar (MCF10A media containing 0.5% Noble agar) in a 6-well plate. One milliliter of medium was added on the top agar and changed once per week to compensate for evaporation. Colonies greater than 100 cells were scored after 21 days.
Three-dimensional Basement Membrane Culture
MCF10A cells were infected with adenovirus-RNAi at m.o.i. 20. Twenty-four hours post infection, cells were transfected with or without HMGA2 siRNA for 60 h. Approximately 8 × 103 cells per well were seeded in four-well Nunc chamber slides coated with Growth Factor Reduced Matrigel (BD Biosciences, San Jose, CA) and covered with growth medium supplemented with 2% Matrigel as previously described (19). The diameter of acinar structures was measured using Spot Advanced software. Fluorescence imaging was performed with Phase I/4′,6-diamidino-2-phenylindole filters on a Zeiss Axiovert 200M equipped with a Hamamatsu Photonics K.K. Deep Cooled digital camera using Axiovision 4.4 software. For cell number counting, cells were recovered from Matrigel after 6, 9, and 12 days by digestion with dispase (BD Biosciences), following the manufacturer's instructions. A 10-μl aliquot of the single cell suspension was loaded onto a hemocytometer and counted in triplicate.
RESULTS
HMGA2 Expression Is Co-repressed by ZBRK1, BRCA1, and CtIP in MECs
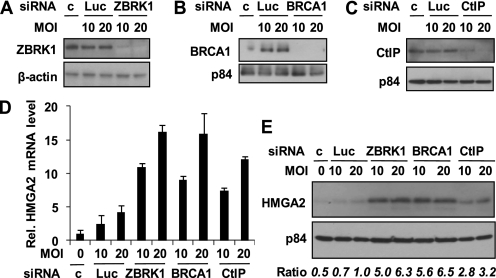
We previously identified 11 genes, including ANG1 and HMGA2, that are co-regulated by BRCA1, CtIP (18), and ZBRK13 through microarray analyses on MCF10A cells depleted of BRCA1, CtIP, or ZBRK1 by adenoviral RNAi. The ZBRK1/BRCA1/CtIP complex co-represses ANG1 (a secreted angiogenic factor) via a ZBRK1 recognition site in the ANG1 promoter. Disruption of this complex up-regulates ANG1, which modulates tumor microenvironment and promotes breast tumor progression (18). How HMGA2 is regulated, however, remains unclear. Based on our microarray data showing a significant increase of HMGA2 expression (4.55- and 2.55-fold by BRCA1-KD and CtIP-KD, respectively) (18), we performed qRT-PCR to confirm HMGA2 expression following adenovirus RNAi-mediated depletion of ZBRK1, BRCA1, or CtIP in MCF10A cells. As shown in Fig. 1, we successfully depleted protein expression of each of these factors (Fig. 1, A–C), which correlates with increased HMGA2 mRNA expression (Fig. 1D). Similarly, HMGA2 protein expression was increased in those cells depleted of any one of those factors (Fig. 1E), further suggesting that a deficiency of any member of the ZBRK1/BRCA1/CtIP complex up-regulates HMGA2 expression.
FIGURE 1.
HMGA2 is repressed by ZBRK1, BRCA1, and CtIP in MCF10A cells. A–C, Western blot analysis on the expression of ZBRK1 (A), BRCA1 (B), and CtIP (C) in cells infected with 10 or 20 m.o.i. of adenoviral luciferase-, ZBRK1-, BRCA1-, or CtIP-RNAi, respectively. c represents no treatment control. β-Actin or p84 serves as an internal loading control. D, quantitative RT-PCR analysis of HMGA2 mRNA expression in cells treated as above. Relative expression levels of HMGA2 in the treated cells were plotted against the control cells. E, Western blot analysis of HMGA2 protein expression in cells treated as above. The lysates were also blotted for p84 as an internal loading control. Relative expression levels of HMGA2 normalized to the expression levels of p84 (Ratio) are also indicated.
A Single ZBRK1 Recognition Site in HMGA2 Promoter Mediates the BRCA1, ZBRK1, and CtIP Repression Complex
To determine whether transcription of HMGA2 is regulated by the ZBRK1/BRCA1/CtIP complex, we performed a reporter assay measuring the luciferase activity of 5.133 kb of the HMGA2 promoter (−5003 to +114; construct H1), as well as two deletion mutants, i.e. −2994 to +114 (construct H2) and −5003 to −2994 (construct H3) in MCF10A cells (Fig. 2A). Upon depletion of ZBRK1 by adenoviral RNAi, reporter activity was significantly increased in constructs H1 and H2 (Fig. 2B), but not construct H3. Similar results were obtained after depletion of BRCA1 or CtIP (Fig. 2B), suggesting that the −2994 to +114 region of the HMGA2 promoter contains the essential element(s) for transcriptional repression by the ZBRK1/BRCA1/CtIP complex.
FIGURE 2.
ZBRK1, BRCA1, and CtIP co-repress HMGA2 expression via a single ZBRK1 recognition element in the promoter. A, schematics of HMGA2 reporter constructs in the promoter region. From the first initiation codon, H1 contains −5003 to +114; H2, from −2994 to +114; and H3, from −5003 to −2994. B, relative luciferase activity of reporter constructs from MCF10A cells infected with adenoviral ZBRK1-, BRCA-, and CtIP-RNAi. Error bar, mean ± S.D. C, schematics of H2 reporter constructs with mutated ZBRK1 recognition site. The sequences of two putative ZBRK1 binding sites are GTGtcaCAGataATT (Site 1, −2523/−2509, sense) and AAGaagCTGgggCGC (Site 2, −820/−806, antisense). H2-1, site 1 is mutated; H2-2, site 2 is mutated; H2-3, both site 1 and 2 are mutated as described under “Experimental Procedures.” D, relative luciferase activity of reporter constructs of H2-1, H2-2, and H2-3 from MCF10A cells infected with adenoviral ZBRK1-, BRCA1-, and CtIP-RNAi. Error bar, mean ± S.D. E, ChIP analysis on a 264-bp fragment (−2574/−2311) encompassing the ZBRK1 binding site 1 in HMGA2 promoter. The association of ZBRK1, BRCA1, and CtIP with the HMGA2 promoter was analyzed by ChIP using MCF10A cells infected with adenoviral luciferase-, ZBRK1-, BRCA1-, or CtIP-RNAi at 20 m.o.i. for 24 h. The immunoprecipitated chromatin DNA was amplified by PCR using ZBRK1 binding site-specific primers. The promoter of the ANG1 gene served as a positive control. The right panel shows the quantities of the immunoprecipitated chromatin DNA from MCF10A cells treated as above by quantitative PCR using the same specific primers as in the left panel.
Sequence analysis revealed that construct H2 (−2994 to +114) contains two potential ZBRK1 consensus sites, i.e. site 1 (−2523/−2509, sense): GTGtcaCAGataATT, and site 2 (−820/−806, antisense): AAGaagCTGgggCGC. We then generated mutant constructs by inactivating either site 1 or 2, or both sites (Fig. 2C), for further investigation. Constructs with mutations in site 1 or both sites, but not site 2 alone, abolished HMGA2 repression by ZBRK1, BRCA1, or CtIP (Fig. 2D), suggesting that site 1 (−2523/−2509) is a strong candidate for a ZBRK1 binding site.
Next, to demonstrate that the ZBRK1/BRCA1/CtIP complex is associated with the HMGA2 promoter in vivo, we performed a ChIP assay on a 264-bp fragment around the ZBRK1 site 1 (−2523/−2509). As shown in Fig. 2E, antibodies against ZBRK1, BRCA1, or CtIP immunoprecipitated the same DNA fragment of the HMGA2 promoter. Using RNAi to deplete any of these proteins completely abolished the association of all the three proteins with the promoter, suggesting that the three participants are all essential for the complex formation. Taken together, these data substantiate a notion that ZBRK1, BRCA1, and CtIP coordinately form a repressor complex tethered at the ZBRK1 recognition site in the HMGA2 promoter.
Depletion of ZBRK1, BRCA1, or CtIP Induces Anchorage-independent Growth
Overexpression of HMGA2 has been shown to lead to a transformed phenotype in cultured lung cells derived from normal tissue (21). To test whether increased expression of HMGA2 in MCF10A mammary epithelial cells leads to a transformed phenotype, we performed soft agar colony formation assays, which is an in vitro assay used to mimic tumorigenicity in a mouse model. Because HMGA2 mRNA and protein were up-regulated in MCF10A cells after ZBRK1, BRCA1, or CtIP depletion by adenoviral RNAi, those cells were subjected to soft agar colony formation assay. As shown in Fig. 3A, the colony formation efficiency of MECs depleted of ZBRK1, BRCA1, or CtIP was higher (7- to 9-fold) than those formed by control green fluorescent protein RNAi-infected cells. Consistently, MCF10A cells with HMGA2 overexpression form significantly more colonies than the vector control cells (Fig. 3B, right panel). To confirm that HMGA2 plays as a main effector of ZBRK1, we assayed the colony formation of cells depleted both HMGA2 and ZBRK1 simultaneously with each individual RNAi. It appeared that double knockdown of HMGA2 and ZBRK1 in MCF10A cells significantly reduced the number of colonies compared with ZBRK1 inactivation alone (Fig. 3C, right panel). Based on these observations, HMGA2 up-regulation in MECs, either by ectopic overexpression or by depletion of ZBRK1, BRCA1, or CtIP, contributes to the transformed phenotype as assayed by soft agar colony formation.
FIGURE 3.
Inhibition of ZBRK1, BRCA1, CtIP, or overexpression of HMGA2 induces anchorage-independent growth of MCF10A cells. A and B, colony formation of MCF10A cells infected with adenoviral luciferase-, ZBRK1-, BRCA1-, or CtIP-RNAi at 20 m.o.i. for 24 h (A) and of MCF10A/HMGA2 or vector alone (B, right panel) cell lines in semisolid medium. Cells on soft agar plates were grown for 3 weeks before colonies were stained and visualized microscopically. Colonies having >100 cells were counted. Results from one representative experiment are shown. Error bars, mean ± S.D. Lines, difference in paired comparison. **, p < 0.01; *, p < 0.05. The left panel of B shows Western analysis of HMGA2 expression in MCF10A/HMGA2 cells in comparison to the vector control cells. β-Actin served as a loading control. ◀, indicates HA-HMGA2; ◁, indicates endogenous HMGA2; and the arrow indicates β-actin. C, colony formation of MCF10A cells infected with adenoviral luciferase-, or ZBRK1-RNAi at 20 m.o.i. for 24 h, followed by transfection with or without HMGA2 siRNA for 60 h, in semisolid medium as described in A (right panel). The left panel shows Western analysis of HMGA2 expression upon siRNA treatment using p84 as internal reference (c1, transfection reagent; c2, scramble siRNA (200 μm).
Reduction of ZBRK1 Stimulates Overproliferation and Increases Acinar Size of MCF10A Cells
Although in vitro transformation assays provide models for investigating certain aspects of the cellular processes associated with tumor initiation and progression, they do not model alterations in tissue architecture that are critically involved in tumor development. To determine whether HMGA2 overexpression interferes with growth and morphogenesis of MECs, MCF10A cells were seeded in Matrigel to examine the process of acini formation. Upon ZBRK1 depletion by adenoviral RNAi, the acini formed were significantly larger than the control acini after 9 days in three-dimensional culture. The maximal size difference was reached at day 12, when the increase in diameter was ∼40% larger than control cells. However, knockdown of ZBRK1 and HMGA2 significantly reduced the acinar sizes compared with ZBRK1 depletion alone, suggesting that an increase in acinar size caused by ZBRK1 inactivation requires HMGA2 (Fig. 4A). Consistently, the acinar sizes of MCF10A/HMGA2 cells were significantly larger than those formed from vector control cells. At day 6, there was a 28% increase in acinar diameter in MCF10A/HMGA2 cells relative to controls, and reached a maximum increase of 50% at day 12 (Fig. 4C). Consistent with the observed size increase, the acini with up-regulated HMGA2, either by reduction of ZBRK1 (Fig. 4B) or ectopic expression (Fig. 4D), also exhibited a significant increase in cell numbers at days 9 and 12 in culture. These data show a link between excess HMGA2 and increased proliferation of mammary epithelial cells, resulting in a change in acinar size. Such deviations from normal epithelial behavior in three-dimensional culture after ZBRK1 inhibition revealed significant insights into the mechanisms involved in the development and progression of breast cancer.
FIGURE 4.
Depletion of ZBRK1 or overexpression of HMGA2 results in increased acinar size and cell proliferation. A and C, acinar size of MCF10A cells either infected with 20 m.o.i. of adenoviral luciferase-, or ZBRK1 RNAi, followed by transfection with or without HMGA2 siRNA (A) or overexpressing HMGA2 or none as control (C). These cells were cultured in three-dimensional Matrigel, and the acinar diameter was assessed at days 3, 6, 9, and 12. Each point represents the mean acinar diameter derived from three independent experiments. In each experiment, at least 200 structures were analyzed per time point. The bars indicate the standard error and *, p < 0.05, highlighting a significant statistical difference. B and D, relative cell number of MCF10A cells from (A and C). A single cell suspension was prepared at days 6, 9, and 12 by enzyme digestion and counted with a hemocytometer. Each point represents the mean cell number derived from three independent experiments. The bars indicate the standard error and **, p < 0.01, highlighting a significant difference.
Adenoviral RNAi-mediated Depletion of ZBRK1 Results in Reduction of Luminal Cell Death
Suppression of luminal cell death may be an important early step in the progression to breast cancer (22). We therefore further characterized the functional consequences of HMGA2 overexpression in MCF10A acini by investigating cell death in the lumen. MCF10A cells with ZBRK1 depletion (Fig. 5A) or HMGA2 overexpression (Fig. 5B) were stained with Hoechst 33342 at days 3, 6, 9, and 12 in Matrigel culture. MCF10A cells with green fluorescent protein adenovirus or MCF10A/vector control cells formed 73% or 84%, respectively, normal acini with a hollow lumen at day 12. In contrast, only 33% of cells with ZBRK1 depletion and 30.5% of cells with HMGA2 overexpression formed normal acini, with the remaining acini containing a filled lumen (Fig. 5, A and B, lower panel). These results suggest that the cell death in the luminal space of HMGA2-overexpressing acini is inhibited, explaining the enhancement of acinar size and increase of total cell numbers.
FIGURE 5.
Frequency of normal acinar formation of MCF10A cells either depleting ZBRK1 expression (A) or ectopic expression of HMGA2 (B). Upper panel: representative images of normal acinar structures or atypical acini with cells filled in lumen after 12 days in culture. All cells were cultured at the same passage number and imaged at the same magnification. Bars represent 25 μm. Arrowheads indicate cells in the lumen. Lower panel: number of normal acini formation of MCF10A cells infected with adenoviral luciferase- or ZBRK1 RNAi (A), and overexpressed HMGA2 or none as control (B). These MCF10A cells were allowed to grow in Matrigel for 3, 6, 9, and 12 days, then fixed and stained with Hoechst 33342 to visualize nuclei. Each point represents the mean number of hollow lumen acini from three independent experiments. In each experiment, at least 200 acini were analyzed per time point. The bars indicate the standard error and *, p < 0.05, highlighting a significant difference.
BRCA1-deficient Mouse Mammary Tumors and Human Breast Cancer Cells Express High Levels of HMGA2
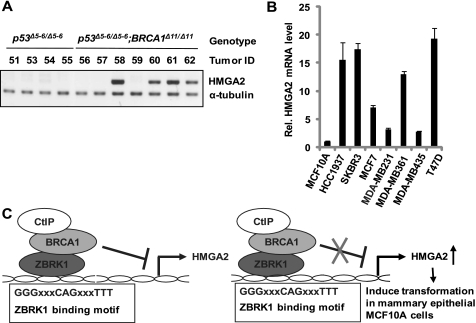
To further confirm the above-mentioned notion that the induction of HMGA2 expression occurs via inactivation of ZBRK1/BRCA1/CtIP complex formation, we compared HMGA2 expression in seven mammary tumors derived from genetic knockout mice with BRCA1 and p53 gene inactivation (BRCA1Δ11/Δ11; p53Δ5–6/Δ5–6) with four tumors derived from p53 gene inactivation (p53Δ5–6/Δ5–6) alone (23, 24). HMGA2 expression, as determined by RT-PCR, was highly up-regulated in 57% of BRCA1-deficient tumors in comparison with none of the control tumors (Fig. 6A). Because BRCA1 expression was frequently reduced in human breast tumors (25–27), the expression of HMGA2 would be expected to be elevated in many of them. To verify this possibility, we examined HMGA2 mRNA expression in seven human breast cancer cell lines, including a BRCA1-deficient HCC1937 cell line, and found that five out of seven (HCC1937 included) have significantly high HMGA2 expression (Fig. 6B). Taken together, these results support the notion that BRCA1 participates in HMGA2 regulation and the elevated HMGA2 expression has a critical role in BRCA1 deficiency-related tumorigenesis.
FIGURE 6.
Overexpression of HMGA2 in BRCA1-deficient mouse mammary tumors and human breast cancer cell lines. A and B, HMGA2 expression in p53Δ5–6/Δ5–6 and BRCA1Δ11/Δ11; p53Δ5–6/Δ5–6 mouse mammary tumors (A) and human breast cancer cells (B) detected by RT-PCR and qRT-PCR, respectively. α-Tubulin served as an internal control. C, schematic model for HMGA2 transcriptional regulation by ZBRK1/BRCA1/CtIP complex via a ZBRK1 recognition site in HMGA2 promoter.
DISCUSSION
Deficiency of BRCA1 leads to mammary gland and ovarian tumorigenesis (28, 29). The molecular mechanism of how BRCA1 inactivation leads to carcinogenesis remains unsubstantiated, although it has been linked to DNA double strand break repair (30). In this communication, we show that BRCA1, by forming a repression complex with ZBRK1 and CtIP, coordinately regulates HMGA2 expression via a ZBRK1 recognition site in the HMGA2 promoter. Depletion of any protein in this complex via adenoviral RNAi in MCF10A mammary epithelial cells activates HMGA2 expression, resulting in increased colony formation in soft agar. Similarly, depletion of ZBRK1, or ectopic overexpression of HMGA2, enhances abnormal acinar size with increased cell number and reduces normal acinar formation. These results indicate that inactivation of the BRCA1/ZBRK1/CtIP repression complex leads to activation of HMGA2 expression, which confers a neoplastic phenotype to mammary epithelial cells. The results therefore offer a potential mechanism to explain how BRCA1 deficiency contributes to breast tumorigenesis.
In our previous study using a three-dimensional morphogenesis assay, MECS depleted of BRCA1 undergo vigorous proliferation but failed to differentiate into acinar structure (17). This suggests that BRCA1 has a role in directly regulating cell proliferation and differentiation separate from its activity in DNA double strand break repair. However, the mechanisms underlying how BRCA1 directly contributes such functions are largely unclear. BRCA1 mediates transcriptional repression through binding to the CtIP co-repressor and transcriptional repressor ZBRK1, which physically tethers to a given gene promoter. To identify genes regulated by this complex, we performed microarray analysis on cells infected with BRCA1, CtIP, or ZBRK1 RNAi and found a dozen genes to be up-regulated upon depletion of this repressor complex. One of these, ANG1, participates in blood vessel formation, and was recently demonstrated to undergo direct repression by BRCA1. Furthermore, BRCA1 was found to form a repressor complex with ZBRK1 and CtIP on the ANG1 promoter (18). Despite its function in remodeling the tumor microenvironment, ANG1 may play a minor role in transforming MECs into neoplastic cells. Untimely expression of HMGA2 is observed in many benign and malignant tumors, suggesting its potential as an oncogene (31). Thus, demonstration of a similar regulation of HMGA2 by BRCA1 will offer a plausible mechanism addressing how BRCA1 inactivation contributes to breast carcinogenesis, specifically through HMGA2 activation.
As we demonstrate in this report, the ZBRK1/BRCA1/CtIP complex repressed HMGA2 gene expression, and removal of any one of the repressor components by adenovirus RNAi induced the expression of HMGA2 (Fig. 1). The HMGA2 promoter contains one ZBRK1 consensus motif at the −2523/−2509 region upstream of exon 1. When ZBRK1, BRCA1, or CtIP was depleted in cells transfected with HMGA2 promoter constructs, a significant induction in reporter activity was observed in cells harboring the wild-type promoter but not the promoter with the ZBRK1 binding site mutated (Fig. 2, B and D). This supports the notion that HMGA2 is subject to negative regulation by ZBRK1/BRCA1/CtIP via a consensus ZBRK1 binding motif (−2523/−2509) on the HMGA2 promoter. The direct and physical localization of ZBRK1/BRCA1/CtIP repressor complex on HMGA2 promoter at this ZBRK1 consensus motif was also determined by using a ChIP assay (Fig. 2E). This repression mode appears to be similar to that of ANG1 promoter as we previously described (18).
Given the oncogenic potential of HMGA2, and its common overexpression in breast cancers, an important but unresolved question is the functional consequence of elevated HMGA2 expression in normal mammary epithelial cells. To address this question, we have exploited the characteristics of the spontaneously immortalized human mammary epithelial cell line MCF10A. In three-dimensional culture in reconstituted basement membrane (Matrigel), these cells form acinar structures that retain important characteristics of glandular epithelium in vivo, such as apico-basal polarization, suppression of proliferation, and acinar cavitation through apoptosis of cells located within the inner cell mass (32). This three-dimensional culture system provides a powerful model for characterizing the biological activities of proteins implicated in breast cancer development and progression. Effects observed upon overexpression and/or constitutive activation of particular oncoproteins include lack of proliferative suppression (cyclin D1), lumenal filling, and development of multiacinar structures (ErbB2) and disruption of cell-cell adhesion in acini (colony-stimulating factor receptor) (22, 33, 34). Consistently, increased expression of HMGA2, either by the overexpression construct or by depletion of ZBRK1, results in increased acinar size, overproliferation, and inhibition of lumenal cell death, supporting a role for HMGA2 in breast carcinogenesis.
Furthermore, MCF10A cells with ZBRK1 depletion or HMGA2 overexpression are capable of forming colonies in soft agar with high efficiency, the best characteristic of transformed cells (Fig. 3), suggesting that derepression of HMGA2 induces mammary epithelial cells transformation. However, how HMGA2 overexpression induces tumorigenicity is not fully understood. In a recent study, Li et al. showed that HMGA2-expressing cells displayed deficiency in DNA end-joining repair and accumulated more endogenous DNA damage, implicating HMGA2 in the promotion of genomic instability and tumorigenesis (35). In another study in transgenic mice, Fedele et al. (36) found that HMGA2-mediated E2F1 activation is a crucial event in pituitary tumorigenesis. Interestingly, Bcl-2 mRNA level was highly elevated in HMGA2-overexpressing MCF10A cells compared with empty vector control.3 Because Bcl-2 is an antiapoptotic member of the Bcl-2 family proteins, it promotes cell survival via inhibiting the mitochondrial release of proapoptotic factors. HMGA2-induced anchorage-independent growth may therefore be mediated, at least in part, through several pathways. Elucidating the downstream targets of HMGA2 and their roles in HMGA2-induced breast tumorigenesis warrants further investigation.
microRNAs have also been shown to regulate HMGA2 expression (37–39). The HMGA2 3′ untranslated region has seven conserved sites complementary to the let-7 microRNA (40), which is expressed in later stages of animal development (41). Disrupting the binding between let-7 and HMGA2 promotes anchorage-independent growth via up-regulation of HMGA2 (42), suggesting that let-7 negatively regulates HMGA2 expression by inhibiting the availability of its mRNA for translation. With our new finding that HMGA2 expression is controlled by the ZBRK1/BRCA1/CtIP repressor complex as demonstrated here (Fig. 6C), it is apparent that normal cells have developed at least two effective control mechanisms to ensure that HMGA2 is silent. Failure in expressing let-7 or forming the BRCA1/ZBRK1/CtIP repressor complex results in activating HMGA2, leading to oncogenic transformation. However, alteration in one of these two regulatory mechanisms may not be sufficient to induce HMGA2 overexpression. Consistently, elevated HMGA2 expression was found in many, but not all, BRCA1-deficient mouse mammary tumors and human breast cancer cells, including HCC1937 (BRCA1-deficient cell) (Fig. 6, A and B) (12). It is likely that other regulatory mechanisms remain effective in some tumors. Therefore, activation of HMGA2 through removal of ZBRK1/BRCA1/CtIP complex on HMGA2 promoter is an important step in breast carcinogenesis. Whether there is any relationship between BRCA1/ZBRK1/CtIP repressor complex and the regulatory mechanisms of let-7 microRNA remains to be explored.
Acknowledgments
We thank Dr. Scott Lowe (Cold Spring Harbor Laboratory, NY) for generously providing us with HMGA2 expression vector, and Drs. Phang-Lang Chen and Erin Goldblatt (University of California at Irvine) for helpful discussion and critical reading, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grant CA94170 (to W. H. L.).
K. M. Ahmed, C. Y. Tsai, and W.-H. Lee, unpublished data.
- HMGA2
- high mobility group AT-hook2
- ZBRK1
- zinc finger and BRCA1-interacting protein with a KRAB domain 1
- BRCA1
- breast cancer-associated gene 1
- CtBP
- C-terminal-binding protein
- CtIP
- CtBP-interacting protein
- ANG1
- angiopoietin-1
- MEC
- mammary epithelial cell
- RNAi
- RNA interference
- m.o.i.
- multiplicity of infection
- ChIP
- chromatin immunoprecipitation
- siRNA
- small interference RNA
- RT
- reverse transcription
- qRT
- quantitative RT.
REFERENCES
- 1.Nagpal S., Ghosn C., DiSepio D., Molina Y., Sutter M., Klein E. S., Chandraratna R. A. (1999) J. Biol. Chem. 274, 22563–22568 [DOI] [PubMed] [Google Scholar]
- 2.Johnson K. R., Disney J. E., Wyatt C. R., Reeves R. (1990) Exp. Cell Res. 187, 69–76 [DOI] [PubMed] [Google Scholar]
- 3.Johnson K. R., Lehn D. A., Reeves R. (1989) Mol. Cell. Biol. 9, 2114–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashar H. R., Cherath L., Przybysz K. M., Chada K. (1996) Genomics 31, 207–214 [DOI] [PubMed] [Google Scholar]
- 5.Reeves R. (2001) Gene 277, 63–81 [DOI] [PubMed] [Google Scholar]
- 6.Huth J. R., Bewley C. A., Nissen M. S., Evans J. N., Reeves R., Gronenborn A. M., Clore G. M. (1997) Nat. Struct. Biol. 4, 657–665 [DOI] [PubMed] [Google Scholar]
- 7.Li L., Yoder K., Hansen M. S., Olvera J., Miller M. D., Bushman F. D. (2000) J. Virol. 74, 10965–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maher J. F., Nathans D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6716–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farnet C. M., Bushman F. D. (1997) Cell 88, 483–492 [DOI] [PubMed] [Google Scholar]
- 10.Sgarra R., Rustighi A., Tessari M. A., Di Bernardo J., Altamura S., Fusco A., Manfioletti G., Giancotti V. (2004) FEBS Lett. 574, 1–8 [DOI] [PubMed] [Google Scholar]
- 11.Wolffe A. P. (1994) Science 264, 1100–1101 [DOI] [PubMed] [Google Scholar]
- 12.Langelotz C., Schmid P., Jakob C., Heider U., Wernecke K. D., Possinger K., Sezer O. (2003) Br J. Cancer 88, 1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouchi T., Monteiro A. N., August A., Aaronson S. A., Hanafusa H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Zhang H., Kajino K., Greene M. I. (1998) Oncogene 17, 1939–1948 [DOI] [PubMed] [Google Scholar]
- 15.Buckley N. E., Hosey A. M., Gorski J. J., Purcell J. W., Mulligan J. M., Harkin D. P., Mullan P. B. (2007) Mol. Cancer Res. 5, 261–270 [DOI] [PubMed] [Google Scholar]
- 16.Zheng L., Pan H., Li S., Flesken-Nikitin A., Chen P. L., Boyer T. G., Lee W. H. (2000) Mol. Cell 6, 757–768 [DOI] [PubMed] [Google Scholar]
- 17.Furuta S., Jiang X., Gu B., Cheng E., Chen P. L., Lee W. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9176–9181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuta S., Wang J. M., Wei S., Jeng Y. M., Jiang X., Gu B., Chen P. L., Lee E. Y., Lee W. H. (2006) Cancer Cell 10, 13–24 [DOI] [PubMed] [Google Scholar]
- 19.Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 20.Liu F., Lee W. H. (2006) Mol. Cell. Biol. 26, 3124–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Cello F., Hillion J., Hristov A., Wood L. J., Mukherjee M., Schuldenfrei A., Kowalski J., Bhattacharya R., Ashfaq R., Resar L. M. (2008) Mol. Cancer Res. 6, 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debnath J., Mills K. R., Collins N. L., Reginato M. J., Muthuswamy S. K., Brugge J. S. (2002) Cell 111, 29–40 [DOI] [PubMed] [Google Scholar]
- 23.Lin S. C., Lee K. F., Nikitin A. Y., Hilsenbeck S. G., Cardiff R. D., Li A., Kang K. W., Frank S. A., Lee W. H., Lee E. Y. (2004) Cancer Res. 64, 3525–3532 [DOI] [PubMed] [Google Scholar]
- 24.Xu X., Wagner K. U., Larson D., Weaver Z., Li C., Ried T., Hennighausen L., Wynshaw-Boris A., Deng C. X. (1999) Nat. Genet. 22, 37–43 [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Chen C. F., Riley D. J., Allred D. C., Chen P. L., Von Hoff D., Osborne C. K., Lee W. H. (1995) Science 270, 789–791 [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., Wang Q., Kajino K., Greene M. I. (2000) DNA Cell Biol. 19, 253–263 [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Silver D. P., Walpita D., Cantor S. B., Gazdar A. F., Tomlinson G., Couch F. J., Weber B. L., Ashley T., Livingston D. M., Scully R. (1998) Mol. Cell 2, 317–328 [DOI] [PubMed] [Google Scholar]
- 28.Couch F. J., DeShano M. L., Blackwood M. A., Calzone K., Stopfer J., Campeau L., Ganguly A., Rebbeck T., Weber B. L. (1997) N. Engl. J. Med. 336, 1409–1415 [DOI] [PubMed] [Google Scholar]
- 29.Zheng L., Li S., Boyer T. G., Lee W. H. (2000) Oncogene 19, 6159–6175 [DOI] [PubMed] [Google Scholar]
- 30.Zhong Q., Chen C. F., Li S., Chen Y., Wang C. C., Xiao J., Chen P. L., Sharp Z. D., Lee W. H. (1999) Science 285, 747–750 [DOI] [PubMed] [Google Scholar]
- 31.Young A. R., Narita M. (2007) Genes Dev. 21, 1005–1009 [DOI] [PubMed] [Google Scholar]
- 32.Shaw K. R., Wrobel C. N., Brugge J. S. (2004) J. Mammary Gland Biol. Neoplasia 9, 297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muthuswamy S. K., Li D., Lelievre S., Bissell M. J., Brugge J. S. (2001) Nat. Cell Biol. 3, 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrobel C. N., Debnath J., Lin E., Beausoleil S., Roussel M. F., Brugge J. S. (2004) J. Cell Biol. 165, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li A. Y., Boo L. M., Wang S. Y., Lin H. H., Wang C. C., Yen Y., Chen B. P., Chen D. J., Ann D. K. (2009) Cancer Res. 69, 5699–5706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedele M., Visone R., De Martino I., Troncone G., Palmieri D., Battista S., Ciarmiello A., Pallante P., Arra C., Melillo R. M., Helin K., Croce C. M., Fusco A. (2006) Cancer Cell 9, 459–471 [DOI] [PubMed] [Google Scholar]
- 37.Lee Y. S., Dutta A. (2007) Genes Dev. 21, 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dröge P., Davey C. A. (2008) Cell Stem Cell 2, 8–9 [DOI] [PubMed] [Google Scholar]
- 39.Park S. M., Shell S., Radjabi A. R., Schickel R., Feig C., Boyerinas B., Dinulescu D. M., Lengyel E., Peter M. E. (2007) Cell Cycle 6, 2585–2590 [DOI] [PubMed] [Google Scholar]
- 40.Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 41.Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., Maller B., Hayward D. C., Ball E. E., Degnan B., Müller P., Spring J., Srinivasan A., Fishman M., Finnerty J., Corbo J., Levine M., Leahy P., Davidson E., Ruvkun G. (2000) Nature 408, 86–89 [DOI] [PubMed] [Google Scholar]
- 42.Mayr C., Hemann M. T., Bartel D. P. (2007) Science 315, 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]