Abstract
GSK-3 is active in the absence of growth factor stimulation and generally acts to induce apoptosis or inhibit cell proliferation. We previously identified a subset of growth factor-inducible genes that can also be induced in quiescent T98G cells solely by inhibition of GSK-3 in the absence of growth factor stimulation. Computational predictions verified by chromatin immunoprecipitation assays identified NF-κB binding sites in the upstream regions of 75% of the genes regulated by GSK-3. p50 bound to most of these sites in quiescent cells, and for one-third of the genes, binding of p65 to the predicted sites increased upon inhibition of GSK-3. The functional role of p65 in gene induction following inhibition of GSK-3 was demonstrated by RNA interference experiments. Furthermore, inhibition of GSK-3 in quiescent cells resulted in activation of IκB kinase, leading to phosphorylation and degradation of IκBα and nuclear translocation of p65 and p50. Taken together, these results indicate that the high levels of GSK-3 activity in quiescent cells repress gene expression by negatively regulating NF-κB through inhibition of IκB kinase. This inhibition of NF-κB is consistent with the role of GSK-3 in the induction of apoptosis or cell cycle arrest in cells deprived of growth factors.
Keywords: Cell/Cycle, Chromatin/Immunoprecipitation/ChIP, Phosphorylation/Transcription Factors, Signal Transduction/Protein Kinases, Signal Transduction/Protein Kinases/Serine/Threonine, Transcription/NF-κB, GSK-3, Phosphatidylinositol 3-Kinase
Introduction
Glycogen synthase kinase-3 (GSK-3)2 is a serine/threonine kinase that plays a key role in regulating cell differentiation, proliferation, and survival (1–3). Although initially identified as a metabolic control enzyme that phosphorylated glycogen-synthase in the absence of insulin, GSK-3 is now known to target multiple cell regulatory proteins and to be controlled by both Wnt signaling and the PI 3-kinase/Akt pathway. Responsible for a diverse range of functions, defects in GSK-3 signaling have been implicated in various diseases including cancer, heart disease, Alzheimer disease, and diabetes.
There are two highly homologous mammalian isoforms, GSK-3α and GSK-3β, which are both ubiquitously expressed. Unlike most protein kinases, GSK-3 is active in the absence of growth factor stimulation and generally acts to inhibit cell proliferation or induce apoptosis (1–5). Wnt signaling disrupts the complex of GSK-3, axin, APC, and β-catenin, inhibiting GSK-3 and leading to the stabilization of β-catenin and transcriptional activation of β-catenin/TCF target genes (6). Growth factor stimulation and activation of PI 3-kinase/Akt signaling also results in inhibition of GSK-3, caused by phosphorylation by Akt (7). In addition to β-catenin, the targets of GSK-3 that have been implicated in the regulation of cell proliferation and survival include cyclin D1 (5), the Bcl-2 family member Mcl-1 (8), eukaryotic translation initiation factor 2B (9, 10), and several transcription factors (1, 2).
The fact that GSK-3 is active in the absence of growth factor stimulation suggests that it may play a role in regulating gene expression in quiescent cells. We have addressed this question by identifying genes whose transcription is regulated by GSK-3 downstream of PI 3-kinase signaling in cells arrested in G0 by growth factor deprivation. These studies initially identified a subset of immediate-early genes whose induction in response to growth factor stimulation was dependent on the activation of PI 3-kinase (11). Twelve of these PI 3-kinase-dependent genes (∼40%) were further shown to be inducible solely by inhibition of GSK-3 in the absence of growth factor stimulation, indicating that the activity of GSK-3 was required to maintain repression of these genes during quiescence (12).
Computational analysis identified binding sites for the transcription factor CREB as highly over-represented in the upstream regions of the GSK-3-regulated genes, and subsequent chromatin immunoprecipitation and siRNA experiments indicated that inhibition of CREB by GSK-3 plays an important role in repressing transcription of these genes in quiescent cells (12). In the current study, we extended this analysis to identify additional transcription factors through which GSK-3 may be regulating gene expression. A combination of computational and experimental approaches determined that inhibition of NF-κB by GSK-3 is similarly involved in maintaining repression of gene expression during quiescence. Furthermore, we have shown that the inhibition of NF-κB by GSK-3 affects activation of p65 (RelA) and occurs upstream of the phosphorylation and degradation of IκB. Previous studies have shown that GSK-3 similarly inhibits NF-κB in some cell types (13–15), although it activates NF-κB in others (16–23). In this context, our results indicate that GSK-3 plays a dual role in the regulation of NF-κB depending on the physiological state of the cell. NF-κB is inhibited by the high intracellular activity of GSK-3 expressed either during quiescence or during apoptosis induced by growth factor deprivation, whereas GSK-3 may be required for the activation of NF-κB in response to cytokine stimulation or in some tumor cells where the NF-κB pathway is constitutively active.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
T98G human glioblastoma cells were grown in minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone). To induce quiescence, the cells were incubated in serum-free medium for 72 h and then either left untreated or treated with DMSO vehicle control, 20 ng/ml of TNFα (R & D Systems), 50 ng/ml of human PDGF-BB (Sigma), 5 μm of SB-216763 (BioMol), or 1 μm of 6-bromoindirubin-3′-oxime (BIO) (Calbiochem) for the times indicated in the text.
Prediction of Transcription Factor Binding Sites
Computational analysis to identify over-represented transcription factor binding sites was done as previously described (11, 12, 24). Briefly, the 1-, 3-, and 5-kb upstream regions of the 12 genes that were up-regulated by inhibition of GSK-3 (12) were obtained from the University of California Santa Cruz Genome Browser along with the corresponding mouse orthologous sequences (human version hg18 and mouse version mm8) (25). These sequences were analyzed with the Match program using the MinSUM threshold and the 588 position weight matrices from TRANSFAC professional version 11.1 (26, 27). For each matrix, a permutation test was used to compare the frequencies of the predicted sites in the GSK-3-regulated genes with those in a background set of 165 genes that were expressed but not induced by PDGF stimulation. The p values were adjusted with a false discovery rate correction (28).
Chromatin Immunoprecipitation
Chromatin immunoprecipitations (ChIPs) were performed as previously described (12), using 5 μg of the following antibodies: anti-p65 (sc-372), anti-RelB (sc-226), anti-c-Rel (sc-71), anti-p50 (sc-114), anti-Bcl-3 (sc-185), anti-HDAC-1 (H-51) (all from Santa Cruz), or anti-p52 (Upstate; 06-413). Protein A-agarose beads were successively washed with low salt, high salt, and LiCl buffers and washed twice with 1× Tris-EDTA. Immunoprecipitated chromatin was quantified with real time PCR using primers located within 250 bp of the predicted transcription factor binding sites. Primer sequences are listed in supplemental Table S1.
Real Time RT-PCR
RNA extraction and real time RT-PCR were performed as described (12), except that 1 μg of total RNA was used in the reverse transcription reactions. Primer sequences for the real time PCR are listed in supplemental Table S1.
RNA Interference
Transfections were performed using predesigned siRNAs against p65 (Ambion; S11915) or a nonspecific negative control (Ambion; 4390843). Shortly before transfection, 105 cells/ml were seeded on 60-mm plates in 4 ml of medium containing 10% fetal bovine serum. Transfection reactions containing 5 nm of siRNA, 20 μl of HiPerfect (Qiagen), and 100 μl of serum-free media were incubated for 10 min at room temperature and added dropwise onto the cells. The cells were then incubated at 37 °C for 24 h and then serum-starved for 48 h to induce quiescence. The quiescent cells were then appropriately treated, after which RNA was extracted and analyzed by real time RT-PCR.
Immunoblot Analysis
Whole cell extracts were prepared by lysing cells with 2× Laemmli buffer (29). Cytoplasmic and nuclear fractions were isolated as described elsewhere (30). The proteins were separated by electrophoresis in 12% SDS-polyacrylamide gels, transferred to a nitrocellulose membrane, and incubated with appropriate primary antibody: anti-IκBα (Cell Signaling; 9242), anti-phospho-IκBα (Ser32) (Cell Signaling; 2859), anti-p65 (Santa Cruz; sc-372), anti-phospho-p65 (Ser468) (Cell Signaling; 3039), anti-p50 (Santa Cruz; sc-114), anti-GSK-3β (BD Transduction Laboratories; 610202), anti-phosopho-GSK-3β (Tyr216) (BD Transduction Laboratories; 612312), anti-PARP (Cell Signaling; 9542), anti-14-3-3 (Upstate; 06-511), or anti-β-actin (Sigma). The immunoblots were visualized using species-specific horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) and chemiluminescence (PerkinElmer Life Sciences).
IκB Kinase Assay
In vitro assays of IκB kinase (IKK) were performed according to the procedure of Liang et al. (31) but using anti-IKKγ antiserum (BD Pharmingen; 559675). Briefly, the T98G cells were incubated in serum-free medium for 72 h prior to treatment with SB-216763 or DMSO. IKK complexes were immunoprecipitated from whole cell extracts by using anti-IKKγ antiserum. The complexes were resuspended in kinase assay buffer containing [γ-32P]ATP. Half the sample was incubated in the presence of GST-IκBα (residues 1–55, containing the IKK phosphorylation site), and the other half was incubated in the presence of GST alone as a control (GST fusion protein constructs were kindly provided by Dr. Thomas Gilmore, Boston University). The reactions were incubated at 30 °C for 20 min and then terminated by adding 4× Laemmli sample buffer and boiling for 5 min. The proteins were separated by SDS-PAGE, dried, and exposed to either film for visualization or a phosphor screen for quantification. Phosphorimages were quantified using Image J analysis software.
RESULTS
Identification of NF-κB Sites in Promoter Regions of Genes Regulated by GSK-3
We previously identified 12 growth factor-inducible genes whose repression in quiescent T98G human glioblastoma cells required GSK-3, which is highly active in these cells in the absence of growth factors (12). These genes were induced not only by PDGF stimulation in a PI 3-kinase-dependent manner but also by inhibition of GSK-3 in the absence of growth factor stimulation. Because genes that participate in the same pathway may be regulated by the same transcription factors, we analyzed the upstream sequences of this set of genes to determine whether there were over-represented transcription factor binding sites in their promoter regions. Functional transcription factor binding sites are generally more likely to be evolutionarily conserved (32); therefore we focused on sites that were conserved between human and mouse orthologs. A similar analysis of the 5-kb upstream regions of these genes previously identified conserved binding sites for CREB as being statistically over-represented in this gene set (12). By analyzing the more proximal promoter regions and using updated versions of both the human and mouse genomes as well as the TRANSFAC data base, we found that NF-κB binding sites were also significantly enriched upstream of the transcription start sites of these genes. Comparisons of the 12 GSK-3-regulated genes with a background set of 165 genes that were expressed in quiescent T98G cells but not induced by growth factor stimulation indicated that binding sites for NF-κB were statistically over-represented in the 1-, 3-, and 5-kb regions upstream of the GSK-3-regulated genes with p values of 4.7 × 10−4, 1.5 × 10−5, and 1.4 × 10−4, respectively. Computational analysis predicted 19 NF-κB sites in the 5-kb upstream regions of the GSK-3-regulated genes. Of the 12 genes regulated by GSK-3, 9 contained at least one predicted NF-κB binding site, suggesting the possible involvement of NF-κB in regulating gene expression downstream of GSK-3.
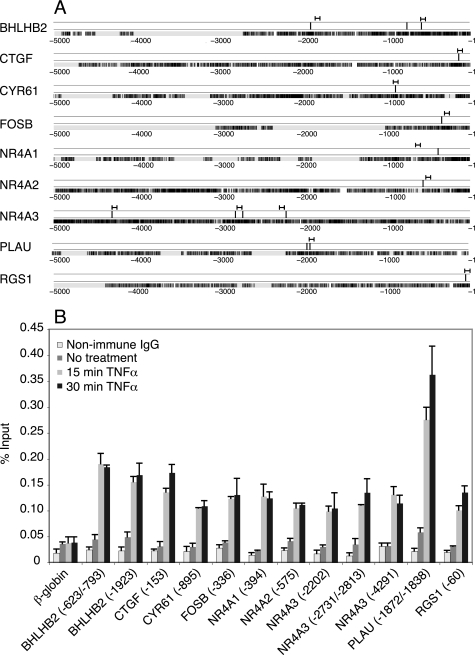
The NF-κB family is comprised of five different members (p65, c-Rel, RelB, p50, and p52), all of which share a conserved Rel homology domain that binds to DNA (33, 34). To initially test whether NF-κB was physiologically able to bind to the predicted sites, ChIP assays were performed following treatment of cells with TNFα, which produces a rapid and robust activation of p65 (33, 34). ChIP assays performed on T98G cells treated for 15 or 30 min with TNFα demonstrated that at least 12 sites upstream of 9 genes exhibited significantly increased p65 binding in response to TNFα stimulation (Fig. 1). Thus, 9 of the 12 genes that are induced in response to inhibition of GSK-3 contain functional NF-κB binding sites in their upstream regulatory regions.
FIGURE 1.
NF-κB sites upstream of GSK-3-regulated genes. A, NF-κB sites located in the 5-kb upstream region relative to the transcription start site are represented by vertical black lines, with the position of the primers used for ChIP assays indicated above. Only sites that were confirmed by ChIP are shown. Displayed below is a depiction of the human sequence aligned to the corresponding mouse ortholog. Black, exact match; gray, mismatch; white, gap. B, T98G cells were either untreated or treated for 15 or 30 min with TNFα. Chromatin fragments were precipitated with either anti-p65 antibody or nonimmune IgG and quantified by real time PCR. The data shown for nonimmune IgG are averaged from cells treated for 15 and 30 min with TNFα. Indicated within parentheses are the positions of the first nucleotide of the binding sites for each gene relative to the transcription start site. If the sites were too close together to be distinguished by a single primer, both sites are listed. β-globin was used as a negative control. The data are presented as the percentages of input and are the means of two independent experiments ± S.E.
Activation of p65 Contributes to Gene Induction in Response to Inhibition of GSK-3
Once the relevant binding sites had been established, we next sought to determine whether NF-κB was bound to these promoters following either growth factor stimulation of quiescent cells or inhibition of GSK-3 in the absence of growth factor stimulation. NF-κB binding was therefore measured after treating quiescent cells with either PDGF or SB-216763, a highly specific GSK-3 inhibitor (35, 36) that we have shown inhibits GSK-3 in quiescent T98G cells without activating the Akt or extracellular signal-regulated kinase (ERK) signaling pathways (12). To comprehensively assess binding of different NF-κB family members, ChIP assays were performed with antibodies against all five family members as well as Bcl-3, which is an IκBα-like protein that can regulate transcription by associating with either p50 or p52 homodimers (37).
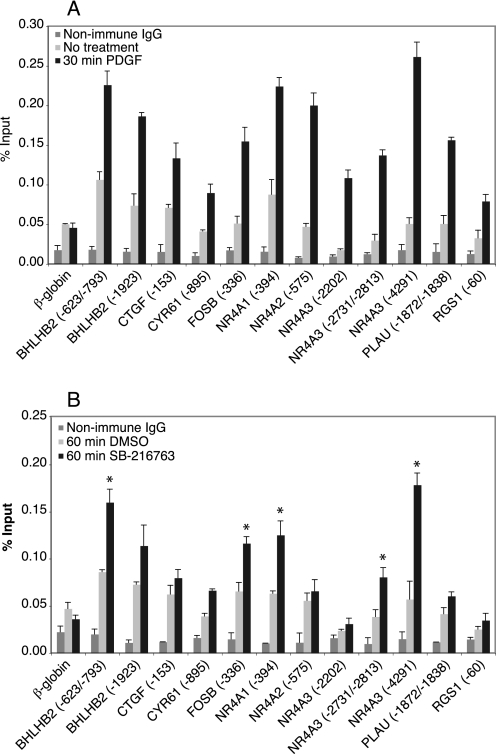
We did not observe any significant binding of RelB, c-Rel, p52, or Bcl-3 to the promoter regions in either untreated quiescent cells or following treatment with PDGF or SB-216763 (data not shown). In contrast, PDGF treatment significantly increased p65 binding to the same 12 sites that responded to TNFα (Fig. 2A). Of note, some sites (e.g. the sites upstream of BHLHB2 and NR4A1) also exhibited higher p65 binding in the untreated cells as compared with the nonimmune IgG or β-globin negative controls, suggesting that there may be a low level of p65 binding in quiescent cells. Importantly, inhibition of GSK-3 by treatment of quiescent cells with SB-216763 in the absence of growth factor stimulation also induced a significant increase in p65 binding to five sites in four genes (Fig. 2B), suggesting that GSK-3 may regulate expression of these genes via p65.
FIGURE 2.
Binding of p65 increases in response to PDGF stimulation or GSK-3 inhibition. Quiescent T98G cells were either untreated or treated for 30 min with PDGF (A) or treated for 60 min with either DMSO or SB-216763 (B). Chromatin fragments were precipitated with either anti-p65 antibody or nonimmune IgG and quantified by real time PCR. The values for nonimmune IgG are from treated cells. The data are presented as the percentage of input and are the means of three independent experiments, ± S.E. In B, * represents sites that showed >1.8-fold increase in binding upon SB-216763 treatment, in addition to statistically significant binding (p < 0.01) as compared with both β-globin and to DMSO control samples, as assessed by t test.
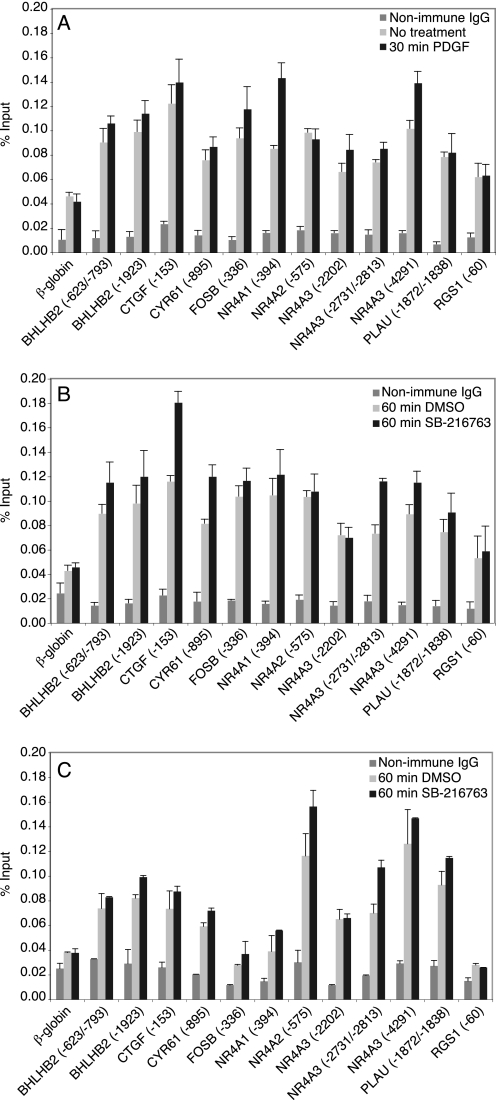
We found that p50, the most common heterodimerization partner of p65, bound to most of the predicted sites in both unstimulated quiescent cells and those treated with either PDGF or SB-216763 (Fig. 3, A and B). Although p50 binding to most of the predicted sites was significantly higher than binding to the β-globin control, we did not observe a significant increase upon growth factor stimulation or inhibition of GSK-3. This suggests that p50 is constitutively bound, which is consistent with a previous report that showed that p50 binds to a majority of its target genes in the absence of stimulation (38).
FIGURE 3.
Binding of p50 and HDAC-1 to predicted NF-κB sites in quiescent and stimulated cells. Quiescent T98G cells were either untreated or treated for 30 min with PDGF (A) or treated for 60 min with either DMSO or SB-216763 (B and C). Chromatin fragments were precipitated with either anti-p50 antibody (A and B), anti-HDAC-1 antibody (C) or nonimmune IgG and quantified by real time PCR. The values for nonimmune IgG are from treated cells. The data are presented as the percentage of input and are the means of two (C) or three (A and B) independent experiments ± S.E.
In unstimulated cells, p50 has been reported to bind to NF-κB sites in a complex with HDAC-1, which represses transcription (39). HDAC-1 was also bound to most of the predicted NF-κB sites in quiescent T98G cells (Fig. 3C), consistent with repression by p50 homodimers. Treatment with SB-216763 did not affect the association of HDAC-1 with these promoters, indicating that gene induction resulted primarily from recruitment of p65 rather than dissociation of HDAC-1 from promoters following inhibition of GSK-3.
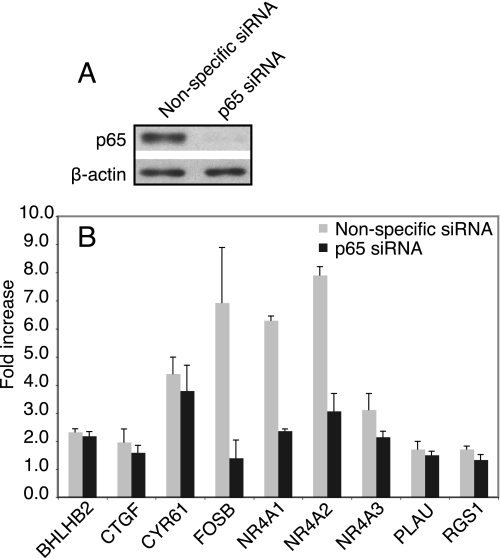
To investigate the functional role of p65 in the induction of GSK-3-regulated genes, siRNA experiments were performed. Transfection with p65 siRNA followed by 48 h of serum withdrawal reduced p65 levels by almost 90% in quiescent T98G cells (Fig. 4A). Induction of the nine genes with confirmed NF-κB binding sites was not affected by knockdown of p65 in quiescent cells treated with PDGF, indicating that there are additional factors inducing these genes upon growth factor stimulation (data not shown). Upon inhibition of GSK-3 by treatment with SB-216763, however, there was at least a 60% decrease in the induction of three genes (FOSB, NR4A1, and NR4A2) when p65 was knocked down (Fig. 4B). It is noteworthy that the induction of NR4A2 markedly decreased upon p65 knockdown, even though ChIP assays did not show an increase in p65 binding to the NR4A2 promoter after inhibition of GSK-3. This suggests that although GSK-3 inhibition did not induce a sufficient increase in p65 binding to be detected by ChIP, activation of p65 still contributes significantly to NR4A2 induction.
FIGURE 4.
Inhibition of gene induction by p65 siRNA. T98G cells were transfected with p65 siRNA or a nonspecific control siRNA for 24 h and serum-starved for 48 h to induce quiescence. Immunoblots were used to measure p65 knockdown (A). Gene induction after treatment for 60 min with either DMSO or SB-216763 was measured by real time RT-PCR (B). The data represent the fold increase compared with the vehicle control and are the means of three independent experiments ± S.E.
Taken together, these results indicate that inhibition of GSK-3 in quiescent cells increased binding of p65 to the promoter regions of a third of the GSK-3-regulated genes. Moreover, p65 is required for efficient induction of at least three genes in response to GSK-3 inhibition.
GSK-3 Regulates p65 at the Level of IκBα Phosphorylation
GSK-3 has previously been reported to inhibit NF-κB either by phosphorylating p65 on Ser468 (40) or by inhibiting the phosphorylation of IκB by IKK (14). We first investigated Ser468 phosphorylation by immunoblotting with an anti-Ser(P)468 antibody. Phosphorylation of Ser468 was undetectable in quiescent T98G cells, although stimulation with TNFα induced robust Ser468 phosphorylation (data not shown), as was previously reported to occur via IKK (41). We therefore concluded that GSK-3 does not inhibit p65 by phosphorylating Ser468 in quiescent cells.
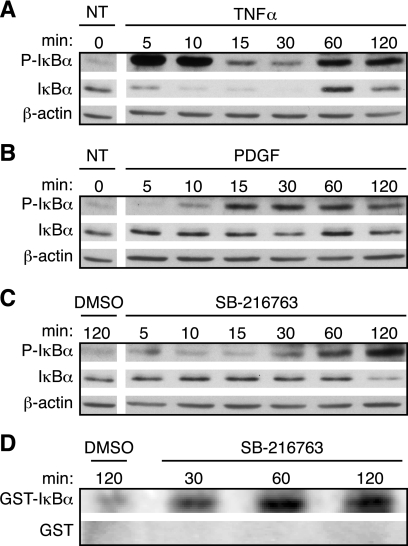
We next investigated whether inhibiting GSK-3 in quiescent cells stimulates p65 by activating IKK. In most cells, p65 is mainly located within the cytoplasm in an inhibitory complex with IκBα. Activated IKK phosphorylates IκBα at serines 32 and 36, causing its proteasomal degradation, which liberates p65 and allows it to translocate to the nucleus (34). Stimulation of T98G cells with TNFα resulted in phosphorylation of IκBα at Ser32 within 5 min (Fig. 5A). This was followed by a marked decrease in the levels of IκBα, indicating that IκBα is quickly phosphorylated and then degraded (Fig. 5A). Resynthesis of IκBα occurred by 60 min, which is expected, because the IκBα gene is activated by NF-κB as part of a negative feedback loop (42). A low level of IκBα phosphorylation was also detected in the untreated cells, consistent with the ChIP results, suggesting that a small pool of p65 is active in quiescent cells. Growth factor stimulation by treatment with PDGF resulted in phosphorylation of IκBα within 15 min, followed by a modest degradation of IκBα at 30 min (Fig. 5B). Inhibition of GSK-3 by treatment with SB-216763 also resulted in an increase in IκBα phosphorylation, with maximum effects seen within 60–120 min, accompanied by partial degradation of IκBα at 120 min (Fig. 5C).
FIGURE 5.
Inhibition of GSK-3 induces phosphorylation of IκBα by increasing IKK activity. Quiescent T98G cells were either not treated (NT) or stimulated with TNFα (A) or PDGF (B) or treated with either DMSO or SB-216763 (C and D) for the indicated times. A–C, cell extracts were analyzed by immunoblotting to detect both phospho-IκBα and total IκBα, with β-actin used as a loading control. The untreated cells were used as controls for both TNFα and PDGF and are repeated in A and B. The data are representative of three independent experiments. D, IKK complexes were immunoprecipitated from cell extracts and in vitro kinase assays were performed as described under “Experimental Procedures.” Parallel SDS-PAGE gels were run and stained with Coomassie Blue to confirm equal loading of GST-IκBα (residues 1–55) and GST-only substrates (data not shown). The kinase assay data are representative of four independent experiments.
To directly test whether inhibition of GSK-3 resulted in the activation of IKK, in vitro kinase assays were performed (Fig. 5D). Endogenous IKK complexes were immunoprecipitated from extracts of T98G cells that had been treated with SB-216763, and kinase activity was measured by the ability of the IKK complexes to phosphorylate an exogenous GST-IκBα (1–55) substrate. Consistent with the IκBα phosphorylation kinetics in vivo (Fig. 5C), treatment with SB-216763 resulted in an increase of IKK activity by 30 min continuing to an increase of 2-fold by 60–120 min as compared with the DMSO control (averaged from four independent experiments). GST-only controls that were run in parallel showed no incorporation of phosphate under identical conditions.
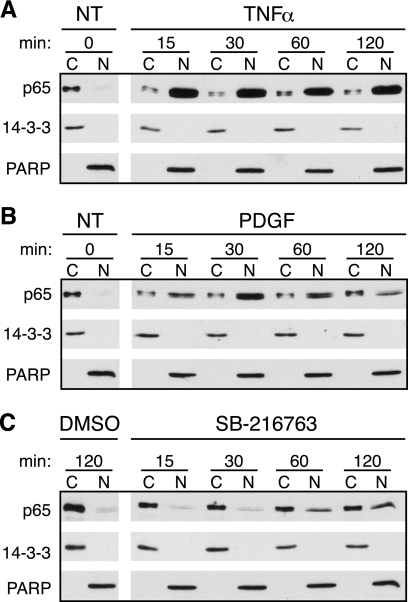
Because activation of IKK and the subsequent degradation of IκBα are followed by nuclear translocation of p65, we further determined whether growth factor stimulation and inhibition of GSK-3 resulted in an increase of p65 in the nucleus. To measure translocation, the cells were fractionated into cytoplasmic and nuclear extracts, and p65 levels were analyzed by immunoblots. Quiescent T98G cells were initially treated with TNFα, which induced a rapid translocation of p65, shown by high levels of p65 in the nucleus with little protein remaining in the cytoplasm (Fig. 6A). Stimulation with PDGF induced translocation as well, with peak accumulation of p65 in the nucleus occurring by 30 min (Fig. 6B). Although less robust, GSK-3 inhibition by treatment with SB-216763 also resulted in translocation of p65, with the highest nuclear levels found after 60–120 min (Fig. 6C). These results show that inhibition of GSK-3 is sufficient to induce p65 activation, with the kinetics of p65 nuclear translocation correlating with the kinetics of IκBα phosphorylation.
FIGURE 6.
Nuclear translocation of p65 is stimulated by inhibition of GSK-3. Quiescent T98G cells were fractionated into cytoplasmic (∼1 μg of protein) or nuclear extracts (∼2 μg of protein) and were either not treated (NT) or stimulated with either TNFα (A) or PDGF (B) or treated with DMSO or SB-216763 (C) for the indicated times. The untreated samples were used as controls for both TNFα and PDGF and are repeated in A and B. Immunoblots were used to analyze the level of p65. 14-3-3 and PARP confirmed the cytoplasmic and nuclear fractions, respectively. Representative data from three independent experiments are presented.
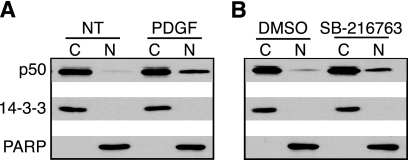
Because the most abundant form of NF-κB consists of the p50/p65 heterodimer, and this complex is presumed to be the primary target of IκBα (34), we thought it likely that this heterodimer was being activated upon both growth factor stimulation and inhibition of GSK-3. To confirm this, we measured translocation of p50 after treatment with either PDGF or SB-216763 (Fig. 7). Although p50 is present in the nuclei of quiescent cells, the amount substantially increased upon either growth factor stimulation or GSK-3 inhibition. This is consistent with the idea that p50 homodimers are constitutively present within the nucleus, and upon stimulation, p50 levels increase because of translocation of the p50/p65 heterodimer.
FIGURE 7.
Inhibition of GSK-3 increases nuclear translocation of p50. Quiescent T98G cells were fractionated into cytoplasmic (∼2 μg of protein) or nuclear extracts (∼2 μg of protein) and were either not treated (NT) or stimulated with PDGF for 30 min (A) or treated with DMSO or SB-216763 for 60 min (B). Immunoblots were used to analyze the level of p50. 14-3-3 and PARP confirmed the cytoplasmic and nuclear fractions, respectively. Representative data from two independent experiments are presented.
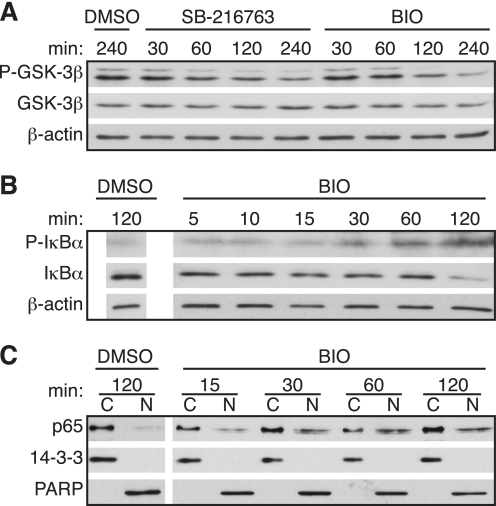
Although SB-216763 is a potent and highly specific inhibitor of GSK-3 (35, 36), we wanted to rule out the possibility that activation of p65 following GSK-3 inhibition was due to off target effects. Therefore, we also investigated the effects of BIO, an indirubin that is a highly selective inhibitor of GSK-3 compared with cyclin-dependent kinases (43). We verified that BIO inhibits GSK-3 to a similar extent as SB-21676 by confirming that both inhibitors decreased the stimulatory tyrosine 216 phosphorylation on GSK-3 (Fig. 8A). Treatment with BIO also resulted in IκBα phosphorylation and degradation, as well as nuclear translocation of p65 (Fig. 8, B and C). Both the extent and kinetics of IκBα phosphorylation and p65 translocation following treatment of cells with BIO were similar to those observed following treatment with SB-216763.
FIGURE 8.
Inhibition of GSK-3 with BIO induces phosphorylation of IκBα and translocation of p65. A, quiescent T98G cells were treated with DMSO, SB-216763, or BIO for the indicated times. Phospho-GSK-3β (Tyr216) and total GSK-3β levels in whole cell extracts were measured by immunoblots, with β-actin used as a loading control. B, levels of phospho-IκBα or IκBα in whole cell extracts were measured by immunoblots, with β-actin used as a loading control. C, cells were fractionated into cytoplasmic (∼1 μg of protein) or nuclear extracts (∼2 μg of protein), and immunoblots were used to analyze the level of p65, with 14-3-3 and PARP confirming the cytoplasmic and nuclear fractions, respectively. The data are representative of two (A) or three (B and C) independent experiments.
These results confirm that inhibition of GSK-3 in quiescent cells stimulates the phosphorylation of IκBα by IKK, resulting in degradation of IκBα and nuclear translocation of p65. Although inhibiting GSK-3 did not cause as rapid or as strong effects as those observed after PDGF- or TNFα-induced activation, the relative levels of p65 translocation and IκBα phosphorylation and degradation were proportional. It thus appears that GSK-3 acts to suppress p65 activity in quiescent cells via inhibition of IKK.
DISCUSSION
GSK-3 plays a critical role in regulating cell proliferation and survival downstream of both Wnt and PI 3-kinase signaling. Unlike most protein kinases involved in signal transduction cascades, GSK-3 is active in the absence of growth factor stimulation and generally functions to induce apoptosis or inhibit cell proliferation (1–5). Because the targets of GSK-3 include multiple transcription factors, we have investigated the role of GSK-3 in regulating gene expression in quiescent cells arrested in G0 by growth factor deprivation, which have a high level of GSK-3 activity (4, 7, 12, 44–46). A subset of the genes induced by growth factor stimulation of such cells are also inducible solely by inhibiting GSK-3, indicating that the continual activity of GSK-3 is required to maintain repression of these genes in the quiescent state (12). In the present study, we have identified inhibition of NF-κB as one of the targets of GSK-3 that mediates repression of gene expression.
Previous studies identified CREB binding sites as over-represented in the promoter regions of GSK-3-regulated genes and demonstrated by ChIP assays and RNA interference that inhibition of CREB by GSK-3 is involved in repressing the expression of these genes in quiescent cells (12). In the present study, utilization of updated computational tools indicated that evolutionarily conserved binding sites for NF-κB were also over-represented in the promoter regions of the GSK-3-regulated genes. These computational predictions were confirmed by ChIP assays demonstrating the binding of p65 and p50, but not other NF-κB family members, to predicted sites in the upstream regions of 9 of 12 of these genes. p50, together with HDAC-1, was bound to promoter regions in both unstimulated and stimulated cells, consistent with previous data indicating that p50 is constitutively bound as a repressor to the promoters of most target genes (38). In contrast, the binding of p65 was stimulated by both TNFα and PDGF, as well as by inhibition of GSK-3 in the absence of growth factor stimulation. Nuclear translocation of both p50 and p65 were also stimulated by inhibition of GSK-3, so activation of these genes is likely occurring as a result of binding of the p50/p65 heterodimer.
We confirmed the functional role of p65 by showing that the induction of three genes in response to inhibition of GSK-3 was reduced when p65 was depleted by siRNA. Although knockdown of p65 did not significantly affect the induction of other genes, it may still play a role in the expression of these genes. Indeed, we have previously shown that inhibition of CREB by GSK-3 also represses gene expression in quiescent cells (12). In addition, inhibition of c-Jun by GSK-3 plays a similar role.3 Therefore it seems likely that multiple factors may act redundantly to regulate gene expression downstream of GSK-3 signaling. We have further shown that GSK-3 inhibition in quiescent cells resulted in stimulation of IKK, leading to phosphorylation and degradation of IκBα, followed by nuclear translocation of p65. This indicates that the high level of active GSK-3 present in cells arrested by growth factor deprivation inhibits p65 by preventing the phosphorylation of IκBα by IKK. Taken together, these results indicate that inhibition of p65 by GSK-3 is involved in maintaining repression of growth factor-inducible genes in quiescent cells.
Although GSK-3 has been previously reported to regulate NF-κB activity, whether it activates or inhibits NF-κB appears to depend upon the physiological context. In contrast to our results, in some cells GSK-3 activates NF-κB following TNFα stimulation. Hoeflich et al. (16) initially reported that inactivation of the mouse GSK-3β gene resulted in embryonic lethality caused by liver degeneration. In contrast to its usual role as a proapoptotic protein kinase, GSK-3 was instead required to prevent TNFα-induced apoptosis of hepatocytes. GSK3β−/− fibroblasts derived from these mice were also sensitive to TNFα-induced apoptosis caused by a decrease in NF-κB activity, indicating that GSK-3 is required for activation of NF-κB following TNFα stimulation (16). GSK-3 has since been implicated in the TNFα-induced activation of NF-κB in several other cell types, including hepatocytes, renal epithelial cells, and intestinal epithelial cells, in addition to mouse fibroblasts (17, 19–21). Consistent with the role of GSK-3 in inhibiting apoptosis induced by TNFα and in stimulating cytokine activation of NF-κB, inhibition of GSK-3 has also been reported to induce apoptosis and to inhibit NF-κB in some human cancer cell lines, particularly pancreatic carcinomas, in which the NF-κB pathway is constitutively active (18, 22, 23).
The mechanism(s) by which GSK-3 acts to stimulate the activity of NF-κB remain to be fully understood. In most cases, GSK-3 did not affect activation of IKK, degradation of IκB, or nuclear translocation of p65 but instead appeared to affect the DNA binding or transcriptional activity of nuclear p65 (16–20). One possible mechanism for these effects is phosphorylation of p65 by GSK-3 (17, 20). It is also noteworthy that inhibition of GSK-3 can lead to the accumulation of nuclear β-catenin, which directly interacts with p65 to inhibit its transcriptional activity (47, 48). However, GSK-3 has also been reported to affect the stability of IκBα in hepatocytes (21) and to be required for maintenance of constitutive IKK activity in some pancreatic cancer cell lines (23).
In contrast to these stimulatory effects of GSK-3 on the activation of NF-κB by TNFα or in cell lines with constitutive NF-κB signaling, our results indicate that the elevated levels of GSK-3 activity resulting from growth factor deprivation act to inhibit NF-κB in quiescent cells. This is consistent with other studies indicating that the elevated activity of GSK-3 normally associated with arrest of cell proliferation or induction of apoptosis acts to inhibit NF-κB. In PC12 cells, growth factor deprivation induces apoptosis by inhibition of PI 3-kinase signaling, at least in part by activation of GSK-3 (4, 13). Inhibition of GSK-3 leads to activation of NF-κB and inhibition of apoptosis in these cells (13). Conversely, overexpression of GSK-3 by transfection of astrocytes induced apoptosis and inhibited NF-κB (14). The high levels of GSK-3 expressed in serum-deprived epithelial cells have likewise been reported to inhibit NF-κB (15). Consistent with our results, the high levels of GSK-3 that inhibited NF-κB in these studies were found to prevent nuclear translocation of p65 (13–15). Moreover, GSK-3 was shown to affect the stability of IκBα in serum-deprived epithelial cells (15) and to inhibit the phosphorylation of IκBα by IKK in astrocytes in which GSK-3 was overexpressed by transfection (14).
Taken together, these findings and the present study indicate that GSK-3 plays distinct roles in regulation of NF-κB depending on the physiology of the cell. GSK-3 can act to promote cell survival and stimulate the activity of NF-κB in cells treated with TNFα or in some tumor cells in which the NF-κB pathway is constitutively active. In contrast, the more common role of GSK-3 as a kinase activated by growth factor deprivation is to induce apoptosis or cell cycle arrest. Under these conditions, GSK-3 acts as a negative regulator of the NF-κB pathway by inhibiting the phosphorylation of IκBα by IKK. In addition to contributing to the induction of apoptosis (13, 14), we have shown that this inhibition of NF-κB by GSK-3 plays a role in suppressing the expression of growth factor-inducible genes in quiescent cells.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA18689.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
J. W. Tullai, J. R. Graham, S. Tacheva, L. Owens, and G. M. Cooper, manuscript in preparation.
- GSK
- glycogen-synthase kinase
- NF-κB
- nuclear factor κB
- PI
- phosphoinositide
- CREB
- cyclic AMP response element-binding protein
- TNF
- tumor necrosis factor
- PDGF
- platelet-derived growth factor
- BIO
- 6-bromoindirubin-3′-oxime
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription
- PARP
- poly(ADP-ribose) polymerase
- IKK
- IκB kinase
- siRNA
- small interfering RNA
- DMSO
- dimethyl sulfoxide
- GST
- glutathione S-transferase
- HDAC
- histone deacetylase.
REFERENCES
- 1.Jope R. S., Johnson G. V. (2004) Trends Biochem. Sci 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 2.Doble B. W., Woodgett J. R. (2003) J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frame S., Cohen P. (2001) Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pap M., Cooper G. M. (1998) J. Biol. Chem. 273, 19929–19932 [DOI] [PubMed] [Google Scholar]
- 5.Diehl J. A., Cheng M., Roussel M. F., Sherr C. J. (1998) Genes Dev. 12, 3499–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nusse R. (2005) Cell Res. 15, 28–32 [DOI] [PubMed] [Google Scholar]
- 7.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 8.Maurer U., Charvet C., Wagman A. S., Dejardin E., Green D. R. (2006) Mol. Cell 21, 749–760 [DOI] [PubMed] [Google Scholar]
- 9.Welsh G. I., Miller C. M., Loughlin A. J., Price N. T., Proud C. G. (1998) FEBS Lett. 421, 125–130 [DOI] [PubMed] [Google Scholar]
- 10.Pap M., Cooper G. M. (2002) Mol. Cell. Biol. 22, 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tullai J. W., Schaffer M. E., Mullenbrock S., Kasif S., Cooper G. M. (2004) J. Biol. Chem. 279, 20167–20177 [DOI] [PubMed] [Google Scholar]
- 12.Tullai J. W., Chen J., Schaffer M. E., Kamenetsky E., Kasif S., Cooper G. M. (2007) J. Biol. Chem. 282, 9482–9491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bournat J. C., Brown A. M., Soler A. P. (2000) J. Neurosci. Res. 61, 21–32 [DOI] [PubMed] [Google Scholar]
- 14.Sanchez J. F., Sniderhan L. F., Williamson A. L., Fan S., Chakraborty-Sett S., Maggirwar S. B. (2003) Mol. Cell. Biol. 23, 4649–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachelder R. E., Yoon S. O., Franci C., de Herreros A. G., Mercurio A. M. (2005) J. Cell Biol. 168, 29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoeflich K. P., Luo J., Rubie E. A., Tsao M. S., Jin O., Woodgett J. R. (2000) Nature 406, 86–90 [DOI] [PubMed] [Google Scholar]
- 17.Schwabe R. F., Brenner D. A. (2002) Am. J. Physiol. Gastrointest Liver Physiol 283, G204–G211 [DOI] [PubMed] [Google Scholar]
- 18.Ougolkov A. V., Fernandez-Zapico M. E., Savoy D. N., Urrutia R. A., Billadeau D. D. (2005) Cancer Res. 65, 2076–2081 [DOI] [PubMed] [Google Scholar]
- 19.Steinbrecher K. A., Wilson W., 3rd, Cogswell P. C., Baldwin A. S. (2005) Mol. Cell. Biol. 25, 8444–8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong R., Rifai A., Ge Y., Chen S., Dworkin L. D. (2008) J. Biol. Chem. 283, 7401–7410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Götschel F., Kern C., Lang S., Sparna T., Markmann C., Schwager J., McNelly S., von Weizsäcker F., Laufer S., Hecht A., Merfort I. (2008) Exp. Cell Res. 314, 1351–1366 [DOI] [PubMed] [Google Scholar]
- 22.Kotliarova S., Pastorino S., Kovell L. C., Kotliarov Y., Song H., Zhang W., Bailey R., Maric D., Zenklusen J. C., Lee J., Fine H. A. (2008) Cancer Res. 68, 6643–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson W., 3rd, Baldwin A. S. (2008) Cancer Res. 68, 8156–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terragni J., Graham J. R., Adams K. W., Schaffer M. E., Tullai J. W., Cooper G. M. (2008) BMC Cell Biol. 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn R. M., Karolchik D., Zweig A. S., Wang T., Smith K. E., Rosenbloom K. R., Rhead B., Raney B. J., Pohl A., Pheasant M., Meyer L., Hsu F., Hinrichs A. S., Harte R. A., Giardine B., Fujita P., Diekhans M., Dreszer T., Clawson H., Barber G. P., Haussler D., Kent W. J. (2009) Nucleic Acids Res. 37, D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kel A. E., Gössling E., Reuter I., Cheremushkin E., Kel-Margoulis O. V., Wingender E. (2003) Nucleic Acids Res. 31, 3576–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) Nucleic Acids Res. 34, D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. B. 57, 289–300 [Google Scholar]
- 29.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30.Meares G. P., Jope R. S. (2007) J. Biol. Chem. 282, 16989–17001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang M. C., Bardhan S., Pace E. A., Rosman D., Beutler J. A., Porco J. A., Jr., Gilmore T. D. (2006) Biochem. Pharmacol. 71, 634–645 [DOI] [PubMed] [Google Scholar]
- 32.Sauer T., Shelest E., Wingender E. (2006) Bioinformatics 22, 430–437 [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S., Karin M. (2002) Cell 109, (Suppl.) S81–S96 [DOI] [PubMed] [Google Scholar]
- 34.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 35.Coghlan M. P., Culbert A. A., Cross D. A., Corcoran S. L., Yates J. W., Pearce N. J., Rausch O. L., Murphy G. J., Carter P. S., Roxbee Cox L., Mills D., Brown M. J., Haigh D., Ward R. W., Smith D. G., Murray K. J., Reith A. D., Holder J. C. (2000) Chem. Biol. 7, 793–803 [DOI] [PubMed] [Google Scholar]
- 36.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer S., Chen Y. H. (2008) Immunol. Res. 42, 210–218 [DOI] [PubMed] [Google Scholar]
- 38.Schreiber J., Jenner R. G., Murray H. L., Gerber G. K., Gifford D. K., Young R. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5899–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong H., May M. J., Jimi E., Ghosh S. (2002) Mol. Cell 9, 625–636 [DOI] [PubMed] [Google Scholar]
- 40.Buss H., Dörrie A., Schmitz M. L., Frank R., Livingstone M., Resch K., Kracht M. (2004) J. Biol. Chem. 279, 49571–49574 [DOI] [PubMed] [Google Scholar]
- 41.Schwabe R. F., Sakurai H. (2005) FASEB J. 19, 1758–1760 [DOI] [PubMed] [Google Scholar]
- 42.Ghosh S., May M. J., Kopp E. B. (1998) Annu. Rev. Immunol. 16, 225–260 [DOI] [PubMed] [Google Scholar]
- 43.Meijer L., Skaltsounis A. L., Magiatis P., Polychronopoulos P., Knockaert M., Leost M., Ryan X. P., Vonica C. A., Brivanlou A., Dajani R., Crovace C., Tarricone C., Musacchio A., Roe S. M., Pearl L., Greengard P. (2003) Chem. Biol. 10, 1255–1266 [DOI] [PubMed] [Google Scholar]
- 44.Saito Y., Vandenheede J. R., Cohen P. (1994) Biochem. J. 303, 27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurel S. J., Rochford J. J., Borthwick A. C., Wells A. M., Vandenheede J. R., Turnbull D. M., Yeaman S. J. (1996) Biochem. J. 320, 871–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cross D. A., Alessi D. R., Vandenheede J. R., McDowell H. E., Hundal H. S., Cohen P. (1994) Biochem. J. 303, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng J., Miller S. A., Wang H. Y., Xia W., Wen Y., Zhou B. P., Li Y., Lin S. Y., Hung M. C. (2002) Cancer Cell 2, 323–334 [DOI] [PubMed] [Google Scholar]
- 48.Deng J., Xia W., Miller S. A., Wen Y., Wang H. Y., Hung M. C. (2004) Mol. Carcinog. 39, 139–146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.