Abstract
Major histocompatibility complex (MHC) class I molecules are ligands for T-cell receptors of CD8+ T cells and inhibitory receptors of natural killer cells. Assembly of the heavy chain, light chain, and peptide components of MHC class I molecules occurs in the endoplasmic reticulum (ER). Specific assembly factors and generic ER chaperones, collectively called the MHC class I peptide loading complex (PLC), are required for MHC class I assembly. Calreticulin has an important role within the PLC and induces MHC class I cell surface expression, but the interactions and mechanisms involved are incompletely understood. We show that interactions with the thiol oxidoreductase ERp57 and substrate glycans are important for the recruitment of calreticulin into the PLC and for its functional activities in MHC class I assembly. The glycan and ERp57 binding sites of calreticulin contribute directly or indirectly to complexes between calreticulin and the MHC class I assembly factor tapasin and are important for maintaining steady-state levels of both tapasin and MHC class I heavy chains. A number of destabilizing conditions and mutations induce generic polypeptide binding sites on calreticulin and contribute to calreticulin-mediated suppression of misfolded protein aggregation in vitro. We show that generic polypeptide binding sites per se are insufficient for stable recruitment of calreticulin to PLC substrates in cells. However, such binding sites could contribute to substrate stabilization in a step that follows the glycan and ERp57-dependent recruitment of calreticulin to the PLC.
Keywords: Antigen Presentation, Chaperone/Chaperonin, Immunochemistry, Immunology, MHC, MHC Class I, TAP Transporter, Calreticulin, Peptide Loading Complex, Tapasin
Introduction
Calreticulin is a soluble protein of the endoplasmic reticulum (ER),3 where it functions in calcium binding and as a molecular chaperone (1). Calreticulin binds to monoglucoslyated N-linked oligosaccharides on newly synthesized glycoproteins and maintains quality control of glycoprotein folding in the ER. A structural model for calreticulin was derived (supplemental Fig. S1A), using the crystal structure of the closely related protein, calnexin (2). Calreticulin is thought to contain three structural domains. The first is a large globular domain comprising the N- and C-terminal regions of the protein that form a β-stranded sandwich and a C-terminal helix (supplemental Fig. S1A, orange, light blue, and green). The glycan binding site is located within this domain (supplemental Fig. S1B). A second hook-shaped P-domain forms a β-stranded hairpin structure inserted in the middle of the globular domain (supplemental Fig. S1A, dark blue). The tip of this domain forms a binding site for ERp57 (3), an ER oxidoreductase that works cooperatively with calreticulin and calnexin in glycoprotein folding (4). A third C-terminal domain, rich in acidic amino acids, is functional as a low affinity/high capacity calcium coordination site (1). This region is not present in the lumenal domain of calnexin.
In vitro studies have shown that calreticulin can bind to misfolded non-glycosylated polypeptides and suppress their irreversible aggregation (5). This activity is induced by various conditions associated with ER stress, including calcium depletion and heat shock (6). These conditions also induce calreticulin oligomerization (6, 7). Much remains to be understood about the one or more binding sites on calreticulin that are used to suppress substrate aggregation, as well as the relevance of this activity to calreticulin-mediated protein folding under physiological non-stress conditions.
Calreticulin is a key player in the MHC class I assembly pathway (8). The MHC class I-dedicated assembly factors, transporter associated with antigen processing (TAP) and tapasin, as well as the generic ER-folding factors ERp57 and calreticulin, form a large complex with MHC class I molecules, collectively called the PLC. TAP provides a major source of peptides for MHC class I molecules, whereas tapasin, ERp57, and calreticulin facilitate assembly of MHC class I molecules with peptides (reviewed in Ref. 9). Calreticulin is a component of the PLC, and calreticulin-deficient cells express reduced cell surface MHC class I molecules (8). The mechanisms by which calreticulin contributes to enhanced MHC class I assembly are not well understood.
Early studies with glycosylation inhibitors, MHC class I mutants, and in vitro binding analyses suggested that glycan-based interactions with MHC class I molecules recruit calreticulin into the PLC (10–12). More recent studies with calreticulin mutants that are defective for glycan or ERp57 binding have suggested that calreticulin can be recruited into the PLC in the absence of interactions with both ERp57 and substrate glycans and that polypeptide-based interactions are important for calreticulin recruitment (13, 14). However, partial truncation of the P domain of calreticulin (including residues mediating ERp57 binding) impacted calreticulin recruitment to the PLC (14), and ERp57- and tapasin-deficient cells have impaired recruitment of calreticulin into the PLC (15, 16). Thus, although MHC class I molecules are one of the best characterized substrates of calreticulin, the precise mechanism by which calreticulin is recruited into the PLC remains unclear. Furthermore, whereas numerous studies involving glycosylation inhibitors and substrates lacking glycans have shown that the presence of monoglucosylated glycans on substrate glycoproteins are important for calreticulin binding and ER quality control, whether or not substrate glycans alone are sufficient for calreticulin recruitment is not well understood, and neither is the molecular basis for differences in substrate profiles between calnexin and calreticulin. Additionally the roles of ERp57, polypeptide-based, and other interactions in substrate recruitment to calreticulin have not been well studied.
To address some of these questions, various truncation mutants targeting different domains of calreticulin and point mutants targeting glycan and ERp57 binding residues were generated. These constructs were used to understand the impacts of truncations and mutations on calreticulin structure and stability, to investigate modes of calreticulin binding to PLC components, and to examine reconstitution of MHC class I assembly in calreticulin-deficient cells.
EXPERIMENTAL PROCEDURES
DNA Constructs
Expression of mCRT in Escherichia coli
Truncation mutants of mouse calreticulin (mCRT) were amplified from the pCMV-SPORT6 (ATCC, MGC-6209) vector using primers that allowed for subsequent ligation-independent cloning (LIC) into the pMCSG7 vector (17). The following oligonucleotide primers were used to terminate CRT at the indicated C-terminal amino acid positions: 399, 5′-TTA TCC ACT TCC AAT GTT ACA GCT CAT CCT TGG CTT-3′; 362, 5′-TTA TCC ACT TCC AAT GTT ATT CCT CTT TAC GCT TCT TGT-3′; 339, 5′-TTA TCC ACT TCC AAT GTT ACT GCT TCT CGG CAG CCT TGG TTA CAC CCC-3′; and 318, 5′-TTA TCC ACT TCC AAT GTT AAT CAT TAG TGA TGA GGA AAT TGT C-3′. The following oligonucleotide primers were used to generate constructs at the N-terminal start sites: 1, 5′-TAC TTC CAA TCC AAT GCT GCC GCA GAC CCT GCC ATC-3′; and 33, 5′-TAC TTC CAA TCC AAT GCT GTC CTC AGT TCT GGC AAG TTT TAC GGG-3′. Underlined bases represent those that are complementary to the sequence encoding mCRT, and additional 5′ sequences were introduced for LIC.
Deletion of the P domain amino acids 187–283 and insertion of two glycine residues (mCRT(ΔP)) was achieved with the Finnzymes Phusion site-directed mutagenesis kit (New England Biolabs). The following primers were used to generate mCRT(ΔP): forward, 5′-GGA CCC GAT GCA AAT ATC TAT GCC TAT-3′ and reverse, 5′-TCC CAG AAA GTC CCA ATC ATC CTC CAA-3′.
DNA constructs were sequenced and transformed into BL21 (DE3) cells for protein expression. All bacterially expressed mCRT constructs lacked the signal sequence and contained an N-terminal MHHHHHHSSGVDLGTENLYFQSNA fusion sequence for nickel affinity chromatography.
Cloning of mCRT Truncation Mutants into Retroviral Vectors
mCRT mutants were cloned into pMSCV-ires-GFP and pMSCV-puro retroviral vectors. pMSCV-puro constructs were created by ligation-dependent cloning, and LIC was used to generate pMSCV-ires-GFP vectors. The mCRT templates used were the corresponding mCRT in the pMSCV-ires-GFP vector (for ΔC × pMSCV-puro) or pCMV-SPORT6 (all other constructs). All constructs retained the mCRT signal sequence and KDEL ER retention motifs.
Generation of LIC-competent Version of pMSCV-ires-GFP
The plasmid pMCSV-ires-GFP (18) was used as the parent vector for construction of a LIC-competent variant. This was accomplished by inserting a unique sequence with a PmeI restriction site. LIC overhangs were then generated by treatment of the linearized vector with T4 DNA polymerase exonuclease activity. The inserted sequence was constructed by annealing two oligonucleotides to generate a fragment with overhangs complimentary to an EcoRI restriction site. The oligonucleotide sequences used were 5′-AATTAGAGAGTTTAAACTTCCAC-3′ and 5′-AATTGTGGAAGTTTAAACTCTCT-3′. The lyophilized oligonucleotides were resuspended in distilled water at a concentration of 20 μm. Prior to annealing these were phosphorylated using T4 polynucleotide kinase (New England Biolabs). The 20-μl reactions contained 2 μl of 10 mm ATP, 12.5 μl of 20 μm oligonucleotide, 2 μl of 10× reaction buffer, 1 μl of enzyme, and 2.5 μl of distilled water. Reactions were incubated for 1 h at 37 °C then shifted to 95 °C for an additional 10 min. The two oligonucleotide reactions were then mixed together at 95 °C and slow cooled to 37 °C. A 2.5-μl aliquot was then added to a 20-μl ligation reaction composed of 1 μl of 45 ng/μl EcoRI-cut pMCSV-ires-GFP, previously treated with calf intestinal phosphatase, 2 μl of 10× reaction buffer, and 13.5 μl of distilled water. The reaction was slow cooled from 37 °C to 16 °C, and 1 μl of 1 unit/μl of T4 DNA ligase (New England Biolabs) was added. Incubation was continued at 16 °C for ∼16 h.
A 2-μl aliquot of the ligation reaction was used to transform 20 μl of competent XL1-Blue cells. These were plated on Lysogeny Broth (LB) agar with ampicillin (Amp) selection and incubated at 37 °C overnight. Colonies were picked and grown in 5 ml of LB-Amp, and plasmid DNA was isolated using Qiagen minispin columns. Positive clones were identified by restriction digestion with PmeI and ClaI and confirmed by DNA sequencing.
Primers for LIC of mCRT(WT), mCRT(ΔC) and mCRT(ΔP) into pMSCV-ires-GFP
The following primers were used: forward, 5′-GAA TTA GAG AGT TTC ACC ATG CTC CTT TCG GTG CCG CTC CTG CTT GGC-3′, reverse for WT and ΔP: 5′-GG AAT TGT GGA AGT TTC TTA CAG CTC ATC CTT GGC TTG GCC AGG GGA TTC-3′, and reverse for ΔC, 5′-GG AAT TGT GGA AGT TTC TTA CAG CTC ATC CTT CG CTT CTC TGC AGC CTT GGT A-3′.
Procedures for LIC
LIC was performed to introduce mCRT sequences into the pMCSG7 and pMSCV-ires-GFP vectors. Overhangs of 15 bp were generated in the insert DNA by processing 0.2 pmol of the PCR product with 0.5 unit of LIC-qualified T4 DNA polymerase (Novagen) in the presence of 4 mm dCTP (for ligation with pMCSG7) or 4 mm dGTP (for ligation with pMSCV-ires-GFP) and 5 mm dithiothreitol, at 20-μl final reaction volume. Reactions were incubated for 30 min at 22 °C followed by 20 min at 75 °C. The pMCSG7 or pMSCV-ires-GFP vectors were linearized by restriction digestion with SspI (for pMCSG7) or PmeI (for pMSCV-ires-GFP) (New England Biolabs) at a concentration of 5 units/μg of DNA at 37 °C for 1.5 h. The protein was removed using a Qiagen PCR minispin kit. Linearized vector DNA was then processed by T4 DNA polymerase to yield single-stranded ends for annealing. Reactions, 60-μl total, were set up with 1.6–2.0 μg of linear DNA and 3.75 units of LIC-qualified T4 polymerase (Novagen) in the presence of 4 mm dGTP (for pMCSG7) or 4 mm dCTP (for pMSCV-ires-GFP) and 5 mm dithiothreitol. Reactions were mixed on ice and incubated as described above for inserts. Insert and vector DNA were annealed in 96-well plates by combining 1 μl of processed vector and 2 μl of processed insert per well, followed by incubation in a PCR machine at 22 °C for 15 min with addition of 1 μl of 25 mm EDTA after 10 min of incubation. Annealed DNA was used to transform competent XL1-Blue cells. Positive clones were identified by PCR analysis. DNA sequences were verified at the University of Michigan DNA sequencing core. Purified plasmid DNA from positive clones was used to transform competent Rosetta cells (Novagen) or BL21 (DE3) cells for protein expression.
Primers for Cloning mCRT into pMSCV-puro
The following primers were used to clone untagged mCRT into the pMSCV-puro vector: forward, 5′-GA CTC G AG ATG CTC CTT TCG GTG-3′ and reverse, 5′-GA GTT AAC CTA CAG CTC ATC CTT-3′, which introduce 5′ XhoI and 3′ HpaI restriction sites. A FLAG-tagged CRT containing a C-terminal FLAG epitope tag was also generated in pMSCV-puro. The following primers were used: forward, 5′-GA CTC GAG ATG CTC CTT TCG GTG-3′ and reverse 5′-GA GTT AAC CTA CAG CTC ATC CTT CTT ATC ATC GTC ATC CTT ATA ATC GGC TTG GCC AGG GGA TTC TTC-3′. Underlined bases represent those that are complementary to the sequence encoding mCRT, with additional 5′ sequences added to insert an XhoI restriction site upstream of the protein sequence, and a FLAG epitope tag inserted before the KDEL ER retention motif, and finally an HpaI restriction site downstream of the protein sequence. Following amplifications from appropriate templates, PCR products were digested with XhoI and HpaI (New England Biolabs). PCR reactions were cleaned using MinElute 96 UF PCR purification kits by Qiagen. The digested products were ligated into the pMSCV-puro vector digested with the same enzymes. Both mCRT retroviral constructs retained the mCRT signal sequence and KDEL ER retention motif.
Generation of Y92A, W244A, W302A, and Y92A/W244A Mutants of mCRT
mCRT(W244A) was obtained by site-directed mutagenesis of mCRT(WT) in pMSCV-puro (Finnzymes Phusion, New England Biolabs). mCRT(Y92A) and mCRT(Y92A/W244A) were obtained by site-directed ligation-independent mutagenesis (19) of mCRT(WT) and mCRT(W244A) constructs in pMSCV-puro. mCRT(W302A) was obtained by site-directed mutagenesis of mCRT in pCMV-SPORT6 (QuikChange II site-directed mutagenesis kit, Stratagene). mCRT(W302A) in pMSCV-puro was subsequently cloned using a 3′ primer that introduced a FLAG epitope tag as described above. A corresponding FLAG-tagged mCRT(WT) was also generated. mCRT(Y92A), mCRT(W244A), mCRT(Y92A/W244A), and mCRT(W302A) were inserted into pMCSG7 from pMSCV-puro or pCMV-SPORT6 templates by LIC using the primers for an N-terminal start site at residue 1, and C-terminal end site at residue 399.
Site-directed Ligation-independent Mutagenesis to Create mCRT(Y92A) and mCRT(Y92A/W244A)
Procedures for site-directed ligation-independent mutagenesis were similar to those described in Ref. 19. To create Y92A, full-length mCRT in the retroviral vector pMSCV-puro was used as a template for the following primers in two separate PCR reactions to create DNA fragments with complementary 5′ overhangs, which are underlined: forward tailed, 5′-GGG GGC GGC GCC GTG AAG CTG TTT CCG AGT GGC TTG-3′; reverse short, 5′-ACA GTC GAT ATT CTG CTC ATG CTT CAC CG-3′; reverse tailed, 5′-CTT CAC GGC GCC GCC CCC ACA GTC GAT ATT CTG CTC-3′; and forward short, 5′-CTG TTT CCG AGT GGC TTG GAC CAG AAG-3′. The PCR products were then incubated with DpnI (New England Biolabs) to digest template DNA, and this reaction was quenched with the addition of the following 2× site-directed ligation-independent mutagenesis buffer: 50 mm Tris, pH 8.8, 300 mm NaCl, 20 mm EDTA. The two products were then mixed and incubated for two cycles of 65 °C for 5 min and 30 °C for 15 min to hybridize the two fragments, and the product was transformed into competent XLI-Blue cells. Positive clones were identified through sequencing performed by the University of Michigan DNA Sequencing Core. The Y92A/W244A mutant was created with the same method as Y92A, except that the original template used was full-length mCRT containing the W244A mutation in pMSCV-puro.
Generation of mCRT(W244A) by Finnzymes Phusion Site-directed Mutagenesis Kit
The following primers were used to generate mCRT(W244A): forward, 5′-GCT GAA CCA CCA GTG ATT CAA AAT CCT GAA TAC-3′ and reverse, 5′-CTC TCC ATC CAT CTC TTC ATC CCA GTC CTC-3′.
Generation of the W302A Mutant Using the QuikChange II Site-directed Mutagenesis Kit
The following primers were used to generate mCRT(W302A): forward, 5′-G GGC CTA GAT CTC GCG CAG GTC AAG TCC GG-3′ and reverse, 5′-CC GGA CTT GAC CTG CGC GAG ATC TAG GCC C-3′.
Cloning Human ERp57 into pET33b
ERp57 was PCR-amplified from the pVL1392 vector (20) using primers that introduced 5′ NcoI and 3′ XhoI sites and deleted the signal sequence. The vector pET33b, which allows for insertion of a C-terminal His tag, and ERp57 inserts were both prepared by XhoI and NcoI digestion and ligated (Invitrogen). The following primers were used: forward, 5′-A TCC ATG GCC TCC GAC GTG CTA GAA CTC-3′ and reverse, 5′-AT CTC GAG GAG ATC CTC CTG TGC CTT C-3′.
Expression of LLO in E. coli
A listeriolysin O (LLO) W492A mutant LLO, which is described to have reduced cytotoxic activity (21), was used as a non-glycosylated calreticulin substrate. Wild-type LLO with a C-terminal histidine tag was mutated by PCR using the primer 5′-GCG AGA ACG GTA ATT GAT GAC C-3′ (Integrated DNA Technologies, Coralville, IA) to change Trp-492 to Ala. The PCR product was ligated into the pET29b plasmid (Novagen, WI), sequenced, transformed into BL21 (DE3, Novagen), and purified as described below.
Protein Purifications
Purification of Calreticulin
Glycerol stocks of BL21 (DE3) cells expressing the calreticulin constructs were inoculated into a 25-ml terrific broth culture (with 50 μg/ml ampicillin) and incubated at 37 °C overnight. The starter culture was added to 1 liter of terrific broth with 50 μg/ml ampicillin and incubated at 37 °C until cell density measured by A600 was 0.8–0.9. Cultures were then incubated at room temperature for 1 h before inducing calreticulin protein expression with 200 μm isopropyl 1-thio-β-d-galactopyranoside. Bacterial cultures were incubated at room temperature for 16–20 h before harvesting cells by centrifugation. Cell pellets were resuspended in 50 ml of 50 mm Tris with 0.33 mg/ml lysozyme and EDTA-free complete protease inhibitors (Roche Applied Science). Cells were lysed by sonication. Subsequently, 10 μg/ml DNase, 1% Triton X-100, 10 mm MgCl2, and 1 mm CaCl2 were added, and the cell lysis suspension was incubated at room temperature for 30 min. Cell debris was removed from samples by centrifugation, followed by vacuum filtration of the supernatant with a Steriflip having a 0.22-μm pore membrane (Millipore). The filtrate was then incubated with nickel-nitrilotriacetic acid-agarose beads (Qiagen) for 2–4 h at 4 °C. The beads were washed with 10 mm imidazole in wash buffer (50 mm Tris, 150 mm NaCl, 1 mm CaCl2, pH 7.5). Murine calreticulin was eluted from beads with 75 and 100 mm imidazole in wash buffer. Protein was concentrated to 0.5–4.0 ml, by centrifugation using Centriplus centrifugal filter devices (Millipore) with molecular weight cut-offs of 10 or 30 kDa, and analyzed by gel-filtration chromatography.
Gel-filtration Analyses of Calreticulin
Purified concentrated calreticulin constructs were analyzed by gel filtration at 4 °C using a Superdex 200 10/300 GL column or Highload 16/60 Superdex 200 (Amersham Biosciences). Buffer used was 20 mm Hepes, 150 mm NaCl, 10% glycerol, 1 mm CaCl2, pH 7.5. Following gel filtration, fractions corresponding to the various monomeric and oligomeric calreticulin species for each construct were pooled and concentrated by centrifugation using Centricon centrifugal filter devices (Millipore) with molecular weight cut-offs of 10 or 30 kDa. Protein concentration was determined measuring absorbance at 280 nm. Extinction coefficients were calculated from the protein amino acid sequence using ProtParam (www.expasy.ch) and are as follows (units are in m−1 cm−1): mCRT1–399, 82,975; mCRT1–362, 82,975; mCRT1–339(ΔC), 82,975; mCRT1–318, 75,985; mCRT33–362, 70485; and mCRT(ΔP), 44,015.
Purification of LLO
BL21 E. coli cells expressing LLO were inoculated into LB broth with 60 μg/ml kanamycin. Cell cultures were expanded to log phase (A600 = 0.8–0.9), induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 16–20 h at room temperature, and the recombinant LLO was purified from bacterial lysates by nickel affinity chromatography. Buffers used throughout the purification procedure were pH 8.0 and contained 10% glycerol to minimize LLO aggregation. LLO was eluted from the nickel beads with 250 mm imidazole in wash buffer, and then stored in aliquots at −80 °C. Relative purity was consistently >95% by SDS-PAGE and SYPRO Red gel staining (Invitrogen). Prior to aggregation assays (described below), small aliquots of LLO were further purified by gel-filtration chromatography using a Superdex 200 10/300 column. Buffer used was 50 mm Hepes, 140 mm NaCl, 10% glycerol, pH 8.0. LLO concentration and extinction coefficient (77,240 m−1 cm−1) were calculated as described above for mCRT.
Purification of ERp57
LB broth containing 50 μg/ml kanamycin was inoculated with BL21 E. coli cells expressing His-tagged ERp57. Cell cultures were expanded to log phase, induced with 250 μm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. ERp57 was purified by resuspending bacterial pellets in lysis buffer containing: 50 mm NaH2PO4, 150 mm NaCl, 0.2 mg/ml lysozyme, 0.1% Triton X-100, 10 mm β-mercaptoethanol, 2.7 mm benzamidine, 5 μg/ml leupeptin, 7 μg/ml pepstatin, and 1.5 mm phenylmethylsulfonyl fluoride, pH 8.0. Cells were disrupted by sonication in lysis buffer. Cell debris was removed from samples by centrifugation, followed by vacuum filtration of the supernatant with a Steriflip having a 0.22-μm pore membrane (Millipore). The filtrate was then incubated with nickel-nitrilotriacetic acid-agarose beads (Qiagen) for 3 h at 4 °C. The beads were washed with wash buffer (50 mm Tris, 300 mm NaCl, pH 8.0). ERp57 was eluted from beads with 10 and 20 mm imidazole in wash buffer. Protein was concentrated to ∼6.5 ml, by centrifugation using Centriplus centrifugal filter devices (Millipore) with molecular weight cut-offs of 10 or 30 kDa, and further purified by Mono Q ion-exchange chromatography. A linear gradient from 150 mm to 1 m NaCl in 50 mm Tris (pH 7.4) was used to elute ERp57 from the column. The fractions of eluted protein containing ERp57 were dialyzed into 10 mm Tris, 150, mm NaCl (pH 7.4), concentrated, and stored at −80 °C after adding 10% glycerol.
Thermostability Analyses by Sypro Orange Binding
These analyses were undertaken as previously described (22, 23). Proteins (16 μm) were incubated in buffer (20 mm Hepes, 150 mm NaCl, and 1 mm CaCl2, pH 7.5) and 1× Sypro Orange Stain (Invitrogen) diluted from a 5000× stock solution in the presence or absence of 48 μm Glcα1–3Manα1–2Manα1–2Man-OH (G1M3, Alberta Research Council) in a total reaction volume of 10 μl. Thermal scans were performed using an ABI PRISM 7900HT Sequence Detection System using temperature increments of 1 °C starting at 25 °C. Within an experiment, each condition was analyzed in triplicate wells. Fluorescence emission was measured across different wavelength bins, and the bin with maximum fluorescence was chosen for further analysis. Fluorescence was normalized within wells as percent maximum fluorescence ((Fobs − Fmin)/(Fmax − Fmin)*100) and plotted against the sample temperature.
Biacore-based Assays
CRT-ERp57 binding was assayed by surface plasmon resonance-based assays using a Biacore 2000 instrument, as previously described (24). Briefly, mCRT(WT) or mutants or the control protein bovine serum albumin were coupled to a Biacore CM5 chip (Amersham Biosciences) using amine-based coupling chemistry. ERp57 binding to each surface was monitored in 10 mm Hepes, 150 mm NaCl, 0.5 mm CaCl2, 0.005% surfactant P-20, pH 7.5, at a flow rate of 5 μl/min at 25 °C over a 30-min period. Signals obtained from the bovine serum albumin surface were subtracted from corresponding signals for each mCRT surface. Specific signals for mCRT binding were plotted as a function of ERp57 concentration, and non-linear curve fitting (1:1 specific binding model) was performed to derive KD values using PRISM software.
Aggregation Assays
In gel-based assays, 2 or 4 μm LLO were incubated with purified mCRT constructs (4–32 μm), or ovalbumin as a negative control, at 37 °C for 1 h. Assays were conducted in the presence of 0.2–0.5 mm CaCl2 with or without 5 mm EDTA or 3 mm ATP. Following incubation, aggregated proteins were separated from solution by microcentrifugation at maximum speed for 30 min at 4 °C. Supernatants containing non-aggregated protein were removed, and the pellets containing the aggregated protein were re-suspended in an equal volume of buffer. Proteins present in the supernatants and pellets were separated by SDS-PAGE and visualized with Coomassie Blue stain. Protein band intensity was quantified using ImageQuant (Amersham Biosciences) and used as a measure of aggregation suppression activity of calreticulin. A light scattering assay was also performed to compare the aggregation suppression activities of different calreticulin mutants. In this assay, 1 μm LLO was incubated in a 96-well plate with 1–16 μm of different mCRT in the presence of 0.5 mm CaCl2. Plates were incubated in a BIO-TEK spectrophotometer pre-heated to 37 °C, and absorbance at 360 nm was monitored over a 60-min period (KC4 software). The absorbance signal for LLO alone was set to 100%.
Cell Cultures
Bosc cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 4.5 g/liter glucose, l-glutamine, and 110 mg/liter sodium pyruvate, supplemented with 10% (v/v) fetal bovine serum (Invitrogen), 100 μg/ml streptomycin, and 100 units/ml penicillin (Invitrogen). Calreticulin-deficient (K42) cells (8) were maintained in RPMI medium 1640 (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 μg/ml streptomycin, and 100 units/ml penicillin.
Generation of Retroviral Supernatants
5.5 μg of pMSCV-ires-GFP or pMSCV-puro vector (encoding different mCRT or control vector lacking mCRT) DNA was mixed with 4 μg of pCL-EcoDNA and 0.5 μg of VSV-G encoding plasmid, and added to a mixture of Opti-MEM (Invitrogen) and FuGENE 6 (Roche Applied Science). Following incubation for 20 min at room temperature, the mixture was added to Bosc cells that had been grown to 70% confluency in a 10-cm tissue culture dish. Medium was changed after 24 h, and after 48 h, supernatants containing retroviruses were harvested and used to infect K42 cells.
Flow Cytometry
For MHC class I surface analysis by flow cytometry, infected K42 cells were resuspended and washed in phosphate-buffered saline containing 1% fetal bovine serum (flow cytometry buffer). 2 × 105 cells were resuspended in 100 μl of flow cytometry buffer containing the MHC class I ascites fluid Y3 (1:100, anti-H2-Kb, ATCC HB-176), AF6 (anti-H2-Kb, ATCC HB-158, 1:50), or 28-14-8S (1:50, anti-H2-Db, ATCC HB-27), and incubated for 30 min on ice. Cells were washed once with buffer, then resuspended in 100 μl of buffer containing goat anti-mouse antibody conjugated to phycoerythrin (1:500), and incubated for 15 min on ice. Cells were washed three times, and data for each sample were collected on the FACSCanto flow cytometer (BD Biosciences). For pMSCV-ires-GFP-infected cells, levels of mCRT expression were normalized by gating on a subset of the GFP-positive population. For pMSCV-ires-GFP-derived cells, flow cytometric analyses were undertaken 37 and 71 h post-infection. For pMSCV-puro-derived cells, following selection and maintenance in media containing 1–2.5 μg/ml puromycin, cells were analyzed no earlier than 3 weeks post-infection. Analyses were performed using WinMDI version 2.8.
Immunoprecipitations
K42 cells or K42 cells expressing mCRT constructs were harvested and lysed in digitonin lysis buffer (10 mm Na2HPO4, 10 mm Tris, 130 mm NaCl, 1% digitonin, complete EDTA-free protease inhibitors, pH 7.5). Indicated immunoprecipitations additionally contained 20 mm N-ethylmaleimide in the lysis buffer. In some analyses, cells were incubated with 10 μg/ml tunicamycin to induce accumulation of de-glycosylated tapasin. Cells were lysed on ice for 1 h, followed by a 30-min centrifugation to remove cell debris. Supernatants were incubated with or without antibodies overnight at 4 °C. Samples were then centrifuged to remove precipitated proteins and incubated for 2 h with Protein G beads (Amersham Biosciences). Beads were washed three times with lysis buffer containing 0.1% digitonin. Samples were separated by 10% SDS-PAGE and transferred to Immobilon membranes (Millipore) for immunoblotting. Membranes were blocked in 5% milk in TBS for 1 h at room temperature, followed by an overnight incubation with primary antibody in TBS plus 0.05% Tween 20 (TTBS) at 4 °C. Membranes were washed for 2 h in TTBS, incubated for 30 min with secondary antibody, and washed again for 2 h at room temperature. Chemiluminescence was detected using the Amersham Biosciences ECL Plus kit.
The following antibodies were used to immunoisolate relevant proteins: rabbit anti-mouse TAP1 serum (1:15, kindly provided by Dr. Ted Hansen), rabbit anti-mCRT antibody (1:400, Abcam, catalog number ab2907), and rabbit anti-Kb antiserum (EX8, 1:100, kindly provided by Dr. Jonathan Yewdell). The following antibodies were used in immunoblotting analyses: goat anti-TAP1 antibody (1:2000, Santa Cruz Biotechnology, Inc., catalog number sc-11465), goat antibody specific to the N terminus of mCRT (1:2000, Santa Cruz Biotechnology, Inc., catalog number sc-7431), hamster anti-tapasin (1:3000, kindly provided by Dr. Ted Hansen), EX8 (1:7500), and rabbit anti-ERp57 (1:3000, Santa Cruz Biotechnology, Inc., catalog number 28823). Secondary antibodies (Jackson ImmunoResearch) were all conjugated to horseradish peroxidase: mouse anti-rabbit (light chain specific), bovine anti-goat, and goat anti-hamster.
RESULTS
C- and N-terminal Truncations Destabilize Monomers of Calreticulin
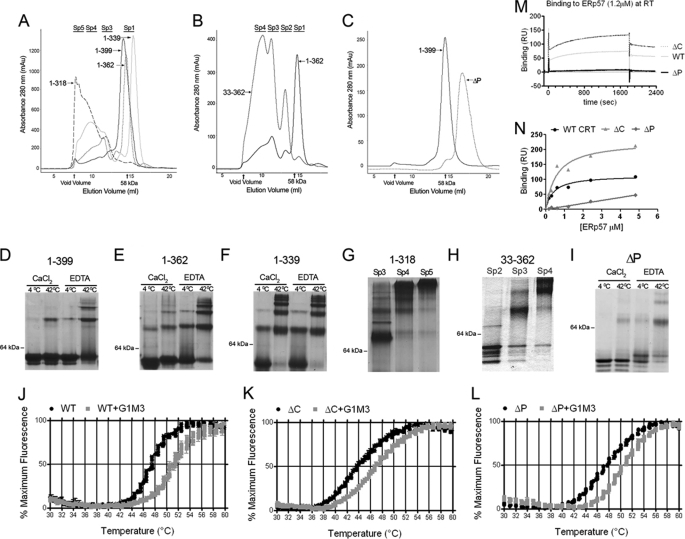
The first 32 residues of the calreticulin globular domain lack significant predicted secondary structure, with the exception of a short β-strand. Near the calreticulin C terminus, preceding the acidic domain is an α-helix, the major predicted helix in the calreticulin structure (supplemental Fig. S1). Based on these predicted structural features, we made truncated versions of calreticulin using Ala-1 and Val-33 as start sites, and Asp-318, Gln-339, Glu-362, and Leu-399 as stop sites. Asp-318 truncates the protein prior to the C-terminal helix, Gln-339 truncates the protein following the C-terminal helix and just prior to the acidic region, whereas Glu-362 extends the calreticulin sequence several residues into the C-terminal acidic domain. A construct in which the entire P-domain of calreticulin was deleted (amino acids 187–283) and replaced by two glycine residues was also generated.
Following protein expression in E. coli and purification on a nickel column, gel-filtration analysis indicated that full-length calreticulin, mCRT1–399 (mCRT(WT)), was present largely in the monomeric form (Sp1, Fig. 1A), as expected. Native PAGE analysis of purified protein verified that mCRT(WT) (12 μm) was largely monomeric at 4 °C, in the presence of 1 mm CaCl2 (Fig. 1D, lane 1). As previously described (6, 7), the formation of dimers and oligomers of mCRT(WT) was induced by heat treatment or calcium depletion (Fig. 1D, lane 1 compared with lanes 2–4). C-terminal deletion constructs mCRT1–362 and mCRT1–339 (mCRT(ΔC)) were also largely monomeric, although the extent of oligomeric forms of these proteins was enhanced relative to mCRT(WT) (Fig. 1A). In response to heat treatment or calcium depletion, dimeric and oligomeric structures were enhanced for mCRT1–362 and mCRT1–339 (mCRT(ΔC)), relative to mCRT(WT) (Fig. 1, compare D–F). Additional deletion of the C-terminal helix (mCRT1–318) dramatically affected calreticulin structure (Fig. 1A). No monomers were recovered from the purification (Fig. 1, A and G), and the major peak migrated in the column void volume. This construct was not used further in this study.
FIGURE 1.
Impacts of calreticulin domains on structure and functional activities. Analytical gel-filtration chromatography of C-terminal (A), N-terminal (B), and P-domain (C) truncation mutants of mCRT. All constructs were directly analyzed following nickel affinity chromatography, except mCRT(ΔP) for which monomers were first isolated by gel filtration and then re-analyzed by a second analytical gel-filtration step shown. Following gel-filtration chromatography, peak fractions were analyzed by native-PAGE following protein (12 μm) incubations for 1 h in 1 mm CaCl2 or 10 mm EDTA at 4 or 42 °C (D–F and I). Fractions corresponding to the indicated oligomeric species (Sp2–Sp5) of mCRT1–318 or mCRT33–362 were directly analyzed by native PAGE (1 mm CaCl2, 4 °C) (G and H). One representative analysis is shown of two or more independent analyses. J–L, monomeric mCRT constructs were incubated with the fluorophore SYPRO Orange and subjected to a thermal stability analysis using a real-time PCR machine. Normalized fluorescence data (mean of triplicate wells ± S.E.) for one of three independent experiments are shown. Compiled Tm values for all constructs discussed in this report are shown in supplemental Table S1. M and N, mCRT constructs were immobilized on a Biacore CM5 chip to response unit (RU) values of 438, 426, and 480 (respectively, for WT, ΔP, and ΔC), and human ERp57 was injected over each surface or a control bovine serum albumin surface. Representative sensorgrams following subtraction of corresponding sensorgrams from the bovine serum albumin surface are shown in M. Dose-response graphs for binding constant calculations are shown in N. One representative of two (ΔC and ΔP) or five (WT) independent analyses of N are shown.
N-terminal truncation of calreticulin (mCRT33–362) also impacted calreticulin structure (Fig. 1B). Oligomeric species were recovered, and monomers were absent. Different oligomeric forms were classified based on the gel-filtration chromatograms and native PAGE analyses as: Sp3, consisting largely of the dimeric form of the protein and Sp4 and Sp5, consisting of higher order oligomeric forms (Fig. 1, A, B, G, and H).
The calreticulin ΔP mutant (mCRT(ΔP)) was recoverable as monomers following purification, as were mCRT(ΔC), mCRT1–362, and mCRT(WT) (Fig. 1C). The oligomerization phenotype for mCRT(ΔP) resembled that of mCRT(WT) (Fig. 1I).
A thermostability assay was next used to compare heat-induced unfolding of mCRT constructs. A fluorescent dye (Sypro Orange) was used that displays enhanced binding to proteins following thermal unfolding. CD studies at 280 nm have previously revealed the occurrence of a conformational change in human calreticulin, which has a transition midpoint (Tm) value of 46.4 °C in the presence of 1 mm CaCl2 (25). A similar Tm value was measured for mCRT(WT) in the Sypro Orange-based thermostability assay (Tm value 47.96 ± 0.21 °C) (Fig. 1J). In the presence of the calreticulin-binding glycan Glcα1–3Manα1–2Manα1–2Man-OH (G1M3), the Tm value was significantly right-shifted to 51.08 ± 0.18 °C. Both mCRT(ΔP) and mCRT(ΔC) displayed fluorescence profiles similar to those obtained with mCRT(WT), and both were right-shifted by G1M3 (Fig. 1, K and L). However, the Tm value measured for mCRT(ΔC) was 44.29 ± 0.44 °C in the absence of G1M3, and 46.91 ± 0.17 °C in the presence of G1M3. These findings indicate that mCRT(ΔC) is less resistant to heat-induced structural changes than mCRT(WT). However, mCRT(ΔC) was able to bind to and be stabilized by G1M3, as was mCRT(ΔP) (supplemental Table S1). Different species of the oligomeric mCRT33–362 construct did not display thermostability profiles resembling those of mCRT(WT) and did not bind G1M3 as indicated by the thermostability-shift assays (data not shown) and, therefore, are not described further in this study.
A Biacore-based assay was used to measure ERp57 binding to the monomeric truncation constructs. mCRT(WT) and mCRT(ΔC) bound to ERp57, whereas binding was essentially undetectable with mCRT(ΔP), as expected (3) (Fig. 1M). Calculated KD values were 0.55 ± 0.26 μm and 1.2 ± 0.8 μm, respectively, for mCRT(WT) and mCRT(ΔC) (Fig. 1N).
Compared with mCRT(WT), mCRT1–339 (ΔC) and mCRT(ΔP) Have Reduced Abilities to Mediate MHC Class I Assembly in CRT−/− Cells
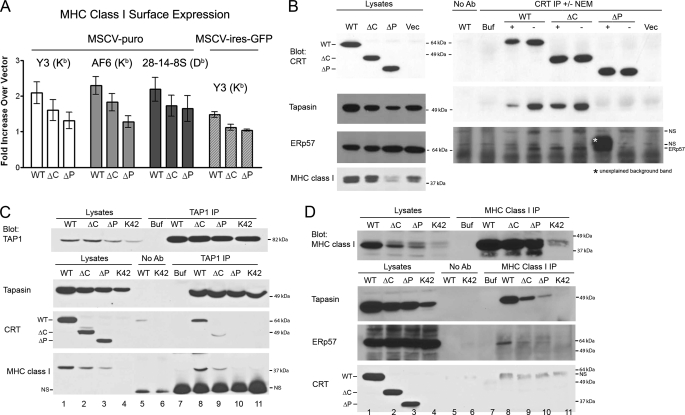
Calreticulin-deficient cells (K42) express reduced cell surface MHC class I, which can be restored by expression of wild-type calreticulin (8). Impacts of mCRT(ΔC) and mCRT(ΔP) truncations upon this activity were next assessed. The pMSCV-puro and pMSCV-ires-GFP viral vectors were utilized for stable and transient expressions, respectively. A small but reproducible induction of surface MHC class I (Kb) molecules was observed when K42 cells were infected with viruses encoding mCRT(WT) compared with those infected with viruses lacking the calreticulin DNA (Fig. 2A). Using both vector systems, mCRT(ΔP) was impaired in the ability to induce surface MHC class I relative to mCRT(WT) (Fig. 2A). A reduction in MHC class I cell-surface expression was also observed in the context of mCRT(ΔC), although less significant than that of mCRT(ΔP) (Fig. 2A).
FIGURE 2.
mCRT(ΔC) and mCRT(ΔP) have reduced abilities to mediate MHC class I assembly in calreticulin-deficient (K42) cells. A, flow cytometric analyses of cell surface expression of MHC class I on K42 cells infected with retroviruses generated with pMSCV-puro (left three sets of bar graphs) or pMSCV-ires-GFP (bar graph set on the extreme right) encoding indicated mCRT constructs or control virus lacking CRT. The -fold induction of mean fluorescence relative to parallel control infections with vector alone is indicated. Y3 (anti-H2-Kb), AF6 (anti-H2-Kb), or 28-14-8S (anti-H2-Db) antibodies were used in the analyses as indicated. Data are averaged over seven independent analyses from four separate infection (Y3, left (MSCV-puro) panel), six independent analyses from two separate infection (AF6 and 28-14-8S panels), and three independent analyses from two separate infections (Y3, right (MSCV-IRES-GFP) panel). B–D, immunoprecipitations from cells expressing indicated mCRT constructs or control vector-infected cells with anti-calreticulin (B) anti-TAP1 (C) or anti-MHC class I (D). Immunoblotting analyses of indicated cell lysates or immunoprecipitates were undertaken with antibodies directed against various PLC components. Lysates were generated in the presence or absence of NEM (B). Data are representative of five, two, or three independent analyses, respectively, for B–D. No antibody (No Ab) controls were performed by incubating indicated lysates with beads, and Buffer (Buf) indicates signals obtained with antibody alone (without lysates). NS indicates nonspecific bands.
Additional assays were developed to compare functional activities of the different calreticulin constructs. We noted that steady-state levels of tapasin and MHC class I heavy chains were significantly reduced in cells lacking mCRT compared with cells expressing mCRT(WT) (Fig. 2B, lysates panels). This was a consistent phenotype in the lysate analyses. Cells expressing mCRT(ΔP) and mCRT(ΔC) also displayed lower steady-state levels of tapasin and MHC class I heavy chains, a phenotype that was more pronounced for mCRT(ΔP) compared with mCRT(ΔC) (Fig. 2B, lysates panels and tapasin and MHC class I blots). Lower steady-state protein level was not a general phenotype of all ER proteins in K42 cells that lacked calreticulin. For example, ERp57 levels appeared enhanced rather than reduced in the absence of calreticulin (Fig. 2B, lysates panels and ERp57 blots).
To assess interactions between PLC components, immunoprecipitations were first performed with a calreticulin-specific antibody, and immunoblotting analyses were undertaken with antibodies directed against different PLC components (Fig. 2B, CRT IP). A strong signal for tapasin was observed with mCRT(WT) and mCRT(ΔC), whereas tapasin binding to mCRT(ΔP) was impaired (Fig. 2B, CRT IP and tapasin blot). ERp57 binding to mCRT(WT) and mCRT(ΔC) was weakly detected in some assays (for example, Fig. 2B, CRT IP and ERp57 blot), and undetectable in other assays (supplemental Fig. S2, ERp57 blot), whereas ERp57 binding to mCRT(ΔP) was undetectable in all analyses. These results suggest that the known P domain-dependent calreticulin-ERp57 interaction (3) was at the detection threshold of these immunoprecipitation reactions. Tapasin forms a disulfide-linked heterodimer with ERp57, an interaction that is observable only if thiol-modifying agents are included during cell lysis (26). A thiol-modifying agent was not required to observe the calreticulin-tapasin interaction (Fig. 2B, tapasin blot, compare signals ± NEM). MHC class I molecules were not detectable in the anti-calreticulin immunoprecipitations (supplemental Fig. S2, MHC class I blot), indicating that glycan-based interactions between MHC class I and calreticulin per se are insufficient for stable detection of complex formation between these proteins.
Anti-TAP1 immunoprecipitations revealed incorporation of mCRT(ΔC) into the MHC class I peptide-loading complex and a deficiency in mCRT(ΔP) incorporation, when expressed at levels similar to mCRT(ΔC) (Fig. 2C, CRT blot and lanes 2, 3, 9, and 10). Tapasin was incorporated into the PLC of all cells (Fig. 2C, tapasin blot and lanes 8–11). Levels of MHC class I detectable within the PLC were reflective of steady-state levels of MHC class I, most readily detectable with mCRT(WT), and very weakly detectable with mCRT(ΔP) or cells expressing the control vector (Fig. 2C, MHC class I blot and lanes 8–11). Thus, although PLCs are observable in calreticulin-deficient cells as previously noted (8), the efficiency of incorporation of MHC class I molecules is significantly impacted by calreticulin deficiency.
Immunoprecipitations with an anti-MHC class I antibody revealed that MHC class I binding to tapasin was most readily detectable in the context of mCRT(WT), with significant reductions noted in other cells (Fig. 2D, tapasin blot and lane 8 compared with lanes 9–11), reflective of alterations in steady-state levels of tapasin. Additionally, ERp57 binding to MHC class I was also most readily detectable in the context of mCRT(WT) (Fig. 2D, ERp57 blot and lane 8 compared with lanes 9–11). A very weak signal for mCRT(WT) was observed in the anti-MHC class I immunoprecipitations in two of four experiments (Fig. 2D, CRT blot and lane 8; a nonspecific band is visualized just below this band), indicating this was also a weak interaction at the detection threshold. Neither of the two mutant calreticulin constructs was observable in these assays.
Together the results in Fig. 2 indicated that calreticulin deficiency impacted steady-state levels of tapasin and MHC class I heavy chains and cell surface expression of MHC class I. Tapasin-calreticulin binding was readily detectable and impacted by the mCRT(ΔP) mutation, which also impacted calreticulin and MHC class I incorporation into the PLC. Furthermore, the mCRT(ΔC) mutation impacted steady-state levels of PLC components.
Point Mutations Targeting the ERp57 and Glycan Binding Sites of Calreticulin Impact the Efficiencies of Calreticulin-Tapasin Binding, Stabilization of Tapasin and MHC Class I, and Calreticulin Recruitment to the PLC
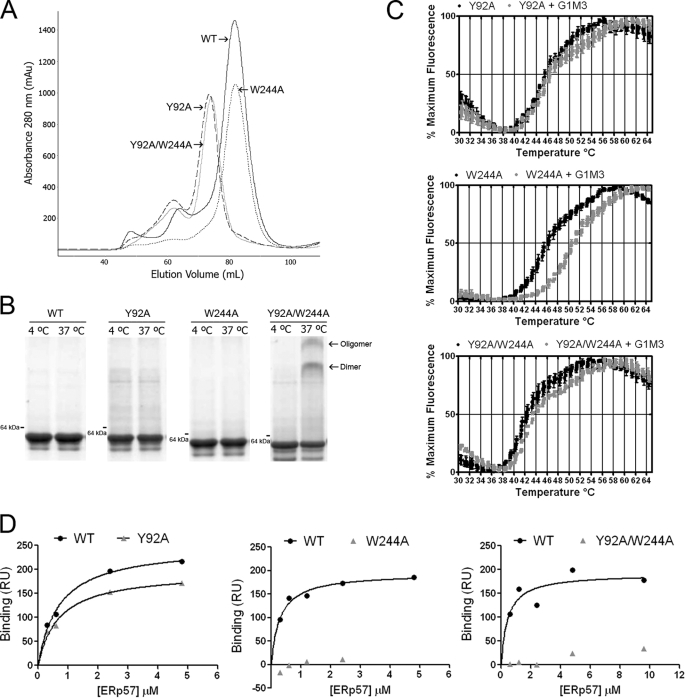
Mutations of calreticulin at residues 92 and 244 have previously been shown to strongly impact glycan and ERp57 binding, respectively (24, 27); we assessed impacts of mutations at these sites upon MHC class I assembly. Following purification, mCRT(Y92A) and mCRT(Y92A/W244A) each eluted as a single major peak that was left-shifted compared with mCRT(WT) and mCRT(W244A) (Fig. 3A). Native-PAGE revealed that the peak fraction of each protein was largely monomeric at 8 μm (37 °C, 0.5 mm CaCl2). Under these conditions, the double mutant showed a small induction of a dimer and oligomeric species (Fig. 3B). These findings indicate that mCRT(Y92A) and mCRT(Y92A/W244A) equilibrate between monomeric and dimeric/higher order forms during purification (likely due to the high concentrations achieved during purification), but that the equilibriums shift predominantly to monomers at a concentration of 8 μm. The Tm values of mCRT(Y92A) and mCRT(W244A), 45.90 ± 0.31 °C and 46.59 ± 0.46 °C, respectively, were slightly left-shifted relative to mCRT(WT) (47.96 ± 0.21 °C), and that of mCRT(Y92A/W244A) was more strongly left-shifted (42.96 ± 0.55 °C), indicating slightly lower resistance of the mutants to heat-induced structural changes (Fig. 3C and supplemental Table S1). In the presence of G1M3, the Tm values for mCRT(Y92A) and mCRT(Y92A/W244A) did not significantly change, indicating these mutants are in fact deficient in glycan binding (Fig. 3C and supplemental Table S1). Similar to mCRT(WT), the Tm value for mCRT(W244A) increased in the presence of G1M3 (50.74 ± 0.03 °C) (Fig. 3C and supplemental Table S1), verifying that this mutant binds glycans. ERp57 binding was strongly reduced with mCRT(W244A) and mCRT(Y92A/W244A) (Fig. 3D; binding signals too low to allow for accurate KD value estimation), whereas mCRT(Y92A) interacted with a KD value of 0.67 μm, similar to that for mCRT(WT).
FIGURE 3.
Y92A, W244A, and Y92A/W244A mutants of mCRT display expected impairments in glycan and/or ERp57 binding. A, representative gel-filtration chromatograms of mCRT(WT), mCRT(Y92A), mCRT(W244A), and mCRT(Y92A/W244A) following nickel affinity purification. B, native PAGE analyses of pooled peak fractions from A, following incubation at 4 or 37 °C for 1 h (8 μm protein in 1 mm CaCl2). C and D, thermostability and glycan and ERp57 binding analyses of indicated mCRT constructs were performed as described in Fig. 1. Data are representative of two independent sets of analyses, except ERp57 binding to the mCRT(Y92A/W244A), for which one analysis was performed.
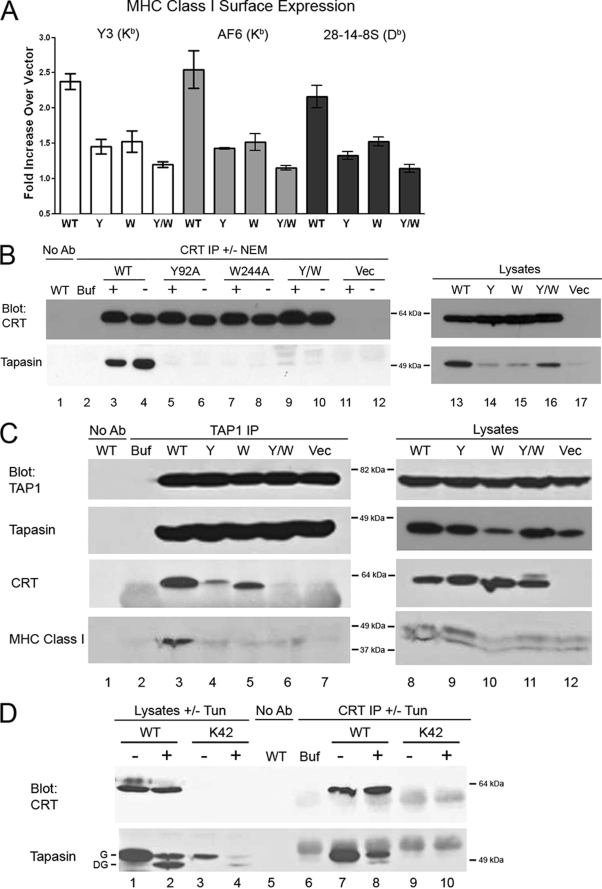
With this knowledge of the conformational integrity and glycan and ERp57 binding phenotypes, mutants were expressed in K42 cells for further analyses. None of the mutants induced surface MHC class I to the same extent as mCRT(WT) (Fig. 4A). Correlating with this result, and similar to the phenotypes seen with mCRT(ΔP), tapasin levels were reduced in the lysates of cells expressing mCRT(Y92A), mCRT(W244A), mCRT(Y92A/W244A), or the vector control (Fig. 4B, lysate panels), despite the expression of the mutant calreticulin molecules at levels equal to or greater than mCRT(WT).
FIGURE 4.
Y92A (Y), W244A (W), and Y92A/W244A (Y/W) mutants of mCRT are impaired in mediating MHC class I assembly. A, flow cytometric analyses were performed with the three indicated MHC class I-specific antibodies. The -fold induction of mean fluorescence by each mCRT relative to parallel control vector infections is indicated. Data are the averages of three independent experiments from a single infection. B–D, immunoprecipitated proteins (IP) (anti-calreticulin or anti-TAP1) or lysates were analyzed by immunoblotting with the indicated antibodies. Lysates were generated in the presence or absence of NEM (B) and from untreated cells or cells pre-treated with tunicamycin (Tun) (D). Data are representative of two independent analyses. In D, G = glycosylated tapasin, DG = deglycosylated tapasin. Control lanes are labeled as in Fig. 2.
In anti-calreticulin-based immunoprecipitations, tapasin co-immunoprecipitation was essentially undetectable with each mutant, either in the presence or in the absence of NEM (Fig. 4B, CRT IP, tapasin blot, and lanes 5–10). Anti-TAP1 immunoprecipitations revealed reduced incorporation of mCRT(Y92A) and mCRT(W244A) and low or undetectable mCRT(Y92A/W244A) incorporation (Fig. 4C, CRT blot, and lanes 3–6) despite expression of each mutant at a level similar to or greater than mCRT(WT). Correlating with lower steady-state MHC class I levels particularly in the context of mCRT(W244A) and mCRT(Y92A/W244A), and despite efficient tapasin incorporation into the PLC, reduced MHC class I incorporation into the PLC was also observed in K42 cells expressing each mutant (Fig. 4C, MHC class I blot and lanes 3–6).
Tyr-92 may be important for calreticulin binding to glycans of tapasin or MHC class I, as both proteins are glycosylated. To further examine the former possibility, K42 cells expressing or lacking mCRT(WT) were pre-treated with the drug tunicamycin to block protein glycosylation to a point at which deglycosylated tapasin was the major observable species (Fig. 4D, tapasin blot and lanes 2 and 4). Under these conditions, the major species that co-immunoprecipitated with calreticulin was the glycosylated form of tapasin, whereas de-glycosylated tapasin was the minor species (Fig. 4D, tapasin blot and lane 8). These results suggest that the tapasin glycan contributes at least in part to the observed interactions between calreticulin and tapasin. However, the observation of the de-glycosylated tapasin band as a minor species in the anti-calreticulin immunoprecipitation suggests that there are additional modes of direct or indirect calreticulin-tapasin interactions that are independent of the tapasin glycan.
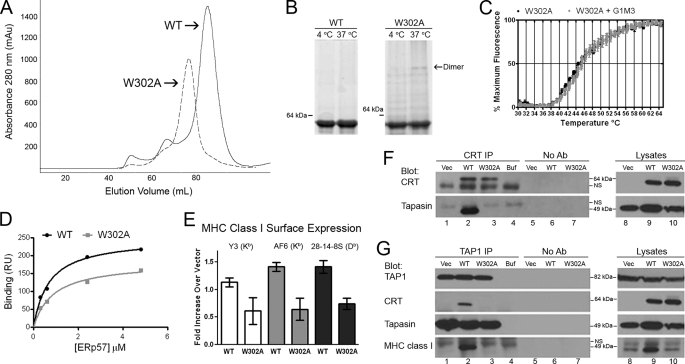
Trp-302 of calreticulin is also thought to comprise a residue in the glycan binding site (supplemental Fig. S1B) (28) and has additionally been implicated in the in vitro chaperone activity of calreticulin (24). Similar to mCRT(Y92A), mCRT(W302A) protein displayed a gel-filtration profile whose major peak was slightly left-shifted relative to that of mCRT(WT) (Fig. 5A), while remaining largely monomeric at 4 and 37 °C at a concentration of 8 μm (Fig. 5B, right panel). In a thermostability assay, mCRT(W302A) displayed a Tm value of 44.82 ± 0.55 °C, lower than that of mCRT(WT) (Tm = 47.96 ± 0.21 °C), and the Tm value of mCRT(W302A) did not right-shift in the presence of G1M3 (Fig. 5C and supplemental Table S1), indicating that the mutant has the expected deficiency in glycan binding. In Biacore assays, mCRT(W302A) was able to bind to ERp57 with a KD value of 0.88 μm, similar to mCRT(WT) (Fig. 5D).
FIGURE 5.
mCRT(W302A) is impaired in mediating MHC class I assembly. A, representative gel-filtration chromatograms of mCRT(WT) and mCRT(W302A) proteins following nickel affinity chromatography. B, native PAGE analysis of pooled peak fractions from A, following indicated incubations (in 0.5 mm CaCl2). C and D, thermostability, glycan, and ERp57 binding analyses for mCRT(W302A) performed as described in Fig. 1. Thermostability data are representative of three independent analyses, and ERp57 binding data is based on a single analysis. E, flow cytometric analyses of the ability of mCRT(WT-FLAG) and mCRT(W302A-FLAG) to restore MHC class I expression in K42 cells were performed as described in Fig. 4A. Data are representative of three experiments from two infections. F and G, immunoprecipitated proteins (anti-calreticulin and anti-TAP1) or lysates were analyzed by immunoblotting with the indicated antibodies. Data are representative of two immunoprecipitations. Control lanes are labeled as indicated in Fig. 2.
In K42 cells, mCRT(W302A) was unable to restore surface MHC class I (Fig. 5E). Steady-state levels of tapasin and MHC class I in the lysates of cells expressing mCRT(W302A) were reduced compared with those found in K42 cells expressing mCRT(WT) (Fig. 5, F and G, lysate panels and lanes 8–10). Immunoprecipitations with anti-calreticulin revealed defects in tapasin binding by mCRT(W302A) (Fig. 5F, CRT IP, tapasin blot, and compare lane 2 with lane 3). Immunoprecipitation with anti-TAP1 revealed impaired incorporation of mCRT(W302A) into the PLC (Fig. 5G, TAP IP, CRT blot, and compare lane 2 with lane 3), and impaired MHC class I incorporation into the PLC in the context of mCRT(W302A) (Fig. 5G, TAP IP, MHC class I blot, and compare lane 2 with lane 3).
Various mCRT Mutants Induce the Activity of Calreticulin in Suppressing Protein Aggregation
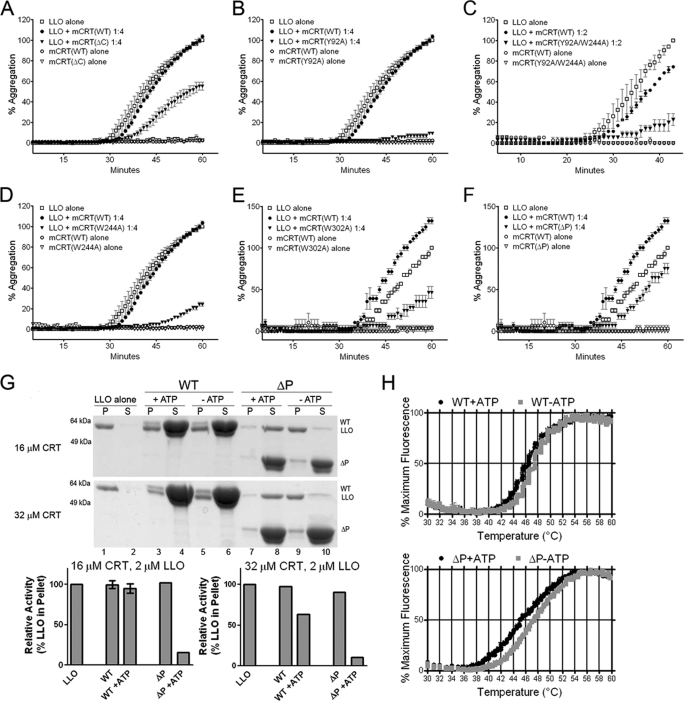
Calreticulin has previously been shown to inhibit thermal aggregation of various non-glycosylated substrate proteins (5, 6, 24), a functional assay indicative of the polypeptide-binding activity of calreticulin. To assess this activity at 37 °C, LLO was used as a substrate. LLO has been shown to aggregate at 37 °C and neutral pH (29), which was reproduced in the present study by gel-based aggregation assays and light scattering assays. By the gel-based assay, in the presence of 0.5 mm CaCl2, mCRT(WT) was unable to significantly rescue LLO from precipitating at LLO:mCRT ratios up to 1:4 (supplemental Fig. S3). Calcium depletion induced activity of mCRT(WT), as did the mCRT(ΔC) truncation, consistent with our previous findings with different substrates (6).
By the more sensitive light scattering assay, in 0.5 mm CaCl2, mCRT(WT) had low activity until high stoichiometric ratios were achieved (1:16 LLO:mCRT(WT); data not shown). mCRT(ΔC) displayed induced activity relative to mCRT(WT) (for example, Fig. 6A). mCRT(Y92A) and mCRT(Y92A/W244A) displayed strongly induced activities relative to mCRT(WT) in all analyses undertaken (for example, Fig. 6, B and C). mCRT(W244A) and mCRT(W302A) also displayed higher or equal activities relative to mCRT(WT) in the majority of conditions tested (14 of 18 conditions analyzed over 4 independent experiments for W244A, and 19 of 24 conditions analyzed over 6 independent experiments for W302A; representative analyses shown in Fig. 6, D and E). The W302A and W244A mutants of calreticulin have both previously been implicated as being impaired for rescue of aggregation of other substrates at 44–45 °C (24); our findings reflect different phenotypes of these two mutants in aggregation assays performed under a physiological non-stress condition.
FIGURE 6.
Aggregation suppression activities of calreticulin mutants in vitro. A–F, light scattering-based aggregation assays of the abilities of indicated mCRT constructs to suppress LLO (1 μm) aggregation at 37 °C in the presence of 0.5 mm CaCl2. G, gel-based aggregation assays. LLO alone (4 μm) or LLO with indicated amounts mCRT(WT) or mCRT(ΔP) were incubated in the presence of 0.5 mm CaCl2 ± 3 mm ATP at 37 °C for 1 h. Proteins present in supernatants (S) and pellets (P) were resolved by SDS-PAGE and visualized by staining with Coomassie Blue dye (top and middle panels). Lower panels, band intensities of LLO in the pellet were quantified and are represented as a percentage relative to LLO observed in the pellet in the absence of any mCRT. H, thermostability of mCRT(WT) or mCRT(ΔP) in the presence of 0.5 mm CaCl2 ± 3 mm ATP as indicated, were measured as described in Fig. 1.
Compared with mCRT(WT), lower activity of mCRT(ΔP) was observed in 9 analyses, and equal or higher activity was observed in 11 analyses of a total of 21 comparisons over 4 independent experiments (for example, Fig. 6F). These variable results taken together with previous findings that the P domain of calnexin facilitates suppression of substrate aggregation (30) prompted us to use gel-based aggregation assays to further compare mCRT(ΔP) and mCRT(WT) activities. Because ATP is a factor that modulates the chaperone activity of calreticulin (5), aggregation suppression activities of mCRT(WT) and mCRT(ΔP) were measured in 0.5 mm CaCl2 ± 3 mm ATP. Both ATP and a 1:16 ratio of LLO to mCRT(WT) were required to partially inhibit LLO aggregation at 37 °C (in 0.5 mm CaCl2) (Fig. 6G, middle panel, and compare lane 3 with lanes 1 and 5). The addition of 3 mm ATP also markedly induced the aggregation suppression activity of mCRT(ΔP) (Fig. 6G, top and middle panels, and compare lane 7 with lanes 1 and 9). This impact of ATP was readily apparent with mCRT(ΔP) even at a 1:8 ratio (Fig. 6G, upper panel, and compare lane 7 with lanes 1 and 9), and ATP induced a more marked left-shift of the Tm value for mCRT(ΔP) compared with mCRT(WT) (Fig. 6H and supplemental Table S1, Tm values of 45.49 ± 0.13 and 46.24 ± 0.02 °C, respectively, in the presence of ATP). Thus, the removal of the P-domain rendered calreticulin more susceptible to ATP-induced structural changes, thereby enhancing its ability to suppress aggregation of a misfolded protein under a physiological condition. Furthermore, several mutations that impaired mCRT function in MHC class I assembly induced its ability to suppress aggregation of a substrate protein in vitro under a physiological condition.
DISCUSSION
In the N-terminally truncated calreticulin construct mCRT33–362, essentially all protein was purified as dimers and higher order oligomers in the presence of 1 mm CaCl2 (Fig. 1, B and H), suggesting that structural changes involving this region of calreticulin may be responsible for inducing oligomer formation under heat-shock or calcium-depleting conditions. The C-terminal helix was also critical for maintaining calreticulin monomers (Fig. 1, A and G); the predominant species induced by this truncation migrated in the void volume. Conformational alterations and self-association interfered with the ability of the truncated variants to bind glycan substrates. A loss of just 20 residues internal to the acidic domain or 33 N-terminal residues is sufficient to induce profound conformational alterations. A number of functional studies of calreticulin described in the literature that used calreticulin truncations lacking all or a subset of residues N- and C-terminal to the P domain (or truncation constructs containing only the regions N- or C-terminal to the P domain), may have to be re-interpreted in light of these findings.
Truncation and point mutations of calreticulin that disrupted glycan binding (mCRT(Y92A) and mCRT(W302A)) or ERp57 binding (mCRT(ΔP) and mCRT(W244A)) were defective for binding tapasin, being recruited into the TAP complex, stabilizing tapasin and MHC class I heavy chains, and inducing MHC class I surface expression. For the Y92A and W244A mutations, binding deficiencies were more strongly apparent in the anti-CRT immunoprecipitations than in the anti-TAP1 immunoprecipitations, indicating that binding defects with these mutants may be partial rather than complete. Nevertheless, strong binding defects were apparent with these mutants as well as with mCRT(ΔP), mCRT(W302A), and mCRT(Y92A/W244A), mutants for which tapasin or TAP binding were essentially undetectable. The Williams laboratory used a different set of mutants, Y111A for a glycan binding-deficient mutant, D241N for an ERp57 binding-deficient mutant (Y128A and D258N, respectively, in protein numbering that includes the signal sequence), and the double mutant Y111A/D241A, analyses of which suggested that calreticulin could be recruited into the PLC in the absence of interactions with both ERp57 and substrate oligosaccharides (14). Previous studies have defined that trisaccharide interactions with calreticulin were essentially undetectable with the Y92F mutant (Y109F in mature protein numbering), whereas binding was detectable with Y111F (Y128F in mature protein numbering) (KB = 11.21 × 104 m−1 for Y128F compared with a KB of 58.8 × 104 m−1 for mCRT(WT)) (27). Thus, it is likely that the choice of mutant impacts the extent of the binding deficiency and forms the basis for the discrepancies in conclusions from the two studies. Additionally, antibodies used in the interactions analyses and the levels of calreticulin expression achieved are likely to determine the sensitivity with which binding differences can be elucidated.
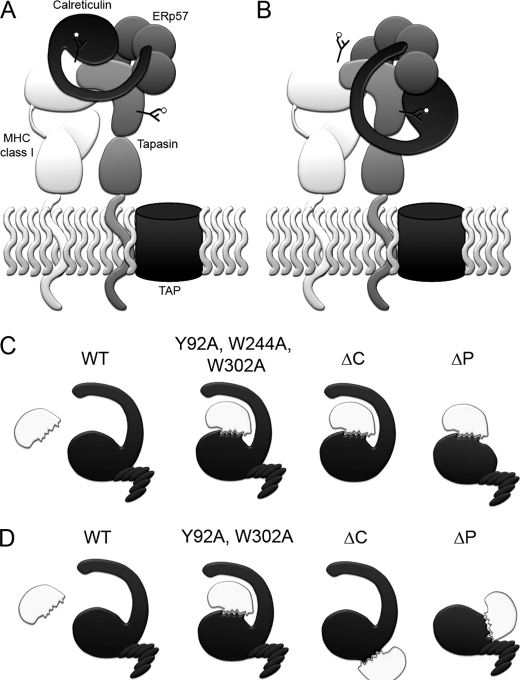
Our mutational data are strongly supportive of a model in which calreticulin-ERp57 and calreticulin-glycan interactions are both important for the recruitment of calreticulin into the PLC and for the functional activity of calreticulin in stabilizing tapasin and MHC class I heavy chains. Although previous studies have suggested a role for MHC class I glycan-dependent interactions for the recruitment of calreticulin to the PLC, support for such a mode of binding comes from studies with glycosylation inhibitors, in vitro binding studies with purified MHC class I molecules and calreticulin, and glycan-deficient mutants of MHC class I molecules (10–12). Support for an ERp57-dependent mode of calreticulin recruitment comes from the known tapasin-ERp57 interaction and its impact on calreticulin recruitment (15, 31, 32). The studies described here represent the first direct evidence for the involvement of the glycan and ERp57 binding sites of calreticulin for its recruitment into the PLC. Furthermore, based on the strong Tyr-92 and Trp-302 dependence of the tapasin-calreticulin binding (Figs. 4 and 5), it remains an open question whether calreticulin recruitment to the PLC involves the glycans of tapasin, or MHC class I molecules, or both proteins (Fig. 7). Two models for the recruitment of calreticulin into the PLC are shown in Fig. 7 that are both consistent with the current data. In both models, the tip of the P domain of calreticulin engages the b′ domain of ERp57. In model A, glycans of MHC class I are bound by calreticulin, and a specific contact interface for tapasin binding is also present on calreticulin. This model is similar to that proposed following structural studies of tapasin-ERp57 complexes (33). Current support for model 7A comes from inhibitor-based data, which showed reduced calreticulin-MHC class I binding in castanospermine-treated cells, and from cells expressing mutant MHC class I molecules lacking an appropriate glycan, which displayed reduced calreticulin binding (10, 11). The treatments or mutations also impacted MHC class I-TAP binding, which leaves open the possibility of indirect impacts upon MHC class I-calreticulin binding. In the additional/alternate model 7B, which is favored by the data in Fig. 4D, calreticulin engages the glycan of tapasin. Further studies with tapasin and calreticulin mutants will be required to address the relevance of one or both models shown in Fig. 7 (A and B) to the recruitment of calreticulin to the PLC.
FIGURE 7.
Modes of substrate recognition by calreticulin. A and B, two mechanisms to explain the central roles of tapasin, glycans, and ERp57 in calreticulin recruitment to the PLC and function within the PLC. In A, calreticulin binds to a MHC class I glycan, to ERp57 via the tip of its P domain, and to tapasin via a specific polypeptide binding site. In B, calreticulin binds to the glycan of tapasin, and to ERp57 via the tip of its P domain. A specific polypeptide-based interaction with MHC class I may also be relevant to this mode of recruitment. Interactions mediated by the acidic domain of calreticulin, which is also important for function, have to be further defined, and are not shown for simplicity. C and D, polypeptide binding sites on calreticulin are induced by mutations targeting distinct domains and residues on calreticulin, likely relating to the structural plasticity of calreticulin, which exposes single (C) or distinct hydrophobic sites (D) upon different truncations/mutations. Because mutations that induce the ability of calreticulin to suppress protein aggregation in vitro inhibit calreticulin recruitment to the PLC in cells, generic polypeptide binding sites per se are insufficient to recruit calreticulin to the PLC under non-stress conditions.
It is certainly possible that the observed glycan-dependent calreticulin-tapasin association (Figs. 4 and 5) reflects a role for calreticulin in the folding and maturation of tapasin, via a mechanism that is independent of the mode by which calreticulin is recruited into the PLC. Similarly, calreticulin could have a function in the maturation of MHC class I outside of the PLC, and such a possibility has very recently been described (34). However, PLC-independent interactions of calreticulin with MHC class I cannot explain the impact of calreticulin upon steady-state levels of tapasin (Figs. 2, 4, and 5). Steady-state levels of tapasin are expected to impact steady-state levels of MHC class I (35) and TAP (36, 37), as well as the functional activity of tapasin upon MHC class I assembly. Thus, the tapasin-calreticulin interactions occurring within the PLC or preceding the formation of the PLC are of functional significance in MHC class I assembly, and do not exclude the additional involvement of calreticulin at a subsequent step in MHC class I assembly.
Despite the ability of calreticulin to bind to monoglucosylated substrates in vitro (12), the data shown here indicate that glycan binding alone is insufficient for stable recruitment of calreticulin to a glycosylated substrate in cells, or for substrate stabilization. Rather, the glycan and ERp57 binding sites and the C-terminal acidic domain function cooperatively in substrate stabilization. Furthermore, ERp57-calreticulin binding influences the functional outcome of the calreticulin interaction with PLC substrates. Are such mechanisms of recruitment and function specific to calreticulin functions in the PLC, owing to the presence of tapasin, an unusual ERp57 substrate that can be isolated as a stable disulfide-linked conjugate with ERp57? We suggest that the observed modes of calreticulin recruitment may be more generally relevant for the following reasons. First, observation of tapasin-calreticulin binding or calreticulin incorporation into the PLC did not require conditions necessary for maintaining disulfide linkage between tapasin and ERp57 (±NEM, Figs. 2 and 4), and calreticulin binding per se could stabilize more transient substrate-ERp57 interactions if the substrate provides a suitable glycan site for cooperative binding. Second, the occurrence of multiple interactions would increase contact probability of the relatively low affinity calreticulin-glycan interactions (micromolar affinity range). Thus, as originally suggested (38), the calreticulin-ERp57 complex may be designed for a partnership in the oxidative folding of glycoproteins. It is noteworthy that the P domains of calnexin and calreticulin differ in length (2); differences in substrate specificities of calnexin and calreticulin could also relate to this difference. Likewise, the acidic domain of calreticulin is absent within the ER luminal domain of calnexin. Functional differences between calnexin and calreticulin may also partly be attributed to this domain, a possibility suggested by the data of Fig. 2.
Calreticulin is able to interact with MHC class I molecules in a glycan-independent manner under conditions of ER stress both in cells and in vitro (6), but whether such interactions also occur under physiological non-stress conditions is unclear. Mutations targeting diverse regions of calreticulin induced its ability to interact with a non-glycosylated polypeptide (Fig. 6), likely by providing increased exposure (at 37 °C in 0.5 mm CaCl2) of either one or several hydrophobic surfaces that can participate in substrate binding (Figs. 7, C and D), as suggested by the slight reduction in Tm values of the mutants (Figs. 1, 3, and 5). However, calreticulin mutants with induced polypeptide binding in vitro were impaired in their binding to tapasin and in their recruitment into the PLC under non-stress conditions (Figs. 2, 4, 5, and 6). Thus, generic polypeptide-based associations per se are insufficient for stable recruitment of calreticulin to PLC components in cells under non-stress conditions. Rather, more significant conformational changes induced in calreticulin by certain types of ER stress may be required to sufficiently populate conformations of calreticulin that are capable of substrate recognition via glycan-independent polypeptide-based associations.
Williams et al. have noted that calreticulin is much more efficient at suppressing aggregation of glycosylated substrates than of non-glycosylated substrates (5) and that mutations of glycan binding site residues specifically impair its ability to suppress aggregation of glycosylated substrates (39). These findings suggest that distinct mechanisms (binding sites) could be involved in calreticulin-mediated suppression of aggregation of appropriately glycosylated substrate versus non-glycosylated substrates. Further studies are needed to define the location(s) of polypeptide binding sites functional for the suppression of aggregation of glycosylated substrates to explore their relevance to substrate stabilization following initial recruitment via glycan and ERp57-dependent interactions.
Supplementary Material
Acknowledgments
We are grateful to Drs. Marek Michalak and Tim Elliott for providing calreticulin-deficient K42 cells and to Drs. Ted Hansen and Jon Yewdell for antibodies used in these studies. We thank James Delproposto of the University of Michigan high through-put protein laboratory for assistance with expression of recombinant truncated proteins, the University of Michigan DNA sequencing core for DNA sequence data, the University of Michigan Hybridoma Core for ascites production, the University of Michigan Microarray core for Sypro Orange binding analyses, and the University of Michigan Diabetes Research and Training Center for financial support. We thank Ramkumar Nandkumar and Kalpana Krishnaswamy (Metaome) supplemental Fig. S1, and Ben Schwartz, Rachael Sturtevant, and Laura Weiser for assistance with the project.
This work was supported, in whole or in part, by National Institutes of Health Grant AI066131 (to M. R.) and by a diversity supplement to National Institutes of Health Grant AI066131 (awarded to N. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- ER
- endoplasmic reticulum
- MHC
- major histocompatibility complex
- TAP
- transporter associated with antigen processing
- PLC
- MHC class I peptide loading complex
- mCRT
- mouse calreticulin
- CMV
- cytomegalovirus
- LIC
- ligation-independent cloning
- GFP
- green fluorescent protein
- LLO
- listeriolysin O
- G1M3
- Glcα1–3Manα1–2Manα1–2Man-OH
- TBS
- Tris-buffered saline
- NEM
- N-ethylmaleimide.
REFERENCES
- 1.Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M. (2009) Biochem. J. 417, 651–666 [DOI] [PubMed] [Google Scholar]
- 2.Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 3.Frickel E. M., Riek R., Jelesarov I., Helenius A., Wuthrich K., Ellgaard L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver J. D., Roderick H. L., Llewellyn D. H., High S. (1999) Mol. Biol. Cell 10, 2573–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito Y., Ihara Y., Leach M. R., Cohen-Doyle M. F., Williams D. B. (1999) EMBO J. 18, 6718–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizvi S. M., Mancino L., Thammavongsa V., Cantley R. L., Raghavan M. (2004) Mol. Cell 15, 913–923 [DOI] [PubMed] [Google Scholar]
- 7.Jørgensen C. S., Ryder L. R., Steinø A., Højrup P., Hansen J., Beyer N. H., Heegaard N. H., Houen G. (2003) Eur. J. Biochem. 270, 4140–4148 [DOI] [PubMed] [Google Scholar]
- 8.Gao B., Adhikari R., Howarth M., Nakamura K., Gold M. C., Hill A. B., Knee R., Michalak M., Elliott T. (2002) Immunity 16, 99–109 [DOI] [PubMed] [Google Scholar]
- 9.Raghavan M., Del Cid N., Rizvi S. M., Peters L. R. (2008) Trends Immunol. 29, 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadasivan B., Lehner P. J., Ortmann B., Spies T., Cresswell P. (1996) Immunity 5, 103–114 [DOI] [PubMed] [Google Scholar]
- 11.Harris M. R., Yu Y. Y., Kindle C. S., Hansen T. H., Solheim J. C. (1998) J. Immunol. 160, 5404–5409 [PubMed] [Google Scholar]
- 12.Wearsch P. A., Jakob C. A., Vallin A., Dwek R. A., Rudd P. M., Cresswell P. (2004) J. Biol. Chem. 279, 25112–25121 [DOI] [PubMed] [Google Scholar]
- 13.Ireland B. S., Brockmeier U., Howe C. M., Elliott T., Williams D. B. (2008) Mol. Biol. Cell 19, 2413–2423 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Zhang Y., Kozlov G., Pocanschi C. L., Brockmeier U., Ireland B. S., Maattanen P., Howe C., Elliott T., Gehring K., Williams D. B. (2009) J. Biol. Chem. 284, 10160–10173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbi N., Tanaka S., Momburg F., Hämmerling G. J. (2006) Nat. Immunol. 7, 93–102 [DOI] [PubMed] [Google Scholar]
- 16.Grandea A. G., 3rd, Golovina T. N., Hamilton S. E., Sriram V., Spies T., Brutkiewicz R. R., Harty J. T., Eisenlohr L. C., Van Kaer L. (2000) Immunity 13, 213–222 [DOI] [PubMed] [Google Scholar]
- 17.Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 18.Van Parijs L., Refaeli Y., Lord J. D., Nelson B. H., Abbas A. K., Baltimore D. (1999) Immunity 11, 281–288; retracted in Immunity (2009) 30, 611 [DOI] [PubMed] [Google Scholar]
- 19.Chiu J., March P. E., Lee R., Tillett D. (2004) Nucleic Acids Res. 32, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silvennoinen L., Myllyharju J., Ruoppolo M., Orrùu S., Caterino M., Kivirikko K. I., Koivunen P. (2004) J. Biol. Chem. 279, 13607–13615 [DOI] [PubMed] [Google Scholar]
- 21.Michel E., Reich K. A., Favier R., Berche P., Cossart P. (1990) Mol. Microbiol. 4, 2167–2178 [DOI] [PubMed] [Google Scholar]
- 22.Lo M. C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., Ellestad G. (2004) Anal. Biochem. 332, 153–159 [DOI] [PubMed] [Google Scholar]
- 23.Malawski G. A., Hillig R. C., Monteclaro F., Eberspaecher U., Schmitz A. A., Crusius K., Huber M., Egner U., Donner P., Müller-Tiemann B. (2006) Protein Sci. 15, 2718–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin V., Groenendyk J., Steiner S. S., Guo L., Dabrowska M., Parker J. M., Müller-Esterl W., Opas M., Michalak M. (2006) J. Biol. Chem. 281, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 25.Li Z., Stafford W. F., Bouvier M. (2001) Biochemistry 40, 11193–11201 [DOI] [PubMed] [Google Scholar]
- 26.Peaper D. R., Wearsch P. A., Cresswell P. (2005) EMBO J. 24, 3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor M., Ellgaard L., Gopalakrishnapai J., Schirra C., Gemma E., Oscarson S., Helenius A., Surolia A. (2004) Biochemistry 43, 97–106 [DOI] [PubMed] [Google Scholar]
- 28.Gopalakrishnapai J., Gupta G., Karthikeyan T., Sinha S., Kandiah E., Gemma E., Oscarson S., Surolia A. (2006) Biochem. Biophys. Res. Commun. 351, 14–20 [DOI] [PubMed] [Google Scholar]
- 29.Schuerch D. W., Wilson-Kubalek E. M., Tweten R. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12537–12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockmeier A., Brockmeier U., Williams D. B. (2009) J. Biol. Chem. 284, 3433–3444 [DOI] [PubMed] [Google Scholar]
- 31.Peaper D. R., Cresswell P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10477–10482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wearsch P. A., Cresswell P. (2007) Nat. Immunol. 8, 873–881 [DOI] [PubMed] [Google Scholar]
- 33.Dong G., Wearsch P. A., Peaper D. R., Cresswell P., Reinisch K. M. (2009) Immunity 30, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howe C., Garstka M., Al-Balushi M., Ghanem E., Antoniou A. N., Fritzsche S., Jankevicius G., Kontouli N., Schneeweiss C., Williams A., Elliott T., Springer S. (2009) EMBO J. 28, 3730–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thammavongsa V., Schaefer M., Filzen T., Collins K. L., Carrington M., Bangia N., Raghavan M. (2009) Immunogenetics 61, 703–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garbi N., Tiwari N., Momburg F., Hämmerling G. J. (2003) Eur. J. Immunol. 33, 264–273 [DOI] [PubMed] [Google Scholar]
- 37.Lehner P. J., Surman M. J., Cresswell P. (1998) Immunity 8, 221–231 [DOI] [PubMed] [Google Scholar]
- 38.Molinari M., Helenius A. (1999) Nature 402, 90–93 [DOI] [PubMed] [Google Scholar]
- 39.Thomson S. P., Williams D. B. (2005) Cell Stress Chaperones 10, 242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.