Abstract
Calreticulin is an abundant endoplasmic reticulum resident protein that fulfills at least two basic functions. Firstly, due to its ability to bind monoglucosylated high mannose oligosaccharides, calreticulin is a central component of the folding quality control system of glycoproteins. On the other hand, thanks to its capacity to bind high amounts of calcium, calreticulin is one of the main calcium buffers in the endoplasmic reticulum. This last activity resides on a highly negatively charged domain located at the C terminus. Interestingly, this domain has been proposed to regulate the intracellular localization of calreticulin. Structural information for this domain is currently scarce. Here we address this issue by employing a combination of biophysical techniques and molecular dynamics simulation. We found that calreticulin C-terminal domain at low calcium concentration displays a disordered structure, whereas calcium addition induces a more rigid and compact conformation. Remarkably, this change develops when calcium concentration varies within a range similar to that taking place in the endoplasmic reticulum upon physiological fluctuations. In addition, a much higher calcium concentration is necessary to attain similar responses in a peptide displaying a randomized sequence of calreticulin C-terminal domain, illustrating the sequence specificity of this effect. Molecular dynamics simulation reveals that this ordering effect is a consequence of the ability of calcium to bring into close proximity residues that lie apart in the primary structure. These results place calreticulin in a new setting in which the protein behaves not only as a calcium-binding protein but as a finely tuned calcium sensor.
Keywords: Calcium, Carbohydrate/Binding Protein, Methods/Circular Dichroism, Methods/Fluorescence, Protein/Molecular Dynamics, Protein/Conformation, Calreticulin, Natively Unfolded
Introduction
Calreticulin (CRT)2 is an endoplasmic reticulum (ER) calcium-binding chaperone that has been associated with several cellular functions both inside (1–3) and outside the ER (4–7). Within the ER, CRT fulfills at least two important roles. Firstly, it is one of the main calcium-binding proteins of the ER, accounting for near one-half of the total calcium-loading capacity of this organelle. On the other hand, due to its lectin activity, CRT retains misfolded glycoproteins, folding intermediates, or partially assembled oligomers, preventing their aggregation and assisting their conformational maturation (8–10). This function is shared by its membrane-bound paralog, calnexin (CNX). These lectins display 39% sequence identity and have indistinguishable binding specificity for monoglucosylated high mannose glycans (9, 11). In addition, CNX and CRT collaborate with ERp57, a protein disulfide isomerase that promotes correct disulfide bonding along the folding of glycoproteins (12, 13).
Eukaryotic cells in culture can survive in the absence of CNX and/or CRT (14, 15). Experimental evidence points to a functional redundancy between both chaperones. For instance, loss of CRT has only marginal consequences on folding efficiency, and retention of incompletely folded glycoproteins is impaired only after abolishing their association with both lectins (16). By contrast, CRT deficiency in mice is embryonically lethal, and cells derived from CRT knockout embryos have impaired Ca2+ homeostasis (17). Surprisingly, CRT-deficient mice can be rescued by overexpression of activated calcineurin (18), suggesting that lethality is a consequence of calcium signaling impairment.
CRT may be divided into three structural domains: N-terminal domain (residues 1–180), proline-rich or P-domain (residues 181–290), and the C-terminal domain (residues 291–400) (19). The N-terminal domain contains the monoglucosylated oligosaccharide binding site, and it is predicted to display eight anti-parallel β-strands. The crystal structure of CNX reveals a single calcium ion located on this domain (20). An analogous site is expected to be present in CRT, accounting for its high affinity low capacity calcium-binding activity. The three-dimensional structure of the P-domain has been solved by NMR (21, 22) and comprises an unusual extended hairpin fold bearing the ERp57 binding site. This arm structure is most likely curved toward the N-domain as it was observed in CNX, shaping a partially solvent-shielded cavity where the incompletely folded glycoprotein would be lodged. The C-terminal domain presents a strikingly high content of acidic residues and accounts for the high capacity (∼20 mol of calcium/mol of CRT) and low affinity (Kd ∼ 2 mm) calcium-binding activity of CRT. This domain is dispensable for CRT lectin activity (23, 24) but is critical for the calcium-buffering properties of the protein (25). On the other hand, it has been suggested that this domain regulates the intracellular localization of CRT. For instance, CRT retention within the ER is not only accomplished by its KDEL C-terminal signal, but also by its C-terminal domain (26). Besides, the presence of CRT in the cytoplasm (4–7) seems to be the consequence of a transport process triggered only in the presence of the C-terminal domain (27).
The only available structural information regarding the C-terminal domain has been recently provided by small angle x-ray scattering, which suggested a globular fold (28). Here we explored the structural effects exerted by calcium upon the C-terminal part (CRT C-t) of this domain. Noticeably, conformational rearrangements in CRT C-t occur within a range of calcium concentrations similar to that spanned during ER physiological calcium fluctuations. Moreover, the structural modulation exerted by calcium on a peptide displaying a randomized sequence of CRT C-t is markedly different, illustrating the sequence specificity of the effects herein observed. Our results place CRT in a new scenario in which the protein behaves not only as a calcium-binding protein but as a finely tuned ER low affinity calcium sensor.
EXPERIMENTAL PROCEDURES
Materials
Most chemicals were purchased from Sigma. Polyglutamic acid (Sigma product P4636) is a homopolymer with an average molecular mass of 3–15 kDa. Alexa Fluor 350 maleimide was obtained from Invitrogen. Full-length mature CRT from rabbit endowed with a His tag at its C terminus cloned in pBAD vector was used as the starting material for further constructions. The sequence of mature CRT devoid of signal peptide was used for residue numbering. CRT mutants in which a tryptophan (CRT-Trp-402) or cysteine (CRT-Cys-402) residues were incorporated beyond the KDEL ER retention signal were generated by the inverse PCR method with primers 5′-CATCATCATCATCATCATTGAGTTTAAACGGTCTCCAGC-3′ and 5′-CCACAGCTCGTCCTTGGCCTGGCC-3′ or 5′-GCACAGCTCGTCCTTGGCCTGGCC-3′, respectively. Glutamine at position 344 was mutated to cysteine (CRT-Q344C) with primers 5′-GAAAGACAAGTGCGACGAGGAGCAGCGG-3′ and 5′-ATCTGCTTCTCGGCCGTCTTGGTGACGCC-3′, whereas cysteine at position 146 (CRT-C146A) was mutated to alanine using primers 5′-AAGGACGACGAGTTCACACACCTG-3′ and 5′-GGCACGGATGTCCTTGTTGATCAG-3′. Wild-type and mutant CRT proteins were expressed in Escherichia coli and purified as previously described (11). Peptides used to assess sequence specificity were custom-synthesized (Life Tein LLC). Peptide Rd (EDECEAEEEKLEKEEKQEEEEEDDKDKADGKEDRLEEREEDDKKDEDAEEKEQDEAEADKEW) consists of a random sequence of identical amino acid composition as CRT C-t. Peptide Rd-N (EDECEAEEEKLEKEEKQEEEEEDDKDW) corresponds to the N-terminal half of Rd, whereas peptide Wt-N (KDKCDEEQRLKEEEEEKKRKEEEEAEW) and Wt-C (EDECDKDDKEDEDEDEEDKDEEEEEAW) correspond to the N- and C-terminal halves of CRT C-t, respectively. To make these peptides suitable for RET experiments a cysteine residue was placed at the fourth position and a tryptophan residue at the end of each peptide. The His tag present at the C-terminal end of CRT was not included on the synthetic peptides.
Chemical Cleavage
Wild-type or mutant CRT was dissolved in 70% formic acid at a concentration of 10 mg/ml. A 100-fold molar excess of crystalline cyanogen bromide was added, and the mixture was incubated in the dark for 24 h at room temperature. The reaction mixture was diluted 10-fold in water and lyophilized. Proteolytic fragments containing the C terminus were isolated through an immobilized Ni2+ affinity chromatography. The fragment containing the sequence KDKQDEEQRLKEEEEEKKRKEEEEAEEDEEDKDDKEDEDEDEEDKDEEEEEAAAGQAKDELHHHHHH was further purified using a Superdex 75 column developed with 20 mm Tris-HCl, 150 mm NaCl, pH 7.5 (buffer A). Its identity was confirmed by N-terminal sequencing and mass spectrometry.
Protein Labeling
Conjugation reaction of Alexa Fluor 350 maleimide (A30505, Invitrogen) was performed according to the instructions of the manufacturer. Briefly, 10 μl of a freshly prepared aqueous stock solution of Alexa Fluor 350 (10 mg/ml) was added to previously reduced protein (20 μm) with 1 mm dithiothreitol dissolved in 25 mm Tris-HCl. The reaction was allowed to proceed in the dark for 2 h at room temperature. Excess probe was quenched by adding a 100-fold molar excess of dithiothreitol. The labeled protein was further purified using a gel filtration Superdex 75 column developed with buffer A.
Spectroscopic Measurements
Far-UV CD measurements were carried out at 25 °C on a Jasco J-815 spectropolarimeter. All measurements were done with protein samples dissolved in buffer A with the addition of CaCl2 at the indicated concentration. The path length of the cell employed was 1 cm, and protein concentration was 10 μm (0.1 mg/ml). Spectra shown are averages of at least 8 scans, which were acquired over the wavelength range of 190–260 nm. Single value decomposition analysis (29, 30) of the CD spectra was carried out with Matlab by building a matrix with the set of CD spectra in the range of 203–260 nm at 0.1 nm intervals. This matrix can be decomposed into the product of three matrices, A = USVT, where S represents a matrix of singular values, U is the matrix of basis vectors, and V is the matrix of amplitude coefficients. Basis spectra were obtained from the US matrix. Significant components were selected as those showing an autocorrelation value above 0.3 and taking into account the magnitude of their singular values. Fluorescence emission spectra were measured at 25 °C in a Jasco FP-6500 spectrofluorometer. The protein concentration used was 3 μm, and the sample was dissolved in buffer A with the addition of the indicated CaCl2 concentration. Excitation wavelength was set to 280 nm, and spectra were recorded between 290 and 500 nm. An average of at least six scans was used for final calculations. Spectra were corrected for dilution effects, and the final dilution of the sample was always <10%. R0 for the tryptophan-Alexa Fluor 350 pair was calculated by using the equation, R0 = 0.211(κ−2n−4QDJ(λ))⅙, where κ is a factor describing the relative orientation of the transition dipoles between the donor and the acceptor (a random orientation was assumed for this calculation, κ2 = 2/3), n is the refractive index of the medium, QD is the quantum yield of the donor in the absence of the acceptor, and J(λ) is the overlap integral between the donor emission and acceptor absorption spectra.
Stokes Radius Measurements
The dependence of CRT hydrodynamic radius with calcium was studied using size-exclusion chromatography (SEC) and dynamic light scattering (DLS). In both experiments the protein was dissolved in 30 mm Tris-HCl, pH 7.5, 300 mm NaCl with the addition of the indicated concentration of CaCl2. SEC was performed in a Superdex 200 column (Amersham Biosciences Pharmacia) at a flow rate of 0.5 ml/min. Protein elution was monitored by absorbance at 280 nm, and the column was calibrated using globular protein standards of known Stokes radii. DLS was measured with a Malvern Nanos S instrument (model ZEN 1600). Diameter distribution was calculated from the average of 20 measurements.
TFE Titrations
2,2,2-Trifluoroethanol (TFE) titrations were followed by monitoring the CD spectra at 25 °C. Protein concentration employed was 10 μm, and the final calcium concentration was achieved by a one-step addition of a CaCl2 stock solution. CD spectra were corrected for protein concentration and expressed in molar ellipticity (per residue) [θ]MRW = θ/(10 × C × l × n), where θ is the observed ellipticity in millidegrees, C is the molar concentration, l the path length in centimeters, and n the number of peptide bonds.
Molecular Modeling
Molecular dynamics simulations of a twelve-residue-long peptide representative of the complete domain (EEDEEDKDDKED) or a scrambled version of this peptide (EDEEEKKDDEDD) were performed using the GROMACS program and the ffG53a6 force field (31). Both systems include 8950 water molecules, 4 Ca2+ cations, or 12 Na+ and 4 Cl−; both are electroneutral, and they display the same ionic strength. Each peptide was located in a cubic box (6.45 nm side) with periodic boundary conditions, and we employed the smooth particle mesh Ewald method to treat the long range electrostatic interactions (32). In these simulations Ca2+ and Na+ concentrations were 24.8 mm and 74.4 mm, respectively, while the Cl− concentration was 24.8 mm. The temperature and pressure were set at 300 K and 1 atm, respectively. This was achieved with the Berendsen thermostat and barostat (33, 34). A time step of 1 fs was employed along the simulations. The initial configuration was generated by adding water molecules and ions to the peptide arranged in an extended geometry. Statistical information was collected for 15 ns, following an initial equilibration period of 5 ns.
Fourier Transform Analysis of Amino Acid Sequences
Amino acid sequences of the C-terminal domain of CRT of different species were converted into numerical strings, where +1, −1, and 0 correspond to basic (RK), acidic (DE), and other amino acids, respectively. These strings in the sequence length domain were then Fourier-transformed into the frequency domain (the abscissa represents frequency = 1/period of the amino acid repeat) with a macro implemented in MS Excel. Finally, to allow for a straightforward comparison among amino acid sequences of different length, the frequency axis was normalized for the total number of amino acids in each sequence. Randomized peptides of the same amino acid composition were run as control samples.
RESULTS
CRT C-t Secondary Structure Is Affected by Calcium
To study the effect of calcium on CRT C-t, we first employed CD spectroscopy. The far-UV CD spectrum of full-length CRT is shaped mainly by the signals arising from the β-sheets in the N-terminal domain, eclipsing any signal from the C-terminal domain (23). This prompted us to measure the spectrum of the isolated domain. Several attempts to express this polypeptide in E. coli failed, probably due to intracellular proteolysis. Because the last methionine residue (Met-340) of CRT lies at the beginning of CRT C-t, we used chemical proteolysis with CNBr to isolate it. The resulting fragment was purified by immobilized metal affinity and SEC and characterized by N-terminal sequencing and mass spectroscopy.
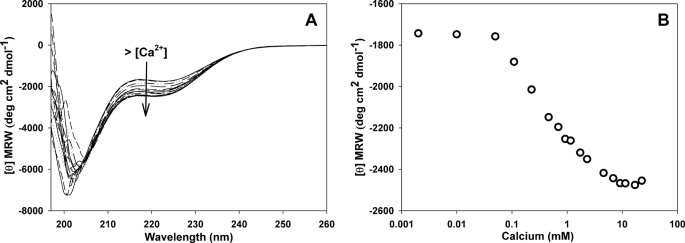
The spectrum of the domain recorded in the absence of calcium shows a negative band near 200 nm (Fig. 1A), indicative of a strong contribution from disordered structural elements and a negative band in the 217–227 range that suggests the presence of α-helical components. Interestingly, calcium addition enhances the intensity of this band and causes a signal reduction of the band centered at 200 nm. Ellipticity values at 222 nm measured for each calcium concentration tested are shown in Fig. 1B. This result shows that the CRT C-t structure is sensitive to calcium, shifting from a disordered structure at low Ca2+ concentrations to a more folded one when calcium is raised. Remarkably, the midpoint of this transition is 400 μm Ca2+, which is near the average calcium level within the ER.
FIGURE 1.
Effect of calcium on CRT C-domain secondary structure. A, far-UV CD spectra at increasing calcium concentrations, ranging from 5 μm to 20 mm. B, molar ellipticity at 222 nm measured at increasing calcium concentrations.
TFE is an extensively employed structure-stabilizing solvent (35, 36). Upon TFE addition, a dramatic change in the CD spectra was observed, pointing at the induction of an α-helical conformation (supplemental Fig. S1A). To further probe the structural effects of calcium, far-UV CD spectra were recorded at increasing concentrations of TFE in the presence of 20 mm calcium. Under this condition, structural changes induced by TFE on the C-domain are enhanced by the presence of calcium. The absolute value of mean residue ellipticity measured at 222 nm is higher for every TFE concentration tested (supplemental Fig. S1B), pointing to a cumulative effect of both added substances.
Secondary Structure Effects Are Sequence Dependent
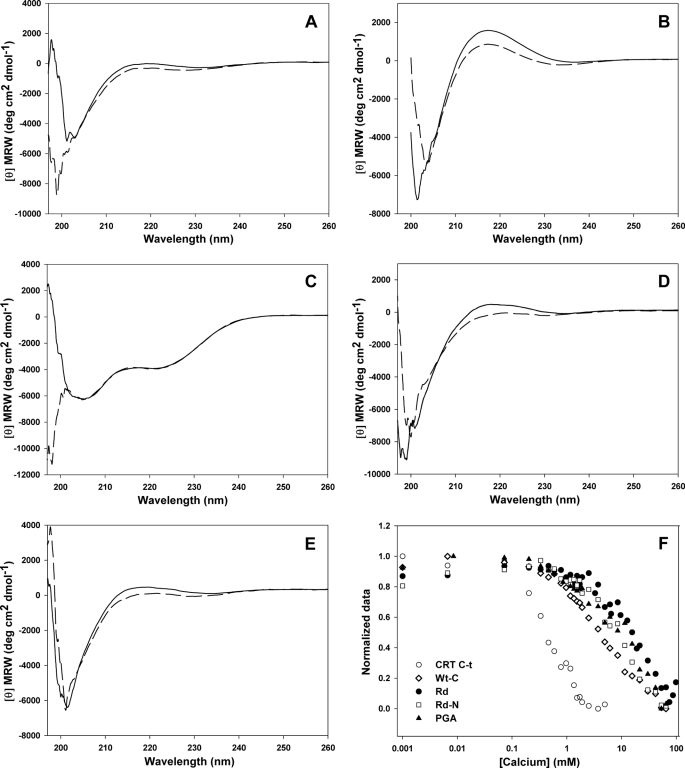
If the behavior of CRT C-t were solely a consequence of its high negative charge, any similar peptide would experience an analogous transition. To address this issue we employed two synthetic polypeptides. First we used a peptide displaying a scrambled sequence of CRT C-t (Rd, see sequence in Table 1). Upon calcium addition, Rd showed a small wavelength shift of the negative band centered at ∼200 nm (Fig. 2A). Although this behavior was similar to that observed with CRT C-t (Fig. 1A), the calcium concentration needed to attain a comparable response was ∼20 times higher (Fig. 2F). Upon calcium binding, the polyglutamic acid (PGA) spectrum also showed a small shift of the 200 nm negative band and a decreased intensity of the positive band centered at 216 nm (Fig. 2B). Similarly to Rd, the conformational transition of this last peptide needed a higher calcium concentration than CRT C-t (Fig. 2F). Therefore, both Rd and PGA, although being equally or more negatively charged than CRT C-t, are expected to be permanently unstructured in the ER environment.
TABLE 1.
Sequence of the peptides employed
| Peptide | Sequence |
|---|---|
| CRT C-t | KDKCDEEQRLKEEEEEKKRKEEEEAEEDEEDKDDKEDEDEDEEDKDEEEEEAAAGQAKDELWHHHHHH |
| WT-N | KDKCDEEQRLKEEEEEKKRKEEEEAEW |
| WT-C | EDECDKDDKEDEDEDEEDKDEEEEEAW |
| Rd | EDECEAEEEKLEKEEKQEEEEEDDKDKADGKEDRLEEREEDDKKDEDAEEKEQDEAEADKEW |
| Rd-N | EDECEAEEEKLEKEEKQEEEEEDDKDW |
FIGURE 2.
Effect of calcium on the secondary structure of C-t-related peptides. Continuous line: CaCl2 5 μm; dashed line: CaCl2 50 mm. A, peptide Rd, displaying a randomized sequence of CRT C-t. B, pGA. C, peptide Wt-N, displaying the N-terminal half of CRT C-t. D, peptide Wt-C, displaying the C-terminal half of CRT C-t. E, peptide Rd-N, displaying the N-terminal half of Rd. F, calcium effect measured at 222 nm from the spectra shown in panels A–E.
To study the contributions from different CRT C-t regions on its secondary structure we employed the peptides Wt-N and Wt-C (Table 1). These peptides encompass the N-terminal and C-terminal halves of the parent molecule, respectively. Interestingly, Wt-N displayed an α-helical conformation, which was not affected by calcium (Fig. 2C). On the other hand, Wt-C displayed a random conformation, which, upon calcium addition, underwent a transition similar to that observed with the complete domain but at higher concentrations (Fig. 2, D and F). By contrast, the N-terminal half of Rd (named Rd-N, see Table 1) showed a behavior similar to that of its parent molecule (Fig. 2, E and F). The CRT C-t spectrum at low calcium concentration could be recreated by adding the spectra of its constituent fragments (supplemental Fig. S2A), whereas an analogous procedure using the spectra recorded at high calcium concentration did not fully reconstitute the spectrum of the domain (supplemental Fig. S2B). This observation indicates that at high calcium concentration long range interactions within CRT C-t occur, with consequences on the ion-binding activity.
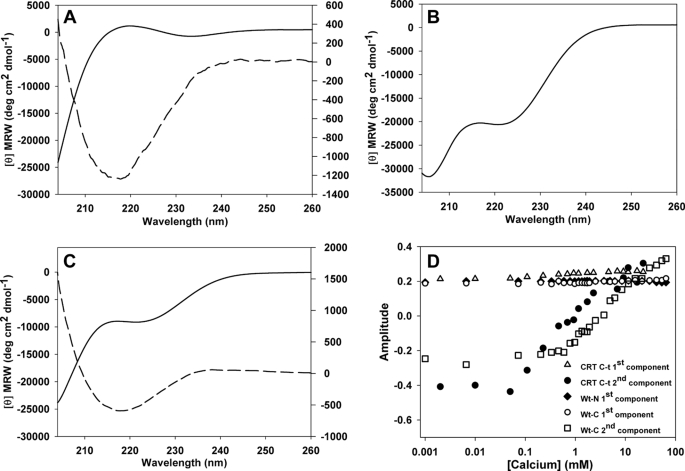
The calcium dependence of Wt-C, Wt-N, and CRT C-t CD spectra was studied using singular value decomposition analysis. This procedure revealed that the Wt-C spectrum results from two basic components (Fig. 3A). The main component displays the typical shape of the spectrum generated by a disordered conformation, whereas the second component is similar to that produced by a β-sheet. Besides, the amplitude of this last component increases with calcium concentration, suggesting that the observed change is due to the emergence of a more ordered conformation (Fig. 3D). On the other hand, the spectra of Wt-N can be ascribed to a single invariant component typical of an α-helix (Fig. 3B). In this case, the other components make little or no contribution to the overall spectrum. Finally, the spectrum of CRT C-t can be mainly explained by two components (Fig. 3C). The first one remained constant along the experiment, and its shape is the result of a mixture of an α-helix conformation with disordered regions, while the second one was similar to a β-sheet component and its contribution increased upon calcium addition (Fig. 3D). Taken together, the analysis suggests that the structural change suffered by CRT C-t is mainly due to its C-terminal half, which starts displaying a β-sheet-like structure upon Ca2+ binding.
FIGURE 3.
Analysis of calcium effect on far-UV CD spectra of CRT C-t and its constituent fragments. Basic spectra of the main component (continuous line, left axis) and of the second most prominent component (dashed line, right axis) of CRT-Ct (A), Wt-N (B), and Wt-C (C). D, amplitude vectors at increasing calcium concentrations for the components shown in A–C.
The CRT C-terminal Domain Becomes More Compact in the Presence of Calcium
To further study the effect of calcium, we developed an intramolecular RET assay. To this end, fluorophore-flanked CRT C-t was engineered by adding a Trp residue at the C terminus of the KDEL retention signal and chemically attaching the probe Alexa Fluor 350 to a cysteine residue introduced at position 344. With this setup, RET experiments were performed using the Trp as the energy donor and Alexa Fluor 350 as the acceptor. Fluorescence emission maximum of this construct was at 350 nm, in agreement with a solvent-exposed position of its Trp residue. Fluorescence emission intensity at 350 nm was measured for the construct at increasing calcium concentrations either in the presence or in the absence of the acceptor (Fig. 4). This experiment indicates the induction of a more compact structure. The efficiency of the energy transfer process was calculated using the equation, E = (1 − (FDA/FD)) (1/fA), where FDA and FD are the fluorescence intensities of the donor in the presence or absence of acceptor, respectively, and fA is the fraction of labeled protein. Considering the maximum value obtained for RET efficiency and the calculated R0 for the two fluorophores (21 Å), the end-to-end distance of CRT C-t at 10 mm Ca2+ was ∼25 Å, whereas in the absence of the ion it was ∼35 Å.
FIGURE 4.
RET measured for the isolated C-t domain and its related peptides. Normalized data are the ratio between emission at 350 nm of a Trp residue located at the C terminus with Alexa Fluor 350 chemically attached to a Cys residue at the N terminus and its emission in the absence of acceptor.
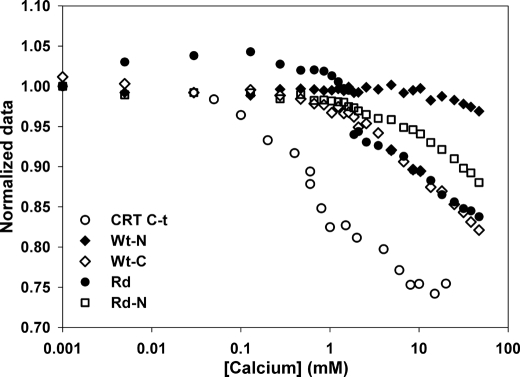
A similar RET experiment performed with the scrambled peptide Rd and the shorter versions Wt-C and Rd-N showed a calcium-induced compaction that occurred at a higher concentration than with CRT C-t, in agreement with the CD measurements. On the other hand, calcium addition did not affect the RET efficiency of Wt-N, probably because it was already structured in the absence of the cation (Fig. 4).
C-terminal Domain Conformation in Full-length CRT
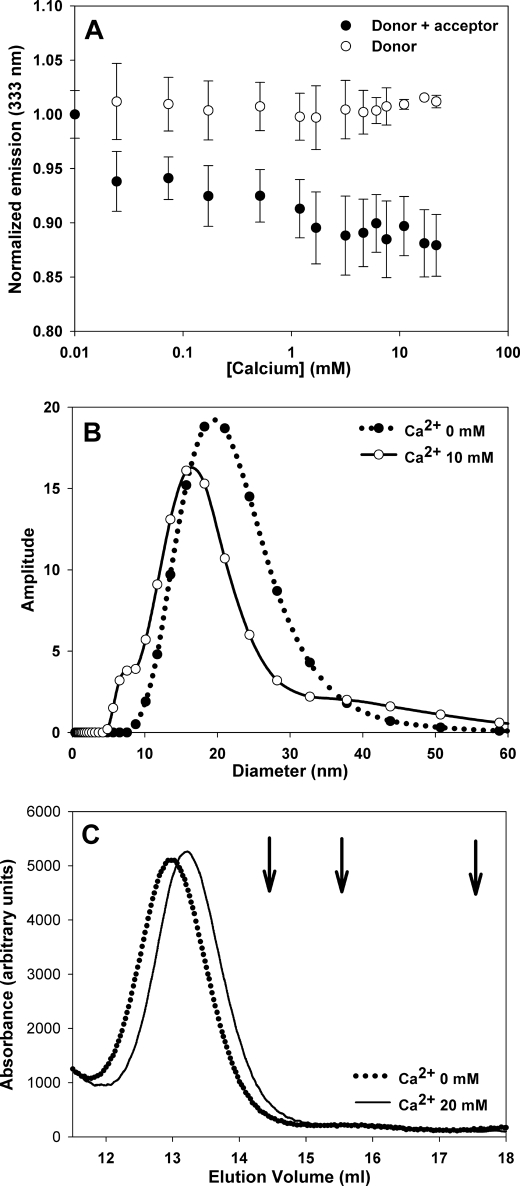
Although CD and RET experiments using the isolated C-terminal domain support a similar view, its validity in the context of the full-length protein is arguable. To address this issue we employed a similar approach based on intramolecular RET. To this end, a CRT endowed with a Cys residue at position 402 and carrying a C146A mutation was engineered. This last residue is the only free cysteine present in CRT, and its mutation ensured that only Cys-402 is chemically modified with Alexa Fluor 350. Fluorescence emission intensity at 333 nm was measured in the presence or in the absence of the acceptor for increasing calcium concentrations (Fig. 5A). Interestingly, the overall behavior is similar to that observed with the isolated domain, pointing to a more compact structure of CRT in the presence of calcium. It is worth mentioning that in this construct only a fraction of the tryptophan residues is expected to be located at a distance suitable for energy transfer, thus explaining the lower RET efficiencies observed by comparison with the previous experiment. Besides, the varied distances of Trp residues to the acceptor preclude any quantitative conclusion regarding the degree of compaction.
FIGURE 5.
Effect of calcium on full-length CRT. A, RET measured for C-domain in the full-length CRT. Normalized emission at 333 nm with increasing calcium concentrations for CRT with (full circles) or without (empty circles) Alexa Fluor 350 chemically attached to a Cys residue located at the C-terminal end of the protein. B and C, diameter distribution measured by DLS and SEC elution profile of CRT in the presence of 5 μm or 10 mm CaCl2 (C). The arrows indicate the elution volume of molecular mass standards. From left to right: aldolase (138 kDa), bovine seroalbumin (67 kDa), and ovalbumin (43 kDa).
In addition, the effect of calcium on the hydrodynamic properties of full-length CRT was studied by DLS and SEC. The diameter distribution measured by DLS was centered at 20.5 and 17.8 nm in the absence and in the presence of 10 mm CaCl2, respectively (Fig. 5B). In addition, the size distribution in the absence of calcium was broader, suggesting a higher flexibility. A similar behavior was observed with SEC, in which protein elution was delayed in the presence of calcium, suggesting a more compact conformation (Fig. 5C). This effect is specific for CRT, because the elution volume of proteins used as standards remained unchanged in the presence of calcium.
Molecular Dynamics
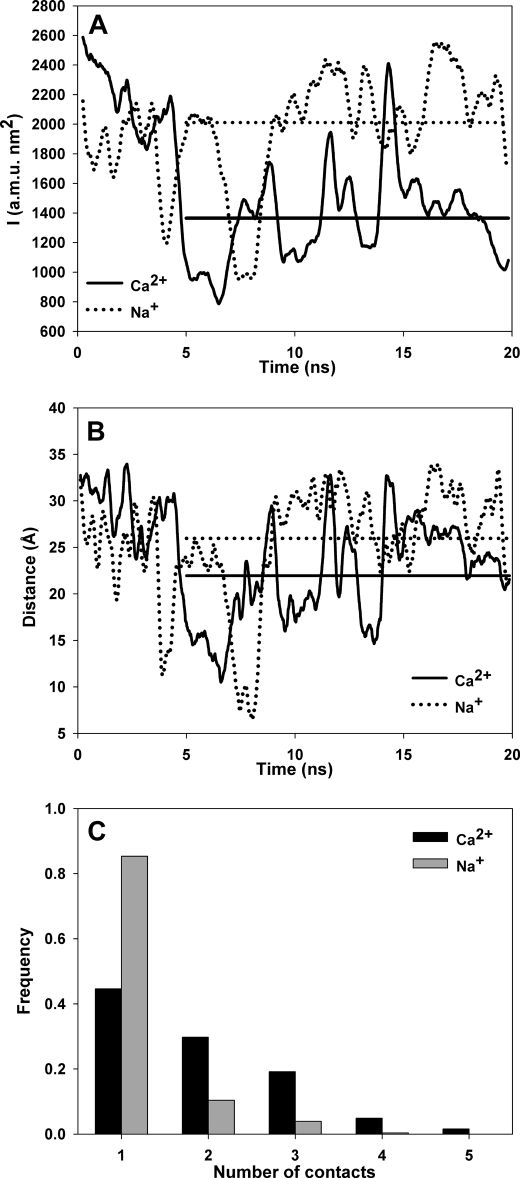
The spectroscopic experiments showed that calcium induces a more rigid and compact conformation on CRT C-t. To gain insight into the mechanism underlying this process we employed molecular dynamic simulations. For this purpose we simulated the behavior of a 12-residue-long peptide that is representative of a complete domain (EEDEEDKDDKED). A 20-ns simulation was performed in the presence of either 12 Na+ plus 4 Cl− or 4 Ca2+. The ionic strength was identical in both simulations (99.2 mm), and the calcium concentration was similar to the highest concentration employed in the spectroscopic measurements (24.8 mm). The simulations started from an extended conformation, and statistical analyses of the trajectories were done using data produced after an equilibration time of 5 ns. A visual inspection reveals differences between the simulations in the presence of calcium or sodium (see supplemental movies 1 and 2, respectively). In both cases many events of ion binding and detaching were observed, assuring that the statistics drawn from the trajectories were reliable. The simulation in the presence of calcium reveals on average a more compact conformation than that obtained in the presence of sodium. A quantitative analysis showed that the average moments of inertia were 1896 and 1365 atomic mass units nm2 for the simulations performed in the presence of sodium or calcium, respectively (Fig. 6A). Moreover, the average end-to-end distance of the peptide diminishes from 26.2 Å in the presence of sodium to 22.0 Å in the presence of calcium (Fig. 6B). These results qualitatively agree with the spectroscopic experiments using the complete domain. The distinctive effect of calcium originates from its ability to bind several residues simultaneously, thus bridging residues distant in the primary structure. By contrast, sodium usually makes single contacts with the peptide and, therefore, has a lesser effect on its conformation (Fig. 6C). In addition, analogous simulations performed with a peptide of identical composition but with different sequence (EDEEEKKDDEDD) showed no major differences in either compaction or moments of inertia between Ca2+ and Na+ (supplemental Fig. S3, A–C).
FIGURE 6.
Molecular dynamics simulation of a peptide representative of CRT C-t. A, moment of inertia and B, end-to-end distance in the presence of calcium (solid line) or sodium (dotted line). Straight horizontal lines are the calculated average values for each condition after 5 ns of equilibration. C, number of ion-peptide contacts for calcium (black bars) and sodium (gray bars).
DISCUSSION
The effect of calcium on CRT has been studied using different approaches. In general, calcium binding increases CRT stability, as revealed by chaotrope-induced denaturation, thermal stability, and protease susceptibility experiments (23, 37–39). These effects are probably a consequence of calcium binding to the N-terminal domain, because, in general, they are observed when calcium concentration varies within the micromolar range. Interestingly, it has been shown that calcium depletion or C-terminal domain removal induces a conformation that exhibits increased affinity for hydrophobic peptides (40). Regarding the role of the C-terminal domain on the lectin activity of CRT, we recently showed that its deletion does not affect the binding constant for Glc1Man9GlcNAc2 oligosaccharide nor its chaperone activity (23). This implies that the lectin-chaperone and Ca2+-buffering activities of CRT are mutually independent.
Calcium concentration in the ER lumen can vary over a wide range, from 1–5 mm when it is full to 10–50 μm after the opening of the ER membrane calcium channels (41, 42). The calcium dissociation constant of CRT C-terminal domain is ∼2 mm (25), bringing about the possibility that in vivo physiological fluctuations could affect its structure and dynamics. The only available structural information regarding this domain was obtained using small angle x-ray scattering, which revealed a globular form (28). Given the high content of negatively charged residues, a more extended conformation would be expected. However, the presence of several intervening positively charged amino acids, which partly dampen the internal electrostatic repulsion, could favor a more compact conformation. Interestingly, this pattern of negatively and positively charged residues is conserved among different species (supplemental Table 1). In this vein, the presence of calcium could further neutralize the remaining negative charge, inducing an even more compact structure.
The far-UV CD spectrum of CRT C-t showed the distinctive pattern of a natively unfolded domain, although the negative band centered at 220 nm points also to a contribution from α-helical elements. We should mention that this experiment had to be performed with the isolated domain, because the spectrum of full-length CRT is mainly shaped by the strong signals arising from the β-sheets of the N-domain (23). Calcium addition induced a shift to a more ordered structure, in which contributions from β-sheet-like components became evident, as assessed by singular value decomposition analysis. This observation does not necessarily imply that the C-terminal domain adopts a bona fide β-sheet structure. Our interpretation is that some residues begin to populate conformations with the characteristic dihedral angles of a β-sheet, in a very dynamic process. Regarding the sequence specificity of this effect, it may be argued that any domain displaying an equivalent negative charge would behave similarly. Nevertheless, the fact that PGA and Rd needed a much higher calcium concentration to become structured shows that sequence plays an important role in the fine tuning of this phenomenon. In this sense, the distribution of charged residues along CRT C-t seems to favor a disorder-to-order transition at lower calcium concentration, as if the domain would be primed to be easily structured. Moreover, a Fourier transform of the charge distribution along sequences of CRT C-t from different species reveals a pattern with a period of ∼7 residues, a feature absent in Rd (supplemental Fig. S4). This analysis shows that charge distribution along CRT C-t is not random.
By splitting CRT C-t we could address the role played by each half of the domain on its behavior. The secondary structures of Wt-N and Wt-C in the absence of calcium are markedly different, the former being an α-helix and the latter a disordered structure. This behavior is in good agreement with a prediction of α-helix propensity made with Agadir (supplemental Fig. S5 (43)). At low calcium the CD spectrum of CRT C-t can be reconstructed by adding the spectra of its constituent parts, suggesting that each half behaves independently from the other. Interestingly, this behavior no longer holds at high calcium concentration, probably due to long range interactions within the domain. Moreover, calcium induced the emergence of β-sheet-like elements in both CRT C-t and Wt-C, although the former required a lower concentration than the latter. By contrast, the α-helical structure of Wt-N was not affected by calcium. These observations show that the structural response of CRT C-t is mainly due to changes on its C-terminal half, whereas the N-terminal part would adjust its sensitivity to a specific range of calcium concentrations.
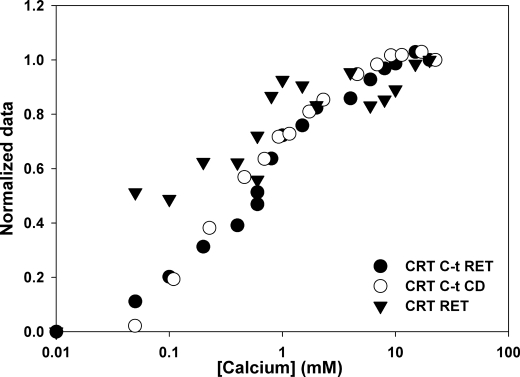
RET experiments show that calcium stabilizes a more compact structure. Using a Förster distance (R0) of 21 Å for the Trp-Alexa Fluor 350 pair, it can be estimated that the end-to-end distance of the domain evolves from around 35 Å at low calcium to 25 Å as calcium concentration increases to 5 mm. Interestingly, Ca2+ concentration needed to compact Wt-C, Rd, or Rd-N was much higher than that needed for CRT C-t, in agreement with the CD experiments. In addition, Wt-N did not show a significant degree of compaction even at Ca2+ concentrations as high as 100 mm. These observations illustrate the importance of both sequence and long range interactions in the behavior of CRT C-t. The specificity of the RET procedure prompted us to set up an experiment to study the behavior of the C-terminal domain within the context of the complete protein. When a full-length CRT bearing only one free cysteine located at the C terminus was labeled with Alexa Fluor 350, a quenching effect on Trp fluorescence could be observed with increasing calcium concentration. This result shows that the C-terminal domain becomes more compact. Nevertheless, we should mention that the RET efficiency with the full-length protein is lower than that observed with the isolated domain. This is an expected consequence of CRT displaying 11 Trp residues, many of them located too far from the acceptor for noticeable RET to occur. This heterogeneous population precludes any quantitative conclusion about the degree of calcium-induced compaction in the full-length protein. As additional tools to study the size of CRT we employed DLS and SEC. Both techniques revealed that calcium induces a more compact structure on CRT, in agreement with the RET experiment. Nevertheless, the hydrodynamic radius and the degree of compaction induced by calcium estimated from both techniques did not fully agree. SEC renders a hydrodynamic radius similar to that measured by others (∼5 nm (44)) and a 2% decrease in radius, whereas DLS renders an average value of ∼9 nm for the hydrodynamic radius and a 10% compaction. SEC and DLS measure molecular volume and diffusion coefficient, respectively. Although these parameters are related, given the dissimilar nature of these experiments and the highly asymmetric structure of CRT, a quantitative difference between both methodologies is not surprising. Comparison of the results obtained with CRT using the different spectroscopic techniques shows that the conformational changes detected either by CD or RET occur over the same range of calcium concentrations (Fig. 7). This indicates that the ordering effect observed on the secondary structure parallels the induction of a more compact structure. Besides, the agreement between the data obtained using full-length CRT and the isolated peptide points to a similar process taking place in both molecules.
FIGURE 7.
Normalized data obtained from CD and RET experiments using the isolated C-domain or full-length CRT.
The mechanism underlying the structural effects exerted by calcium was explored using molecular dynamics simulation. Overall, results obtained with this approach are in good agreement with the spectroscopic measurements. In the simulations performed in the presence of calcium the peptide adopts on average a more compact conformation. The reason for this behavior rests on the ability of calcium to simultaneously coordinate several acidic residues of the peptide, bringing into close proximity residues that are distant in the primary sequence. Although other divalent cations could be expected to induce similar effects, this would be irrelevant in the biological system addressed here. We should point out that the degree of calcium-induced compaction observed in the simulation (∼14%) is smaller than that measured by RET on the complete domain (∼29%). This discrepancy is probably a consequence of long range effects that are missing in the short peptide and the different length of the molecules used in each experiment.
Remarkably, the disorder-to-order transition occurs when calcium concentration varies within a range similar to that spanned during physiological fluctuations in the ER. As we mentioned above, this response is highly dependent on the primary structure. In this sense, it is likely that the insertion of positive residues at strategic positions played a major role in the fine tuning of this phenomenon. This brings about the possibility that signals that mobilize ER calcium could trigger a conformational change for the CRT C-terminal domain. As we mentioned above, the main roles of CRT in the ER are as lectin-chaperone and calcium buffer. It is noteworthy that both activities are mutually independent (23), opening the question about the biological significance of maintaining these two functions on the same polypeptide. We speculate that the structural response of this domain to calcium fluctuations influences CRT subcellular localization. Some lines of evidence support this hypothesis. CRT retention in the ER is accomplished by a KDEL retrieval signal. However, a protein lacking this signal is also located in the ER, whereas deleting the C-terminal domain (26, 27) or perturbing ER calcium induces CRT relocalization (45). In the light of the results presented here, it will be interesting to study the effect of calcium concentration on these processes and its relevance for cell physiology. Experiments to address these issues are currently underway in our laboratory.
Supplementary Material
Acknowledgments
cDNA encoding rabbit CRT was a generous gift from Dr. Marek Michalak. Spectroscopic experiments were performed with equipment provided by Dr. Gonzalo Prat Gay. Technical assistance by Susana Raffo and Mariángeles González is greatly appreciated.
This work was supported, in whole or in part, by National Institutes of Health Grant GM044500. This work was also supported by the Howard Hughes Medical Institute, by the University of Buenos Aires, and by the National Agency for the Promotion of Science and Technology (Argentina).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, Table S1, and Movies 1 and 2.
- CRT
- calreticulin
- DLS
- dynamic light scattering
- CNX
- calnexin
- ER
- endoplasmic reticulum
- PGA
- polyglutamic acid
- Rd
- a scrambled version of CRT C-terminal domain
- RET
- resonance energy transfer
- SEC
- size-exclusion chromatography
- TFE
- 2,2,2-trifluoroethanol
- C-t
- C-terminal.
REFERENCES
- 1.Mery L., Mesaeli N., Michalak M., Opas M., Lew D. P., Krause K. H. (1996) J. Biol. Chem. 271, 9332–9339 [DOI] [PubMed] [Google Scholar]
- 2.Labriola C., Cazzulo J. J., Parodi A. J. (1999) Mol. Biol. Cell 10, 1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michalak M., Corbett E. F., Mesaeli N., Nakamura K., Opas M. (1999) Biochem. J. 344, 281–292 [PMC free article] [PubMed] [Google Scholar]
- 4.Burns K., Duggan B., Atkinson E. A., Famulski K. S., Nemer M., Bleackley R. C., Michalak M. (1994) Nature 367, 476–480 [DOI] [PubMed] [Google Scholar]
- 5.Dedhar S., Rennie P. S., Shago M., Hagesteijn C. Y., Yang H., Filmus J., Hawley R. G., Bruchovsky N., Cheng H., Matusik R. J., Giguère V. (1994) Nature 367, 480–483 [DOI] [PubMed] [Google Scholar]
- 6.Coppolino M. G., Woodside M. J., Demaurex N., Grinstein S., St-Arnaud R., Dedhar S. (1997) Nature 386, 843–847 [DOI] [PubMed] [Google Scholar]
- 7.Holaska J. M., Black B. E., Rastinejad F., Paschal B. M. (2002) Mol. Cell. Biol. 22, 6286–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nauseef W. M., McCormick S. J., Clark R. A. (1995) J. Biol. Chem. 270, 4741–4747 [DOI] [PubMed] [Google Scholar]
- 9.Spiro R. G., Zhu Q., Bhoyroo V., Söling H. D. (1996) J. Biol. Chem. 271, 11588–11594 [DOI] [PubMed] [Google Scholar]
- 10.Caramelo J. J., Parodi A. J. (2008) J. Biol. Chem. 283, 10221–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassilakos A., Michalak M., Lehrman M. A., Williams D. B. (1998) Biochemistry 37, 3480–3490 [DOI] [PubMed] [Google Scholar]
- 12.Van der Wal F. J., Oliver J. D., High S. (1998) Eur. J. Biochem. 256, 51–59 [DOI] [PubMed] [Google Scholar]
- 13.Molinari M., Helenius A. (1999) Nature 402, 90–93 [DOI] [PubMed] [Google Scholar]
- 14.Jannatipour M., Rokeach L. A. (1995) J. Biol. Chem. 270, 4845–4853 [DOI] [PubMed] [Google Scholar]
- 15.Parlati F., Dignard D., Bergeron J. J., Thomas D. Y. (1995) EMBO J. 14, 3064–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molinari M., Eriksson K. K., Calanca V., Galli C., Cresswell P., Michalak M., Helenius A. (2004) Mol. Cell 13, 125–135 [DOI] [PubMed] [Google Scholar]
- 17.Nakamura K., Zuppini A., Arnaudeau S., Lynch J., Ahsan I., Krause R., Papp S., De Smedt H., Parys J. B., Muller-Esterl W., Lew D. P., Krause K. H., Demaurex N., Opas M., Michalak M. (2001) J. Cell Biol. 154, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L., Nakamura K., Lynch J., Opas M., Olson E. N., Agellon L. B., Michalak M. (2002) J. Biol. Chem. 277, 50776–50779 [DOI] [PubMed] [Google Scholar]
- 19.Smith M. J., Koch G. L. (1989) EMBO J. 8, 3581–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 21.Ellgaard L., Riek R., Herrmann T., Güntert P., Braun D., Helenius A., Wüthrich K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3133–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frickel E. M., Riek R., Jelesarov I., Helenius A., Wuthrich K., Ellgaard L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conte I. L., Keith N., Gutiérrez-Gonzalez C., Parodi A. J., Caramelo J. J. (2007) Biochemistry 46, 4671–4680 [DOI] [PubMed] [Google Scholar]
- 24.Leach M. R., Cohen-Doyle M. F., Thomas D. Y., Williams D. B. (2002) J. Biol. Chem. 277, 29686–29697 [DOI] [PubMed] [Google Scholar]
- 25.Baksh S., Michalak M. (1991) J. Biol. Chem. 266, 21458–21465 [PubMed] [Google Scholar]
- 26.Sönnichsen B., Füllekrug J., Nguyen Van P., Diekmann W., Robinson D. G., Mieskes G. (1994) J. Cell Sci. 107, 2705–2717 [DOI] [PubMed] [Google Scholar]
- 27.Afshar N., Black B. E., Paschal B. M. (2005) Mol. Cell. Biol. 25, 8844–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nørgaard Toft K., Larsen N., Steen Jørgensen F., Højrup P., Houen G., Vestergaard B. (2008) Biochim. Biophys. Acta 1784, 1265–1270 [DOI] [PubMed] [Google Scholar]
- 29.Henry E. R., Hofrichter J. (1992) Methods Enzymol. 210, 129–192 [Google Scholar]
- 30.Mukhopadhyay K., Basak S. (1998) Biophys. Chem. 74, 175–186 [DOI] [PubMed] [Google Scholar]
- 31.van der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C. (2005) J. Comp. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 32.Essman U., Perela L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) J. Chem. Phys. 103, 8577–8592 [Google Scholar]
- 33.Gunsteren W. F., Berendsen H. J. (1990) Angew. Chem. Int. Ed. Engl. 29, 992–1023 [Google Scholar]
- 34.Berendsen H. J., Postma J. V. (1984) J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 35.Nelson J. W., Kallenbach N. R. (1986) Proteins 1, 211–217 [DOI] [PubMed] [Google Scholar]
- 36.Storrs R. W., Truckses D., Wemmer D. E. (1992) Biopolymers 32, 1695–1702 [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Stafford W. F., Bouvier M. (2001) Biochemistry 40, 11193–11201 [DOI] [PubMed] [Google Scholar]
- 38.Tan Y., Chen M., Li Z., Mabuchi K., Bouvier M. (2006) Biochim. Biophys. Acta 1760, 745–753 [DOI] [PubMed] [Google Scholar]
- 39.Corbett E. F., Michalak K. M., Oikawa K., Johnson S., Campbell I. D., Eggleton P., Kay C., Michalak M. (2000) J. Biol. Chem. 275, 27177–27185 [DOI] [PubMed] [Google Scholar]
- 40.Rizvi S. M., Mancino L., Thammavongsa V., Cantley R. L., Raghavan M. (2004) Mol. Cell 15, 913–923 [DOI] [PubMed] [Google Scholar]
- 41.Miyawaki A., Llopis J., Heim R., McCaffery J. M., Adams J. A., Ikura M., Tsien R. Y. (1997) Nature 388, 882–887 [DOI] [PubMed] [Google Scholar]
- 42.Yu R., Hinkle P. M. (2000) J. Biol. Chem. 275, 23648–23653 [DOI] [PubMed] [Google Scholar]
- 43.Muñoz V., Serrano L. (1994) Nat. Struct. Biol. 1, 399–409 [DOI] [PubMed] [Google Scholar]
- 44.Bouvier M., Stafford W. F. (2000) Biochemistry 39, 14950–14959 [DOI] [PubMed] [Google Scholar]
- 45.Tufi R., Panaretakis T., Bianchi K., Criollo A., Fazi B., Di Sano F., Tesniere A., Kepp O., Paterlini-Brechot P., Zitvogel L., Piacentini M., Szabadkai G., Kroemer G. (2008) Cell Death Differ. 15, 274–282 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.