Abstract
SSeCKS/Gravin/AKAP12 (“SSeCKS”) encodes a cytoskeletal protein that regulates G1 → S progression by scaffolding cyclins, protein kinase C (PKC) and PKA. SSeCKS is down-regulated in many tumor types including prostate, and when re-expressed in MAT-LyLu (MLL) prostate cancer cells, SSeCKS selectively inhibits metastasis by suppressing neovascularization at distal sites, correlating with its ability to down-regulate proangiogenic genes including Vegfa. However, the forced re-expression of VEGF only rescues partial lung metastasis formation. Here, we show that SSeCKS potently inhibits chemotaxis and Matrigel invasion, motility parameters contributing to metastasis formation. SSeCKS suppressed serum-induced activation of the Raf/MEK/ERK pathway, resulting in down-regulation of matrix metalloproteinase-2 expression. In contrast, SSeCKS had no effect on serum-induced phosphorylation of the Src substrate, Shc, in agreement with our previous data that SSeCKS does not inhibit Src kinase activity in cells. Invasiveness and chemotaxis could be restored by the forced expression of constitutively active MEK1, MEK2, ERK1, or PKCα. SSeCKS suppressed phorbol ester-induced ERK1/2 activity only if it encoded its PKC binding domain (amino acids 553–900), suggesting that SSeCKS attenuates ERK activation through a direct scaffolding of conventional and/or novel PKC isozymes. Finally, control of MLL invasiveness by SSeCKS is influenced by the actin cytoskeleton: the ability of SSeCKS to inhibit podosome formation is unaffected by cytochalasin D or jasplakinolide, whereas its ability to inhibit MEK1/2 and ERK1/2 activation is nullified by jasplakinolide. Our findings suggest that SSeCKS suppresses metastatic motility by disengaging activated Src and then inhibiting the PKC-Raf/MEK/ERK pathways controlling matrix metalloproteinase-2 expression and podosome formation.
Keywords: Cell/Migration, Chemotaxis, Cytoskeleton/Actin, Signal Transduction/Protein Kinases, Signal Transduction/Protein Kinases/MAP, Tumor/Metastases, SSeCKS/Gravin/AKAP12
Introduction
Metastasis is the most frequent cause of death for patients with cancer. However, the molecular mechanisms of metastasis are poorly understood as a result of their apparent complexity.
The Src-suppressed protein kinase C substrate (SSeCKS),2 the rodent orthologue of human GRAVIN, has recently emerged as an important regulator of cell signaling and cytoskeletal dynamics (1–4). Multiple studies have shown that SSeCKS can suppress growth rates and promote reorganization of the actin-based cytoskeleton in v-Src-transformed fibroblasts, thus leading to classification of SSeCKS as a class II tumor suppressor gene (5, 6). The expression of SSeCKS is down-regulated by several oncogenes and is suppressed strongly in various cancers, including prostate, ovary, and breast (1, 7–9).
SSeCKS re-expression can suppress all aspects of Src-induced oncogenic growth in vitro including anchorage- and growth factor-independent proliferation and Matrigel invasiveness (10). The regulated re-expression of SSeCKS in rat prostatic cancer MAT-LyLu (MLL) cells decreases anchorage-independent growth in vitro; in nude mouse models, SSeCKS re-expression slightly decreased the growth of primary subcutaneous tumors although it resulted in severely decreased levels of macroscopic lung metastases. However, SSeCKS re-expression neither inhibited wound healing motility in vitro nor metastatic lung colonization in vivo. Formation of avascular microscopic metastases by SSeCKS re-expressing MLL cells correlated with decreased levels of pro-angiogenic factors such as hypoxia-inducible factor-1α, VEGF, fibroblast growth factor-7, angiopoietin, tenascin C, platelet-derived growth factor-Rβ, and osteopontin, and increased levels of anti-angiogenic factors such as vasostatin and Col18α1 (endostatin precursor). Indeed, the finding that macroscopic lung metastasis formation could only be partially rescued by the forced re-expression of VEGF 165 or 121 isoforms in SSeCKS-expressing MLL cells strongly suggests that additional SSeCKS-regulated factors contribute to its metastasis suppressing activity (7, 11).
Given the importance of invasion enzymes such as metalloproteinases (MMP) in the metastatic process (12), we addressed here whether SSeCKS re-expression could affect invasiveness through control of MMP expression and secretion. We show that SSeCKS significantly decreases Matrigel invasiveness and chemotaxis in several cancer cell types, correlating with decreased MMP-2 expression and a suppression of serum-induced activation of a PKC-Raf/MEK/ERK pathway. That SSeCKS disengages Src signaling, but not activation, is based on the continued serum-induced phosphorylation of Shc after SSeCKS re-expression in MLL cells. Finally, SSeCKS inhibits podosome formation independent of the actin cytoskeleton, whereas inhibition of MEK/ERK activation requires actin cytoskeletal remodeling. The results suggest that SSeCKS partly suppresses metastasis through regulation of oncogenic motility parameters in both actin cytoskeleton-dependent and -independent manners.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following primary antibodies (Ab) were used: rabbit polyclonals specific for MEK1, PKCα, PKCδ, GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA), GFP (Invitrogen), phospho-c-RafSer-338, phospho-MEK1/ 2Ser-217/221, phospho-ERK1/2Thr-202/Tyr-204, phospho- ShcTyr-239/240, c-Raf, Shc, HA tag (6E2) mouse mAb, MEK2 and ERK1/2 (Cell Signaling Technology, Beverly, MA). The MEK1/2 inhibitor (U0126), MMP inhibitor (GM6001), and actin polymerization inhibitor (cytochalasin D) and jasplakinolide were from Calbiochem. Recombinant mouse MMP-9 and MMP-2 were from R&D Systems, Inc. (Minneapolis, MN). Phorbol 12-myristate 13-acetate (PMA) was from Avanti Polar Lipids Inc. (Alabaster, AL).
Cell Culture
MLL cell lines expressing tetracycline-regulated (tet-OFF) SSeCKS (MLL/tet-SSeCKS) were described previously (7). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine serum (BS) and 0.7 μg/ml of tetracycline.
Chemotaxis and Invasion Assays
Modified Boyden chamber assays were performed to assess chemotaxis and invasion by using a polyethylene terephthalate tissue culture insert with 8-μm pores (BD Biosciences). For chemotaxis, 3 × 104 cells in 100 μl of serum-free DMEM were added to Boyden chambers. For invasion, 5 × 104 cells in 100 μl of serum-free DMEM were added to the wells of a MATRIGELTM Invasion Chamber (growth factor reduced Matrigel Matrix, BD Biosciences). The lower chambers contained 5% BS or 20 μg/ml of platelet-derived growth factor-BB (BD Biosciences) in 600 μl of DMEM as a chemoattractant. The cells were allowed to migrate over the course of 12 (chemotaxis) and 24 h (invasion), and cells that had not penetrated the filters were removed by scrubbing with cotton swabs. Chambers were fixed and stained using Diff-Quik Stain Set (Dade Behring Inc., Newark, DE), and examined under a bright-field microscope. Values for migration were obtained by counting 6 fields per membrane (×20 objective) and represent the average of three independent experiments. For U0126 treatment, the drug (final concentration, 10 μm) was added for 18 h to MLL/tet-SSeCKS cells with DMEM, 5% BS prior to and then for the duration of the migration assays.
Zymography
Equal numbers of cells were plated onto 6-well culture dishes and allowed to reach confluence over 24 h. Cells were washed in phosphate-buffered saline and then incubated for 20 h in 1 ml of serum-free medium. Conditioned medium samples harvested from duplicate wells were pooled (2 ml) and concentrated to 40 μl using Centricon YM-10 centrifugal filter devices (Millipore Corp., Bedford, MA). Concentrated conditioned medium samples (10 μl) were then loaded onto 10% SDS-PAGE gels that had been co-polymerized with 1 mg/ml of gelatin (Sigma). Electrophoresis was performed under non-reducing conditions at 125 volts for 2 h. Gels were washed four times, 20 min each, successively in washing buffers 1, 2, 3, and 4 (13) at room temperature, and incubated in wash buffer 4 overnight at 37 °C. Gels were stained for 90 min with 0.5% Coomassie Blue R-250 and destained with methanol:acetic acid:water (50:10:40). Areas of protease activity appear as clear bands where the protease digested the substrate against a dark blue background.
Western Blotting (IB)
MLL/tet-SSeCKS cells grown in the presence or absence of tet (0.7 μg/ml) were plated in 10-cm dishes and serum-starved overnight by incubation with serum-free DMEM. The cells were stimulated with 10% BS in DMEM for 10 min (or the times indicated in specific figure legends) and immediately lysed in RIPA buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 5 mm EDTA, 8% glycerol, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 10 mm Na3VO4, 1 mm sodium fluoride, Complete Protease Inhibitor Mixture (Roche Diagnostics)). 40 μg of total protein per sample was separated by SDS-PAGE, blotted onto polyvinylidene fluoride membranes that were blocked for 30 min with 3% bovine serum albumin (Sigma) in 1× TBS/T (0.1% Tween 20 in Tris-buffered saline), and then probed as described previously (11). Digital imaging and signal quantification were performed on a Chemi-Genius2 Bio-Imager (Syngene, Frederick, MD) using GeneTools software. In some cases, cells were treated with PMA (100 nm) for various lengths of time, or CytD (10 μm) or jasplakinolide (100 nm) for 60 min prior to serum stimulation.
Transient Expression
SSeCKS-EGFP plasmid (kindly provided by Dr. Joseph M. Miano, University of Rochester School of Medicine and Dentistry) was transiently transfected into HT29 (colon cancer), MDA-MB231 (breast cancer), and CWR22Rv1 (prostate cancer) cell lines. After fluorescence-activated cell sorter sorting, GFP positive cells were used for Western blotting and invasion assays. Constitutively active MEK1 (CA-MEK1) plasmids (ΔN3/S218E/S222D (14), kindly provided by Dr. Natalie G. Ahn, University of Colorado at Boulder), CA-MEK2 plasmid (S222E (15), kindly provided by A. Bakin, RPCI), or CA-hPKCα-CAT (kindly provided by Dr. Jae-Won Soh, Inha University, Korea) were co-transfected transiently with pEGFP DNA (Clontech/Takara, Mountain View, CA) into MLL/tet-SSeCKS cells using Lipofectamine reagent (Invitrogen) according to the manufacturer's recommendations, and then incubated for 40 h. GFP positive cells were used for chemotaxis, invasion, and zymography. 293T cells were co-transfected with expression vectors encoding GFP fusions of either full-length SSeCKS or SSeCKS deleted of its PKC-binding domain (amino acids 553–900), and after serum starvation overnight, the cells were treated with 100 nm PMA for 15 min followed by IB analysis.
Cell Morphology
MLL/tet-SSeCKS cells plated overnight in 6-well dishes in the presence or absence of tet, and in some cases with U0126, CytD, or jasplakinolide, were photographed digitally using a Nikon TE2000-E inverted phase-contrast microscope (Melville, NY) equipped with a Roper CoolSnap HQ CCD camera (Ottbrunn, Germany). Texas Red phalloidin solutions (Invitrogen) were applied to the cells for F-actin staining.
Immunofluorescence Analysis
MLL/tet-SSeCKS cells grown on glass coverslips (22 mm2) for 24 h in the presence or absence of tet, and in some cases with U0126, CytD, or jasplakinolide, were fixed with cold 60% acetone, 3.7% paraformaldehyde in phosphate-buffered saline, and blocked with 3% nonfat dry milk in phosphate-buffered saline for 30 min at room temperature. Actin filaments were stained with rhodamine-labeled phalloidin (Sigma; 1:500), nuclei were stained with 4′,6-diamidino-2-phenylindole (Invitrogen; 1:1000), and vinculin was stained using a primary mAb (clone hVIN-1; 1:250; Sigma) followed by FITC-conjugated goat anti-mouse IgG (1:250; Chemicon, Temecula, CA). Fluorescent images were captured using Nikon TE2000-E inverted microscope equipped with Roper CoolSnap HQ CCD camera.
PKC Activity Assay
PKC kinase activity was measured using the PepTagR Nonradioactive Protein Kinase Assays (Promega Corp., Madison, WI) according to the manufacturer's instructions. Briefly, MLL/tet-SSeCKS cells were plated in 6-well dishes for 24 h in the presence or absence of tet, lysed in cold PKC extraction buffer, and then aliquots of lysates were assessed for PKC activity. Quantification of the PKC-phosphorylated substrate peptide, separated by electrophoresis through 1% agarose gels, was accomplished with a Chemi-Genius2 Bio-Imager (Syngene, Frederick, MD) using GeneTools software.
Co-immunoprecipitation Assays
293T cells were transfected with SSeCKS-EGFP and/or HA-MEK1 (kindly provided by Dr. Michael J. Weber, University of Virginia Health System). Cells were lysed in RIPA buffer after 48 h and immunoprecipitation was performed using anti-MEK1 antibody, followed by IB with the indicated antibodies. Lysates of 293T cells transfected with HA-MEK1 were incubated with Ni2+ beads bound to the His-TAT-SSeCKS protein. After washing three times in RIPA buffer, the beads were subjected to IB analysis and probed with anti-HA mouse mAb.
Semiquantitative Reverse Transcriptase-PCR
1 μg of total cellular RNA from MLL/tet-SSeCKS cells grown in the presence or absence of tet for 2 days was isolated using TRIzol reagent (Invitrogen) and converted to cDNA as described previously (5) using Superscript II reverse transcriptase (Invitrogen). One microliter of the resulting cDNA was used as template for each PCR. The PCR conditions were optimized for each primer pair: MMP2, forward, 5′-GGAGAAGGCTGTGTTCTTCG-3′, reverse, 5′-CGGGTCCATTTTCTTCTTCA-3′ (214 bp product); MMP14 (MT1-MMP), forward, 5′-GTACCCCAAGTCAGCTCTGC-3′, reverse, 5′-CAGTGAACGCTGGCAGTAAA-3′ (247 bp product); TIMP2, forward, 5′-TGGACGTTGGAGGAAAGAAG-3′, reverse, 5′-GTCCATCCAGAGGCACTCAT-3′ (224 bp product) using an MJ Research PTC-200 DNA thermal cycler (Bio-Rad). Cycle numbers were used that produced mid-log phase amplification (21–26 cycles). β-Actin primers were used as an internal control for cDNA normalization as described previously (16). The PCR products were analyzed on 2% agarose gels and the products were digitally imaged and quantified on a Chemi-Genius2 Bio-Imager (Syngene) using GeneTools software.
RESULTS
SSeCKS Inhibits Matrigel Invasion
We showed previously that the tet-regulated re-expression of SSeCKS in MLL cells to physiologic levels found in untransformed prostatic epithelial cells inhibited neither their wound healing motility in vitro nor their ability to colonize peripheral sites from subcutaneous primary tumor sites in vivo (7, 11), indicating that SSeCKS does not inhibit certain parameters of motility involved in metastasis. However, the finding that SSeCKS re-expression produced only micrometastases with little histological evidence of local invasiveness suggests that SSeCKS antagonizes specialized parameters of metastatic motility such as invasiveness or chemotaxis. To investigate these more specialized functions, we assessed the motility of MLL/tet-SSeCKS cells in the presence (− SSeCKS) or absence (+ SSeCKS) of tet in in vitro assays for chemotaxis or invasiveness.
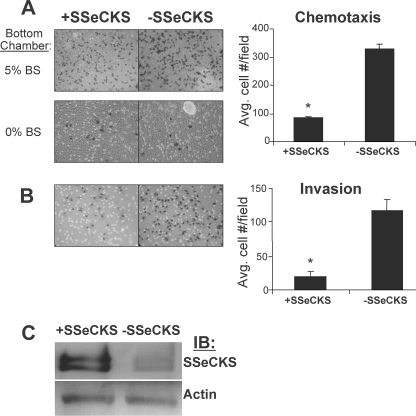
To determine the effect of SSeCKS on chemotaxis, MLL/tet-SSeCKS cells were placed in serum-free medium with or without tet on 8-μm pore membranes of Boyden chambers, using 5% BS as a chemoattractant in the lower wells. Fig. 1A shows that in triplicate experiments, SSeCKS re-expression decreased chemotaxis roughly 4-fold (p < 0.01). Similar results were obtained using platelet-derived growth factor-BB (20 μg/ml) as the chemoattractant (data not shown). In the absence of serum in the bottom chamber (Fig. 1A, 0% BS), there was no significant cell migration either in the presence or absence of tet, indicating that the motility measured toward 5% BS was indeed chemotaxis. We performed an immunoblot to show that SSeCKS protein levels are induced in the absence of tet (Fig. 1C). It is important to note that we previously showed that these induced levels reflect SSeCKS re-expression rather than an overexpression because they are comparable with ”normal“ SSeCKS levels in untransformed, immortalized rat prostate epithelial EP12 cells (7). To explore invasive activity, similar Boyden chamber assays were performed except that the MLL/tet-SSeCKS cells were placed atop membranes coated with 1-mm layers of growth factor-reduced Matrigel, and then assayed for growth toward the bottom chamber containing 5% BS. SSeCKS re-expression inhibited invasiveness 6.2-fold (p < 0.01; Fig. 1B). Importantly, the invasiveness of MLL cells is dependent on MMP- and -9 because treatment with the MMP inhibitor, GM6001, reduced MLL invasive potential and MMP-2/9 activity at similar rates (supplemental Fig. S1, A and B). Taken together, these data indicate that SSeCKS can inhibit specialized in vitro parameters of motility associated with metastasis such as chemotaxis and Matrigel invasiveness. To address whether SSeCKS can inhibit invasiveness of other cancer cell types, human HT29 colon cancer, MDA-MB-231 breast cancer, and CWR22Rv1 prostate cancer cells were transiently transfected with SSeCKS-EGFP or GFP expressing plasmids, and then GFP-positive invasive cells were scored. SSeCKS decreased the relative invasive potentials of all these cell lines 60–70% compared with controls (supplemental Fig. S2A), strongly suggesting that SSeCKS has a more generic role in suppressing oncogenic invasiveness.
FIGURE 1.
SSeCKS suppresses chemotaxis and Matrigel invasion of metastatic prostate cancer cells. A, chemotaxis. MLL/tet-SSeCKS cells were seeded atop Boyden chamber membranes in the presence or absence of tet (− or + SSeCKS, respectively) with 5% BS in the bottom well as the chemoattractant (top left panels) or no serum, as a negative control (bottom left panels). After 12 h, the cells were fixed, stained, and counted as described under “Experimental Procedures.” Six separate microscopic fields on the stained membranes from duplicate experiments were counted to determine the average number of cells/field (error bar = S.D.). *, p < 0.005. B, invasion. The ability of MLL/tet-SSeCKS cells to invade after 24 h through 1-mm growth factor-reduced Matrigel barriers atop Boyden chamber membranes was assessed. Six separate microscopic fields on the stained membranes from duplicate experiments were counted to determine the average number of cells/field (error bar = S.D.). *, p < 0.01. C, immunoblot (IB) of SSeCKS and actin protein levels from MLL/tet-SSeCKS cells grown in the presence or absence of tet (− or + SSeCKS, respectively).
SSeCKS Inhibits MMP-2 Expression/Secretion
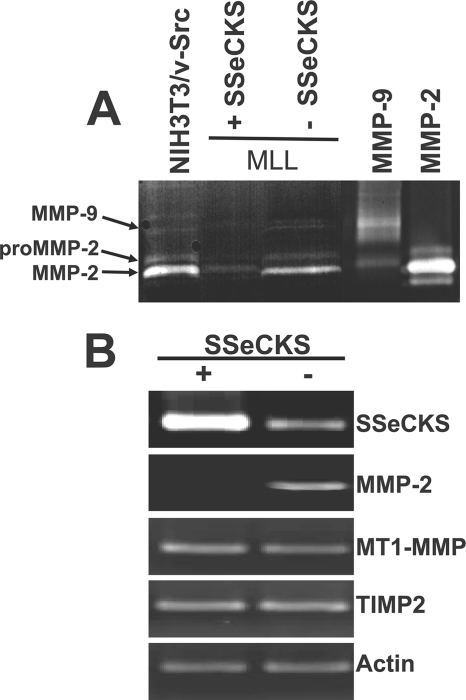
Tumor cell invasion associated with metastasis requires both specialized cell migration and the ability to degrade basement membranes by secreted or membrane-bound proteases (17, 18). We sought to determine whether alterations in the expression and/or secretion of MMP, known to be up-regulated in many metastatic tumors, might be responsible for the SSeCKS-mediated decreases in invasion. Metastatic prostate cancer cell lines and cancer tissues exhibit increased MMP-2 and MMP-9 expression levels, and mice deficient in MMP-2 or MMP-9 are much less susceptible to experimental metastasis (19). To assess MMP activity, conditioned medium from equal numbers of MLL/tet-SSeCKS cells incubated with or without tet overnight in serum-free DMEM were subjected to gelatin zymography. Active, recombinant MMP-2 and MMP-9 controls are visualized as clear 67- and 95-kDa bands, respectively, in Coomassie Blue-stained gels as a result of gelatinolytic activity. Given that MLL cells are known to express very low relative levels of MMP-9 (confirmed in our zymography gels (Fig. 2A)) and that invasive activity is driven by MMP-2 (20), we focused on MMP-2. The re-expression of SSeCKS resulted in a significant reduction in MMP-2-associated gelatinolytic activity (Fig. 2A). We confirmed that the decrease in secreted, active MMP-2 reflected regulation at the transcriptional level by subjecting total RNA from the experiment in Fig. 2A to semi-quantitative reverse transcriptase-PCR. SSeCKS suppressed MMP-2 transcript levels comparable with the decrease in secreted MMP-2 enzyme (compare Fig. 2, A with B). In contrast, SSeCKS re-expression had no effect on the transcript levels of MT1-MMP (MMP-14), a membrane-tethered (MT) MMP known to activate pro-MMP-2 through direct cleavage (21). In addition, SSeCKS did not affect the RNA levels of TIMP-2 (Fig. 2B), which is required for MMP-2 activation by binding to MT1-MMP and which also serves as a major MMP antagonist (22). These data suggest that SSeCKS modulates the invasive activity of MLL cells by decreasing MMP-2 expression.
FIGURE 2.
SSeCKS inhibits MMP-2 expression and secretion. A, conditioned medium obtained from an overnight incubation of equal numbers of MLL/tet-SSeCKS cells grown in serum-free DMEM +/− tet was subjected to zymography as described under “Experimental Procedures.” Controls include overnight medium from NIH3T3/v-Src cells and purified MMP-2 and -9. The 95- and 67-kDa MMP-9 and -2, respectively, are identified by arrows as well as the unactivated form of MMP-2 (pro-MMP-2). Aliquots of purified, enzymatically active MMP-2 and -9 were run as controls. B, RNA derived from MLL/tet-SSeCKS cells grown in serum-free DMEM +/− tet was subjected to semi-quantitative reverse transcriptase-PCR using primer sets for MMP-2, MT1-MMP, TIMP2, and β-actin as described under “Experimental Procedures.” The results are typical of at least three independent experiments.
SSeCKS Modulates Growth Factor-induced MEK/ERK Activity
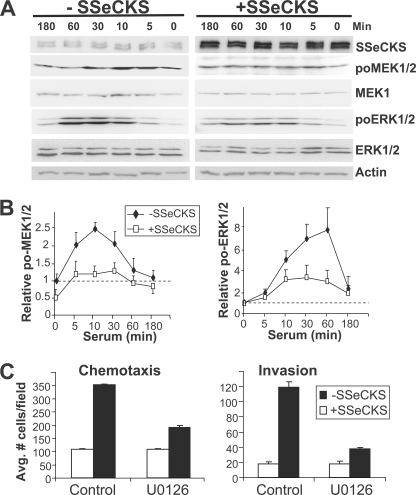
The expression of MMPs and thus, the invasive potential of CaP cells is dependent on growth factor-induced activation of the MEK/ERK signaling pathway (23, 24). Given our previous demonstration that SSeCKS could suppress serum-induced ERK1/2 activation in NIH3T3 fibroblasts (25), we addressed whether SSeCKS could suppress MEK/ERK signaling in MLL CaP cells. Thus, MLL/tet-SSeCKS cells were serum starved overnight in either the presence or absence of tet, stimulated with 10% serum for various times, and then cell lysates were probed for phospho-MEK or -ERK relative to total MEK or ERK protein levels as an indicator of activated MEK/ERK signaling. As shown in Fig. 3A, cells grown in the absence of tet exhibited a serum-independent induction of SSeCKS protein levels. In the presence of tet, the addition of serum induced a rapid increase in phospho-MEK levels between 5 and 30 min, whereas SSeCKS re-expression decreased the basal phospho-MEK levels by half and then suppressed serum-induced phospho-MEK levels over the same period (Fig. 3, A and B). The effect of SSeCKS on phospho-ERK1/2 levels was even more pronounced: whereas serum induced a 5–8-fold rise in relative phospho-ERK1/2 levels in the presence of tet, SSeCKS re-expression resulted in only a 2–3-fold increase over the same period. In contrast to phospho-MEK, SSeCKS did not affect basal phospho-ERK1/2 levels. A similar decrease in serum-stimulated MEK and ERK activity by SSeCKS was also found in other tumor cell lines (supplemental Fig. S2B). These data agree with our previous demonstration in NIH3T3 showing that SSeCKS re-expression could attenuate serum-induced ERK2 activation (25).
FIGURE 3.
Inhibition of chemotaxis and invasiveness by SSeCKS correlates with suppression of serum-induced MEK-1/2 and ERK-1/2 activation. A, re-expression of SSeCKS attenuates MEK/ERK. Serum-starved MLL/tet-SSeCKS cells (+/− tet) were stimulated with 10% BS in DMEM for various times, and then RIPA lysates were analyzed by IB (40 μg of protein/lane) for SSeCKS (to show inducibility in the absence of tet), phospho- and apo-forms of MEK1/2 and ERK1/2, and actin (protein loading control). B, graphic representation of the relative MEK1/2 and ERK1/2 activation levels (phospho-protein levels normalized to total MEK1/2 or ERK1/2 protein levels, with basal levels set arbitrarily at 1, as shown by the dotted lines). Similar effects by serum and SSeCKS were identified in two other independent experiments (error bar = S.D.). C, Boyden chamber chemotaxis and invasion assays as described in the legend to Fig. 1 were preformed in the presence of U0126 (10 μm) or vehicle in the presence (black columns) or absence (white columns) of tet (error bar = S.D. from two independent experiments).
Given that chemotaxis and invasiveness require MEK activity (26, 27) and our data that SSeCKS re-expression inhibits serum-induced MEK activation, we argued that the metastatic motility of parental MLL (i.e. which suffer SSeCKS down-regulation) would be much more sensitive to MEK inhibitory drugs. Indeed, MLL/tet-SSeCKS cells incubated in the presence of tet showed 2- and 3-fold decreases in chemotactic and invasive motility, respectively, when treated with the MEK inhibitor U0126 (compared with vehicle alone; Fig. 3C). In contrast, U0126 had little effect on the already decreased chemotactic and invasive motility induced by SSeCKS re-expression, presumably because SSeCKS had already suppressed MEK activation. Interestingly, U0126 induced a level of cell flattening in MLL/tet-SSeCKS cells grown in the presence of tet (− SSeCKS) similar to that induced by SSeCKS re-expression alone in the absence of U0126 (Fig. 3D). This suggests that part of the cytoskeletal remodeling induced by SSeCKS is through the inhibition of MEK activation.
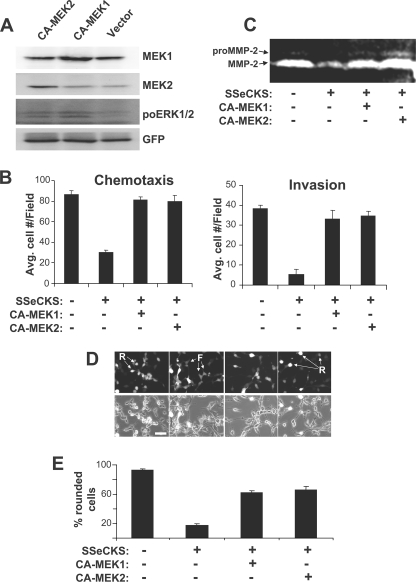
To determine whether activated MEK is sufficient to induce metastatic motility in the MLL cells, we transiently expressed constitutively active (CA) alleles of MEK1 or MEK2 in MLL/tet-SSeCKS cells. Expression of each CA-MEK isoform was first verified by immunoblotting (Fig. 4A), and indeed, both CA-MEK1 and -2 induced ERK activity (as measured by ERKThr-202/Tyr-204 IB) under conditions of constant growth in serum-containing medium. Expression of CA-MEK1 or CA-MEK2 rescued SSeCKS-suppressed chemotaxis and invasiveness (Fig. 4B). The ectopic expression of a CA-ERK2 allele (28) also induced increased chemotactic activity (supplemental Fig. S2) indicating that the inhibitory effects of SSeCKS on oncogenic motility probably are mediated through MEK to ERK. Consistent with our concept that MEK activity is required for MMP-2 expression, the expression of CA-MEK1 or CA-MEK2 in SSeCKS re-expressor cells induced a significant increase in secreted MMP-2 (Fig. 4C). Consistent with our previous result that MEK inhibition induced MLL cell flattening (Fig. 3D), CA-MEK1 or CA-MEK2 restored the parental, round cell morphology in SSeCKS re-expressing cells (Fig. 4, D and E). To address whether SSeCKS inhibits MEK through a direction interaction, we determined whether SSeCKS-GFP could be co-immunoprecipitated with HA-MEK1 when co-transfected into 293T cells (supplemental Fig. S4A) or whether the His-TAT-SSeCKS protein loaded onto Ni2+ beads could pull down HA-MEK1 overexpressed in 293T cell lysates (supplemental Fig. S4B). Neither scenario showed evidence of protein association, suggesting that SSeCKS exerted its regulation upstream of MEK.
FIGURE 4.
Activated MEK rescues SSeCKS-suppressed chemotaxis and invasion. A, CA-MEK1 or -MEK2 were transiently co-transfected along with pEGFP into MLL/tet-SSeCKS cells. The IB analysis shows increased MEK1 or MEK2 protein levels in cells expressing CA-MEK1 or CA-MEK2 (versus vector alone), respectively. The CA-MEK alleles induced increased phosphorylation of ERK1/2 as measured by the relative pro-ERK1/2 levels. GFP protein levels are shown as a loading control. (Note that the cell lysates expressing CA-MEK alleles could not be probed for changes in phospho-MEK levels because the CA-MEK products contain amino acid substitutions at the site recognized by phospho-specific MEK Abs (Ser217/221).) B, chemotaxis and invasion assays were performed as described in the legend to Fig. 1 comparing MLL/tet-SSeCKS/vector (−) versus MLL/tet-SSeCKS/CA-MEK (+) grown in the presence or absence of tet (error bar = S.D. from two independent experiments). C, CA-MEK1 or CA-MEK2 rescues MMP-2 expression in SSeCKS re-expressing MLL cells. Conditioned medium from serum-starved MLL/tet-SSeCKS/vector (−) versus MLL/tet-SSeCKS/CA-MEK (+) grown in the presence or absence of tet were assessed for MMP-2 activity by zymography as described in the legend to Fig. 2. D, SSeCKS-induced cell flattening is suppressed by the forced expression of CA-MEK1 or CA-MEK2. Round (R) or flat (F) transfected cells (GFP-positive) were identified by fluorescence (panel D, top row) plus phase-contrast microscopy (panel D, bottom row). Size bar = 5 μm. E, the percentage of round cells in at least three independent fields from panel D (>50 cells/field). Error bars = S.D.
SSeCKS-mediated Suppression of MEK and ERK Is Reversed by Preventing Actin Cytoskeletal Remodeling
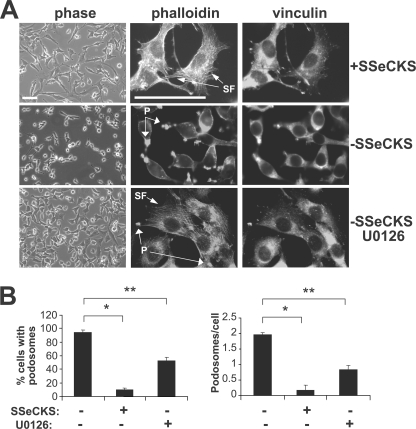
SSeCKS is associated with the actin-based cytoskeleton as well as the plasma membrane via an N-terminal myristyl modification (1). We showed previously that SSeCKS re-expression in Src- or Ras-transformed cancer cells re-establishes normal cytoskeletal structures such as actin stress fibers and mature focal adhesion plaques (10, 29). Indeed, SSeCKS re-expression in MLL/tet-SSeCKS cells results in cell flattening marked by the re-establishment of actin stress fibers (Fig. 5A). This was accompanied by a dramatic increase in the size and number of vinculin-associated focal adhesions (Fig. 5A). Similar to the findings we previously reported (36), SSeCKS re-expression decreased the percentage of cells with podosomes roughly 10-fold in MLL/tet-SSeCKS cells (Fig. 5B). U0126 exhibited a similar effect as SSeCKS by inducing cell flattening and the formation of stress fibers and focal adhesions (Fig. 5A), but it only decreased the percentage of cells with podosomes roughly 3-fold (Fig. 5B), suggesting that SSeCKS likely inhibited podosome formation through MEK-dependent and -independent mechanisms.
FIGURE 5.
SSeCKS inhibits podosome formation and induces stress fiber formation. A, MLL/tet-SSeCKS cells grown in the presence or absence of tet, and in the presence of tet with U0126 cells were imaged by phase-contrast microscopy (left column) or fixed and stained for F-actin (rhodamine-phalloidin) stress fibers (SF) (middle column), and focal adhesions (vinculin, right column). Podosomes (P) are defined as enrichments of F-actin on the cell surface, often exhibiting a “ring” structure. Size bars = 5 μm. B, SSeCKS re-expression decreases total podosome formation. The percentage of cells exhibiting podosomes (left) or the frequency of podosomes per cell (right) was quantified as the average of three separate visual fields containing roughly 30 cells per field; error bars = S.E. *, p < 0.01; **, p < 0.05.
The literature has conflicting data in different cell types on the dependence of growth factor-stimulated ERK activation on an intact cytoskeleton, with some studies showing no effect by CytD (30), with others showing an inhibitory effect (31–33). We addressed whether the ability of SSeCKS to suppress MEK and ERK activation is affected by CytD, an interrupter of polymerized actin. Treatment of MLL/tet-SSeCKS cells with CytD reversed SSeCKS-induced cell flattening and disrupted SSeCKS-induced formation of stress fibers (supplemental Fig. S5, A and B). This phenomenon is similar to the effect of CytD on NIH3T3 cells overexpressing SSeCKS (3). In contrast, treatment with jasplakinolide, which stabilizes actin stress fibers primarily by decreasing the dissociation rate of actin subunits in low concentration (34), induced flattening in the absence of SSeCKS, and in SSeCKS re-expressor cells, induced additional flattening over that induced by SSeCKS (supplemental Fig. S5, A and B). Interestingly, neither CytD nor jasplakinolide suppressed podosome formation in the absence of SSeCKS, whereas SSeCKS re-expression suppressed podosome formation regardless of the effects of these drugs on the actin cytoskeleton (supplemental Fig. S5B). CytD treatment also inhibited serum-induced MEK1/2 and ERK1/2 phosphorylation in the absence of SSeCKS (supplemental Fig. S5, C and D, compare lanes b to c). Consistent with our findings in Fig. 3A, SSeCKS re-expression suppressed MEK and ERK activation, however, CytD did not change these outcomes (supplemental Fig. S5, C and D, compare lanes f to g). In contrast, MEK1/2 and ERK1/2 could be activated by serum in the presence or absence of SSeCKS following actin stress fiber stabilization by jasplakinolide (supplemental Fig. S5, C and D, compare lanes a to d and e to h). This indicates that SSeCKS requires actin cytoskeletal remodeling capability to inhibit MEK/ERK. Unlike the results in Fig. 5a, which suggest that activated MEK is sufficient to induce cell rounding and stress fiber loss, the activation of MEK or ERK1/2 by serum in the presence of jasplakinolide is not sufficient to induce these cytoskeletal changes (supplemental Fig. S5, A and B). This strongly suggests that the cell flattening induced by SSeCKS and jasplakinolide are inherently different in that only SSeCKS suppresses podosome formation and serum-induced MEK/ERK activation.
SSeCKS Inhibits PKCα-triggered Ras-independent Raf/MEK/ERK Activation by Scaffolding PKC
Having shown that SSeCKS likely inhibits MEK activation through upstream mediators, we investigated whether activation of the MEK kinase, Raf, was also inhibited. Indeed, SSeCKS delayed serum-induced c-Raf Ser-338 phosphorylation without affecting total c-Raf protein levels (Fig. 6). Growth factor receptors are known to induce Raf activation through Src-Ras pathways (35), however, we previously showed that SSeCKS could inhibit Src-induced oncogenic growth parameters without affecting innate Src kinase activity or phosphorylation of most Src substrates in cells (10, 36). Consistent with this, SSeCKS re-expression did not affect serum-induced Shc phosphorylation or total Shc protein levels (Fig. 6). This suggests that SSeCKS disengages serum-induced Src activation from the Raf/MEK/ERK pathways, possibly by inducing cytoskeletal remodeling associated with cell flattening.
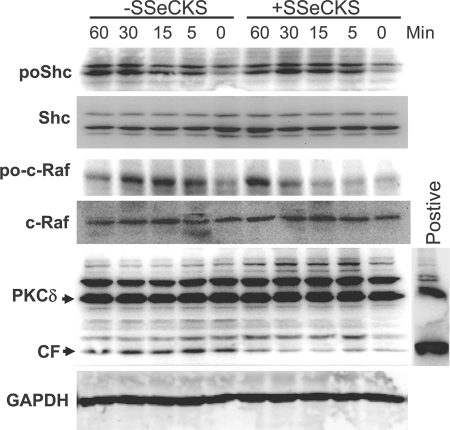
FIGURE 6.
Inhibition of serum-induced c-Raf and PKCδ activation by SSeCKS. Serum-starved MLL/tet-SSeCKS cells (+/− tet) were stimulated with 10% BS in DMEM for various times, and then RIPA lysates were analyzed by IB (30 μg of protein/lane) for phospho- and apo-forms of c-Raf and Shc, PKCδ and GAPDH. CF, cleaved fragment identifying active PKCδ.
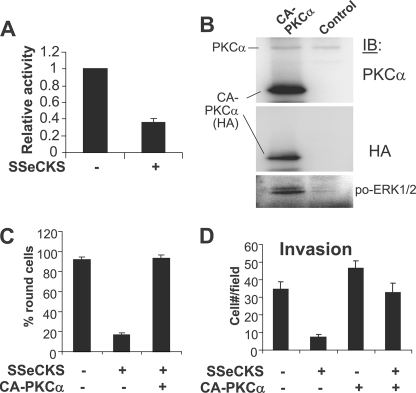
Another mechanism to activate Raf/MEK/ERK pathways is through the direct phosphorylation of Raf by PKC isozymes (37, 38). Indeed, SSeCKS is known to bind to and inhibit the kinase activity of PKC (1, 2, 39–41). As shown in Fig. 6, SSeCKS re-expressing MLL cells have 2–3-fold lower activation levels of PKCδ, as assessed by the relative levels of so-called 40-kDa cleaved fragments, generated by the caspase-3-mediated cleavage of activated PKCδ forms (42). The low level of cleaved fragments under these conditions most likely represents background apoptosis for MLL cells (8 ± 1.8%). As a control, lysates of 293T cells transfected with a PKCδ-cleaved fragment expression vector were blotted with PKCδ-specific Ab to identify the 40-kDa isoform. We confirmed that SSeCKS re-expressing MLL cells have 50–65% reductions in total PKC activity (Fig. 7A), yet have no changes in total PKCα or -δ (Fig. 6, data not shown). To address whether PKC was sufficient to reverse the effects of SSeCKS on cell flattening, invasiveness, and ERK activation, SSeCKS re-expressing MLL cells were transiently transfected with HA-tagged CA-PKCα plus pEGFP, and the GFP-positive cells were assessed. Fig. 7B shows that expression of CA-PKCα (containing a deletion of its regulatory domain) induced a 2.5-fold increase in relative phospho-ERK1/2 levels. Moreover, CA-PKCα induced cell rounding to levels found in the MLL cells not re-expressing SSeCKS (Fig. 7C). Although CA-PKCα could marginally increase Matrigel invasiveness in the absence of SSeCKS, it rescued the ability of SSeCKS re-expressing MLL to invade (Fig. 7D).
FIGURE 7.
Activated PKCα rescues SSeCKS-suppressed invasion and -induced cell flattening. A, re-expression of SSeCKS decreases PKC activity. Lysates from MLL/tet-SSeCKS cells grown overnight in +/− tet conditions in the absence of serum were analyzed for total PKC kinase activity as described under “Experimental Procedures.” Error bars = S.D. of triplicates, with the mean PKC activity in the absence of SSeCKS re-expression set at 1.0. B, IB analysis of 293T cell lysates transiently co-transfected with CA-PKCα(HA) plus pEGFP, probed for PKCα, HA, or ERK1/2pro-Thr-202/Tyr-204. C, SSeCKS-induced cell flattening is suppressed by forced expression of CA-PKCα. The percentage of round cells in at least three independent fields is shown. Error bars = S.D. D, CA-PKCα rescues Matrigel invasion of SSeCKS re-expressing MLL. MLL/tet-SSeCKS cells transiently transfected with CA-PKCα plus pEGFP were grown in + or − tet conditions and then subjected to Matrigel invasion assays as described in the legend to Fig. 1B except that only GFP-positive invasive cells were scored. Error bars = S.D. of triplicate wells.
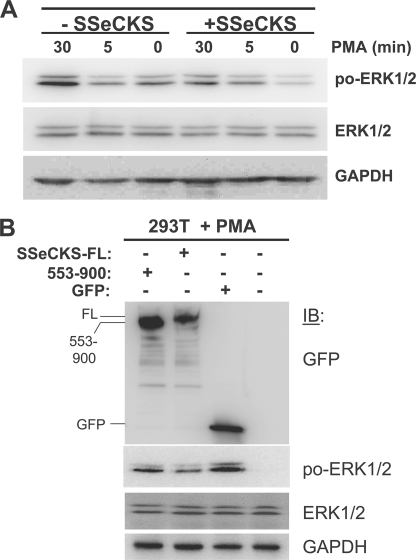
To further investigate whether SSeCKS could antagonize the ability of PKC to activate ERK1/2, serum-starved MLL/tet-SSeCKS cells (+/−tet) were stimulated with 100 nm PMA in DMEM for various times, and then RIPA lysates were analyzed for phospho- versus total ERK1/2. Re-expression of SSeCKS decreased basal and PMA-induced ERK1/2 phosphorylation (Fig. 8A). This suggests that SSeCKS interferes with conventional and/or novel PKC isozymes.
FIGURE 8.
SSeCKS inhibits PMA-induced ERK1/2 activation via scaffolding of PKC. A, re-expression of SSeCKS decreases PMA-induced ERK1/2 activity. Serum-starved MLL/tet-SSeCKS cells (+/− tet) were stimulated with 100 nm PMA in DMEM for various times, and then RIPA lysates were analyzed by IB (30 μg of protein/lane) for phospho-ERK1/2, ERK1/2, and GAPDH. B, full-length (FL), but not Δ553–900 SSeCKS, suppresses PMA-induced ERK1/2 activity. Lysates of 293T cells transiently transfected with pEGFP control vector or expression vectors encoding GFP fusions of FL or Δ553–900 SSeCKS, treated with 100 nm PMA for 15 min, were probed by IB analysis for GFP, phospho-ERK1/2, ERK1/2, and GAPDH.
Given data that SSeCKS binds PKC in cells and via purified proteins (1, 2, 39–41, 43, 44), that binding inhibits PKC activity (40),3 and that PKC binding and inhibitory activity maps SSeCKS amino acids 553–900, we addressed whether SSeCKS scaffolding activity for PKC was responsible the attenuation of PMA-induced ERK1/2 phosphorylation. Thus, 293T cells were co-transfected with expression vectors encoding GFP fusions of either full-length SSeCKS, SSeCKS deleted of its PKC-binding domains (amino acids 553–900),3 or GFP alone (Fig. 8B). After serum starvation overnight, the cells were treated with 100 nm PMA for 15 min, and cell lysates were then assessed by IB for ERK1/2 activity. Full-length SSeCKS could suppress PMA-induced ERK1/2 activity (Fig. 8B). In contrast, the Δ553–900 mutant failed to significantly reduce the PMA effect compared with cells expressing GFP alone. This strongly suggests that SSeCKS attenuates PMA-induced ERK activation through its direct scaffolding of PKC. Taken together with the data in Fig. 6, these findings strongly suggest that SSeCKS suppresses invasiveness by disengaging serum-induced Src activation from downstream Raf/MEK/ERK effectors, and by inhibiting PKC-mediated activation of Raf/MEK/ERK signaling.
DISCUSSION
The complex set of processes required to produce a progressing metastatic lesion includes specialized parameters of cell motility such as chemotaxis and invasion, as well as the ability to initiate neovascularization at peripheral sites. We showed previously that SSeCKS could control metastasis progression by attenuating VEGF-mediated angiogenesis, however, the re-expression of VEGF isoforms could only partially restore macrometastasis forming activity to SSeCKS re-expressing CaP cells (11). The current study shows that SSeCKS can also attenuate chemotaxis and Matrigel invasiveness in in vitro assays, suggesting that the inhibition of these motility parameters also contributes to the metastasis-suppressing activity of SSeCKS. Other factors not yet studied that might contribute to SSeCKS-mediated metastasis suppression include down-regulation of a host of pro-angiogenic factors other than VEGF, or up-regulation of anti-angiogenic factors such as vasostatin and the endostatin precursor, collagen type 18α. It was noteworthy that in our previous study (11), SSeCKS did not induce increased apoptosis in the primary tumor, although it is not clear if SSeCKS affects tumor cell survival in the metastatic niche.
It is likely that control of chemotaxis is less important to the metastasis-suppressing activity of SSeCKS in vivo because there is little decrease in lung colonization after SSeCKS re-expression in MLL cells (7, 11). Thus, SSeCKS does not seem to inhibit MLL motility to metastatic sites. Given that SSeCKS re-expression results in avascular micrometastases, it is likely that the decrease in invasive potential of SSeCKS plays a more important role in metastasis suppression.
Our data indicate that the suppression of chemotaxis and Matrigel invasiveness by SSeCKS are dependent on activation of a MEK/ERK signaling pathway that controls the expression and secretion of MMP-2 and -9. Our result showing that the MEK inhibitor, U0126, can block invasiveness only in the absence of SSeCKS re-expression suggests that SSeCKS effectively blocks the majority of growth factor-induced MEK activation, thereby leaving little MEK to be inhibited by U0126. These data agree with the recent finding that inhibition of ERK1/2 activation suppresses MLL invasiveness, and that the level of activated ERK1/2 correlates with metastatic potential (45). Indeed, CA-MEK1, -MEK2, -ERK1, or -ERK2 can partially rescue the chemotactic and invasive motility in SSeCKS re-expressing MLL cells. This rescue activity correlates with increased MMP-2 production, and given our finding that SSeCKS has no effect on MT1-MMP or TIMP-2 expression, these data strengthen our notion that SSeCKS controls metastasis in part by suppressing MMP-2 expression and secretion, although a lesser role for MMP-9 cannot be ruled out.
We found that the ability of SSeCKS to regulate serum-induced MEK and ERK activation requires the ability to remodel the actin cytoskeleton, correlating with the ability of jasplakinolide to reverse SSeCKS-mediated MEK/ERK inhibition and cell flattening. Indeed, our previous data showed that the ability of SSeCKS to control cytoskeletal remodeling and cellular morphology (cell flattening) in NIH3T3 cells requires an intact actin cytoskeleton (3). Our current data suggest that cytoskeletal remodeling induced by SSeCKS re-expression differs from the cytoskeletal stabilization induced by jasplakinolide.
The mechanism by which SSeCKS regulates MEK/ERK signaling likely involves two concurrent processes: disengagement of activated Src from its downstream Raf/MEK/ERK effectors, and inhibition of PKC-induced activation of Raf, probably through direct scaffolding of PKC by SSeCKS. Recent evidence indicates that human SSeCKS (Gravin/AKAP12) can bind Src directly during the resensitization/recycling process of β2-adrenergic receptors (46). However, our laboratory has shown that SSeCKS re-expression or even overexpression does not inhibit intrinsic Src kinase activity in the cell, which includes Src autophosphorylation as well as phosphorylation of Src substrates (10, 36). We have speculated previously that by re-establishing normalized F-actin stress fibers and mature focal adhesion complexes, SSeCKS re-expression may attenuate oncogenic signaling by simply slowing the rate of signal transduction to that found in untransformed cells (47). Thus, SSeCKS may attenuate MEK/ERK signaling in part by scaffolding growth factor-activated Src away from downstream mediators. Given the opposing effects induced by SSeCKS and activated PKC on the cytoskeleton (SSeCKS, establishment of stress fibers and focal adhesions, leading to cell flattening; PKC, actin cytoskeletal remodeling, leading to cell rounding), it is likely that SSeCKS inhibits PKC-mediated activation of invasion and chemotaxis pathways by both direct binding of PKC and by sequestering it to inactive sites in the cell, away from downstream mediators.
Supplementary Material
Acknowledgments
We thank A. Bakin, J. Miano, M. Weber, and Natalie G. Ahn for sharing plasmid reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant CA94108, Department of Defense Grants PC040256 and PC061246 (to I. H. G.), and National Institutes of Health NCI Cancer Center Support Grant 2P30 CA016056.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
L.-W. Guo, B. Su, L. Gao, and I. H. Gelman, submitted for publication.
- SSeCKS
- Src-suppressed protein kinase C substrate
- 293T
- SV40-immortalized human embryonic kidney cell line 293
- BS
- bovine serum
- CA
- constitutively active
- CaP
- prostate cancer
- CytD
- cytochalasin D
- DMEM
- Dulbecco's modified Eagle's medium
- MLL
- MAT-LyLu
- MMP
- matrix metalloproteinases
- PKC/PKA
- protein kinase C/A
- PMA
- phorbol 12-myristate 13-acetate
- tet
- tetracycline
- VEGF
- vascular endothelial growth factor
- MT
- membrane-tethered
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- ERK
- extracellular signal-regulated kinase
- mAb
- monoclonal antibody
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- EGFP
- enhanced green fluorescent protein
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- IB
- immunoblot.
REFERENCES
- 1.Lin X., Tombler E., Nelson P. J., Ross M., Gelman I. H. (1996) J. Biol. Chem. 271, 28430–28438 [DOI] [PubMed] [Google Scholar]
- 2.Chapline C., Cottom J., Tobin H., Hulmes J., Crabb J., Jaken S. (1998) J. Biol. Chem. 273, 19482–19489 [DOI] [PubMed] [Google Scholar]
- 3.Gelman I. H., Lee K., Tombler E., Gordon R., Lin X. (1998) Cell Motil. Cytoskeleton 41, 1–17 [DOI] [PubMed] [Google Scholar]
- 4.Nelson P. J., Moissoglu K., Vargas J., Jr., Klotman P. E., Gelman I. H. (1999) J. Cell Sci. 112, 361–370 [DOI] [PubMed] [Google Scholar]
- 5.Lin X., Nelson P. J., Frankfort B., Tombler E., Johnson R., Gelman I. H. (1995) Mol. Cell. Biol. 15, 2754–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson P. J., Gelman I. H. (1997) Mol. Cell. Biochem. 175, 233–241 [DOI] [PubMed] [Google Scholar]
- 7.Xia W., Unger P., Miller L., Nelson J., Gelman I. H. (2001) Cancer Res. 61, 5644–5651 [PubMed] [Google Scholar]
- 8.Welsh J. B., Zarrinkar P. P., Sapinoso L. M., Kern S. G., Behling C. A., Monk B. J., Lockhart D. J., Burger R. A., Hampton G. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge O., Pergamenschikov A., Williams C., Zhu S. X., Lønning P. E., Børresen-Dale A. L., Brown P. O., Botstein D. (2000) Nature 406, 747–752 [DOI] [PubMed] [Google Scholar]
- 10.Lin X., Gelman I. H. (1997) Cancer Res. 57, 2304–2312 [PubMed] [Google Scholar]
- 11.Su B., Zheng Q., Vaughan M. M., Bu Y., Gelman I. H. (2006) Cancer Res. 66, 5599–5607 [DOI] [PubMed] [Google Scholar]
- 12.Zetter B. R. (1998) Annu. Rev. Med. 49, 407–424 [DOI] [PubMed] [Google Scholar]
- 13.Kleinman H. K., Jacob K. (2007) Current Protocols in Cell Biology, John Wiley and Sons, Inc., New York [Google Scholar]
- 14.Mansour S. J., Matten W. T., Hermann A. S., Candia J. M., Rong S., Fukasawa K., Vande Woude G. F., Ahn N. G. (1994) Science 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 15.Abbott D. W., Holt J. T. (1999) J. Biol. Chem. 274, 2732–2742 [DOI] [PubMed] [Google Scholar]
- 16.Moissoglu K., Gelman I. H. (2003) J. Biol. Chem. 278, 47946–47959 [DOI] [PubMed] [Google Scholar]
- 17.Boissier S., Ferreras M., Peyruchaud O., Magnetto S., Ebetino F. H., Colombel M., Delmas P., Delaissé J. M., Clézardin P. (2000) Cancer Res. 60, 2949–2954 [PubMed] [Google Scholar]
- 18.Derossi D., Chassaing G., Prochiantz A. (1998) Trends Cell Biol. 8, 84–87 [PubMed] [Google Scholar]
- 19.Itoh T., Tanioka M., Yoshida H., Yoshioka T., Nishimoto H., Itohara S. (1998) Cancer Res. 58, 1048–1051 [PubMed] [Google Scholar]
- 20.Lokeshwar B. L. (1999) Ann. N.Y. Acad. Sci. 878, 271–289 [DOI] [PubMed] [Google Scholar]
- 21.Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. (1995) J. Biol. Chem. 270, 5331–5338 [DOI] [PubMed] [Google Scholar]
- 22.Egeblad M., Werb Z. (2002) Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 23.Suyama K., Shapiro I., Guttman M., Hazan R. B. (2002) Cancer Cell 2, 301–314 [DOI] [PubMed] [Google Scholar]
- 24.Vayalil P. K., Mittal A., Katiyar S. K. (2004) Carcinogenesis 25, 987–995 [DOI] [PubMed] [Google Scholar]
- 25.Lin X., Nelson P., Gelman I. H. (2000) Mol. Cell. Biol. 20, 7259–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinkas J., Leder P. (2002) Cancer Res. 62, 4781–4790 [PubMed] [Google Scholar]
- 27.Guvakova M. A., Adams J. C., Boettiger D. (2002) J. Cell Sci. 115, 4149–4165 [DOI] [PubMed] [Google Scholar]
- 28.Levin-Salomon V., Kogan K., Ahn N. G., Livnah O., Engelberg D. (2008) J. Biol. Chem. 283, 34500–34510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia W., Gelman I. H. (2002) Exp. Cell Res. 277, 139–151 [DOI] [PubMed] [Google Scholar]
- 30.Heuertz R. M., Hamann K. J., Hershenson M. B., Leff A. R. (1997) Am. J. Respir. Cell Mol. Biol. 17, 456–461 [DOI] [PubMed] [Google Scholar]
- 31.Kim B., Feldman E. L. (1998) J. Neurochem. 71, 1333–1336 [DOI] [PubMed] [Google Scholar]
- 32.Rouppe van der Voort C., Kavelaars A., van de Pol M., Heijnen C. J. (2000) J. Neuroimmunol. 108, 82–91 [DOI] [PubMed] [Google Scholar]
- 33.Bijian K., Takano T., Papillon J., Le Berre L., Michaud J. L., Kennedy C. R., Cybulsky A. V. (2005) Am. J. Physiol. Renal Physiol. 289, F1313–F1323 [DOI] [PubMed] [Google Scholar]
- 34.Bubb M. R., Spector I., Beyer B. B., Fosen K. M. (2000) J. Biol. Chem. 275, 5163–5170 [DOI] [PubMed] [Google Scholar]
- 35.Schlessinger J., Bar-Sagi D. (1994) Cold Spring Harbor Symp. Quant. Biol. 59, 173–179 [DOI] [PubMed] [Google Scholar]
- 36.Gelman I. H., Gao L. (2006) Mol. Cancer Res. 4, 151–158 [DOI] [PubMed] [Google Scholar]
- 37.Kolch W., Heidecker G., Kochs G., Hummel R., Vahidi H., Mischak H., Finkenzeller G., Marme D., Rapp U. R. (1993) Nature 364, 249–251 [DOI] [PubMed] [Google Scholar]
- 38.Schönwasser D. C., Marais R. M., Marshall C. J., Parker P. J. (1998) Mol. Cell. Biol. 18, 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapline C., Mousseau B., Ramsay K., Duddy S., Li Y., Kiley S. C., Jaken S. (1996) J. Biol. Chem. 271, 6417–6422 [DOI] [PubMed] [Google Scholar]
- 40.Nauert J. B., Klauck T. M., Langeberg L. K., Scott J. D. (1997) Curr. Biol. 7, 52–62 [DOI] [PubMed] [Google Scholar]
- 41.Piontek J., Brandt R. (2003) J. Biol. Chem. 278, 38970–38979 [DOI] [PubMed] [Google Scholar]
- 42.Ghayur T., Hugunin M., Talanian R. V., Ratnofsky S., Quinlan C., Emoto Y., Pandey P., Datta R., Huang Y., Kharbanda S., Allen H., Kamen R., Wong W., Kufe D. (1996) J. Exp. Med. 184, 2399–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyatt S. L., Liao L., Aderem A., Nairn A. C., Jaken S. (1994) Cell Growth Diff. 5, 495–502 [PubMed] [Google Scholar]
- 44.Yan X., Walkiewicz M., Carlson J., Leiphon L., Grove B. (2009) Exp. Cell Res. 315, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suthiphongchai T., Promyart P., Virochrut S., Tohtong R., Wilairat P. (2003) Oncol. Res. 13, 253–259 [DOI] [PubMed] [Google Scholar]
- 46.Tao J., Wang H. Y., Malbon C. C. (2007) J. Biol. Chem. 282, 6597–6608 [DOI] [PubMed] [Google Scholar]
- 47.Gelman I. H. (2002) Front. Biosci. 7, d1782–d1797 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.