Abstract
Mammalian cytosolic and mitochondrial thioredoxin reductases are essential selenocysteine-containing enzymes that control thioredoxin functions. Thioredoxin/glutathione reductase (TGR) is a third member of this enzyme family. It has an additional glutaredoxin domain and shows highest expression in testes. Herein, we found that human and several other mammalian TGR genes lack any AUG codons that could function in translation initiation. Although mouse and rat TGRs have such codons, we detected protein sequences upstream of them by immunoblot assays and direct proteomic analyses. Further gene engineering and expression analyses demonstrated that a CUG codon, located upstream of the sequences previously thought to initiate translation, is the actual start codon in mouse TGR. The use of this codon relies on the Kozak consensus sequence and ribosome-scanning mechanism. However, CUG serves as an inefficient start codon that allows downstream initiation, thus generating two isoforms of the enzyme in vivo and in vitro. The use of CUG evolved in mammalian TGRs, and in some of these organisms, GUG is used instead. The newly discovered longer TGR form shows cytosolic localization in cultured cells and is expressed in spermatids in mouse testes. This study shows that CUG codon is used as an inefficient start codon to generate protein isoforms in mouse.
Keywords: enzymes/Oxidation-Reduction, evolution/Protein, Sulfhydryls/Disulfide, Translation, Protein Synthesis, Protein Isoforms, Selenium, Thioredoxin Reductase, Translation Initiation
Introduction
Mammalian thioredoxin reductases (TRs)3 are essential enzymes that belong to a pyridine nucleotide disulfide oxidoreductase family (1, 2). In addition to the catalytic site, typical of the entire superfamily, TRs contain a C-terminal penultimate selenocysteine residue encoded by UGA codon (3). This selenocysteine is inserted with the help of selenocysteine insertion sequence element present in the 3′-UTRs of TRs and other selenoprotein genes. The TRs play key roles in the control of cellular redox homeostasis by maintaining thioredoxins (Trxs) in the reduced state, but they are also able to directly reduce certain small molecules such as selenite (4), hydroperoxides (5), dehydroascorbate (6), and NK-lysin (7).
Three TRs exist in mammals: TR1 (also known as TrxR1, TxnRd1, or TrxRα), TR3 (TrxR2, TxnRd2, TrxRβ), and TGR (TR2, TxnRd3). TR1 and TR3 functions are well characterized. The former is a cytosolic enzyme involved in cell growth (8), whereas the latter is mainly localized to mitochondria and is involved in heart development (9). Both proteins are present in all vertebrates and are essential for mouse embryogenesis (8, 9). On the other hand, mammalian TGR is abundant in testis, and its function is not well understood (10–14). It was suggested that TGR promotes disulfide bond isomerization between GPx4 and other proteins. GPx4 is another selenoprotein abundant in testes, which is both an enzyme and a structural protein of the mitochondrial sheath in sperm cells (15).
The main feature that distinguishes TGR from other mammalian TRs is an N-terminal glutaredoxin (Grx) domain. Grx is a Trx-fold protein and a component of another major redox system in mammals: the glutathione system (16–18). Despite an atypical active site motif in the Grx domain (i.e. CXXS instead of CXXC), this domain exhibits Grx activity either in concert with TGR or when expressed alone (11). Thus, this domain allows TGR to participate in both Trx and glutathione systems (13). A Grx-containing form of TR1 is also known, but it does not display activities typical of Grx (19, 20). Prior to TGR discovery, it was thought that Trx and glutathione systems work independently, but increasing evidence suggests cross-talk between these systems. In this regard, several previous observations deserve a particular attention. First, in Drosophila melanogaster, the Trx system substitutes for glutathione reductase (21, 22); second, in Schistosoma mansoni and related platyhelminths, there is neither TR nor glutathione reductase (GR), and TGR alone replaces both major redox systems (23–25).
We previously focused on the mouse TGR as a model protein (10–14). However, examination of its homologs in other mammals revealed a lack of initiation codons in several sequences in the position of the AUG codon previously predicted to serve as the start codon. Translation initiation signals other than AUG are common in viruses; they are also used in bacteria but are extremely rare in eukaryotes. In mammals, non-AUG triplets with the change in one nucleotide in AUG (with the exception of AGG and AAG codons) could direct translation initiation in vitro (26). However, not all of them are able to serve this function in vivo. To date, only about 30 proteins are known that utilize non-canonical initiation sites in mammals (27). The majority of these proteins are regulators of transcription and translation, growth factors, and cation transport channels. In some cases, the utilization of non-AUG codon is driven by IRES structure recognition (28, 29), and in other cases, it is driven by conventional ribosome-scanning mechanism (30, 31). In this work, we found that CUG is used as a start codon in mouse TGR and that this feature evolved to generate isoforms of this protein.
EXPERIMENTAL PROCEDURES
Analysis of TGR Genes
Genomic, non-redundant, and expressed sequence tag databases at the National Center for Biotechnology Information (NCBI) were scanned with tBLASTN using mouse TGR sequence (NM_153162) as a query. TGR sequences were then extended upstream and aligned using ClustalX.
Expression and Purification of Recombinant TGR
To generate a construct for expression of the short form of TGR in Escherichia coli, cDNA of mouse TGR was amplified using primers F1 and R1 (supplemental Table S1). The reverse primer contained a selenocysteine insertion sequence element, derived from E. coli formate dehydrogenase H gene, that was inserted immediately downstream of the TAG stop signal of TGR. This PCR product was cloned into pET28a(+) plasmid (Novagen) in-frame with the preexisting N-terminal His tag using EcoRI and NdeI restriction sites. The construct for expression of the full-length TGR was prepared in two stages. First, the sequence was amplified with primers F2 and R2 and then cloned into pET24(+) using EcoRI and NdeI sites. Second, a PCR procedure was used to add a His tag sequence at the N terminus using primers F3 and R3. The resulting plasmids were co-transformed into E. coli BL21(DE3) cells (New England Biolabs) together with pSUABC plasmid (32). Cells were grown in LB medium supplemented with 20 μm FAD and 10 μm sodium selenite, kanamycin, and chloramphenicol, and induction of protein synthesis was performed by adding 50 μm isopropyl-1-thio-β-d-galactopyranoside at A600 = 1 and incubating cells at 17 °C overnight.
Affinity purification of proteins was carried out using Talon resin (Clontech). 50 mm phosphate buffer, pH 7.5, was used containing 300 mm NaCl and 5 mm imidazole as an equilibration/wash solution, and 50 mm phosphate buffer, pH 7.5, containing 300 mm NaCl and 300 mm imidazole was used as the elution solution. Following elution, proteins were concentrated for further use.
Constructs for Expression in Mammalian Cells
GFP fusion constructs were prepared on the basis of pEGFPN1 (Clontech). The N-terminal part of mouse TGR including the extended longer form (designated extTGR) was cloned using primers F4 and R4 into the EcoRI/BamHI sites of pEGFPN1. The sequence located upstream of the previously predicted AUG start codon was separately fused to a GFP sequence using the same forward primer and R5 as a reverse primer, resulting in the extTGR-GFP construct. Variants of extTGR-GFP plasmids carrying deletions were made as follows: (i) Δ203–256 used primers D1 and D2, (ii) Δ1–92 used primers D3 and D4, and (iii) Δ93–119 used primers D5 and D6 (schematic representation is shown in supplemental Fig. S2). Plasmids carrying point mutations in mouse TGR Kozak sequence were made as follows: (i) mutation of CTG codon at position 146–148 of cDNA into CTC used primers P1 and P2, (ii) mutation of CTG at position 146–148 into ATG used primers P3 and P4, (iii) mutation of CC at position 144–145 into GT used primers P5 and P6, (iv) mutation of CA in position 136–137 into TT used primers P7 and P8, (v) mutation of GAG at position 149–151 into CAT used primers P9 and P10, and (vi) mutation of GCC at position 143–145 into CAT used primers P11 and P12. To examine TGR expression in HEK 293 cells, we removed the AUG start codon in the extTGR-GFP construct using primers P13 and P14. To replace the AUG codon of GFP in pEGFPN1 with the Kozak sequence of CUG codon in mouse TGR, we used primers F6 and R6 and obtained a PCR product from pEGFPN1, which was then inserted into the same vector digested with BamHI/NotI.
Cell Transfection and Lysate Preparation
Transfection of HEK 293 cells was carried out by the calcium chloride method. COS-1 and NIH 3T3 cells were transfected by Lipofectamine 2000 (Invitrogen). After 24–48 h of incubation, cells were collected and lysed in CelLytic M (Sigma). Lysates were directly used for SDS-PAGE analysis on 10% Bis-Tris mini gels (Invitrogen) followed by Western blotting. We prepared rabbit polyclonal antibodies against a shorter version of TGR and separately against peptide sequences coded by sequences upstream of AUG. Monoclonal anti-GFP antibodies were from Sigma. ECLTM donkey anti-rabbit (or anti-mouse in the case of GFP) IgG horseradish peroxidase-linked antibodies were used as secondary antibodies.
Tissue Samples
Testes were taken from C57BL/6 mice fed standard rodent chow (Harlan Teklad, Madison, WI). Tissues were fixed in 10% formalin and processed for paraffin embedding at the Veterinary Diagnostic Center, University of Nebraska, Lincoln, NE. Immunohistochemistry was performed with a Histostain-Plus kit (Zymed Laboratories Inc.) according to the manufacturer's instructions. Briefly, prior to staining, sections of testes were deparaffinized with xylene and passed through a graded series of ethanol. Non-immune goat serum (10%) was used to block nonspecific binding. The slides were incubated with antibodies against a short form of TGR (1:300 dilution) or antibodies against N-terminal sequences of TGR (1:10,000 dilution) for 1 h and washed with phosphate-buffered saline containing 0.05% Tween 20 (PBST). Biotinylated secondary antibodies were applied to the sections for 10 min. The slides were then washed with PBST and incubated with horseradish peroxidase-conjugated streptavidin followed by rinsing in PBST. Staining was performed using 3,3′-diaminobenzene chromogen. In addition, staining by hematoxylin (Invitrogen) was done according to the manual. Images were collected using a light Olympus AX70 microscope at the University of Nebraska-Lincoln Microscopy Core Facility.
RESULTS
Several Mammalian TGR Genes Lack AUG Start Codon
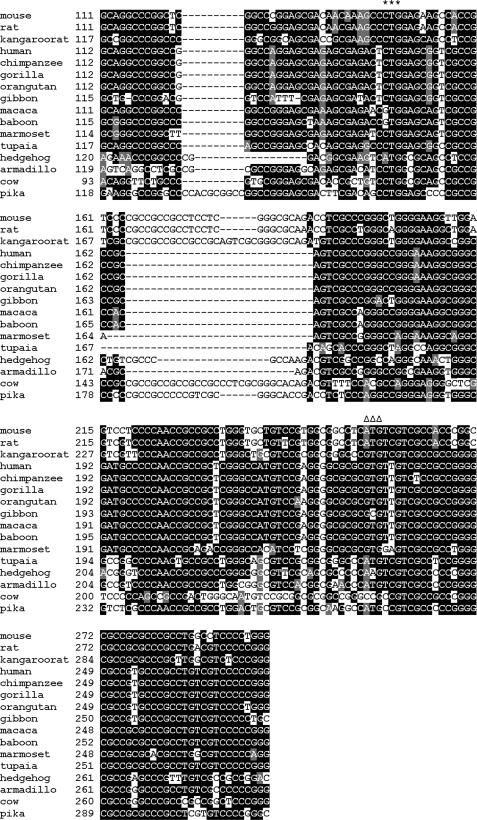
This study began with a surprising observation that human TGR gene lacked an AUG start codon in the position corresponding to the previously predicted start codon in mouse TGR and that upstream sequences in the human gene lacked any AUG at all. Multiple sequence alignment of mammalian TGR genes revealed that the mouse AUG was only present in rodents and several other animals, such as tupaia and armadillo, whereas humans, other primates, and several other mammals replaced AUG with other codons, and all these sequences lacked AUG upstream in the correct open reading frame (Fig. 1).
FIGURE 1.
Alignment of 5′ nucleotide sequences of mammalian TGRs. The alignment of nucleotide sequences of mammalian TGRs corresponding to the 5′-UTR and an initial part of the coding sequence is shown. The newly identified start codon is marked by stars, and the AUG previously thought to serve as the initiation signal is marked by triangles. For sequence accession numbers, see supplemental Table S2.
Mammalian TGRs Have Coding Sequences Upstream of AUG Codon in Mouse TGR
A region upstream of the mouse AUG codon showed high sequence conservation at both nucleotide and protein levels in mammals, and any changes in the nucleotide sequence were multiples of three (i.e. preserving the frame) (Figs. 1 and 2). This arrangement was indicative of coding sequences.
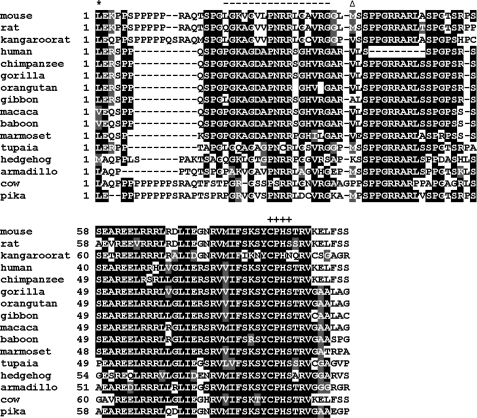
FIGURE 2.
Alignment of N-terminal protein sequences of mammalian TGRs. The alignment of N-terminal sequences of mammalian TGRs, starting from the CUG codon, is shown. First amino acid residue in protein is marked by stars, and the methionine previously thought to be the initial residue is marked by triangles. The active site of the Grx domain of TGR is designated by crosses. The peptide in the longer TGR form that was used as antigen for polyclonal antibodies is marked with dashes.
To directly detect N-terminal coding sequences, we purified TGR from rat testes and subjected it to liquid chromatography-tandem mass spectrometry analyses. In addition to tryptic peptides corresponding to internal sequences of TGR, this procedure identified a peptide that extended 8 amino acids upstream of the AUG codon. Taken together, these observations suggested that non-canonical translation initiation is used in mammalian TGR genes.
Mouse TGR Has an Alternative Start Codon Upstream of the Previously Reported AUG
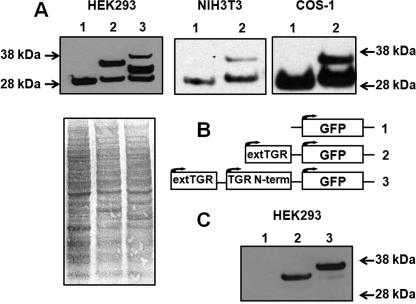
We verified the presence of the actual TGR mRNA sequences in testes of C57BL/6 mice by amplification and cDNA sequencing. Then, we cloned a region of mouse TGR corresponding to 22 amino acids downstream of the AUG together with the entire upstream sequences, or only the sequences upstream of the AUG, and prepared fusion constructs with GFP (including its AUG start codon). These constructs were then transfected into HEK 293, NIH 3T3, or COS-1 cells and examined for translation initiation by subjecting protein extracts to Western blots with anti-GFP antibodies (Fig. 3, A and B). When the sequences located upstream of the AUG were used, two protein forms were detected. One was GFP alone, and the other was a fusion of GFP with sequences coded by the upstream region; these sequences must have corresponded to the N terminus of TGR. The other construct yielded three bands that corresponded to GFP alone, AUG-originated mouse TGR, and the form that began with the natural start codon of TGR. Simultaneous detection of multiple protein forms suggested that the natural upstream start codon is inefficiently used.
FIGURE 3.
Expression of TGR constructs in cell lines. A, immunoblot analysis of expression products. The samples were as follows: lane 1, GFP alone; lane 2, GFP coding sequence fused with the 5′-UTR and sequences coding for the N-terminal region of TGR; lane 3, GFP sequences fused with the 5′-UTR and sequences coding for an additional N-terminal 22 amino acids. The constructs were expressed in the indicated cell lines, and the expressed proteins were probed in immunoblot assays with antibodies specific for GFP. B, schematic representation of constructs used in A. Numbers correspond to those in A. Arrows indicate positions at which translation initiation occurs. extTGR, extended longer form of TGR; N-term, N terminus. C, immunoblot analysis of samples shown in A with antibodies specific for the N-terminal region of TGR-L.
Mammalian TGRs Have a Candidate CUG Start Codon
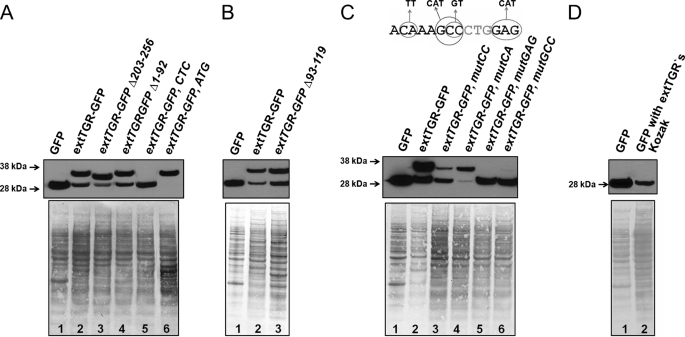
To identify an upstream start codon, we introduced deletions in the mouse TGR cDNA sequence and transfected such constructs into HEK 293 cells. When nucleotides 203–256 were deleted, the upper band, which corresponded to translation from an alternative start codon, appeared lower (Fig. 4A). This observation suggested that the upstream start codon should be closer to the 5′ end. Removing nucleotides 1–92 of TGR cDNA did not affect translation initiation. Nucleotides 93–119 that are conserved in mammals were not required either (Fig. 3B). From these observations and taking into account a 24-nucleotide gap in rat and mouse sequences relative to human TGR, we defined a 40-nucleotide region that contained an alternative start codon (Fig. 1). Based on the sequence alignment, CUG codon at position 146–148 emerged as a promising candidate for translation initiation. This codon was preserved in almost all mammals except hedgehog, macaque, and gibbon, but in those organisms, it was replaced with GUG or AUG codons, the two most common translation start sites in nature.
FIGURE 4.
Analysis of translation initiation function of the CUG codon. A, deletion and point mutation of CUG codon and its flanking areas were used to examine the ability of CUG to initiate translation. Lane 1, GFP control; lane 2, GFP fused with 5′-UTR and sequences coding for the N-terminal region of TGR-L; lane 3, GFP fusion construct carrying deletion at nucleotide positions 203–256; lane 4, GFP fusion construct with deletion at nucleotide positions 1–92; lane 5, GFP fusion construct in which CUG was mutated to CUC in nucleotide position 148; lane 6, GFP fusion construct in which CUG codon was mutated to AUG. extTGR, extended longer form of TGR. B, deletion of conserved sequences upstream of CUG does not affect translation initiation. Lane 1, GFP control; lane 2, GFP fusion construct that contains the 5′-UTR and sequences coding for the N-terminal region of TGR-L; lane 3, same construct, but with deletion in nucleotide positions 93–119. C, Kozak consensus sequences flanking the CUG codon are important for efficient initiation of translation. The upper panel summarizes mutations that were examined. The middle panel shows an immunoblot analysis with anti-GFP antibodies. Lysates of HEK 293 cells were used. Cells were transfected with GFP fusion constructs with mutations in the Kozak consensus sequence of the CUG codon. Lane 1, GFP control; lane 2, GFP fusion construct coding for the N-terminal part of TGR-L; lane 3, the same construct but CC was replaced with GT; lane 4, CA was replaced with TT; lane 5, GAG was replaced with CAT; and lane 6, GCC was replaced with CAT. The lower panel shows the protein loading control. D, Kozak consensus sequence of the longer form of TGR is sufficient to initiate translation at CUG codon in HEK 293 cells. Immunoblot analysis with antibodies specific for GFP is shown. Lane 1, expression of control GFP construct in HEK 293 cells; lane 2, expression of the GFP construct with AUG start codon replaced by CUG and its Kozak sequence (−8 to +6 positions relative to CUG) derived from the longer form of TGR.
To test whether CUG is the natural start codon, we mutated it into CUC. The mutation resulted in the loss of the larger protein form, whereas the forms that started from downstream sequences were intact (Fig. 4A, lane 5). A similar experiment was carried out in NIH 3T3 cells (supplemental Fig. S1). When CUG was changed to AUG, efficiency of translation initiation from this site increased such that only the larger protein form was detected (Fig. 4A, lane 6). Thus, CUG is a natural, albeit inefficient, start codon, and in addition, a downstream AUG codon can serve as a start codon wherein two protein forms are synthesized from the same mRNA. The novel large TGR form is hereafter designated as TGR-L; it differs from the previously known TGR form by a 4-kDa N-terminal extension.
Taking into account the data shown in Figs. 3 and 4, we developed polyclonal antibodies against the LGKVGVLPNRRLGAVRG peptide, which is part of the N-terminal extension and is unique to the long TGR form. Western blot analysis of the same samples as those shown in Fig. 3A with these antibodies provided additional evidence for TGR-L existence (Fig. 3C). In Fig. 3A, the upper band corresponded to the translation of the N-terminal part of TGR-L. Despite excellent Kozak sequence of the CUG codon, it serves as a weak initiator of translation and is subject to leaky scanning. This explains the observation of a middle band in lane 3 that originates from AUG start codon of the short TGR form. Moreover, this AUG has a weak Kozak consensus; thus, ribosome may also initiate translation at the downstream GFP sequence.
Mechanism of Translation Initiation from the CUG Codon
All cases of non-canonical start codon usage in mammals can be separated into two groups: IRES-dependent and IRES-independent. We carried out site-directed mutagenesis studies to determine the mechanism of CUG-initiated translation; specifically, we determined whether it utilizes IRES or is based on a typical ribosome-scanning mechanism. As discussed above, deletion of sequences upstream or downstream of the CUG codon and its 25 flanking nucleotides at the 5′ end had no influence on translation initiation. Because the shortest experimentally verified viral IRES has a length of 56 nucleotides, whereas the average size in mammals is about 300 nucleotides according to the IRESite data base (33), the functional sequences flanking CUG could not accommodate IRES. We further examined this mRNA region by Mfold and did not identify a stable mRNA structure that could function as IRES. Thus, IRES-dependent mechanism is not likely. We also made a set of constructs with point mutations in the consensus sequence that flanks the CUG (Fig. 4C). This region is referred to as the Kozak sequence for alternative initiation in a recent bioinformatics study (27); positions −7, −6, and −4 are particularly important in addition to the classical Kozak. These mutations either completely blocked or severely inhibited translation initiation. On the other hand, certain changes in nucleotide sequences facilitated it (e.g. replacement of GA in positions −7 and −6 with TT-increased fidelity of the CUG start codon). To further exclude a role of possible vicinal sequences in the CUG-driven translation initiation, we replaced the native AUG start codon in the control GFP construct by the Kozak region (from −8 to +6 positions) of TGR-L, including the CUG codon, and expressed this construct in HEK 293 cells. A clear and sharp band was observed by Western blotting with anti-GFP antibodies. Thus, the Kozak sequence of mouse TGR is sufficient for translation initiation at CUG codon (Fig. 4D).
Tissue Distribution of TGR and TGR-L
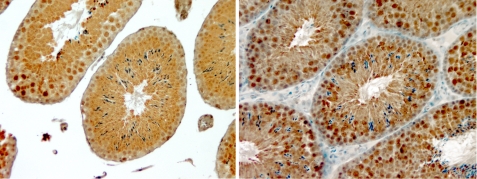
Previous studies showed that TGR is abundant in testis and is expressed in seminiferous tubuli. To examine TGR-L expression in vivo, we employed polyclonal antibodies against the unique N-terminal part of TGR-L. As a control, we used antibodies prepared against a recombinant TGR that lacked the N-terminal 4-kDa region; these antibodies recognized all TGR forms (total TGR). Immunohistochemical analyses revealed that the total TGR was evenly distributed among seminiferous tubuli cells, whereas TGR-L was less abundant on the outer edge of tubuli (Fig. 5). Thus, the long TGR form was expressed in mouse testes, and it showed an expression pattern that differed from that of total TGR. Overall, both TGR forms were apparently present in mouse testes.
FIGURE 5.
Localization of TGR in mouse seminiferous tubuli. The left panel shows staining of a mouse testis section with antibodies specific for the N-terminal part of TGR-L (brown) and hematoxylin (blue). The right panel shows staining of a mouse testis section with antibodies specific for the recombinant TGR and hematoxylin.
Localization of TGR in Cultured Cells
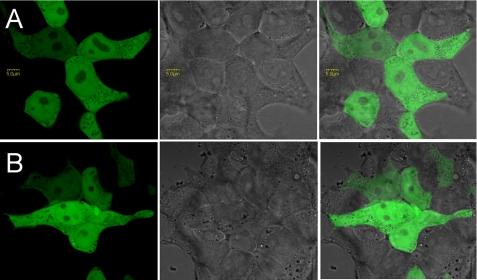
We examined the N-terminal sequence of TGR-L for being a localization signal. Computational analyses by PSORT II and other programs did not identify signal sequences in this region of TGR. We transfected HEK 293 cells with the construct coding for the 4-kDa sequence of TGR-L (designated as extTGR in Figs. 3 and 4) followed by GFP. Because the CUG start codon was not stringent in translation initiation resulting in background from unfused GFP, we mutated the natural AUG start codon of GFP. The resulting protein product was expressed and localized to cytosol (Fig. 6). As a control, we used a similar vector with the N-terminal part of regular TGR. Some nuclear staining was also detected, but it was not different from the expression of GFP alone (which has an inherent ability to pass through the nuclear membrane). The data suggest that the N-terminal sequence of TGR-L is not involved in targeting the protein to cellular compartments when expressed in transfected cells.
FIGURE 6.
Localization of the N-terminal region of TGR-L and a shorter form of TGR fused to GFP in HEK 293 cells. A, expression construct coding for the N-terminal part of TGR-L (up to the nucleotide position 256) fused to GFP. B, expression construct coding for the N-terminal part of short TGR (nucleotides 256–323) fused to GFP.
The Two Forms of TGR Occur in Mammals
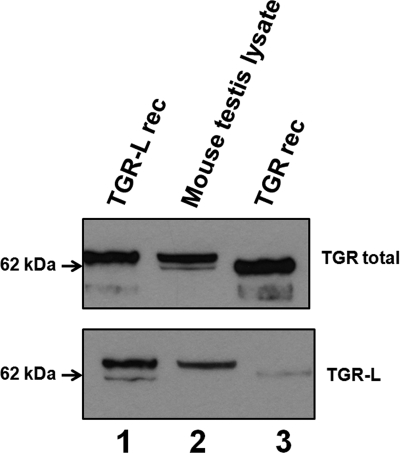
We examined whether the two forms of TGR occur in vivo. Western blot analysis of mouse testes with antibodies specific for recombinant TGR revealed two bands, which corresponded to TGR-L and a shorter form (Fig. 7). The assignment of the upper band to TGR-L was further verified by Western blotting with antibodies specific for the synthetic peptide in the N-terminal portion of TGR-L. We also observed heterogeneity of TGR forms in rat testes (data not shown). Overall, the two TGR forms were both generated upon expression of the gene in cell culture experiments and existed in vivo.
FIGURE 7.
TGR isoforms in mouse testes. Mouse testis lysate and recombinant TGR forms were subjected to immunoblot analyses. The upper panel represents samples analyzed with anti-TGR antibodies (lane 2). Recombinant TGR (lane 3) and TGR-L (lane 1) were purified from E. coli. The lower panel shows the same membrane probed with antibodies specific for the N-terminal part of TGR-L.
CUG Usage Evolved in Mammals
We traced the use of the CUG codon in TGRs by analyzing evolution of this protein in vertebrates and examining translation initiation signals in these sequences. Placing this information on the tree of life revealed that the ancestral form of TGR had a AUG start codon. This codon is still used in fish, birds, and amphibians. Most mammals, however, contain extension at the 5′ end of TGR mRNA with a conserved CUG codon, and some primates (baboon, macaque) use GUG instead. The acquired use of CUG and its conservation in mammals indicate that translation initiation is best served by a non-canonical codon in TGRs.
DISCUSSION
Numerous isoforms of mammalian thioredoxin reductases TR1 and TR3 are known. Some of them were predicted from the analyses of expressed sequence tag sequences, and some were experimentally verified (20, 34–38). As a rule, alternative first exon splicing is used in these genes wherein different mRNA forms are transcribed from unique promoters and generate unique N-terminal sequences that converge onto the common TR module. TR1 and TR3 forms may be targeted by their N-terminal sequences to different cellular compartments or to distinct interacting partners. In some cases, an alternative form of TR1, normally a cytosolic protein, localizes to mitochondria, whereas the mitochondrial TR3 can be targeted to the cytosol when alternative first exon splicing skips over the sequences coding for the mitochondrial signal (34). There is also an intriguing isoform of TR1 that has a Grx domain that shows no activity in the Grx assays. However, this domain can be activated by mutating two amino acids in the active site. Further studies linked the function of the Grx-containing TR1 to cytoskeleton rearrangements and cell shape (39, 40).
For TGR, however, no alternative forms were described. Searches in the expressed sequence tag data base did not reveal obvious candidates. However, on Western blots, mouse and rat TGRs appear as two bands (Fig. 7). Through a series of experiments, we found that TGR exists in two forms generated by leakage through a weak translation initiator codon. In experiments involving HEK 293 cells, translation from a AUG codon was more efficient than from a CUG codon. However, in mouse testis, this may not be the case. In addition, the mechanisms and details of translation initiation may vary between various organisms. In any case, the mechanism used to generate TGR forms differs from that in other TRs, although all three enzymes are present during spermatid development (cytosolic and mitochondrial TRs are essential enzymes that are expressed in all cell types). Both TGR isoforms occur in mouse testes; however, the longer form appears to be more abundant. Analysis of the alignment of mammalian TGRs suggests that human and some other organisms may only have a single form, the long form of TGR, due to the absence of the AUG codon downstream the CUG in mouse and rat sequences.
Utilization of non-AUG start codons is highly unusual in mammals. Only several proteins are known to use codons other than AUG, and they include growth factors, kinases, and transcription factors (27). Some of these proteins evolved an IRES structure that facilitates translation initiation, whereas others still utilize cap-dependent, ribosome-scanning mechanisms. For instance, FGF2 (fibroblast growth factor) has as many as five vicinal CUG codons, four of which participate in the IRES-driven translation, whereas the function of the remaining one is cap-dependent (41). In rare cases, regulation of CUG translation by trans-acting factors can occur (42). Recent studies suggest an unknown CUG-specific methionine-independent translational mechanism (43). In the majority of such proteins, a non-AUG codon is auxiliary to the main AUG start signal and is located upstream of it. Several such proteins are transcriptional regulators known to use only non-AUG codons to initiate translation (44–46). One exception is a phosphoribosylpyrophosphate synthase, which is involved in nucleotide synthesis and was found exclusively in testes (47).
The selective usage of CUG codon in TGR generates protein isoforms, and evolutionary analyses suggest that this function evolved specifically in mammals and has been almost uniformly preserved in these organisms. Non-mammalian (e.g. amphibians, fish, birds) TGRs still use AUG as the start codon. However, in the majority of mammals, a conserved CUG is used instead. Some primates replace it with GUG, which perhaps could also serve as an inefficient start signal. Our data suggest that the function of CUG is to provide inefficient translation initiation that allows production of two forms of TGR from a single mRNA species.
This work was supported, in whole or in part, by National Institutes of Health Grant GM065204 (to V. N. G.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- TR
- thioredoxin reductase
- TGR
- thioredoxin/glutathione reductase
- Trx
- thioredoxin
- Grx
- glutaredoxin
- GFP
- green fluorescent protein
- HEK
- human embryonic kidney
- IRES
- internal ribosomal entry site
- UTR
- untranslated region
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Arnér E. S. (2009) Biochim. Biophys. Acta 1790, 495–526 [DOI] [PubMed] [Google Scholar]
- 2.Holmgren A., Björnstedt M. (1995) Methods Enzymol. 252, 199–208 [DOI] [PubMed] [Google Scholar]
- 3.Gladyshev V. N., Jeang K. T., Stadtman T. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6146–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björnstedt M., Kumar S., Holmgren A. (1995) Methods Enzymol. 252, 209–219 [DOI] [PubMed] [Google Scholar]
- 5.Zhong L., Holmgren A. (2000) J. Biol. Chem. 275, 18121–18128 [DOI] [PubMed] [Google Scholar]
- 6.May J. M., Mendiratta S., Hill K. E., Burk R. F. (1997) J. Biol. Chem. 272, 22607–22610 [DOI] [PubMed] [Google Scholar]
- 7.Andersson M., Holmgren A., Spyrou G. (1996) J. Biol. Chem. 271, 10116–10120 [DOI] [PubMed] [Google Scholar]
- 8.Jakupoglu C., Przemeck G. K., Schneider M., Moreno S. G., Mayr N., Hatzopoulos A. K., de Angelis M. H., Wurst W., Bornkamm G. W., Brielmeier M., Conrad M. (2005) Mol. Cell Biol. 25, 1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad M., Jakupoglu C., Moreno S. G., Lippl S., Banjac A., Schneider M., Beck H., Hatzopoulos A. K., Just U., Sinowatz F., Schmahl W., Chien K. R., Wurst W., Bornkamm G. W., Brielmeier M. (2004) Mol. Cell Biol. 24, 9414–9423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q. A., Wu Y., Zappacosta F., Jeang K. T., Lee B. J., Hatfield D. L., Gladyshev V. N. (1999) J. Biol. Chem. 274, 24522–24530 [DOI] [PubMed] [Google Scholar]
- 11.Sun Q. A., Su D., Novoselov S. V., Carlson B. A., Hatfield D. L., Gladyshev V. N. (2005) Biochemistry 44, 14528–14537 [DOI] [PubMed] [Google Scholar]
- 12.Su D., Novoselov S. V., Sun Q. A., Moustafa M. E., Zhou Y., Oko R., Hatfield D. L., Gladyshev V. N. (2005) J. Biol. Chem. 280, 26491–26498 [DOI] [PubMed] [Google Scholar]
- 13.Sun Q. A., Kirnarsky L., Sherman S., Gladyshev V. N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3673–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehr S., Malinouski M., Finney L., Vogt S., Labunskyy V. M., Kasaikina M. V., Carlson B. A., Zhou Y., Hatfield D. L., Gladyshev V. N. (2009) J. Mol. Biol. 389, 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ursini F., Heim S., Kiess M., Maiorino M., Roveri A., Wissing J., Flohé L. (1999) Science 285, 1393–1396 [DOI] [PubMed] [Google Scholar]
- 16.Fernandes A. P., Holmgren A. (2004) Antioxid. Redox. Signal. 6, 63–74 [DOI] [PubMed] [Google Scholar]
- 17.Shelton M. D., Chock P. B., Mieyal J. J. (2005) Antioxid. Redox. Signal. 7, 348–366 [DOI] [PubMed] [Google Scholar]
- 18.Holmgren A. (2000) Antioxid. Redox. Signal. 2, 811–820 [DOI] [PubMed] [Google Scholar]
- 19.Rundlöf A. K., Janard M., Miranda-Vizuete A., Arnér E. S. (2004) Free Radic. Biol. Med. 36, 641–656 [DOI] [PubMed] [Google Scholar]
- 20.Su D., Gladyshev V. N. (2004) Biochemistry 43, 12177–12188 [DOI] [PubMed] [Google Scholar]
- 21.Kanzok S. M., Fechner A., Bauer H., Ulschmid J. K., Müller H. M., Botella-Munoz J., Schneuwly S., Schirmer R., Becker K. (2001) Science 291, 643–646 [DOI] [PubMed] [Google Scholar]
- 22.Cheng Z., Arscott L. D., Ballou D. P., Williams C. H., Jr. (2007) Biochemistry 46, 7875–7885 [DOI] [PubMed] [Google Scholar]
- 23.Alger H. M., Williams D. L. (2002) Mol. Biochem. Parasitol. 121, 129–139 [DOI] [PubMed] [Google Scholar]
- 24.Angelucci F., Miele A. E., Boumis G., Dimastrogiovanni D., Brunori M., Bellelli A. (2008) Proteins 72, 936–945 [DOI] [PubMed] [Google Scholar]
- 25.Bonilla M., Denicola A., Novoselov S. V., Turanov A. A., Protasio A., Izmendi D., Gladyshev V. N., Salinas G. (2008) J. Biol. Chem. 283, 17898–17907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peabody D. S. (1989) J. Biol. Chem. 264, 5031–5035 [PubMed] [Google Scholar]
- 27.Wegrzyn J. L., Drudge T. M., Valafar F., Hook V. (2008) BMC Bioinformatics 9, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieft J. S. (2008) Trends Biochem. Sci. 33, 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baird S. D., Turcotte M., Korneluk R. G., Holcik M. (2006) RNA 12, 1755–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. (1999) Gene 234, 187–208 [DOI] [PubMed] [Google Scholar]
- 32.Arnér E. S., Sarioglu H., Lottspeich F., Holmgren A., Böck A. (1999) J. Mol. Biol. 292, 1003–1016 [DOI] [PubMed] [Google Scholar]
- 33.Mokrejs M., Vopálenský V., Kolenaty O., Masek T., Feketová Z., Sekyrová P., Skaloudová B., Kríz V., Pospísek M. (2006) Nucleic Acids Res. 34, D125–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turanov A. A., Su D., Gladyshev V. N. (2006) J. Biol. Chem. 281, 22953–22963 [DOI] [PubMed] [Google Scholar]
- 35.Rundlöf A. K., Carlsten M., Arnér E. S. (2001) J. Biol. Chem. 276, 30542–30551 [DOI] [PubMed] [Google Scholar]
- 36.Rundlöf A. K., Fernandes A. P., Selenius M., Babic M., Shariatgorji M., Nilsonne G., Ilag L. L., Dobra K., Björnstedt M. (2007) Differentiation 75, 123–132 [DOI] [PubMed] [Google Scholar]
- 37.Chang E. Y., Son S. K., Ko H. S., Baek S. H., Kim J. H., Kim J. R. (2005) Free Radic. Biol. Med. 39, 1666–1675 [DOI] [PubMed] [Google Scholar]
- 38.Miranda-Vizuete A., Spyrou G. (2002) Mol. Cells 13, 488–492 [PubMed] [Google Scholar]
- 39.Dammeyer P., Damdimopoulos A. E., Nordman T., Jiménez A., Miranda-Vizuete A., Arnér E. S. (2008) J. Biol. Chem. 283, 2814–2821 [DOI] [PubMed] [Google Scholar]
- 40.Damdimopoulou P. E., Miranda-Vizuete A., Arnér E. S., Gustafsson J. A., Damdimopoulos A. E. (2009) Biochim. Biophys. Acta 1793, 1588–1596 [DOI] [PubMed] [Google Scholar]
- 41.Arnaud E., Touriol C., Boutonnet C., Gensac M. C., Vagner S., Prats H., Prats A. C. (1999) Mol. Cell Biol. 19, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A. C., Vagner S. (2005) J. Biol. Chem. 280, 4144–4153 [DOI] [PubMed] [Google Scholar]
- 43.Starck S. R., Ow Y., Jiang V., Tokuyama M., Rivera M., Qi X., Roberts R. W., Shastri N. Y. (2008) PloS one 3, e3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao J. H., Davidson I., Matthes H., Garnier J. M., Chambon P. (1991) Cell 65, 551–568 [DOI] [PubMed] [Google Scholar]
- 45.Falvey E., Fleury-Olela F., Schibler U. (1995) EMBO J. 14, 4307–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imataka H., Olsen H. S., Sonenberg N. (1997) EMBO J. 16, 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taira M., Iizasa T., Shimada H., Kudoh J., Shimizu N., Tatibana M. (1990) J. Biol. Chem. 265, 16491–16497 [PubMed] [Google Scholar]