Abstract
Mitochondrial folate-dependent one-carbon (1-C) metabolism converts 1-C donors such as serine and glycine to formate, which is exported and incorporated into the cytoplasmic tetrahydrofolate (THF) 1-C pool. Developing embryos depend on this mitochondrial pathway to provide 1-C units for cytoplasmic process such as de novo purine biosynthesis and the methyl cycle. This pathway is composed of sequential methylene-THF dehydrogenase, methenyl-THF cyclohydrolase, and 10-formyl-THF synthetase activities. In embryonic mitochondria, the bifunctional MTHFD2 enzyme catalyzes the dehydrogenase and cyclohydrolase reactions, but the enzyme responsible for the mitochondrial synthetase reaction has not been identified in embryos. A monofunctional 10-formyl-THF synthetase (MTHFD1L gene product) functions in adult mitochondria and is a likely candidate for the embryonic activity. Here we show that the MTHFD1L enzyme is present in mitochondria from normal embryonic tissues and embryonic fibroblast cell lines, and embryonic mitochondria possess the ability to synthesize formate from glycine. The MTHFD1L transcript was detected at all stages of mouse embryogenesis examined. In situ hybridizations showed that MTHFD1L was expressed ubiquitously throughout the embryo but with localized regions of higher expression. The spatial pattern of MTHFD1L expression was virtually indistinguishable from that of MTHFD2 and MTHFD1 (cytoplasmic C1-THF synthase) in embryonic day 9.5 mouse embryos, suggesting coordinated regulation. Finally, we show using stable isotope labeling that in an embryonic mouse cell line, greater than 75% of 1-C units entering the cytoplasmic methyl cycle are mitochondrially derived. Thus, a complete pathway of enzymes for supplying 1-C units from the mitochondria to the methyl cycle in embryonic tissues is established.
Keywords: Development Differentiation, Enzymes/Multifunctional, Metabolism/Folate, Organisms/Mammal, Subcellular Organelles/Mitochondria, Transcription/Development
Introduction
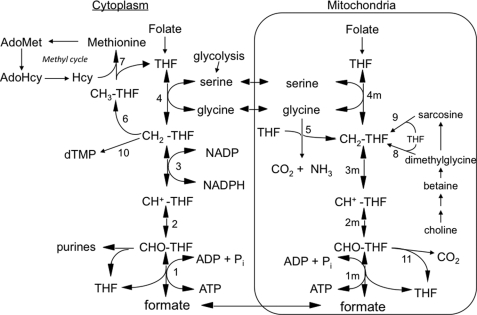
Tetrahydrofolate (THF)4-dependent one-carbon (1-C) metabolism is highly compartmentalized in eukaryotes, with THF-dependent enzymes found in mitochondria, cytoplasm, and nuclei (1, 2). The 3-carbon of serine is the major 1-C donor in most organisms, including humans (3), and THF can be charged with this one-carbon unit in both the cytoplasmic and mitochondrial compartments via serine hydroxymethyltransferase (SHMT) (Fig. 1, reactions 4 and 4m), resulting in the formation of 5,10-methylene-THF (CH2-THF). Cytoplasmic CH2-THF can be reduced to 5-methyl-THF (see Fig. 1, reaction 6) for entry into the methyl cycle, oxidized to 10-formyl-THF (10-CHO-THF) (Fig. 1, reactions 3 and 2) for purine synthesis, or it can be used for nuclear thymidylate (dTMP) synthesis (Fig. 1, reaction 10) (2). In mitochondria, the other product of the SHMT reaction, glycine, can also serve as a source of one-carbon units (4, 5). It is metabolized by the mitochondrially localized glycine cleavage system (Fig. 1, reaction 5), producing CH2-THF from its 2-carbon. CH2-THF, from either serine or glycine, can be oxidized to 10-CHO-THF by mitochondrial versions of reactions 3 and 2 in Fig. 1. 10-CHO-THF can either be converted to formate and THF by 10-formyl-THF synthetase (Fig. 1, reaction 1m) or oxidized to form CO2 and THF by 10-formyl-THF dehydrogenase (Fig. 1, reaction 11) (6, 7).
FIGURE 1.
Embryonic mammalian one-carbon metabolism. Reactions 1–4 are in both the cytoplasmic and mitochondrial (m) compartments. Reactions 1–3, 10-formyl-THF synthetase, 5,10-methenyl-THF cyclohydrolase, and 5,10-methylene-THF dehydrogenase, respectively, are catalyzed by trifunctional C1-THF synthase in the cytoplasm (MTHFD1). In mammalian mitochondria, reaction 1m is catalyzed by monofunctional MTHFD1L, and reactions 2m and 3m are catalyzed by bifunctional MTHFD2. The other reactions are catalyzed by the following: serine hydroxymethyltransferase (reactions 4 and 4m), glycine cleavage system (reaction 5), 5,10-methylene-THF reductase (reaction 6), methionine synthase (reaction 7), dimethylglycine dehydrogenase (reaction 8), sarcosine dehydrogenase (reaction 9), thymidylate synthase (reaction 10), and 10-formyl-THF dehydrogenase (reaction 11; only the mitochondrial activity of this enzyme is shown, but it has been reported in both compartments in mammals). All of the reactions from choline to sarcosine are mitochondrial except the betaine to dimethylglycine conversion, which is cytoplasmic. Hcy, homocysteine; AdoHcy, S-adenosylhomocysteine.
The cytoplasmic and mitochondrial compartments are metabolically connected by transport of serine, glycine, and formate across the mitochondrial membranes, supporting a mostly unidirectional flow (clockwise in Fig. 1) of 1-C units from serine to formate and onto methionine. In fact, it appears that under most conditions, the majority of 1-C units for cytoplasmic processes are derived from mitochondrial formate (6–17).
In mammals, birds, and yeast, the cytoplasmic activities of CH2-THF dehydrogenase, 5,10-methenyl-THF cyclohydrolase, and 10-CHO-THF synthetase (Fig. 1, reactions 1–3) are present on a trifunctional enzyme called C1-THF synthase (18–22). The CH2-THF dehydrogenase activity of C1-THF synthase is NADP-dependent, and this redox cofactor specificity may explain the preference of the cytoplasmic pathway to operate in the reductive direction in vivo (i.e. from 10-CHO-THF to CH2-THF) (23). The cytoplasmic NADP/NADPH redox couple is maintained in a reduced state (24, 25), greatly favoring the reductive direction of NADP-linked dehydrogenases (26). The mammalian version of this trifunctional enzyme is encoded by the MTHFD1 gene (27–29), and its cytoplasmic protein product will herein be designated as MTHFD1.
The enzymes catalyzing reactions 1m–3m (Fig. 1) in mammalian mitochondria are much less clear. MacKenzie and co-workers (30, 31) characterized a bifunctional NAD-dependent CH2-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase (reactions 3m and 2m), originally isolated from ascites tumor cells. This enzyme was later shown to be a mitochondrial protein (32, 33), encoded by the nuclear MTHFD2 gene. Notably, this enzyme (MTHFD2 protein) is found only in transformed mammalian cells and embryonic or nondifferentiated tissues (30) and is clearly essential during embryonic development (34, 35). The final step in the mammalian mitochondrial pathway to formate (10-CHO-THF synthetase; reaction 1m) is catalyzed by mitochondrial C1-THF synthase, encoded by the MTHFD1L gene (36). This isozyme, herein referred to as MTHFD1L, is a homolog of the cytoplasmic MTHFD1. Unlike MTHFD1, however, MTHFD1L is a monofunctional enzyme, containing only the 10-CHO-THF synthetase activity (37).
To date, the MTHFD1L protein has been confirmed only in mitochondria from adult tissues (36, 38). However, a 10-CHO-THF synthetase activity has been detected in mouse embryonic fibroblasts (MEFs) (28), and the authors suggested MTHFD1L as the most likely candidate enzyme in embryos to explain this activity. However, in normal embryonic tissues, neither the 10-CHO-THF synthetase activity nor an associated protein has been demonstrated. Previous metabolic studies with MEFs support the above model of mitochondrial formate production and release, suggesting a role for 10-CHO-THF synthetase activity in this process (14, 28, 39). Although embryonically derived cells provide a useful experimental system to study the compartmentation of 1-C metabolism, there are limitations of these cultured cells as models for embryogenesis. We thus sought evidence for the participation of MTHFD1L in providing 1-C units to the cytoplasm during normal embryogenesis.
Here we show that embryonic mitochondria contain the MTHFD1L protein and are capable of producing formate from the 1-C donor glycine. We investigate the spatial and temporal expression patterns of the MTHFD1L gene in mouse embryos and compare them to those of MTHFD2 and MTHFD1. Finally, metabolic tracer studies in MEFs show that disruption of mitochondrial 1-C flux creates significant disturbances in the cytoplasmic methyl cycle. These results thus verify and extend the current model of mammalian 1-C cycle metabolism to embryos.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Oligonucleotides were synthesized by IDT (Coralville, IA). The nitrocellulose membranes were obtained from Midwest Scientific (Valley Park, MO). T7, T3, and SP6 polymerases were purchased from Epicentre Biotechnologies (Madison, WI). Restriction enzymes were purchased from either Invitrogen or Fisher/Promega (Madison, WI). Geneticin (G-418 sulfate) was obtained from American Bioanalytical (Natick, MA). Kits for plasmid preparation were from Qiagen.
Animals
All of the study protocols were approved by the Institutional Animal Care and Use Committee of The University of Texas at Austin and conform to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Wild type CF-1® mice (Charles River Laboratories) were housed at the UT-Austin Animal Resource Center. Vaginal plugs observed at noon in female mice were considered as embryonic day 0.5 (E0.5). The embryos were also staged based on somite number.
Isolation of Mitochondria
Mouse embryos between E13.0 and E17.5 were dissected from pregnant females along with the adult livers. Mitochondria were isolated as described previously for adult liver (6, 7) except that in some cases, the pellet from the initial centrifugation was resuspended, Dounce homogenized, and recentrifuged, and the supernatant was pooled with the first supernatant.
Mitochondrial Oxidation of [2-14C]Glycine
Mitochondrial formate production was measured essentially as described previously (6, 7), except that [2-14C]glycine (Morevek Biochemicals, Brea, CA) was used as 1-C donor. Incubations contained 0.4 ml of intact mitochondria, 5.0 mm [2-14C]glycine (1000 dpm/nmol), 50 mm HEPES (pH 7.2), 50 mm KCl, 25 mm KH2PO4, 2 mm MgCl2, and 240 mm sucrose in a total volume of 1 ml. [14C]Formate produced was determined by enzymatic assay using formate dehydrogenase (from Candida boidinii; Sigma-Aldrich) as described previously (6). Formate production was normalized to the mitochondrial protein concentration of each sample as determined by the Bradford assay (40). The p values were calculated utilizing the Student's t test.
SDS-PAGE and Immunoblotting
Extracts of cultured cells and mitochondria isolated from embryos were analyzed by SDS-PAGE and immunoblotting as described (38). The membranes were probed with a polyclonal rabbit antibody specific for MTHFD1L (38) or a rabbit anti-Hsp60 antibody (Stressgen Biotechnologies, San Diego, CA). The secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit IgG (Zymed Laboratories Inc., San Francisco, CA).
Northern Analysis
A Mouse Conceptus full stage Northern blot was obtained from Seegene, Inc. (Seoul, South Korea). Each lane contained 20 μg of total RNA from embryos aged E4.5–18.5. The probes were synthesized with [α-32P]dATP (3000 Ci/mmol; PerkinElmer Life Sciences) by asymmetric PCR using Taq polymerase (Applied Biosystems, Carlsbad, CA) and reagents supplied in the Strip E-Z PCR kit (Ambion, Austin, TX) according to the kit manufacturer's instructions. A cDNA containing the full-length mouse MTHFD1L coding sequence was obtained from the American Type Culture Collection (Image Clone identification number 6311761, GenBankTM access number BQ91728). The SP6 promoter/primer lay at the 5′ end of the insert, whereas the T7 promoter/primer site was at the 3′ end. Mouse MTHFD2 (clone 24FL) and MTHFD1 (clone 14EB) cDNAs, each containing partial sequences, were generously provided by Dr. R. E. MacKenzie (McGill University). For 24FL, the T7 promoter flanked the 5′ end of the insert; the T3 promoter/primer was on the 3′ side. The orientation of the 14EB insert was opposite that of 24FL. The 432-bp MTHFD1L probe was synthesized using forward and reverse primers STP5′ (5′-AAGAACTTGCAGCCGCATCTC-3′) and Mmomito (5′-CCAGACCAGCCTCCTTGGCCAAA-3′), respectively. The 431-bp MTHFD2 probe was synthesized using forward and reverse primers M2F5′ (5′-ACAATCCCGCCAGTCACTCCTAT-3′) and M2F3′ (5′-TTCGTGAGCTCCATCTGTGTGCA-3′), respectively. The 450-bp MTHFD1 probe was synthesized using forward and reverse primers MC5′ (5′-TGAAGGCTGCTGAGGAGATTG-3′) and MC3′ (5′-TTACTTCGACCAACCACCACCG-3′), respectively. Hybridization and visualization were as described previously (36). Densitometric quantification of Northern blot bands was performed using Image J (National Institutes of Health). The bands were normalized to a digitized photographic negative of ribosomal RNA on the gel from which the blot was prepared. The densities of 18 and 28 S bands for each lane were summed and divided by the intensity in the lane of highest density. The relative band densities were determined similarly for the MTHFD1L, MTHFD2, and MTHFD1 blots. The normalized values for each lane were obtained by dividing the individual lane standardized intensities for each blot by the standardized intensities of the ribosomal RNA values in the respective lane. Approximate transcript sizes were estimated by comparison with the mobilities of RNA standards.
In Situ Hybridizations of Whole Mouse Embryos
Antisense and sense probes to MTHFD1L and MTHFD2 transcripts were constructed for in situ hybridizations in whole embryos (E9.5–13.5). For both the 5′- and 3′ end probes to MTHFD1L, the full-length mouse cDNA was shortened to ensure optimal probe length. Shortening for the 5′ end probe used two HindIII restriction sites: one within the insert and the other in the vector. The 3′ end probe was constructed using SmaI sites. Religation of each MTHFD1L plasmid yielded 873 bp of 5′ sequence and 1008 bp of 3′ sequence, respectively. Following linearization, antisense and sense probes were synthesized using T7 and SP6 polymerases, respectively. For MTHFD2 antisense and sense probes, the complete 1045-bp insert was utilized. Following linearization, T7 and T3 polymerases were utilized for synthesis of the sense and antisense probes, respectively. Polymerization reactions included 10 mm dithiothreitol (from the polymerase kits), 4 units of RNase Inhibitor (Ambion), 1× digoxigenin RNA labeling mix (1 mm ATP, CTP, and GTP, 0.65 mm UTP, 0.35 mm digoxigenin-11-UTP (Roche Applied Science)) and diethylpyrocarbonate-treated water (Ambion). Purification of the digoxigenin-labeled RNA probes was carried out by ammonium acetate precipitation. Probe concentration was determined using a NanoDrop Spectrophotometer (Thermo-Fisher Scientific).
RNA in situ hybridization was performed as described previously (41) using BM Purple (Roche Applied Science) as the alkaline phosphatase substrate. Light microscopy was carried out using an Olympus SZH10 (Olympus Optical, Hamburg, Germany), and the images were captured via CCD camera (Optronics, Goleta CA). The images were collated using Adobe Photoshop® (Adobe Systems, San Jose, CA).
Metabolic Tracer Studies
Wild type (IF22) and nullizygous MTHFD2 (IF74) MEFs were donated by Dr. R. E. MacKenzie (McGill University). Both cell lines were grown on Dulbecco's modified Eagle's medium/High medium (Hyclone, Logan, UT) supplemented with 15% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 1× penicillin/streptomycin, 1× nonessential amino acids (Hyclone), and 200 μm l-glutamine. In addition, for IF22 cells, 3.0 μm thymidine and 30 μm hypoxanthine were added; for IF74 cells, 167 μm sodium formate and 154 μm deoxyuridine monophosphate were added (14, 34, 39). The cells were grown at 37 °C under 5% CO2 on 150-mm plates (Nunc) and divided or harvested at 80–95% confluency. The protocol for labeling is a modification of that described previously (13). The labeling medium (a gift from Dr. Patrick Stover, Cornell University) was α-minimum essential medium (Hyclone) lacking sodium bicarbonate, folic acid, nucleotides, nucleosides, histidine, serine, glycine, and methionine. Folic acid (4 mg/liter), methionine (10 μm), sodium bicarbonate (2.6 g/liter), [5,5,5-2H3]leucine (100 μm), and [2,3,3-2H3]serine (500 μm) were added to the medium. Dialyzed fetal bovine serum (15%, 1:10 v/v in water) was also added. The cells were trypsinized, resuspended in labeling medium, and plated. The cells were harvested after two or three cell divisions and either used immediately or frozen at −70 °C.
Crude protein extracts were prepared by suspending cells in 5% trichloroacetic acid and incubating on ice. Precipitated proteins were recovered by centrifugation, the supernatants were discarded, and the pellets were suspended in constant boiling 6 n HCl (Pierce) and placed under vacuum in borosilicate tubes. Hydrolysis of proteins was at 110 °C for 20 h. The amino acids were derivatized using ethylchloroformate and trifluoroethanol (42, 43). The lyophilized hydrolysate pellet was suspended in 20 μl of water and the pH adjusted to ∼7.0 with ammonium bicarbonate. To this hydrolysate suspension, 100 μl of water (60 parts), trifluoroethanol (32 parts), and pyridine (8 parts) and 5 μl of ethylchloroformate were mixed and incubated for 2–3 min at room temperature. 100 μl of 1% ethylchloroformate in chloroform was added to each sample and mixed, and the phases were allowed to separate. The chloroform phase was removed for mass spectrometric analysis.
Mass Spectrometry Analysis
Derivatized amino acids were subjected to gas chromatography (Agilent 6890N with a SGE BP5 25 meter long column, 0.22 mm I.D. and 0.25 μm film thickness) coupled to a Waters Autospec magnetic sector mass spectrometer. Gas chromatography parameters were as previously published (43). Methionine eluted from the column at ∼7.5 min. The natural abundance for fully derivatized ethylchloroformate/trifluoroethanol-methionine (m/z = 302–308) was determined in non-deuterium-enriched IF22 MEFs by dividing the maximum height of each mass spectra peak by the sum of the heights of all seven peaks. Deuterium enrichment for IF22 and IF74 cell lines were calculated as described elsewhere (44), based on the natural abundances and the heights of peaks in the deuterium-enriched sample peaks. The native cellular natural abundances for MEFs were 1.76% for m/z = 302, 4.22% for m/z = 303, 78.73% for m/z = 304, 9.79% for m/z = 305, 4.91% for m/z = 306, 0.52% for m/z = 307, and 0.07% for m/z = 308. For deuterium-enriched samples, the heights of the methionine mass spectra peaks m/z = 304, 305, 306, and 307 were measured. Resolution of the resulting matrix of equations for singly and doubly deuterated and nondeuterated methionine was achieved using the LINEST function in Microsoft Excel (12). Mass spectra data were only considered reliable if the relative intensity of the base peak was above 15,000 and below 65,500. Below 15,000, background levels became problematic. Above 65,500, the mass spectrometer detector was saturated. To eliminate outliers caused by instrument error, multiple individual scans within each mass spectra peak were analyzed. Statistical analysis was performed using the JMP® statistical analysis software (SAS Institute, Cary, NC). All of the p values comparing wild type MEFs (IF22) and MTHFD2-null MEFs (IF74) were calculated using the Student's t test.
RESULTS
Embryonic Mitochondria Synthesize Formate
Intact adult rat liver mitochondria are able to oxidize various 1-C donors to formate without the addition of any other cofactors or substrates (6, 7). Mitochondrial production of formate requires reactions 3m, 2m, and 1m of the folate-dependent pathway (Fig. 1). To determine whether normal embryonic mitochondria possess this pathway, 1-C oxidation assays were performed on mitochondria isolated from mouse embryos at E13.0 and E15.5. In this assay, a radiolabeled 1-C donor is taken up by intact mitochondria and metabolized to formate and CO2 if the complete folate-dependent pathway is functioning. [2-14C]glycine was used because it can only donate 1-C units in mitochondria via the glycine cleavage system (Fig. 1, reaction 5). Formate production was assayed in mitochondria isolated from whole mouse embryos and from the livers of the respective mothers.
Formate production from adult mouse liver mitochondria (1.65 ± 0.12 nmol/mg) agreed well with previous results using serine (6, 7) or glycine.5 E15.5 mouse embryo mitochondria produced significantly more formate (2.03 ± 0.004 nmol/mg; p = 0.026) than adult liver mitochondria. E13.0 embryo mitochondria exhibited even higher formate production (3.6 nmol/mg), but because of the large quantity of mitochondria needed for this assay and the miniscule size of the E13.0 embryos, only a single replicate could be done. These data clearly indicate that normal embryonic mitochondria possess a complete folate-dependent pathway, including the 10-CHO-THF synthetase reaction.
Embryonic Tissues and Cell Lines Express the MTHFD1L Protein
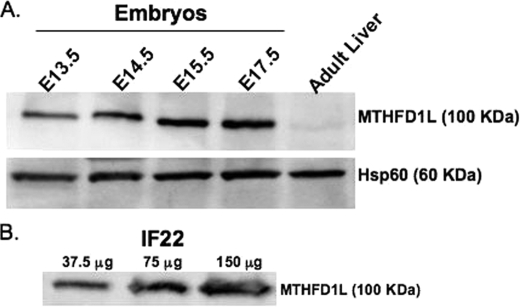
The best candidate protein to contain a 10-CHO-THF synthetase activity in embryonic mitochondria is the MTHFD1L gene product, which has previously been characterized in adult tissues (36, 38). To confirm the presence of the MTHFD1L protein in embryos, isolated mitochondria from E13.5–17.5 embryos were subjected to SDS-PAGE and immunoblotting using antibodies specific for MTHFD1L (38). A duplicate membrane was probed with antibodies against the mitochondrial matrix marker, Hsp60, as a loading control. The MTHFD1L protein is very abundant in mouse embryos on all days examined (Fig. 2A, top panel), compared with that found in the adult liver. The low expression of the MTHFD1L enzyme in adult mouse liver is consistent with previous reports showing low transcript levels in adult human liver (36) and low protein levels in adult rat livers (38).
FIGURE 2.
Mitochondrial C1-THF synthase (MTHFD1L) is expressed in both embryos and MEFs. A, immunoblotting was performed on mitochondria isolated from E13.5 to E17.5 embryos and from adult liver. In each lane, 50 μg of total mitochondrial protein was loaded. The top panel shows MTHFD1L protein intensities. Hsp60 is a mitochondrial matrix marker used as a loading control. B, MTHFD1L protein was observed by immunoblot of increasing amounts of IF22 MEF whole cell extracts.
Mouse embryonic fibroblasts have been used as a model system to study embryonic 1-C flux (14, 28, 39). To better characterize this model, whole cell protein extracts were prepared from wild type MEFs and subjected to immunoblotting for MTHFD1L. Fig. 2B shows that MEFs also express MTHFD1L, further validating these cells as a model of normal embryonic metabolism.
MTHFD1L, MTHFD1, and MTHFD2 Transcripts Exhibit Similar Spatial Expression Patterns but Different Temporal Expression Patterns in Developing Mouse Embryos
Transcripts of MTHFD1 and MTHFD2 have previously been examined in embryos for both their spatial and, to a limited extent, temporal expression patterns (35). Because the 10-CHO-THF synthetase activity of MTHFD1L bridges the activities of mitochondrial MTHFD2 and cytoplasmic MTHFD1 in the 1-C cycle, the temporal and spatial embryonic expression patterns of the MTHFD1L gene were investigated by Northern blot and in situ hybridization and compared with those of MTHFD2 and MTHFD1.
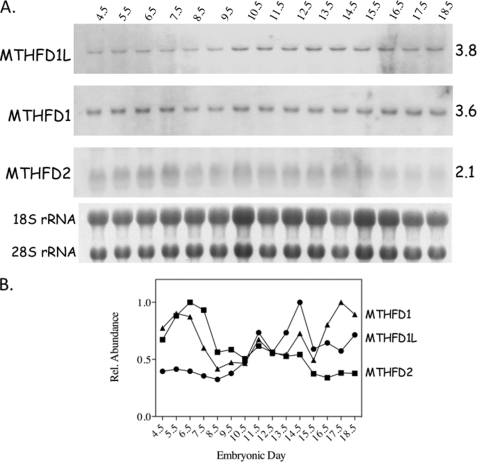
A mouse developmentally staged Northern blot was probed for the transcripts of MTHFD1L, MTHFD2, and MTHFD1. Each lane contained 20 μg of total RNA isolated from mouse embryos of a single age group (E4.5–18.5). All three transcripts were detected at the expected size at all embryonic stages investigated (Fig. 3A), but their temporal expression patterns differed considerably. MTHFD1L expression starts relatively low but rises as birth approaches (between E19 and E21). Conversely, MTHFD2 expression begins relatively high but decreases as the embryos develop. The MTHFD1 transcript peaks around day E6 and again just before birth. The results normalized to the ribosomal RNA in their respective lanes are illustrated graphically in Fig. 3B. The MTHFD1 and MTHFD2 expression profiles shown here are consistent with previously published results (27, 35).
FIGURE 3.
Temporal expression of MTHFD1L, MTHFD1, and MTHFD2 transcripts in mouse embryos. A, developmentally staged Northern blots were hybridized with probes to MTHFD1L, MTHFD1 and MTHFD2 transcripts. The age of the embryos from which the RNA was obtained is indicated above the blots in embryonic days. Approximate transcript sizes, in kilobases, is indicated on the right. The 18 and 28 S ribosomal RNAs from a photographic negative of the gel from which the blot was prepared are shown below the blots. These bands were used as loading controls. B, normalized relative transcript levels from each blot.
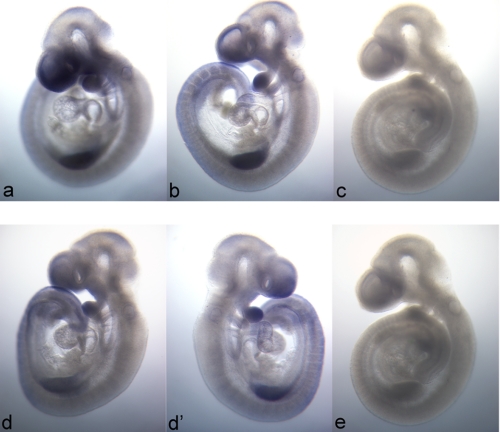
MTHFD1 and MTHFD2 transcripts are reported to stain ubiquitously throughout mouse embryos but share localized regions of elevated expression (35). To examine the spatial expression of the MTHFD1L gene, in situ hybridizations for the MTHFD1L and MTHFD2 mRNAs were performed on E9.5 whole mouse embryos. Fig. 4 illustrates that, like MTHFD1 and MTHFD2, the MTHFD1L transcript is also expressed throughout the embryo but with localized regions of higher expression including the future forebrain, the first branchial arch, and the forelimb bud. Indeed, the spatial pattern of MTHFD1L expression, using either a 5′ end (Fig. 4, panel a) or 3′ end (Fig. 4, panel b) probe, is virtually indistinguishable from that of MTHFD2 (Fig. 4, panels d and d′) in E9.5 mouse embryos. Sense probes to MTHFD1L (Fig. 4, panel c) and MTHFD2 (Fig. 4, panel e) showed no staining.
FIGURE 4.
Spatial distribution of MTHFD1L and MTHFD2 expression in E9.5 mouse embryos. In situ hybridization patterns of MTHFD1L (top row) are identical for probes to the 5′ end (panel a) and 3′ end (panel b). MTHFD1L (panels a and b) and MTHFD2 (panels d and d′) demonstrate nearly identical expression profiles. The left and right sides of the same embryo stained for MTHFD2 are represented in panels d and d′, respectively. Panels c and e are E9.5 embryos stained with sense probes to the 5′ end of MTHFD1L and to MTHFD2, respectively. The images are all 40× magnification.
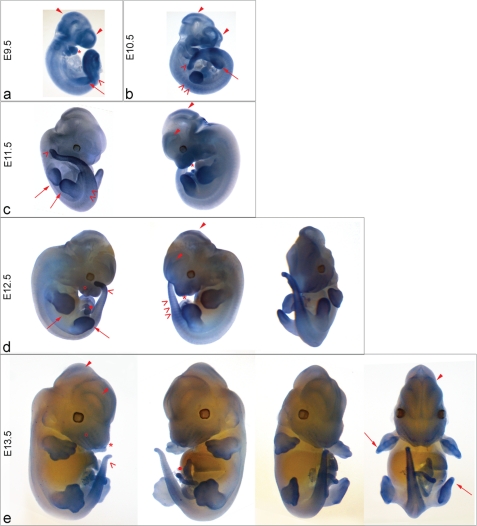
Using the 5′ end probe, the localization of MTHFD1L was examined at different embryonic stages to further characterize which areas exhibit elevated expression and how expression in these areas changes during embryogenesis. Regions of higher expression for all days examined included portions of the neural tube, the developing forebrain and midbrain (Fig. 5, closed arrowheads), the craniofacial region including the first two branchial arches (asterisk), somites (faintly at E10.5), presomitic mesoderm and tail (open arrowheads), limb buds (long arrows), and umbilicus (Fig. 5, panels d and e, closed circles). From E11.5 to E13.5 (Fig. 5, panels c–e), the developing digits (long arrows) are outlined, whereas the vibrissae (open circles) only become defined on E12.5 and E13.5 (Fig. 5, panels d and e). In these older embryos (Fig. 5, panels d and e), it is unlikely that the probe completely penetrates the interior of the embryo. Nevertheless, these panels reveal MTHFD1L expression in surface features such as the whiskers and thin structures such as the umbilical cord, digits, and tail. As for MTHFD2, MTHFD1L expression was not observed in the developing heart. This MTHFD1L expression pattern is essentially indistinguishable from the patterns previously reported for MTHFD2 and MTHFD1 on days E9.5 and E10.5 (35). Thus, despite temporal differences in embryonic transcript levels between MTHFD2, MTHFD1L, and MTHFD1 (as shown by Northern blotting; Fig. 3), their spatial expression patterns are essentially identical.
FIGURE 5.
mRNA Expression of MTHFD1L in Mouse Embryos. In situ hybridization of MTHFD1L (5′ end probe) mRNA was performed on whole mouse embryos ranging from E9.5 to E13.5. Expression was observed throughout the embryos at all of these stages. MTHFD1L is especially prominent in the developing forebrain and midbrain (closed arrowheads); developing limb buds and digits (panels a–e, long arrows); somites, presomitic mesoderm, and developing tail (open arrowheads); and developing branchial arches and embryonic jaw (asterisks). Expression is observed in the developing vibrissae (open circles) at E12.5 (panel d) and E13.5 (panel e) and umbilicus (closed circle). The images were photographed at 40× magnification for E9.5 and E10.5 (panel a and b), 30× for E11.5 and E12.5 (panels c and d), and 12.5× for E13.5 embryos (panel e).
Embryonic Mitochondria Contribute Significant 1-C Units to the Methyl Cycle
Having shown the presence of functional MTHFD1L and a complete mitochondrial folate-dependent 1-C pathway in embryonic cells, studies in MEF cells were performed to determine whether disruption of this pathway impacts cytoplasmic methyl cycle 1-C unit utilization, and if so, to what extent. Immortalized wild type (IF22) and MTHFD2-null (IF74) embryonic fibroblast lines were derived from E9.5–11.5 mouse embryos (39). In this assay, previously used with MCF-7 and SH-SY5Y cells (13, 16), deuterated serine ([2,3,3-2H3]serine) serves as the 1-C donor, and the incorporation of deuterium into methionine is followed. Serine donates its third carbon, along with two deuterium atoms, to THF to form deuterated methylene-THF (CD2-THF). If this occurs in the cytoplasm, catalyzed by cSHMT (Fig. 1, reaction 4), methionine derived from this 1-C unit will retain both deuterium atoms. On the other hand, if the deuterated serine donates its 1-C unit in the mitochondrion, via mitochondrial serine hydroxymethyltransferase, and this mitochondrial CD2-THF is converted to formate, one deuterium will be lost in the CH2-THF dehydrogenase reaction (Fig. 1, reaction 3m). Formate produced from this 1-C unit will be singly deuterated. Incorporation of this deuterated formate into the cytoplasmic THF 1-C pool will produce singly deuterated 5,10-methylene-THF (CDH-THF). Methionine derived from this mitochondrial 1-C unit will retain only one deuterium atom.
[2,3,3-2H3]serine was incubated with the two MEF cell lines. Extracted cellular proteins were hydrolyzed into free amino acids, derivatized, and analyzed by gas chromatography-mass spectrometry to determine the relative quantities of singly and doubly deuterated and nondeuterated methionine in each sample. The data presented in Table 1 indicate that disruption of the mitochondrial 1-C pathway caused a dramatic decrease (p < 0.00001) in the ratio of singly deuterated methionine to total deuterated methionine. For wild type cells (IF22), this ratio ranged from 76 to 92%. In the MTHFD2-null cell line (IF74), only 19 to 27% of deuterated methionine was singly deuterated. The ratios of total deuterated methionine to total methionine, singly deuterated methionine to total methionine, and doubly deuterated methionine to total methionine also differed significantly between the wild type and mutant cell lines. Of particular interest, the doubly deuterated methionine to total methionine ratio indicates that the MTHFD2-null cell line (IF74) exhibited 9-fold greater incorporation of cytoplasmically derived (doubly deuterated) 1-C units relative to the wild type cells. These data clearly demonstrate that disruption of mitochondrial flux causes significant changes in methyl cycle metabolism.
TABLE 1.
Metabolic tracer studies: determination of deuterated 1-C unit species in methionine
Singly deuterated methionine to total deuterated methionine (MetD1/MetTD), total deuterated methionine to total methionine (MetTD/MetTot), singly deuterated methionine to total methionine (MetD1/MetTot), and doubly deuterated methionine to total methionine (MetD2/MetTot) were measured in wild type (IF22) and MTHFD2 nullizygous (IF74) MEFs after incubation with [2,3,3-2H3]serine. The relative intensities of each methionine isotopic species were measured using gas chromatography-mass spectrometry. Total deuterated methionine is the sum of singly and doubly deuterated methionine. Total methionine is total deuterated methionine plus nondeuterated methionine. Each sample calculation gives the percentage ± standard deviation derived from the individual mass spectra of the respective ratio for each biological replicate. Each average calculation reports the average of the biological replicates along with the standard deviation. The p values compare the IF22 and IF74 averages.
| MetD1/MetTD | MetTD/MetTot | MetD1/MetTot | MetD2/MetTot | |
|---|---|---|---|---|
| IF22 | ||||
| Sample 1 | 92.2 ± 7.1 | 4.9 ± 1.7 | 4.6 ± 1.7 | 0.4 ± 0.3 |
| Sample 2 | 89.0 ± 6.8 | 3.9 ± 1.6 | 3.4 ± 1.2 | 0.5 ± 0.4 |
| Sample 3 | 75.9 ± 7.6 | 2.8 ± 1.3 | 2.1 ± 1.0 | 0.7 ± 0.4 |
| Average | 85.7 ± 8.6 | 3.9 ± 1.1 | 3.4 ± 1.3 | 0.5 ± 0.2 |
| IF74 | ||||
| Sample 1 | 26.9 ± 14.4 | 5.5 ± 2.2 | 1.4 ± 0.8 | 4.1 ± 1.7 |
| Sample 2 | 23.5 ± 2.9 | 7.3 ± 1.4 | 1.7 ± 0.5 | 5.6 ± 0.9 |
| Sample 3 | 18.9 ± 7.9 | 5.0 ± 2.6 | 1.1 ± 0.7 | 3.9 ± 1.7 |
| Average | 23.1 ± 4.0 | 5.9 ± 1.2 | 1.4 ± 0.3 | 4.5 ± 0.9 |
| p values for IF22 vs. IF74 | <0.00001 | 0.003 | <0.0001 | <0.0001 |
DISCUSSION
This study establishes a critical role for the 10-formyl-THF synthetase activity of the MTHFD1L protein in mitochondrial formate production during mammalian embryogenesis. We show that intact mitochondria isolated from mouse embryos possess a functional 1-C pathway as evidenced by their ability to synthesize formate from radiolabeled glycine. Moreover, the MTHFD1L protein was detected in both embryonic mitochondria and MEF cells by immunoblotting (Fig. 2). Thus, MTHFD1L catalyzes the final step in the mitochondrial conversion of 1-C units to formate in embryos. Moreover, MTHFD1L levels were substantially higher in embryonic mitochondria than in adult liver mitochondria (Fig. 2A), and embryonic mitochondria exhibited greater formate production. Because embryogenesis is a time of massive cell proliferation as well as cell migration and differentiation, it is not surprising that mitochondrial 1-C flux and MTHFD1L are elevated compared with adult tissues.
Northern blots and in situ hybridizations were performed to examine the temporal and spatial expression profiles of MTHFD2, MTHFD1L, and MTHFD1 during mouse embryogenesis. Although all three genes are expressed at all stages of fetal development, each transcript exhibited a distinct temporal profile (Fig. 3). MTHFD1L is expressed at relatively low levels early but increases in the later stages of embryogenesis. In contrast, MTHFD2 expression is high during early development but decreases as birth approaches, consistent with previous results (35). The MTHFD1 transcript exhibited an early peak of expression around embryonic day 6 and then increased again as birth approached (Fig. 3A). A late increase in MTHFD1 expression was also observed in fetal rat liver (27). Thus, transcriptional control of these genes provides a mechanism to modulate the flux of 1-C units from mitochondria into cytoplasmic processes such as purine and thymidylate biosynthesis and the methyl cycle. De novo purine biosynthesis is essential for cell division, and S-adenosylmethionine (AdoMet) synthesis is critical for chromatin and DNA methylation, which play essential roles during cell differentiation (45, 46) and cell migration (47). Because proper development requires precise interplay of cell proliferation, migration, and differentiation, coordinate control of the transcription of these three genes likely plays an important role during embryogenesis.
In situ hybridization studies were performed on whole mouse embryos to compare the spatial transcript profiles of MTHFD1L and MTHFD2. Previous reports showed that MTHFD2 and MTHFD1 have nearly identical spatial patterns (35). Our results show that the MTHFD1L gene shares this same spatial expression (Fig. 4). Staining was ubiquitous throughout the embryo, but with certain regions exhibiting elevated levels of expression. Regions showing the most intense staining included the developing brain, craniofacial structures, the developing limbs and digits, parts of the neural tube, and the tail bud (Fig. 5). Interestingly, nearly all of these regions are associated with neural crest cells (NCCs). The craniofacial region and neural tube, as well as the peripheral and autonomic nervous system (found in the limbs, digits, and vibrissae) are all of NCC origin (48, 49). Moreover, many of the elevated areas are developing into highly complex and organized tissues, organs, and body regions (i.e. forebrain, limbs, and digits). In either case, these cells would be undergoing high levels of cell division, differentiation, and, in the case of NCCs, migration. In fact, dysregulation of NCCs has previously been suggested to be a factor in folate-associated neural tube defects (50, 51). As seen with MTHFD1 and MTHFD2, the regions of elevated expression for MTHFD1L parallel those reported here (Fig. 4) and elsewhere for MTHFD2 (34) on days E9.5 and E10.5. Thus, in developing mammals, MTHFD1L, MTHFD2, and MTHFD1 appear to have two alternative expression states: a ubiquitous low basal expression level throughout the embryo and a higher expression level in selected tissues. The low basal expression levels are sufficient to support 1-C metabolism in most cells, but certain cells require increased 1-C flux to supply 1-C units for de novo purine and/or AdoMet synthesis.
Of note, at least two folate-responsive mouse mutations causing neural tube defects affect regions that show elevated expression of MTHFD1L, MTHFD1, and MTHFD2. In crooked tail mice (52, 53), the tail develops an abnormal “crook” in heterozygotes, and exencephaly is common in homozygotes. Cart1 mutants develop exencephaly associated with decreased density of mesenchyme cells in the developing forebrain (54). The forebrain and tail bud of mouse embryos are regions where MTHFD2, MTHFD1L, and MTHFD1 transcripts are all elevated (Fig. 5). Because these areas have higher potential rates of mitochondrial 1-C flux, aberrations in folate-mediated 1-C metabolism, whatever the cause, might be more detrimental in these areas compared with regions expressing lower levels of these enzymes. Indeed, a common variant in the human MTHFD1L gene has recently been shown to be associated with increased risk of neural tube defects in an Irish population (55).
Embryonic cells thus possess a complete mitochondrial 1-C pathway capable of oxidizing 1-C units from donors such as serine or glycine to formate. Moreover, this mitochondrial pathway is linked to cytoplasmic de novo purine synthesis and the methyl cycle via the 10-CHO-THF synthetase and the CH2-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase activities of cytoplasmic MTHFD1. This suggests that the mitochondrial 1-C pathway, in conjunction with MTHFD1 in the cytoplasm, modulates the flow of 1-C units not only into purines but also into the methyl cycle. Previous studies with cultured cells confirm that mitochondria supply 1-C units to the cytosol for de novo synthesis of purines (4, 14, 28, 39) and thymidylate (13, 16). Studies in animals and cancer-derived cell lines have suggested that the mitochondrial 1-C pathway can also supply most of the methyl cycle 1-C units as well (12, 13, 16, 17, 29). However, direct evidence that disruption of mitochondrial 1-C flux causes perturbations in the methyl cycle has been lacking. We used metabolic tracer experiments in MEFs to test this hypothesis. Wild type cells directed significantly more mitochondrially derived 1-C units into the methyl cycle than the MTHFD2-null cells (86% versus 23%; Table 1), confirming the participation of the mitochondrial pathway in the production of methyl cycle 1-C units in embryonic cells. These results are consistent with previous reports showing that 70–99% of methyl cycle 1-C units are of mitochondrial origin in normal mammalian cells and tissues (12, 13, 16).
Furthermore, the data from these metabolic tracer studies show that no more than 25–30% of 1-C units in the methyl cycle could be cytoplasmically derived. This number correlates with that seen in adult tissues (12). This residual of singly deuterated 1-C units in the mthfd2-null cells suggests that there is likely an additional mitochondrial 5,10-CH2-THF dehydrogenase active in these cells. We have recently discovered a MTHFD2-like protein with this activity in adult mitochondria.6 Future work will determine whether this enzyme is also present in embryonic cells and tissues.
One potential consequence of this compartmentalized pathway that directs most 1-C units through the mitochondria is that de novo purine synthesis may have priority over dTMP and AdoMet production with respect to the 1-C units. That is, mitochondrial formate passes through the 10-CHO-THF pool (donor for purines) before it reaches the CH2-THF pool (donor for dTMP) and 5-methyl-tetrahydrofolate pool (donor for AdoMet). This might be especially important in rapidly dividing cells. The CH2-THF pool may be further compartmentalized between cytoplasm and nucleus. Previous studies with cultured cells suggested that cSHMT can regulate the partitioning of CH2-THF between thymidylate and AdoMet synthesis (13). It was subsequently shown that during the S phase of the cell cycle, cSHMT translocates into the nucleus and provides CH2-THF for dTMP synthesis (2). Thus, compartmentalization may be an important mechanism by which the limited supply of THF-activated 1-C units is preferentially directed into specific pathways to meet the changing metabolic needs of the cell. Enzyme defects that block or decrease 1-C flux through one compartment may cause increased flux through alternative pathways in other compartments. Indeed, knock-out mice lacking cSHMT exhibit increased uracil incorporation into liver DNA, suggesting that cSHMT normally prevents uracil misincorporation by maintaining adequate nuclear dTMP synthesis (17). Conversely, mice with deficient MTHFD1 expression exhibit decreased uracil incorporation into liver DNA (29). Thus, 1-C unit compartmentalization may play a role in normal developmental processes, as well as disease processes, such as thymidylate deficiency-induced double-stranded DNA breaks that are common in some colorectal cancers (56–58).
Acknowledgments
We acknowledge Margaret Centilli for help with the mice and Mariam Kuruvilla for assisting with the in situ hybridizations. We also thank Dr. Byron Wingerd for assistance with the statistical analysis of the mass spectrometry data and manuscript critique and Dr. Olga de Marcucci, Dr. Gisela Kramer, and Dr. M. Alejandra Manzan for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK61425 and GM086856 (to D. R. A.) and HD10668 (to K. A.).
A. Tibbetts, personal communication.
S. Bolusani, B. Young, N. Cole, A. S. Tibbetts, and D. R. Appling, manuscript in preparation.
- THF
- tetrahydrofolate
- 1-C
- one-carbon
- AdoMet
- S-adenosylmethionine
- CH2-THF
- 5,10-methylene-tetrahydrofolate
- 10-CHO-THF
- 10-formyl-tetrahydrofolate
- cSHMT
- cytoplasmic serine hydroxymethyltransferase
- dTMP
- thymidylate
- E
- embryonic day
- MEF
- mouse embryonic fibroblast
- MTHFD1
- cytoplasmic C1-tetrahydrofolate synthase
- MTHFD1L
- mitochondrial C1-tetrahydrofolate synthase
- MTHFD2
- NAD-dependent 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methenyl-tetrahydrofolate cyclohydrolase
- NCC
- neural crest cell.
REFERENCES
- 1.Appling D. R. (1991) FASEB J. 5, 2645–2651 [DOI] [PubMed] [Google Scholar]
- 2.Woeller C. F., Anderson D. D., Szebenyi D. M., Stover P. J. (2007) J. Biol. Chem. 282, 17623–17631 [DOI] [PubMed] [Google Scholar]
- 3.Davis S. R., Stacpoole P. W., Williamson J., Kick L. S., Quinlivan E. P., Coats B. S., Shane B., Bailey L. B., Gregory J. F., 3rd (2004) Am. J. Physiol. Endocrinol. Metab. 286, E272–E279 [DOI] [PubMed] [Google Scholar]
- 4.Fu T. F., Rife J. P., Schirch V. (2001) Arch. Biochem. Biophys. 393, 42–50 [DOI] [PubMed] [Google Scholar]
- 5.Lamers Y., Williamson J., Theriaque D. W., Shuster J. J., Gilbert L. R., Keeling C., Stacpoole P. W., Gregory J. F., 3rd (2009) J. Nutr. 139, 666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barlowe C. K., Appling D. R. (1988) Biofactors 1, 171–176 [PubMed] [Google Scholar]
- 7.García-Martínez L. F., Appling D. R. (1993) Biochemistry 32, 4671–4676 [DOI] [PubMed] [Google Scholar]
- 8.Barlowe C. K., Appling D. R. (1990) Mol. Cell Biol. 10, 5679–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasternack L. B., Laude D. A., Jr., Appling D. R. (1994) Biochemistry 33, 74–82 [DOI] [PubMed] [Google Scholar]
- 10.Pasternack L. B., Littlepage L. E., Laude D. A., Jr., Appling D. R. (1996) Arch. Biochem. Biophys. 326, 158–165 [DOI] [PubMed] [Google Scholar]
- 11.Kastanos E. K., Woldman Y. Y., Appling D. R. (1997) Biochemistry 36, 14956–14964 [DOI] [PubMed] [Google Scholar]
- 12.Gregory J. F., 3rd, Cuskelly G. J., Shane B., Toth J. P., Baumgartner T. G., Stacpoole P. W. (2000) Am. J. Clin. Nutr. 72, 1535–1541 [DOI] [PubMed] [Google Scholar]
- 13.Herbig K., Chiang E. P., Lee L. R., Hills J., Shane B., Stover P. J. (2002) J. Biol. Chem. 277, 38381–38389 [DOI] [PubMed] [Google Scholar]
- 14.Patel H., Pietro E. D., MacKenzie R. E. (2003) J. Biol. Chem. 278, 19436–19441 [DOI] [PubMed] [Google Scholar]
- 15.Quinlivan E. P., Davis S. R., Shelnutt K. P., Henderson G. N., Ghandour H., Shane B., Selhub J., Bailey L. B., Stacpoole P. W., Gregory J. F., 3rd. (2005) J. Nutr. 135, 389–396 [DOI] [PubMed] [Google Scholar]
- 16.Anguera M. C., Field M. S., Perry C., Ghandour H., Chiang E. P., Selhub J., Shane B., Stover P. J. (2006) J. Biol. Chem. 281, 18335–18342 [DOI] [PubMed] [Google Scholar]
- 17.MacFarlane A. J., Liu X., Perry C. A., Flodby P., Allen R. H., Stabler S. P., Stover P. J. (2008) J. Biol. Chem. 283, 25846–25853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paukert J. L., Williams G. R., Rabinowitz J. C. (1977) Biochem. Biophys. Res. Comm. 77, 147–154 [DOI] [PubMed] [Google Scholar]
- 19.Schirch L. (1978) Arch. Biochem. Biophys. 189, 283–290 [DOI] [PubMed] [Google Scholar]
- 20.Smith G. K., Mueller W. T., Wasserman G. F., Taylor W. D., Benkovic S. J. (1980) Biochemistry 19, 4313–4321 [DOI] [PubMed] [Google Scholar]
- 21.Hum D. W., Bell A. W., Rozen R., MacKenzie R. E. (1988) J. Biol. Chem. 263, 15946–15950 [PubMed] [Google Scholar]
- 22.Thigpen A. E., West M. G., Appling D. R. (1990) J. Biol. Chem. 265, 7907–7913 [PubMed] [Google Scholar]
- 23.West M. G., Horne D. W., Appling D. R. (1996) Biochemistry 35, 3122–3132 [DOI] [PubMed] [Google Scholar]
- 24.Sies H. (1982) in Metabolic Compartmentation (Sies H. ed) pp. 205–231, Academic Press, New York [Google Scholar]
- 25.Veech R. L. (1987) in Pyridine Nucleotide Coenzymes: Chemical, Biochemical, and Medical Aspects, Part B (Dolphin D., Poulson R., Avramovic O. eds) pp. 79–105, John Wiley & Sons, New York [Google Scholar]
- 26.Atkinson D. E. (1977) Cellular Energy Metabolism and Its Regulation, Academic Press, New York [Google Scholar]
- 27.Howard K. M., Muga S. J., Zhang L., Thigpen A. E., Appling D. R. (2003) Gene 319, 85–97 [DOI] [PubMed] [Google Scholar]
- 28.Christensen K. E., Patel H., Kuzmanov U., Mejia N. R., MacKenzie R. E. (2005) J. Biol. Chem. 280, 7597–7602 [DOI] [PubMed] [Google Scholar]
- 29.MacFarlane A. J., Perry C. A., Girnary H. H., Gao D., Allen R. H., Stabler S. P., Shane B., Stover P. J. (2009) J. Biol. Chem. 284, 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mejia N. R., MacKenzie R. E. (1985) J. Biol. Chem. 260, 14616–14620 [PubMed] [Google Scholar]
- 31.Mejia N. R., Rios-Orlandi E. M., MacKenzie R. E. (1986) J. Biol. Chem. 261, 9509–9513 [PubMed] [Google Scholar]
- 32.Mejia N. R., MacKenzie R. E. (1988) Biochem. Biophys. Res. Commun. 155, 1–6 [DOI] [PubMed] [Google Scholar]
- 33.Bélanger C., MacKenzie R. E. (1989) J. Biol. Chem. 264, 4837–4843 [PubMed] [Google Scholar]
- 34.Di Pietro E., Sirois J., Tremblay M. L., MacKenzie R. E. (2002) Mol. Cell Biol. 22, 4158–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Pietro E., Wang X. L., MacKenzie R. E. (2004) Biochim. Biophys. Acta 1674, 78–84 [DOI] [PubMed] [Google Scholar]
- 36.Prasannan P., Pike S., Peng K., Shane B., Appling D. R. (2003) J. Biol. Chem. 278, 43178–43187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walkup A. S., Appling D. R. (2005) Arch. Biochem. Biophys. 442, 196–205 [DOI] [PubMed] [Google Scholar]
- 38.Prasannan P., Appling D. R. (2009) Arch. Biochem. Biophys. 481, 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel H., Di Pietro E., Mejia N., MacKenzie R. E. (2005) Arch. Biochem. Biophys. 442, 133–139 [DOI] [PubMed] [Google Scholar]
- 40.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 41.Conlon R. A., Rossant J. (1992) Development 116, 357–368 [DOI] [PubMed] [Google Scholar]
- 42.Vatankhah M., Moini M. (1994) Biol. Mass Spectrometry 23, 277–282 [Google Scholar]
- 43.Helgadóttir S., Rosas-Sandoval G., Söll D., Graham D. E. (2007) J. Bacteriol. 189, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storch K. J., Wagner D. A., Burke J. F., Young V. R. (1988) Am. J. Physiol. 255, E322–E331 [DOI] [PubMed] [Google Scholar]
- 45.Bai S., Ghoshal K., Datta J., Majumder S., Yoon S. O., Jacob S. T. (2005) Mol. Cell Biol. 25, 751–766 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Kobayakawa S., Miike K., Nakao M., Abe K. (2007) Genes Cells 12, 447–460 [DOI] [PubMed] [Google Scholar]
- 47.Horswill M. A., Narayan M., Warejcka D. J., Cirillo L. A., Twining S. S. (2008) Exp. Eye Res. 86, 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basch M. L., García-Castro M. I., Bronner-Fraser M. (2004) Birth Defects Res. C Embryo Today 72, 109–123 [DOI] [PubMed] [Google Scholar]
- 49.Huang X., Saint-Jeannet J. P. (2004) Dev. Biol. 275, 1–11 [DOI] [PubMed] [Google Scholar]
- 50.Antony A. C., Hansen D. K. (2000) Teratology 62, 42–50 [DOI] [PubMed] [Google Scholar]
- 51.Spiegelstein O., Mitchell L. E., Merriweather M. Y., Wicker N. J., Zhang Q., Lammer E. J., Finnell R. H. (2004) Dev. Dyn. 231, 221–231 [DOI] [PubMed] [Google Scholar]
- 52.Carter M., Ulrich S., Oofuji Y., Williams D. A., Ross M. E. (1999) Hum. Mol. Genet. 8, 2199–2204 [DOI] [PubMed] [Google Scholar]
- 53.Carter M., Chen X., Slowinska B., Minnerath S., Glickstein S., Shi L., Campagne F., Weinstein H., Ross M. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greene N. D., Copp A. J. (2005) Am. J. Med. Genet. C Semin Med. Genet. 135C, 31–41 [DOI] [PubMed] [Google Scholar]
- 55.Parle-McDermott A., Pangilinan F., O'Brien K. K., Mills J. L., Magee A. M., Troendle J., Sutton M., Scott J. M., Kirke P. N., Molloy A. M., Brody L. C. (2009) Hum. Mutat. 30, 1650–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J., Giovannucci E., Kelsey K., Rimm E. B., Stampfer M. J., Colditz G. A., Spiegelman D., Willett W. C., Hunter D. J. (1996) Cancer Res. 56, 4862–4864 [PubMed] [Google Scholar]
- 57.Kawakami K., Ruszkiewicz A., Bennett G., Moore J., Watanabe G., Iacopetta B. (2003) Clin. Cancer Res. 9, 5860–5865 [PubMed] [Google Scholar]
- 58.Sugiura T., Nagano Y., Inoue T., Hirotani K. (2004) Biochem. Biophys. Res. Commun. 315, 204–211 [DOI] [PubMed] [Google Scholar]