Abstract
To study the metabolic activity of NF-κB, we investigated phenotypes of two different mouse models with elevated NF-κB activities. The transcriptional activity of NF-κB is enhanced either by overexpression of NF-κB p65 (RelA) in aP2-p65 mice or inactivation of NF-κB p50 (NF-κB1) through gene knock-out. In these models, energy expenditure was elevated in day and night time without a change in locomotion. The mice were resistant to adulthood obesity and diet-induced obesity without reduction in food intake. The adipose tissue growth and adipogenesis were inhibited by the elevated NF-κB activity. Peroxisome proliferator-activator receptor γ expression was reduced by NF-κB at the transcriptional level. The two models exhibited elevated inflammatory cytokines (tumor necrosis factor-α and interleukin-6) in adipose tissue and serum. However, insulin sensitivity was not reduced by the inflammation in the mice on a chow diet. On a high fat diet, the mice were protected from insulin resistance. The glucose infusion rate was increased more than 30% in the hyperinsulinemic-euglycemic clamp test. Our data suggest that the transcription factor NF-κB promotes energy expenditure and inhibits adipose tissue growth. The two effects lead to prevention of adulthood obesity and dietary obesity. The energy expenditure may lead to disassociation of inflammation with insulin resistance. The study indicates that inflammation may prevent insulin resistance by eliminating lipid accumulation.
Keywords: Adipocyte, Energy Metabolism, Inflammation, Insulin, NF-κB/Transcription Factor, Obesity, Adipogenesis
Introduction
The IKKβ2/NF-κB signaling pathway plays an important role in the control of inflammation, apoptosis, carcinogenesis, and oxidative stress (1). In this pathway, the serine kinase IKKβ (IKK2) activates the transcription factor NF-κB through phosphorylation of NF-κB inhibitor (IκBα). In obesity research, the metabolic activity of IKKβ was tested in the control of insulin sensitivity (2–4) or food intake in transgenic mice (5). In these studies, the IKKβ activity was modified either globally or tissue-specifically in several major tissues/organs, such as the liver (3, 4), skeletal muscle (6), and brain (5). In these studies, the role of IKKβ in the regulation of energy expenditure and adipose tissue growth was not examined. Although IKKβ and NF-κB activities are parallel in most cases, their activities are not identical (7). IKKβ has NF-κB-independent activities (7, 8). We investigated the metabolic activity of NF-κB using the NF-κB transgenic mice in the current study.
NF-κB activation is associated with energy expenditure in cachexia (9, 10). However, the cause/effect relationship has not been tested for NF-κB/energy expenditure in transgenic models. NF-κB is a transcription factor that regulates expression of a broad spectrum of genes. Its activity is found in many types of cells, including adipocytes and macrophages (1, 11). The common form of NF-κB contains two subunits: p65 (RelA) and p50 (NF-κB1). p65 contains the transactivation domain and mediates transcriptional activation of target genes. p50 usually inhibits the transcriptional activity of p65 (12), and the inhibition disappears in the p50-KO mice (13). Expression of NF-κB target genes (TNF-α, interferon-γ, inducible nitric-oxide synthase, etc.) is increased in p50-KO mice (13–15). NF-κB activity is increased by either p65 overexpression or p50 knock-out. In the current study, the NF-κB activity is enhanced in these approaches in two lines of transgenic mice (aP2-p65 mice and p50-KO mice).
In cellular models, NF-κB was shown to inhibit the differentiation and function of adipocytes in the signaling pathway of TNF-α (16, 17). The molecular mechanism is related to inhibition of PPARγ activity by NF-κB. There are several molecular models for the inhibition (18, 19). These include suppression of PPARγ in mRNA expression (16, 20), DNA binding activity (21), and interaction with transcriptional coactivators (22). It is not clear which mechanism plays a major role in the physiological conditions. In this study, we addressed this issue by examining PPARγ activity in the adipose tissue of aP2-p65 mice and p50-KO mice. Our results suggest that an increase in the NF-κB activity leads to transcriptional inhibition of PPARγ expression. This negative regulation may be responsible for the inhibition of adipocyte differentiation and suppression of adipose tissue growth in the transgenic mice. The transgenic mice exhibited an increase in energy expenditure and resistance to diet-induced obesity.
MATERIALS AND METHODS
Animals
The male p50-KO mice and their controls were purchased from the Jackson Laboratory (Bar Harbor, ME). The aP2-p65 mice were made in our laboratory with a plasmid vector (supplemental Fig. 1). Male mice were used in the phenotype studies unless specified. All of the mice were housed in the animal facility at the Pennington Biomedical Research Center with a 12/12-h light-dark cycle and constant temperature (22–24 °C). The mice had free access to water and diet. Chow diet (MF 5015, containing 11% (w/w) or 25% of calories in fat) and the high fat diet (D12331, 36% (w/w) or 58% of calories in fat (Research Diets, New Brunswick, NJ)) were used. All procedures were performed in accordance with the National Institutes of Health guidelines for the care and use of animals and were approved by the Institutional Animal Care and Use Committee at the Pennington Biomedical Research Center. The mice were ear-punched for identification and kept at four per cage.
Adipogenesis in Vitro
Preadipocytes were isolated and differentiated as described elsewhere (23). Differentiated cells were fixed with 10% formalin for 1 h and stained with 300 nm DAPI for DNA and 10 μg/ml BODIPY® 493/503 for triglyceride (Invitrogen). The BODIPY fluorescence signal was quantified and normalized with DAPI signal to reduce variation from cell density.
Western Blot
The whole cell lysate was prepared and the Western blot was conducted according to methods reported earlier (8). Rabbit polyclonal antibodies to NF-κB p65 (sc-8008) and monoclonal antibody to PPARγ (sc-7273x) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies to p50 (ab7971) and tubulin (ab7291) were obtained from Abcam (Cambridge, UK). Tubulin was used as an internal control.
Body Weight and Composition
Body weight and composition were measured after overnight fasting. Body composition was measured using quantitative NMR (Minispec Mn10 NMR scanner) as described previously (24). In the test, conscious and unrestrained mice were individually tested for body composition using a Brucker model mq10 NMR analyzer (Brucker, Milton, Canada). The fat and lean mass were recorded within 1 min. Each group contained 8–9 mice, and a mean value was used to determine the significance of change in the transgenic mice.
Food Intake
Food intake was determined in individually housed mice of 10 weeks of age. The test was conducted daily for 3 days. The intake was determined by the net reduction in diet weight after excluding the spilled diet. The food intake was adjusted for the lean body weight.
Energy Expenditure and Body Temperature
Indirect calorimetry measurement was performed with the Comprehensive Laboratory Animal Monitoring System (Columbus Instruments, Columbus, OH) for individually housed mice (24). After 48 h of adaptation, the data for oxygen consumption (VO2), carbon dioxide production (VCO2), respiratory exchange ratio, and spontaneous physical activity were simultaneously recorded and used in calculating energy expenditure. The energy expenditure (EE; kcal/kg/h) was calculated with the formula, EE = (3.815 + 1.232 × VCO2/VO2) × VO2 × 0.001 (25). The energy expenditure data were normalized with body lean mass in each mouse. The core body temperature was determined in the rectum at 2.5 cm in depth using a Thermalert (model TH-8) temperature monitor (Physitemp (Clifton, NJ)) (26). Each group contained 8–9 mice.
PPARγ-luciferase Vector and Transfection Assay
The PPARγ luciferase reporter plasmid was constructed by inserting the mouse PPARγ2 5′ DNA into the pTAL-luc vector. The γ2 promoter DNA was amplified by PCR from the mouse PPARγ genomic DNA in the BAC-RP23–348 vector, which was obtained from the BACPAC Resource Center at the Children's Hospital Oakland Research Institute. The primer sequences at the 5′-end 5′-ggggtaccacaagtcactgaattatattaggt-3′ for the 1-kb promoter and 5′-ggggtaccctcttcctctatcccgatggttcctga-3′ for the 2-kb promoter. The 3′-end primer is 5′-tcccccgggaacagcataaacagagatttgct-3′. The PCR was conducted using Pfu Ultra high fidelity DNA polymerase (Stratagene). The promoter was cloned at the KpnI/SmalI sites in the multiple cloning region of pTAL vector. Vector DNA was approved by sequencing after construction. The plasmid was named pTAL-PPARγ-luc. The p65 expression vector was described earlier (22). The expression vector of CCAAT/enhancer-binding protein δ was obtained from Addgene (Cambridge, MA) (catalog number 12559). The transfection assay was conducted in 3T3-L1 adipocytes using electroporation with the Nucleofector II electroporator (Amaxa Biosystems, Cologne, Germany) as described elsewhere (27).
Blood Glucose, Insulin Tolerance Test (ITT), and Glucose Tolerance Test (GTT)
The blood glucose test, ITT, and GTT were conducted as described elsewhere (24).
Immunohistostaining
Formalin-fixed epididymal fat pads were processed, embedded in paraffin, and sectioned at 5 μm. The slides were blotted with a monoclonal primary antibody to F4/80 (sc-71087, Santa Cruz Biotechnology, Inc.) at a 1:200 dilution. A biotinylated secondary antibody (mouse IgG) in the ABC kit was used together with the AEC substrate kit (AEC101, Sigma) for a color reaction according to the manufacturer's instructions.
Serum Insulin and Cytokines
Serum insulin and cytokines were measured using a multiplex kit (catalog number MADPK-71k-03, Linco Research Inc. (St. Charles, MO)).
Quantitative Real Time RT-PCR
The TaqMan RT-PCR was used to quantify mRNA in the total RNA prepared with the TRIzol reagent. The signal was normalized with ribosome 18 S RNA. The following probes and primers were obtained from Applied Biosystems. These primers included p65 (Mm00501346_m1), adiponectin (Mm00456425_m1), aP2 (Mm00445880_m1), leptin (Mm00434759_m1), IL-6 (Mm00446190_m1), F4/80 (Mm00802530_m1), TNF-α (Mm00443258_m1), PPARγ (Mm00440945_m1), fatty acid synthase (Mm00662319_m1), sterol response element-binding protein (Mm00550338_m1), lipoprotein lipase (Mm00434770_m1), hormone-sensitive lipase (Mm00495359_m1), mcp-1 (Mm00441242_m1), IκBa (Mm00477798_m1), and pref-1 (Mm00494477_m1).
Electrophoretic Mobility Shift Assay
The nuclear extract was made from cells or adipose tissues using the two-step protocol (28). The DNA-protein reaction and gel shift assay were conducted as described elsewhere (29).
Hyperinsulinemic-Euglycemic Clamp
The hyperinsulinemic-euglycemic clamp studies were performed in conscious mice after 16 weeks on HFD (at 6 months of age) at the University of Massachusetts Mouse Phenotyping Center. Briefly, a 2-h hyperinsulinemic (2.5 milliunits/kg/min with 150 milliunits/kg body weight priming)-euglycemic clamp was conducted in overnight-fasted mice with [3-3H]glucose and 2-deoxy-d-[1-14C]glucose to assess glucose metabolism in individual organs (30).
Statistical Analysis
Statistical analyses for food intake, body weight, body content, indirect calorimetry, fasting blood glucose, ITT, GTT, blood lipid profile, blood cytokine levels, and all real time RT-PCR results were performed using the two-tailed, unpaired, Student's t test for the WT littermates versus transgenic (Tg) mice. The results are presented as means ± S.E.
RESULTS
Generation of aP2-p65 Mice
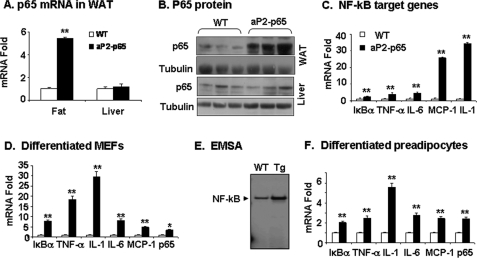
The transgenic mice were generated from C57BL/6 mice using a targeting vector shown in supplemental Fig. 1. To confirm p65 expression in adipose tissue, we examined p65 mRNA and protein in tissues of 6-week-old mice. The results show that p65 activity was significantly elevated in the adipose tissue (but not in liver) of Tg mice (Fig. 1, A and B). Expression of NF-κB target genes was induced by the transgene in adipose tissue (Fig. 1C). To confirm the p65 activity in adipocytes, we examined expression of p65 and NF-κB target genes in aP2-p65 adipocytes differentiated from MEFs or preadipocytes in vitro. The MEF cells were made from embryos of aP2-p65 mice. After differentiation, the MEF cells exhibited a significant increase in mRNA of p65 and NF-κB target genes (Fig. 1D). DNA binding of NF-κB was also increased in the differentiated cells, as indicated by the result of the electrophoretic mobility shift assay (Fig. 1E). Similar gene expression was observed in differentiated preadipocytes of aP2-p65 mice (Fig. 1F). The results confirm that NF-κB p65 overexpression is achieved in adipocytes and involved in elevated inflammation in adipose tissue of Tg mice. The p65 expression was also increased in macrophages (supplemental Fig. 1B), as expected from the aP2 promoter activity.
FIGURE 1.
p65 expression in aP2-p65 mice. A, p65 mRNA in epididymal fat and liver. B, p65 protein in fat and liver. The p65 protein was determined in the whole cell lysate in a Western blot. C, mRNA of NF-κB target genes in epididymal fat of 8-week-old mice. mRNA was determined by quantitative RT-PCR. D, mRNA of p65 and NF-κB target genes in differentiated MEFs of aP2-p65 mice. MEFs were made from embryos of aP2-p65 mice and differentiated into adipocytes in vitro. E, DNA binding activity of NF-κB in differentiated MEFs. The electrophoretic mobility shift assay (EMSA) result is shown. F, mRNA of p65 and NF-κB target genes in differentiated preadipocytes of epididymal fat pad. In the bar graph, values are the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.001 by Student's t test.
Body Weight and Composition
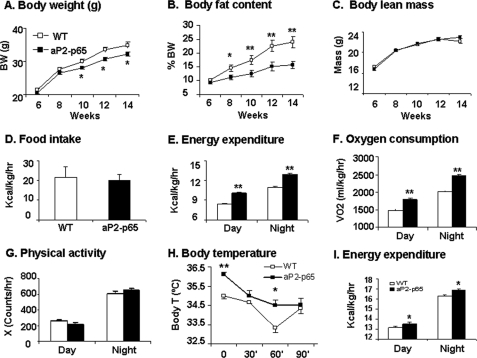
The metabolic phenotype was examined in the aP2-p65 Tg mice by monitoring the body weight, body composition, food intake, and energy expenditure. Compared with the WT mice, the Tg mice had a similar body weight within 8 weeks of age (Fig. 2A). On the chow diet, the Tg mice gained less body weight after 8 weeks of age. At 14 weeks of age, they were 9% lower in body weight (Fig. 2A) and had 68% less fat content than the control mice (Fig. 2B). However, their body lean mass was not changed (Fig. 2C). The data suggest that the reduced gain in fat mass may account for the smaller body weight after 8 weeks in the Tg mice. The genetic modification of NF-κB p65 may protect the Tg mice from adulthood obesity.
FIGURE 2.
Lean phenotype in aP2-p65 mice. Body weight (BW), fat content, food intake, and energy expenditure were monitored in the aP2-p65 mice. A, body weight. B, body fat content (percentage). This was determined by NMR. C, body lean mass. D, food intake. Food intake was monitored daily for 3 days at 10 weeks of age. The average daily food intake (g) was converted into kcal and normalized with body lean mass (kg). E, energy expenditure. The test was conducted at 12 weeks of age, and the unit is kcal/kg of body lean mass/h. F, oxygen consumption. The unit is volume (ml) of oxygen/kg of body lean mass/h. G, physical activity. Interruption of a horizontal laser beam (X) was used to indicate the locomotor activity. H, body temperature and cold response. The test was conducted at 24 weeks in age. I, energy expenditure in female aP2-p65 mice (n = 5). The test was conducted at 16 weeks in age. A–E, n = 8; G and H, n = 10 in WT or Tg group. Values are the mean ± S.E. *, p < 0.05; **, p < 0.001 by Student's t test. RER, respiratory exchange ratio.
Energy Metabolism
Adipose tissue mass reflects energy balance in the body. To understand the mechanism of reduction in body fat, we examined food intake and energy expenditure in the mice. Compared with the WT mice, the Tg mice exhibited no decrease in food intake (Fig. 2D) but had an increase in energy expenditure (Fig. 2E). The increase was observed during both day and night time. Oxygen consumption was elevated (Fig. 2F). The energy expenditure was not due to an increase in physical activity because the locomotor activity was not changed in the Tg mice (Fig. 2G). The disassociation suggests an elevation in the basal metabolic rate. The body temperature was about 1 °C higher in the Tg mice under room temperature (Fig. 2H). In response to cold (4 °C) stimulation, the WT mice exhibited a sharp drop in body temperature at 60 min (Fig. 2H). However, such a drop was not observed in the Tg mice, suggesting a better thermogenesis in the Tg mice. The substrate utilization was not significantly changed in the Tg mice (data not shown). Energy expenditure was also increased in the female Tg mice (Fig. 2I). We examined the second founder of aP2-p65 mice and observed a similar phenotype (supplemental Fig. 2).
White Adipose Tissue (WAT)
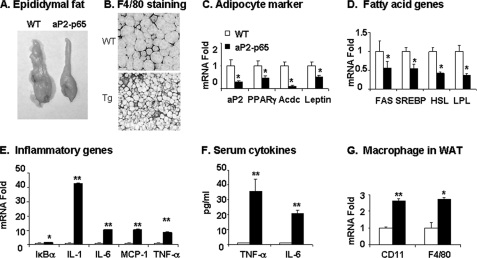
Although energy expenditure may explain the reduced gain in fat mass, a direct impact of NF-κB p65 on fat tissue may also play a role. To test this possibility, we examined the structure and function of WAT by determining the pad size and gene expression. First, we examined the epididymal fat pads. Compared with the WT mice, the Tg mice exhibited 60% reduction in the fat mass and >100% decrease in adipocyte size (Fig. 3, A and B). In the Tg mice, adipocyte genes, such as aP2, PPARγ, adiponectin, and leptin, were all reduced dramatically (>50%) (Fig. 3C), suggesting a deficiency in adipocyte function. A decrease in fatty acid synthesis or increase in lipolysis may induce a reduction in adipocyte size. To test the possibilities, we examined the expression of genes related to fatty acid synthase and lipolysis, such as fatty acid synthase, sterol response element-binding protein, lipoprotein lipase, and hormone-sensitive lipase. Expression of fatty acid synthase and sterol response element-binding protein was significantly reduced in the Tg tissues (>50%) (Fig. 3D). The expression of lipoprotein lipase and hormone-sensitive lipase was also reduced (Fig. 3D). These data suggest that the reduction in adipocyte size may be a result of lipid synthesis inhibition but not elevation of lipolysis. This possibility is supported by the PPARγ deficiency in the Tg mice.
FIGURE 3.
Adipose tissue inflammation. Fat tissue was collected at the age of 20 weeks and used in morphology and gene expression studies. A, epididymal fat pad. B, F4/80 immunostaining in WAT. The macrophages are shown in brown. C, adipocyte-specific genes. mRNA expression was determined by real time RT-PCR and expressed as -fold change. D, fatty acid genes, including genes related to fat synthesis and lipolysis. E, inflammatory genes. F, cytokines in serum. TNF-α and IL-6 were determined in serum using multiplex kits. G, macrophage markers. The mRNA expression was determined by real time RT-PCR. In this figure, values are means ± S.E. (n = 6). *, p < 0.05; **, p < 0.001 by Student's t test. Acdc, adiponectin; FAS, fatty acid synthase; SREBP, sterol response element-binding protein; HSL, hormone-sensitive lipase; LPL, lipoprotein lipase.
Inflammation in Adipose Tissue
An increase in the transcriptional activity of NF-κB may lead to expression of many inflammatory genes, such as TNF-α, IL-1, IL-6, and mcp-1. Expression of these inflammatory genes was examined in the adipose tissue to assess the NF-κB function. All of these genes were significantly increased in the Tg mice (Fig. 3E). NF-κB also induces the IκBα gene, whose product inhibits NF-κB activity (Fig. 3E). TNF-α and IL-6 proteins were determined in the serum, and their concentrations were increased 36- and 21-fold, respectively, in the Tg mice (Fig. 3F). Since macrophages are a dominant source of these cytokines, we examined macrophage infiltration in the adipose tissue by marker gene expression. In immunohistostaining, the number of F4/80-positive cells was increased in the adipose tissue of Tg mice (Fig. 3B). Expression of F4/80 and CD11 was elevated in mRNA (Fig. 3G). These data suggest that macrophage infiltration and activation are enhanced in the adipose tissue of Tg mice. The elevated mRNA expression may lead to the increase in TNF-α and IL-6 in serum.
Adipogenesis in Vitro
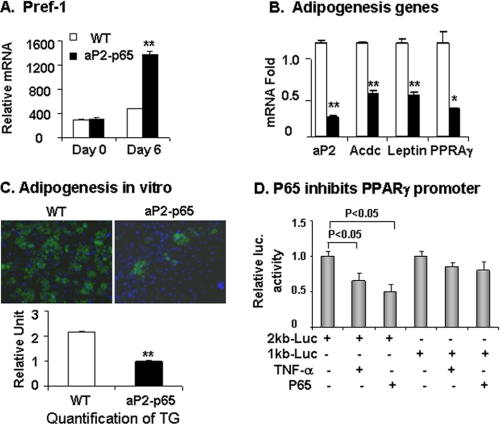
In cellular models, inhibition of PPARγ by NF-κB contributes to suppression of adipogenesis (22). In the aP2-p65 transgenic mice, the reduced fat mass was observed with decreased PPARγ expression. This association supports adipogenic deficiency. To prove it, we isolated preadipocytes from the stromal-vascular fraction of inguinal fat pads and examined their differentiation potential in vitro. The differentiation was monitored by gene expression and triglyceride accumulation. WT and Tg preadipocytes expressed similar levels of preadipocyte marker pref-1 (Fig. 4A). In the Tg cells, pref-1 expression was higher (Fig. 4A), and adipokines (adiponectin and leptin) were lower after differentiation (Fig. 4B). Expression of PPARγ and aP2 was significantly reduced as well (Fig. 3B). triglyceride abundance was 50% lower in the differentiated Tg cells (Fig. 4C). The data suggest that differentiation of preadipocytes is reduced in the Tg mice, and reduced PPARγ may represent the molecular mechanism.
FIGURE 4.
Differentiation of preadipocytes. The intact epididymal fat pad was collected at the age of 5 weeks and used to prepare preadipocytes. The differentiation was induced in the classic adipogenic mixture supplemented with thiazolidinedione (2 μm). A, pref-1 expression in preadipocytes. B, mRNA at day 6 of differentiation. Adiponectin mRNA was examined together with leptin, PPARγ, and aP2. C, triglyceride (TG) content indicated by BODIPY-stained lipid droplets (green fluorescence) at day 8 of differentiation. Blue fluorescence dye of DAPI indicates nucleus. Triglyceride quantification by ratio of BODIBY and DAPI signals is shown. D, the PPARγ promoter activity. The transcription of PPARγ was analyzed in 3T3-L1 adipocytes using the PPARγ2-luciferase reporter system in the transient transfection. p65 was transiently cotransfected with the PPARγ-Luc vector. The cells were serum-starved and treated either with or without TNF-α (20 ng/ml) overnight. In the bar graphs, each point represents mean ± S.E. (n = 3). **, p < 0.001 by Student's t test.
The PPARγ mRNA reduction may be a consequence of transcriptional suppression. To test this possibility, we investigated the PPARγ gene promoter activity in a luciferase reporter that contains the PPARγ2 promoter DNA of 1 or 2 kb in length. In the cotransfection assay, the 2-kb reporter was inhibited by NF-κB p65 in the basal condition in a dose-dependent manner (Fig. 4D). The inhibition was not observed in the 1-kb promoter, suggesting that the NF-κB response element may be located in the PPARγ gene promoter DNA beyond the 1-kb fragment from the transcription start site. These data suggest that p65 inhibits PPARγ expression by targeting the gene promoter.
Resistance to Dietary Obesity
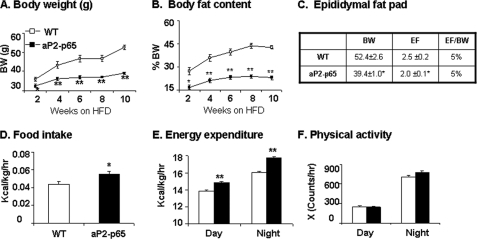
In the aP2-p65 mice, the increased energy expenditure and decreased adipogenesis suggest that the Tg mice may have resistance to obesity. To test this possibility, the dietary obesity model was used. Compared with the WT mice, the Tg mice gained less in body weight and fat content. The reduction was observed at 2 weeks and remained throughout the experiment up to 10 weeks on HFD (Fig. 5A). At the end of 10 weeks, the body weight was 40% lower, and the fat mass (adiposity) was 60% less in the aP2-p65 mice (Fig. 5, A and B). The reduction in fat mass occurs in both subcutaneous and visceral fat. This is supported by the percentage of epididymal fat mass over body mass (Fig. 5C). This group of data suggests that the Tg mice are protected from diet-induced obesity. Food intake and energy expenditure were both significantly increased in the Tg mice on HFD (Fig. 5, D and E). The energy expenditure was enhanced in both day and night time. Physical activity may not contribute to the energy expenditure because the locomotor activity was not increased in the Tg mice (Fig. 5F). These data suggest that the aP2-p65 mice are resistant to diet-induced obesity.
FIGURE 5.
Adiposity and energy metabolism on HFD. Body weight, body composition, food intake, and energy expenditure were examined using the NMR and metabolic chamber. The energy metabolism and food intake were examined at 10–12 weeks of age (5–6 weeks on HFD). A, body weight. B, body fat content (percentage). This was determined by NMR. C, epididymal fat. The epididymal fat (EF) was determined in weight and percentage of body weight (BW). D, food intake. Food intake was monitored daily for 3 days. Average daily food intake (g) was converted into kcal and normalized with the body lean mass (kg) and 24 h. E, energy expenditure. The unit is kcal/kg of body lean mass/h. F, spontaneous physical activity. The frequency of horizontal movement was shown for day and night time. Values are the mean ± S.E. (n = 8). *, p < 0.05; **, p < 0.001 by Student's t test.
p50-KO Mice Share the Same Metabolic Phenotype
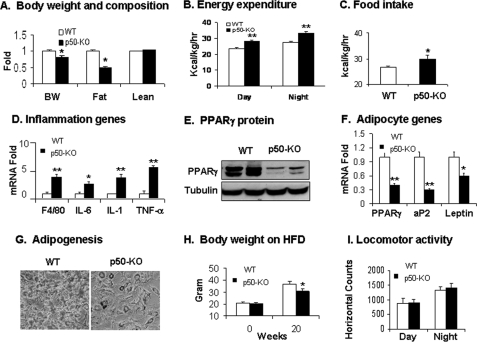
In the p50-KO mice, the NF-κB p50 subunit is inactivated globally, and the transcriptional activity of NF-κB is enhanced in the body (31). The NF-κB p65 activity is enhanced in the nucleus of cells from the p50-KO mice (supplemental Fig. 3). The KO mice represent a model of systemic inflammation. In adulthood, the p50-KO mice were 15% lower in body weight and 50% lower in fat mass at 12 weeks of age (Fig. 6A). There was no difference in body lean mass between the WT and KO mice (Fig. 6A). In the p50-KO mice, energy expenditure and food intake both were increased (Fig. 6, B and C). Inflammation was increased in the adipose tissue, as indicated by the macrophage marker F4/80 and inflammatory cytokines (TNF-α, IL-1, and IL-6) (Fig. 6D). PPARγ mRNA and proteins were reduced in the adipose tissue (Fig. 6, E and F). Expression of aP2 and leptin was also significantly decreased (Fig. 6F). The preadipocytes exhibited a reduced capacity in differentiation, as indicated by triglyceride abundance in the differentiated cells (Fig. 6G). At 20 weeks on HFD, the KO mice had 10% less body weight relative to the WT mice (Fig. 6H). Locomotor activity was not increased in the p50-KO mice (Fig. 6I). These observations suggest that the p50-KO mice have a metabolic phenotype similar to that of the aP2-p65 mice. Their energy expenditure was increased by the elevated NF-κB activity. The phenotype provides the second line of evidence that NF-κB induces energy expenditure.
FIGURE 6.
Lean phenotype of p50-KO mice. The phenotype study was conducted in p50-KO mice at 8–12 weeks of age on chow diet. A, body weight and composition were examined using NMR. The weight (g) of body fat and lean mass was used in the calculation. The results are expressed relative to the control used as a standard. B, energy expenditure was monitored using the metabolic chamber and normalized with body lean mass. C, food intake was determined in mice at 10 weeks of age. The daily average food intake was expressed in kcal normalized by body lean mass and time. D, inflammation gene expression in epididymal fat pads in quantitative RT-PCR. E, PPARγ protein determined in fat tissue in a Western blot. F, mRNA of adipocyte-specific genes was determined in fat tissues by quantitative RT-PCR. G, adipogenesis of preadipocytes of p50-KO mice in vitro. H, the body weight of p50-KO mice before and after HFD feeding at 20 weeks. I, spontaneous physical activity of p50-KO mice monitored in the metabolic chamber. In this figure, values are the mean ± S.E. (n = 9). *, p < 0.05; **, p < 0.001 by Student's t test.
Insulin Sensitivity in aP2-p65 Mice
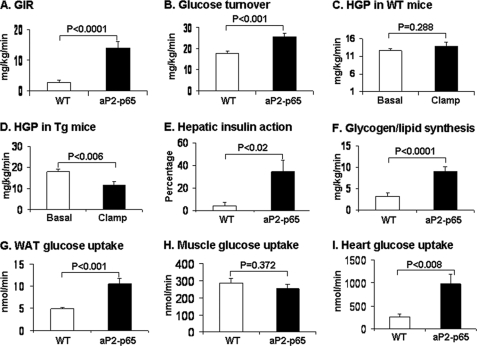
Insulin sensitivity and glucose metabolism were examined in the aP2-p65 mice. On the chow diet, the sensitivity was determined by fasting glucose, insulin, ITT, and GTT. These assays were conducted in mice between 12 and 16 weeks of age. Compared with the WT mice, the Tg mice did not exhibit a significant change in any of the parameters except that their insulin level tended to be lower (supplemental Fig. 4). Insulin sensitivity was further tested using the hyperinsulinemic-euglycemic clamps in conscious mice after 16 weeks on HFD (at 24 weeks of age). Plasma glucose levels were maintained at ∼130 mg/dl during the clamps in both WT and Tg mice. In the Tg mice, steady-state glucose infusion rates were elevated by more than 4-fold compared with the WT mice (Fig. 7A). Insulin-stimulated whole body glucose turnover was increased by ∼50%, indicating enhanced insulin sensitivity in the Tg mice (Fig. 7B). Insulin failed to suppress hepatic glucose production in WT mice, reflecting severe insulin resistance from HFD (Fig. 7C). In contrast, insulin caused a 35% suppression of hepatic glucose production in the Tg mice, showing improved hepatic insulin action in these mice (Fig. 7, D and E). Insulin-stimulated whole body glycogen plus lipid synthesis was increased 2-fold in the Tg mice (Fig. 7F). Insulin-stimulated glucose uptake in white adipose tissue and heart were increased, but muscle glucose uptake was not altered in the Tg mice (Fig. 7, G–I). These data demonstrate that insulin action is enhanced in adipose tissue, heart, and liver of Tg mice following HFD.
FIGURE 7.
Insulin sensitivity in aP2-p65 mice. Hyperinsulinemic-euglycemic clamps were conducted in conscious mice following 16 weeks of HFD (at 24 weeks of age). A, steady-state glucose infusion rates (GIR) during clamps. B, insulin-stimulated whole body glucose turnover. C, hepatic glucose production (HGP) in WT mice. D, hepatic glucose production in Tg mice. E, hepatic insulin action expressed as percentage suppression of basal HGP during clamps. F, whole body glycogen and lipid synthesis. G, insulin-stimulated glucose uptake in WAT. H, insulin-stimulated glucose uptake in skeletal muscle (quadriceps). I, insulin-stimulated glucose uptake in heart. Values are means ± S.E. (n = 6–9).
DISCUSSION
The current study was designed to understand the molecular mechanism of inflammation in the pathogenesis of insulin resistance. The role of NF-κB in insulin resistance was indicated by the phenotypes of IKKβ transgenic mice and the activities of TNF-α (2–4, 22, 32). These studies suggest that NF-κB may inhibit insulin sensitivity by induction of inflammatory cytokines or through inhibition of PPARγ activity (11, 18). However, the NF-κB activity has never been directly examined in animals using the NF-κB transgenic approach. In the current study, this issue is addressed in two lines of NF-κB transgenic mice, in which the transcriptional activity of NF-κB is increased by different mechanisms. The two models share the same metabolic phenotypes in energy expenditure, insulin sensitivity, and resistance to obesity. The phenotypes of aP2-p65 mice suggest that NF-κB activity in adipose tissue is important in the regulation of metabolism.
In the two models, inflammation was observed in adipose tissue with elevated macrophage infiltration and expression of inflammatory cytokines (TNF-α, IL-1, IL-6, and mcp-1). The systemic inflammation is indicated by elevated proteins for TNF-α and IL-6 in the serum (15). The inflammation is a result of increased NF-κB activity from overexpression of the p65 subunit (aP2-p65) or deletion of the p50 subunit (p50-KO) (12). Although NF-κB p65 activity is increased in microphages in the p65 transgenic mice, we did not observe elevated p65 expression in liver. This might be a result of the dilution of Kuffer's cells by hepatocytes. Macrophages produce much more proinflammatory cytokines (especially TNF-α and IL-1) than adipocytes (33).
Induction of energy metabolism by NF-κB is an intriguing observation. This activity is responsible for the prevention of adulthood obesity and dietary obesity in the two models. The mechanism of energy expenditure involves the inflammatory cytokines, such as TNF-α, IL-1, and IL-6, whose levels are positively associated with energy expenditure (10). In transgenic mice with deficiency in these cytokines or their receptors, the body weight gain is enhanced in mouse models for TNF-α (34), IL-1 (35), and IL-6 (36). When their activities are enhanced in transgenic mice, energy expenditure is increased, and body weight gain is attenuated in several models (37–40). The cytokines may act in the central nervous system to regulate energy balance (36, 41–44). They may target the hypothalamus (43, 44). In the peripheral tissues, TNF-α and IL-1 enhance mitochondrial function through activation of PGC-1α by phosphorylation (45). The increased energy expenditure may reflect the combined activities of multiple cytokines in our models because their expression was induced by NF-κB. Our data suggest that NF-κB is a powerful transcription factor in the induction of energy metabolism. It may be a target for drug intervention of energy metabolism (46, 47).
Inhibition of adipose tissue growth may also contribute to the energy expenditure in the two models. PPARγ expression was inhibited by NF-κB, and the inhibition was observed at the transcriptional level in our study. The inhibition was observed in the PPARγ gene promoter, and a NF-κB response element was found within the 1–2 kb region from the transcription start site. Given the master activity of PPARγ in adipocytes, the suppression may be the molecular mechanism of adipose tissue deficiency in the aP2-p65 mice and p50-KO mice. The inhibition may promote energy expenditure by stimulating lipid oxidation because lipid accumulation is inhibited in adipose tissue. The fat-specific PPARγ knock-out mice (PPARγ adipose-KO) exhibited a similar increase in energy expenditure (48). Insulin sensitivity is also protected in the PPARγ adipose-KO mice (48). This phenotype may be related to an increase in the NF-κB activity because PPARγ suppresses the transcriptional activity of NF-κB (49). However, this possibility needs to be tested in the PPARγ adipose-KO mice.
The current study suggests that inflammation may have two different activities in the regulation of metabolism. The first is inhibition of insulin sensitivity (negative effect), as suggested by many studies of low grade inflammation. The second is induction of energy expenditure (positive effect), as indicated by current and other reports (34–40). Our study indicates that energy expenditure may antagonize insulin resistance by eliminating lipid accumulation through elevated lipid oxidation, which prevents lipotoxicity in the pathogenesis of insulin resistance. In both aP2-p65 mice and p50-KO mice (15), insulin sensitivity was examined by the hyperinsulinemic-euglycemic clamp. The systemic insulin sensitivity was enhanced in both models, as indicated by the glucose infusion rate. In terms of tissue specificity, insulin sensitivity was dramatically enhanced in liver and fat but not in the skeletal muscle, suggesting the importance of the two organs in inflammation action. The muscle data suggest that the skeletal muscle is not sensitive to inflammation on insulin sensitivity. In our models, the positive effect of inflammation overrides the negative effect of inflammation, leading to an increase in insulin sensitivity. The energy expenditure represents a new aspect of inflammation that was not reported in the IKKβ transgenic studies (2–4).
In summary, the current study suggests that elevated inflammation from NF-κB has a significant activity in the induction of energy expenditure. The enhanced energy expenditure prevented diet-induced obesity and insulin resistance in the aP2-p65 and p50-KO mice. The energy expenditure may be a result of up-regulated expression of NF-κB target genes and inhibition of lipid accumulation. The energy expenditure may be a key mechanism for the disassociation of inflammation with insulin resistance.
Supplementary Material
Acknowledgment
We greatly appreciate the technical support of Gang Yu in making the aP2-p65 targeting vector.
This work was supported, in whole or in part, by National Institutes of Health Grants DK068036 and R56DK068036-06 (to J. Y.) and DK80756 (to J. K. K.). This work was also supported by American Diabetes Association Research Awards 7-07-RA-189 (to J. Y.) and 7-07-RA-80 (to J. K. K.). The quantitative RT-PCR study was conducted in the genomic core, which is supported by NIDDK, National Institutes of Health, Clinical Nutrition Research Unit Grant 1P30 DK072476.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- IKKβ (IKK2)
- IκBα kinase 2
- NF-κB
- nuclear factor κB
- WAT
- white adipose tissue
- HFD
- high fat diet (58% of calories in fat)
- IκBα
- inhibitor κB α
- p50 (NF-κB1)
- NF-κB p50 subunit
- p65 (RelA)
- NF-κB p65 subunit
- TNF-α
- tumor necrosis factor α
- KO
- knock-out
- DAPI
- 4′,6-diamidino-2-phenylindole
- PPAR
- peroxisome proliferator-activator receptor
- ITT
- insulin tolerance test
- GTT
- glucose tolerance test
- RT
- reverse transcription
- IL
- interleukin
- WT
- wild type
- Tg
- transgenic
- MEF
- mouse embryo fibroblast.
REFERENCES
- 1.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 2.Yuan M., Konstantopoulos N., Lee J., Hansen L., Li Z. W., Karin M., Shoelson S. E. (2001) Science 293, 1673–1677 [DOI] [PubMed] [Google Scholar]
- 3.Cai D., Yuan M., Frantz D. F., Melendez P. A., Hansen L., Lee J., Shoelson S. E. (2005) Nat. Med. 11, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arkan M. C., Hevener A. L., Greten F. R., Maeda S., Li Z. W., Long J. M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M. (2005) Nat. Med. 11, 191–198 [DOI] [PubMed] [Google Scholar]
- 5.Zhang X., Zhang G., Zhang H., Karin M., Bai H., Cai D. (2008) Cell 135, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai D., Frantz J. D., Tawa N. E., Jr., Melendez P. A., Oh B. C., Lidov H. G., Hasselgren P. O., Frontera W. R., Lee J., Glass D. J., Shoelson S. E. (2004) Cell 119, 285–298 [DOI] [PubMed] [Google Scholar]
- 7.Perkins N. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 49–62 [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Gao Z., Yin J., Quon M. J., Ye J. (2008) J. Biol. Chem. 283, 35375–35382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strasser F. (2007) Curr. Opin. Support Palliat. Care 1, 312–316 [DOI] [PubMed] [Google Scholar]
- 10.Tisdale M. J. (1997) J. Natl. Cancer Inst. 89, 1763–1773 [DOI] [PubMed] [Google Scholar]
- 11.Shoelson S. E., Lee J., Goldfine A. B. (2006) J. Clin. Invest. 116, 1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz M. L., Baeuerle P. A. (1991) EMBO J. 10, 3805–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bohuslav J., Kravchenko V. V., Parry G. C., Erlich J. H., Gerondakis S., Mackman N., Ulevitch R. J. (1998) J. Clin. Invest. 102, 1645–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadjeva M., Tomczak M. F., Zhang M., Wang Y. Y., Dull K., Rogers A. B., Erdman S. E., Fox J. G., Carroll M., Horwitz B. H. (2004) J. Immunol. 173, 5786–5793 [DOI] [PubMed] [Google Scholar]
- 15.Gao Z., Yin J., Zhang J., He Q., McGuinness O. P., Ye J. (2009) J. Biol. Chem. 284, 18368–18376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruan H., Hacohen N., Golub T. R., Van Parijs L., Lodish H. F. (2002) Diabetes 51, 1319–1336 [DOI] [PubMed] [Google Scholar]
- 17.Ruan H., Pownall H. J., Lodish H. F. (2003) J. Biol. Chem. 278, 28181–28192 [DOI] [PubMed] [Google Scholar]
- 18.Ye J. (2008) Biochem. Biophys. Res. Commun. 374, 405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilherme A., Virbasius J. V., Puri V., Czech M. P. (2008) Nat. Rev. Mol. Cell Biol. 9, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B., Berger J., Hu E., Szalkowski D., White-Carrington S., Spiegelman B. M., Moller D. E. (1996) Mol. Endocrinol. 10, 1457–1466 [DOI] [PubMed] [Google Scholar]
- 21.Suzawa M., Takada I., Yanagisawa J., Ohtake F., Ogawa S., Yamauchi T., Kadowaki T., Takeuchi Y., Shibuya H., Gotoh Y., Matsumoto K., Kato S. (2003) Nat. Cell Biol. 5, 224–230 [DOI] [PubMed] [Google Scholar]
- 22.Gao Z., He Q., Peng B., Chiao P. J., Ye J. (2006) J. Biol. Chem. 281, 4540–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staszkiewicz J., Gimble J. M., Manuel J. A., Gawronska-Kozak B. (2008) Stem Cells 26, 2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Z., Wang Z., Zhang X., Butler A. A., Zuberi A., Gawronska-Kozak B., Lefevre M., York D., Ravussin E., Berthoud H. R., McGuinness O., Cefalu W. T., Ye J. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E84–E91 [DOI] [PubMed] [Google Scholar]
- 25.Hofmann S. M., Zhou L., Perez-Tilve D., Greer T., Grant E., Wancata L., Thomas A., Pfluger P. T., Basford J. E., Gilham D., Herz J., Tschöp M. H., Hui D. Y. (2007) J. Clin. Invest. 117, 3271–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Z., Yin J., Zhang J., Ward R. E., Martin R. J., Lefevre M., Cefalu W. T., Ye J. (2009) Diabetes 58, 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J., Gao Z., Yin J., He Q. (2007) Am. J. Physiol. Endocrinol. Metab. 293, E1118–E1128 [DOI] [PubMed] [Google Scholar]
- 28.Ye J., Ghosh P., Cippitelli M., Subleski J., Hardy K. J., Ortaldo J. R., Young H. A. (1994) J. Biol. Chem. 269, 25728–25734 [PubMed] [Google Scholar]
- 29.Ye J., Cippitelli M., Dorman L., Ortaldo J. R., Young H. A. (1996) Mol. Cell. Biol. 16, 4744–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H. J., Higashimori T., Park S. Y., Choi H., Dong J., Kim Y. J., Noh H. L., Cho Y. R., Cline G., Kim Y. B., Kim J. K. (2004) Diabetes 53, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 31.Sha W. C., Liou H. C., Tuomanen E. I., Baltimore D. (1995) Cell 80, 321–330 [DOI] [PubMed] [Google Scholar]
- 32.Gao Z., Hwang D., Bataille F., Lefevre M., York D., Quon M. J., Ye J. (2002) J. Biol. Chem. 277, 48115–48121 [DOI] [PubMed] [Google Scholar]
- 33.Fain J. N., Bahouth S. W., Madan A. K. (2004) Int. J. Obes. Relat. Metab. Disord. 28, 616–622 [DOI] [PubMed] [Google Scholar]
- 34.Pamir N., McMillen T. S., Kaiyala K. J., Schwartz M. W., LeBoeuf R. C. (2009) Endocrinology 150, 4124–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chida D., Osaka T., Hashimoto O., Iwakura Y. (2006) Diabetes 55, 971–977 [DOI] [PubMed] [Google Scholar]
- 36.Wallenius V., Wallenius K., Ahrén B., Rudling M., Carlsten H., Dickson S. L., Ohlsson C., Jansson J. O. (2002) Nat. Med. 8, 75–79 [DOI] [PubMed] [Google Scholar]
- 37.Xu H., Hirosumi J., Uysal K. T., Guler A. D., Hotamisligil G. S. (2002) Endocrinology 143, 1502–1511 [DOI] [PubMed] [Google Scholar]
- 38.Matsuki T., Horai R., Sudo K., Iwakura Y. (2003) J. Exp. Med. 198, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somm E., Henrichot E., Pernin A., Juge-Aubry C. E., Muzzin P., Dayer J. M., Nicklin M. J., Meier C. A. (2005) Diabetes 54, 3503–3509 [DOI] [PubMed] [Google Scholar]
- 40.García M. C., Wernstedt I., Berndtsson A., Enge M., Bell M., Hultgren O., Horn M., Ahrén B., Enerback S., Ohlsson C., Wallenius V., Jansson J. O. (2006) Diabetes 55, 1205–1213 [DOI] [PubMed] [Google Scholar]
- 41.Anforth H. R., Bluthe R. M., Bristow A., Hopkins S., Lenczowski M. J., Luheshi G., Lundkvist J., Michaud B., Mistry Y., Van Dam A. M., Zhen C., Dantzer R., Poole S., Rothwell N. J., Tilders F. J., Wollman E. E. (1998) Eur. Cytokine Netw. 9, 279–288 [PubMed] [Google Scholar]
- 42.Luheshi G. N., Gardner J. D., Rushforth D. A., Loudon A. S., Rothwell N. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7047–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klir J. J., Roth J., Szelényi Z., McClellan J. L., Kluger M. J. (1993) Am. J. Physiol. 265, R512–R517 [DOI] [PubMed] [Google Scholar]
- 44.Luheshi G. N. (1998) Ann. N.Y. Acad. Sci. 856, 83–89 [DOI] [PubMed] [Google Scholar]
- 45.Puigserver P., Rhee J., Lin J., Wu Z., Yoon J. C., Zhang C. Y., Krauss S., Mootha V. K., Lowell B. B., Spiegelman B. M. (2001) Mol. Cell 8, 971–982 [DOI] [PubMed] [Google Scholar]
- 46.von Haehling S., Genth-Zotz S., Anker S. D., Volk H. D. (2002) Int. J. Cardiol. 85, 173–183 [DOI] [PubMed] [Google Scholar]
- 47.Guttridge D. C., Mayo M. W., Madrid L. V., Wang C. Y., Baldwin A. S., Jr. (2000) Science 289, 2363–2366 [DOI] [PubMed] [Google Scholar]
- 48.Jones J. R., Barrick C., Kim K. A., Lindner J., Blondeau B., Fujimoto Y., Shiota M., Kesterson R. A., Kahn B. B., Magnuson M. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6207–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pascual G., Fong A. L., Ogawa S., Gamliel A., Li A. C., Perissi V., Rose D. W., Willson T. M., Rosenfeld M. G., Glass C. K. (2005) Nature 437, 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.