Abstract
During development and nerve injury, complex interactions between glial cells and neurons are essential for establishing proper nerve function. Neurotrophins play multiple roles in the developing nervous system, including cell survival, growth, and differentiation. Here we show that migration of Schwann cells, isolated from sciatic nerves, is significantly enhanced by neurotrophin 3, but not by nerve growth factor or brain-derived neurotrophic factor. The neurotrophin-3-induced cell migration was also observed in Schwann cells isolated from sciatic nerves of p75NTR-/- mice, indicating that neurotrophin 3 enhances cell migration through TrkC. This effect was blocked by K252a, an inhibitor of the Trk receptor family. Additionally, the neurotrophin-3-induced cell migration depended on Rho GTPases (Rac1 and Cdc42) and c-Jun N-terminal kinase. We obtained the same results with Cos-7 cells expressing TrkC. Taken together, these results suggest that neurotrophin 3 activation of TrkC induces Schwann cell migration through the c-Jun N-terminal kinase signaling pathway.
Cell migration is thought to be initiated in response to extracellular cues, which can be diffusible factors and signals present on neighboring cells (1). In the developing peripheral nervous system, Schwann cells, the myelin-forming glia, proliferate, migrate along the axonal surface, ensheath individual axons, and eventually form the myelin sheath (2, 3). Schwann cell migration is also essential in response to tissue damage after injury (4). After nerve transection, Schwann cells migrate from the proximal and distal stumps and form a continuous tissue cable, guiding regenerating axons. However, the factors that regulate migration of Schwann cells and their signaling mechanisms remain elusive.
Neurotrophins are involved in almost every aspect of normal and pathological neuronal behavior. These diffusible trophic factors are best known for their ability to support the survival and growth of neuronal cells and for the proper development of nervous-system function (5–7). Neurotrophins consist of nerve growth factor, brain-derived neurotrophic factor (BDNF), neurotrophin 3 (NT3), and neurotrophin 4/5, and they activate two structurally unrelated receptors, the p75 neurotrophin receptor (NTR) and the receptor tyrosine kinases designated (Trk). The p75NTR binds all neurotrophins with a very similar affinity, whereas the Trk receptors show more restricted neurotrophin-binding affinity (7).
Several lines of evidence suggest that small GTPases of the Rho family regulate the organization of the actin cytoskeleton (8, 9). The best-characterized Rho GTPases, RhoA, Rac1, and Cdc42, are implicated in the formation of actin stress fibers, lamellipodia, and filopodia, respectively, and through them the process of cell migration. Rho GTPases act as molecular switches and cycle between active (GTP-bound) and inactive (GDP-bound) states. Their activities are controlled positively by guanine nucleotide-exchange factors (GEFs), which catalyze the replacement of GDP with GTP, and negatively by GTPase-activating proteins, which enhance the endogenous GTPase activity. Rac1 and Cdc42 not only regulate the actin cytoskeleton through their effector molecules, but also activate the signaling cascade of the c-Jun N-terminal kinase (JNK) group of mitogen-activated protein kinases (10).
We demonstrated that BDNF enhances myelination of Schwann cells through p75NTR and that NT3 inhibits the process through TrkC (11, 12). To further investigate the inhibitory role of NT3 in the myelination process, the effects of NT3 on proliferation, differentiation, and migration were assayed on primary Schwann cells. We found that NT3 greatly enhances cell migration solely through TrkC. In addition, we found that this effect depends on Rho GTPases (Rac1 and Cdc42) and JNK. These results suggest that NT3 activation of TrkC inhibits the Schwann cell myelination program by enhancing Schwann cell migration through the Rac1/Cdc42/JNK pathway, providing an example of a functional role of this pathway.
Materials and Methods
Materials. The following antibodies were purchased: anti-JNK and anti-RhoA (Santa Cruz Biotechnology); anti-Rac1 and anti-Cdc42 (BD Biosciences Pharmingen); antiphosphorylated (pThr183/pTyr185) JNK (Cell Signaling Technology, Beverly, MA). The following inhibitors were purchased: K252a and Clostridium difficile Toxin B (Calbiochem-Novabiochem); SP600125 (Biomol, Plymouth Meeting, PA). The anti-TrkC antibody was generously provided by A. Welcher (Amgen Biologicals). Nerve growth factor was purchased from Serotec. BDNF, NT3, and TrkC-Fc were gifts from Regeneron Pharmaceuticals (Tarrytown, NY). GST-tagged mDia1-RBD and αPak-CRIB were purified from Escherichia coli BL21 (DE3) pLysS as described (13, 14).
Plasmids. The coding region of mouse TrkC (GenBank accession no. AY336094) was isolated by 5′ and 3′ rapid amplification of cDNA ends by using mouse brain Marathon-Ready cDNA (BD Biosciences Clontech) and was ligated into mammalian expression vector pCMV. The Dbl homology domain of Vav2 (15), a tyrosine-phosphorylation-dependent GEF for RhoA and Rac1, was amplified from total RNA of 293 cells and was subcloned into pCMV. The mammalian expression plasmids encoding Dbl homology and pleckstrin homology domains of a Cdc42-specific GEF Frg and Rho GTPases were constructed as described (14). The pUSE-constitutively active (CA)-Src plasmid was purchased from Upstate Biotechnology (Charlottesville, VA). The E. coli expression plasmids encoding RhoA·GTP-binding domain (RBD) of mDia1 and Rac1·GTP- and Cdc42·GTP-binding domain (CRIB) of αPak were constructed as described (16, 17). The SRα-HA-JNK1 plasmid was generously provided by M. Karin (University of California at San Diego, La Jolla).
Cell Culture and Transfection. Primary Schwann cells were prepared from sciatic nerves of Sprague–Dawley rats or mice (129S or p75NTR-/-; The Jackson Laboratory) at postnatal day 2 (12). Schwann cells were cultured on poly-l-lysine (100 μg/ml)-coated dishes and were plated for experiments on type I collagen (100 μg/ml)-coated dishes in DMEM containing 10% heat-inactivated FBS and 50 μg/ml gentamicin at 37°C. Before performing experiments, Schwann cells were cultured in Sato medium (18) containing 1 mg/ml BSA for 16 h. Cos-7 cells were cultured in DMEM containing 10% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin. Cos-7 cells were plated on collagen-coated dishes and were transfected by using calcium phosphate precipitation. Transfection efficiency typically exceeded 98% by using an enhanced GFP-expressing plasmid as the control. The final amount of the transfected DNA for a 6-cm dish was adjusted to 15 μg by addition of empty vector pCMV. SRα-HA-JNK1 (1 μg) or pCMV-Rho GTPases (3 μg) were cotransfected with pCMV-TrkC (0.5 μg), pUSE-CA-Src (1 μg), or pCMV-GEFs for Rho GTPases (10 μg) into Cos-7 cells. The medium was replaced 24 h after transfection, and cells were cultured in DMEM containing 1% FBS and 1 mg/ml BSA for 16 h. Unless otherwise indicated, cells were pretreated with or without K252a (100 nM, 45 min), SP600125 (100 nM, 45 min), or Toxin B (1 ng/ml, 16 h) before stimulation with NT3 (10 ng/ml, 0–120 min). To confirm cell viability under these experimental conditions, cells were stained with 0.4% trypan blue. Trypan-blue-incorporating cells numbered <1% in each experiment. Cell Migration Assay. Cell migration was measured by using a 24-well Boyden chamber (BD Biosciences) (19). In brief, polyethylene terephthalate (8-μm pore size) filters were coated with collagen. Cells (1 × 105 cells for Schwann cells or 4 × 105 cells for Cos-7 cells) in 500 μl of medium per well were loaded into the upper chambers, which were inserted into the tissue-culture wells containing neurotrophins at the final concentration of 10 ng/ml in 750 μl of medium per well. After incubation at 37°C for 5–6 h, the filters were stained with Giemsa solution. The number of stained, migrating cells at the bottom surface of the filters was counted at four fields per filter in two to four independent experiments.
Immunoprecipitation and Immunoblotting. Cells were lysed in 200 μl (35-mm dishes) or 600 μl (60-mm dishes) of lysis buffer as described (14, 15). Aliquots of the supernatants were mixed with protein G resin preabsorbed with 0.5 μg of anti-JNK antibody. The immunoprecipitates or the proteins expressed in the cell lysates were denatured and then separated on SDS-polyacrylamide gels. The electrophoretically separated proteins were transferred to nitrocellulose membranes, blocked, and immunoblotted. The bound antibodies were detected by using the ECL system (Amersham Biosciences).
JNK Assay. Immunoprecipitated JNK was immunoblotted with antiphosphorylated JNK antibody, which recognizes the active form (20). To compare the total amount of JNK, the immunoblotting was also performed with anti-JNK antibody. Three to four separate experiments were performed, and images of protein bands were captured by using an Epson GT-7000U scanner (Long Beach, CA). The band intensity in the immunoblot was semiquantified by using NIH IMAGE 1.61 (http://rsb.info.nih.gov/nih-image). Levels of the phosphorylated forms were normalized to the amount of total kinase.
Pull-Down Assays for Rho GTPases. To detect GTP-bound Rho GTPases, we performed pull-down assays using GST-tagged mDia1-RBD or αPak-CRIB (16, 17). The representatives of two to four experiments are shown in the figures.
Statistical Analysis. Statistical analysis was performed with STATVIEW 5.0 (SAS Institute, Cary, NC). Values shown represent the mean ± SD from separate experiments. Student's t test was carried out for intergroup comparisons.
Results
NT3 Enhances Schwann Cell Migration Through TrkC. In an attempt to uncover potential factors involved in the regulation of migration, conditioned media from dorsal root ganglion (DRG) neurons were applied to primary Schwann cells. Migration assays were performed in Boyden chambers, where Schwann cells were plated on the filters in the upper wells and allowed to migrate out onto the lower chamber through 8-μm pores. The presence of conditioned media greatly enhanced Schwann cell migration, whereas addition of the NT3 scavenger TrkC-Fc (1 μg/ml) to conditioned media diminished the effect by ≈50% (data not shown). These studies were consistent with our findings that DRG neurons secrete NT3 (11) and hinted at the physiological effects of NT3, which then drove the following experiments.
To investigate the effects of NT3 on Schwann cell migration, NT3 was placed in the lower wells of the Boyden chambers forming a concentration gradient that extended into the upper wells. As shown in Fig. 1A, stimulation with NT3 enhanced Schwann cell migration by ≈4-fold. This effect was blocked by pretreatment with K252a, an inhibitor of the Trk receptor family (Fig. 1 A and B). Pretreatment with K252a completely abolished the NT3-induced tyrosine phosphorylation of TrkC (data not shown). In contrast, nerve growth factor and BDNF did not promote cell migration (Fig. 1C). These results suggest that NT3 enhances Schwann cell migration through TrkC, consistent with our findings that Schwann cells express full-length TrkC but not TrkA or TrkB (12). Because Schwann cells express high levels of p75NTR (12), we examined whether p75NTR is involved in Schwann cell migration induced by NT3 by using Schwann cells from p75NTR knockout mice. As shown in Fig. 1D, NT3 was able to promote migration of p75NTR-/- Schwann cells at a level similar to wild-type Schwann cells. These results indicate that TrkC but not p75NTR mediates the NT3-induced migration of Schwann cells.
Fig. 1.
NT3 mediates migration of primary Schwann cells and Cos-7 cells through TrkC. Cell migration was measured in Schwann cells (A–D), isolated from sciatic nerves of rats (A–C) and mice (D), and Cos-7 cells transiently transfected with the plasmid encoding TrkC (E–G), by using Boyden chambers. Cells were pretreated with or without K252a (A, B, E, and F). After incubation with (+) or without (-) NT3, the migrating cells were stained and analyzed (A and E), and the number of stained cells was counted (B and F). (C) Rat Schwann cells were incubated with NT3, BDNF, or nerve growth factor, and the number of stained, migrating cells was counted. (D) Schwann cells isolated from sciatic nerves of mice (p75NTR+/+ or p75NTR-/-) were incubated with or without NT3, and the number of stained, migrating cells was counted. (G) Cos-7 cells were transfected with or without TrkC and were incubated with or without NT3. Data were evaluated by using Student's t test. *, P < 0.01.
NT3 Enhances Migration of Cos-7 Cells Expressing TrkC. To confirm that NT3 activation of TrkC stimulates cell migration, a transient transfection system using Cos-7 cells was used. Cos-7 cells do not express any neurotrophin receptors at any detectable level by immunoblotting (data not shown). NT3 enhanced migration of Cos-7 cells transfected with the plasmid encoding TrkC by >2-fold, and this effect was blocked by pretreatment with K252a (Fig. 1 E and F). In contrast, mock-transfected Cos-7 cells did not respond to NT3 (Fig. 1G).
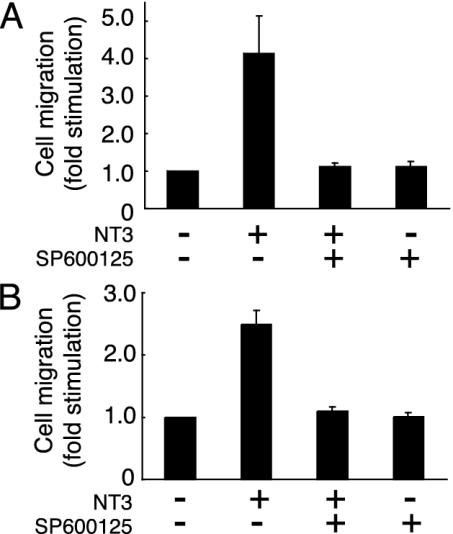
NT3 Enhances Cell Migration Through JNK. JNK is thought to be a key regulator of many cellular processes during development (10). Therefore, we next investigated whether NT3 enhances cell migration through JNK. Pretreatment with SP600125 (21), a specific inhibitor of JNK, blocked the NT3-induced migration of Schwann cells (Fig. 2A) and Cos-7 cells transfected with TrkC (Fig. 2B), suggesting that JNK may mediate cell migration induced by NT3. Specific inhibitors of the extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways had no effect on migration (data not shown).
Fig. 2.
The NT3-induced cell migration involves JNK. Rat Schwann cells (A) and Cos-7 cells transfected with TrkC (B) were pretreated with SP600125. After incubation with NT3, the migrating cells in the Boyden chambers were stained and counted.
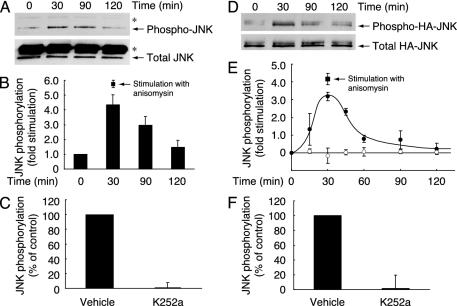
To determine whether NT3 stimulates the intrinsic activity of JNK in primary Schwann cells, endogenous JNK was immunoprecipitated from the lysate of Schwann cells and immunoblotted with antiphosphorylated JNK antibody, which recognizes the active state (Fig. 3 A–C). Stimulation with NT3 activated JNK in a time-dependent manner in Schwann cells at a level similar to the JNK activation by anisomysin, an artificial activator of JNK (Fig. 3 A and B). The JNK activity reached maximum level at 30 min after stimulation with NT3 and remained activated for at least 120 min. In addition, the NT3-induced JNK activation was inhibited by pretreatment with K252a (Fig. 3C). These results demonstrate that NT3 activation of TrkC stimulates JNK activity in Schwann cells.
Fig. 3.
JNK is activated after stimulation with NT3. JNK activity was measured in rat Schwann cells (A–C) and Cos-7 cells cotransfected with TrkC and HA-JNK (D–F). (A) Schwann cells were treated with NT3. Endogenous JNK was immunoprecipitated with anti-JNK antibody and blotted with antiphosphorylated JNK antibody or anti-JNK antibody. Asterisks indicate the heavy chain of IgGs. (B) Levels of JNK phosphorylation were quantified and normalized against the total immunoprecipitated JNK levels. As a positive control, anisomysin (10 μg/ml, 30 min)-induced JNK phosphorylation is also shown. (C) Schwann cells were pretreated with K252a, and the activity of JNK was measured 30 min after stimulation with NT3. (D) Cos-7 cells cotransfected with TrkC and HA-JNK were treated with NT3. Transfected HA-JNK was immunoprecipitated with anti-JNK antibody and blotted with antiphosphorylated JNK antibody or anti-JNK antibody. (E) Levels of HA-JNK phosphorylation were quantified and normalized against the total immunoprecipitated HA-JNK levels (•). Mock transfections are represented (○). As a positive control, anisomysin-induced HA-JNK phosphorylation is also shown (▪). (F) Cos-7 cells cotransfected with TrkC and HA-JNK were pretreated with K252a, and the activity of HA-JNK was measured 30 min after stimulation with NT3.
Similar results were obtained after stimulation with NT3 in Cos-7 cells cotransfected with the plasmids encoding JNK and TrkC (Fig. 3 D–F). Stimulation with NT3 activated JNK in a time-dependent manner and at levels similar to the JNK activation by anisomysin (Fig. 3 D and E). In addition, the NT3-induced JNK activation was inhibited by pretreatment with K252a (Fig. 3F). The dominant-inhibitory variant of JNK kinases MKK4 or MKK7 also blocked the NT3-induced migration (data not shown), indicating that JNK activity is necessary for migration.
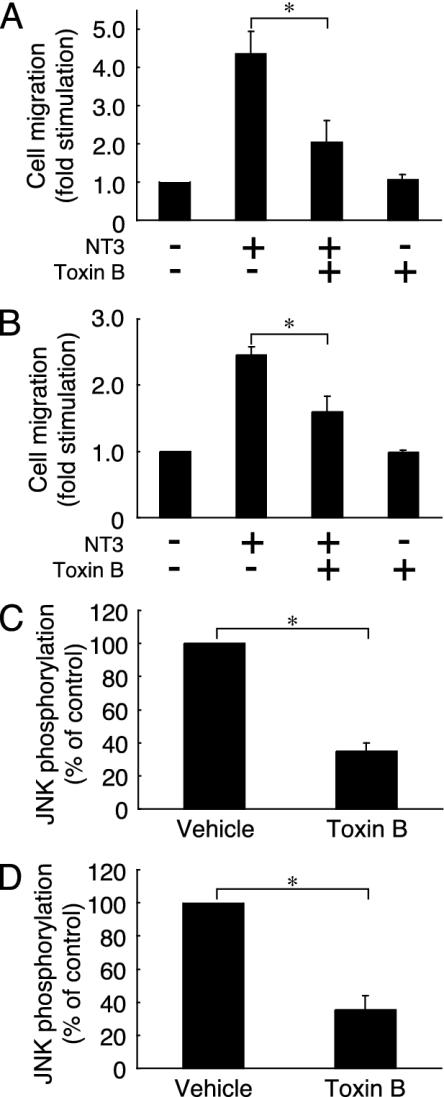
NT3 Enhances Cell Migration Through Rho GTPases. Because the Rho GTPases Rac1 and Cdc42 act upstream of JNK in various signaling pathways (22, 23), we examined the involvement of these Rho GTPases in cell migration induced by NT3. Pretreatment with Toxin B (24), which glycosylates and inhibits RhoA, Rac1, and Cdc42, blocked the NT3-induced migration of Schwann cells (Fig. 4A) and Cos-7 cells transfected with TrkC (Fig. 4B), suggesting that Rho GTPases are also involved in the NT3-induced cell migration. Furthermore, pretreatment with Toxin B inhibited the JNK activation by NT3 in Schwann cells (Fig. 4C) and Cos-7 cells transfected with TrkC (Fig. 4D). These results demonstrate an involvement of Rho GTPases as a regulator of cell migration.
Fig. 4.
Involvement of Rho GTPases in NT3-induced cell migration and JNK activation. Rat Schwann cells (A and C) and Cos-7 cells transfected with TrkC (B) or TrkC and HA-JNK (D) were pretreated with Toxin B. (A and B) After incubation with NT3, the migrating cells in the Boyden chambers were stained and counted. (C and D) The activity of endogenous JNK or HA-JNK was measured 30 min after stimulation with NT3. Data were evaluated by using Student's t test. *, P < 0.01.
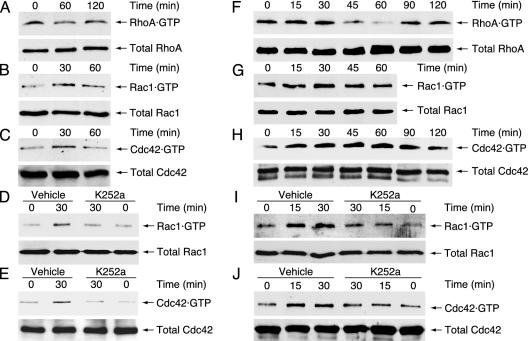
We therefore measured the activities of endogenous Rho GTPases RhoA, Rac1, and Cdc42 in Schwann cells by the pull-down assays with GST-tagged mDia1-RBD and αPak-CRIB. The mDia1-RBD specifically associates with active GTP-bound RhoA, and the αPak-CRIB specifically interacts with active GTP-bound Rac1 and Cdc42. The GTP-bound forms of RhoA and Rac1 were detected in the lysate of Cos-7 cells cotransfected with the plasmids encoding CA-Src and Dbl homology domain of RhoA- and Rac1-GEF Vav2 (13). The GTP-bound Cdc42 was detected in the lysate of Cos-7 cells transfected with Dbl homology and pleckstrin homology domains of Cdc42-GEF Frg (15). As shown in Fig. 5 B and C, Rac1 and Cdc42 were activated in a time-dependent manner after stimulation with NT3 in Schwann cells. The activities of Rac1 and Cdc42 reached maximum levels at 30 min after stimulation with NT3. These effects were inhibited by pretreatment with K252a (Fig. 5 D and E). In contrast, RhoA was down-regulated after stimulation with NT3 in Schwann cells (Fig. 5A). The RhoA activity reached the lowest level at 60 min after stimulation with NT3. Similar to Schwann cells, Rac1 and Cdc42 were activated after stimulation with NT3 in a time-dependent manner, in Cos-7 cells cotransfected with the plasmids encoding TrkC and Rac1 or Cdc42 (Fig. 5 G and H). In addition, these effects were inhibited by pretreatment with K252a (Fig. 5 I and J). In contrast, RhoA was down-regulated after stimulation with NT3 in Cos-7 cells (Fig. 5F). Taken together with the results from Fig. 4, these results suggest that NT3 activation of TrkC stimulates JNK activity and cell migration through activation of Rac1 and Cdc42. Furthermore, the time course of the activation of Rac1 (Fig. 5 B and G) and Cdc42 (Fig. 5 C and H) was very similar to that of JNK activation (Fig. 3 B and E), indicating that Rac1 and Cdc42 can act as activators of the JNK cascade in the NT3/TrkC signaling pathway. Although RhoA was down-regulated following stimulation with NT3, RhoA does not appear to act as an upstream regulator of JNK cascade (22, 23, 25).
Fig. 5.
Rho GTPases Rac1 and Cdc42 are activated after stimulation with NT3. The activities of Rho GTPases were measured in rat Schwann cells (A–E) and Cos-7 cells cotransfected with RhoA (F), Rac1 (G and I), or Cdc42 (H and J), together with TrkC. (A–C) The activities of endogenous Rho GTPases were measured after the addition of NT3 with the pull-down assay by using recombinant mDia1-RBD or αPak-CRIB. (D and E) Cells were pretreated with K252a, and the activities of endogenous Rac1 and Cdc42 were measured after the addition of NT3. (F–H) The activities of transfected Rho GTPases were measured after the addition of NT3. (I and J) Cells were pretreated with K252a, and the activities of transfected Rac1 and Cdc42 were measured after the addition of NT3. The total Rho GTPases in the cell lysates were immunoblotted with anti-Rho GTPases antibodies.
Discussion
The formation of peripheral myelin by Schwann cells can be divided into three major stages: proliferative, premyelinating, and myelinating stages. The proliferative stage is characterized by proliferation and migration of premyelinating Schwann cells. We demonstrated that NT3 inhibits myelination of Schwann cells through TrkC and that expression of NT3 levels correlated with the initiation of the proliferation stage (11, 12). Thereafter, NT3 levels gradually decrease throughout the proliferative and early premyelinating stages. In addition, the expression of TrkC decreases in the early myelinating stage of Schwann cell/DRG neuronal cocultures and in sciatic nerves during the postnatal development (12). This finding suggests a potential role for NT3 in the proliferative stage of Schwann cell development. Here we show that NT3 stimulates Schwann cell migration through TrkC.
Like NT3, glial growth factor (GGF), an isoform of neuregulin 1, enhances Schwann cell migration (26) and inhibits myelination (27). Because the NT3 scavenger TrkC-Fc inhibits Schwann cell migration by ≈50% induced with conditioned media from DRG neurons, it is possible that secreted GGF is responsible for the remaining activity. Therefore, NT3 and GGF may act cooperatively in this process. Future studies to determine the expression and localization of NT3, GGF, and their receptors during development and nerve injury should aid in examining the redundancy of NT3 and GGF in controlling Schwann cell migration and in identifying other possible functions. The levels of neuregulin isoforms (28) and the cognate receptors ErbB2 (29) and ErbB3 (30) are down-regulated in sciatic nerves during the postnatal development. It is conceivable that down-regulation of NT3/TrkC and GGF/ErbB2/ErbB3 is required for the transition into the myelinating stage. This possibility also suggests that NT3/TrkC and GGF/ErbB2/ErbB3 may have the ability to maintain Schwann cells in a nonmyelinating state, an idea supported by the finding that GGF is a mitogen for Schwann cells (31). However, whether NT3 is a mitogen for Schwann cells is not yet known.
Schwann cell migration is not only essential for development of the peripheral nervous system but also for regeneration and remyelination after nerve injury. During regeneration, Schwann cells play a key role in supporting neuronal survival and growth. After nerve injury, NT3 synthesis is up-regulated in satelite cells surrounding neurons in the transected DRG (32). Therefore, if NT3 establishes a concentration gradient, highest at satelite cells near the transected site and decreasing toward the periphery, Schwann cells could potentially migrate out of distal nerve stumps along the NT3 gradient to promote neuronal survival.
It has been widely reported that JNK is responsible for cell migration during Drosophila development, specifically during the process of dorsal closure (10). We show that JNK is required for the NT3/TrkC-induced migration of Schwann cells. In addition, we also demonstrate that Rac1 and Cdc42 are necessary for activation of JNK. Thus, it is possible that Rac1 and Cdc42 associate with JNK upstream kinases such as MEKK1 and MLK2 (25), resulting in up-regulation of the JNK cascade. In contrast, RhoA is down-regulated through TrkC, and it is unlikely that RhoA regulates the JNK cascade (22, 23). Although it remains unclear whether down-regulation of RhoA is involved in Schwann cell migration, RhoA is often regulated oppositely from Rac1 and Cdc42 in the signal transduction cascades of the nervous system (33). RhoA and Rac1/Cdc42 may be oppositely regulated by upstream regulators including GEFs and GTPase-activating proteins for Rho GTPases in the TrkC signaling pathway.
In the present study, we demonstrate that NT3/TrkC uniquely stimulates Schwann cell migration through the Rac1/Cdc42/JNK pathway. This finding provides yet another example of how NT3/TrkC may function during development or after nerve injury by recruiting Schwann cells along axons or to the site of injury before the initiation of myelination. On the basis of these findings, we summarize the proposed signaling pathway in Fig. 6. Recently, paxillin, a focal adhesion protein, has been identified as a target for JNK in the migration of epithelial cells and keratocytes (34). Therefore, paxillin may be a candidate for the JNK substrate involved in Schwann cell migration. Further study is necessary to clarify the detailed mechanism of Schwann cell migration by Rac1/Cdc42/JNK. Such studies would be useful for the development of therapeutic procedures after nerve injury and for the elucidation of the early process of myelination by Schwann cells.
Fig. 6.

Schematic model for the signaling pathway coupling NT3/TrkC to Schwann cell migration. Details are described in Discussion.
Acknowledgments
We thank Drs. D. Shiokawa and L. Chen for helpful discussions. This work was supported by grants from the National Institute of Neurological Disorders and Stroke, the Muscular Dystrophy Association, and the McGowan Charitable Trust (to E.M.S.) and by a National Research Service Award (to J.R.C.).
Abbreviations: BDNF, brain-derived neurotrophic factor; NT3, neurotrophin 3; NTR, neurotrophin receptor; GEF, guanine nucleotide-exchange factor; CRIB, Rac1·GTP- and Cdc42·GTP-binding domain; RBD, RhoA·GTP-binding domain; DRG, dorsal root ganglion; JNK, c-Jun N-terminal kinase; CA, constitutively active.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY336094).
References
- 1.Webb, D. J., Parsons, J. T. & Horwitz, A. F. (2002) Nat. Cell Biol. 4, E97-E100. [DOI] [PubMed] [Google Scholar]
- 2.Bunge, M. B., Williams, A. K. & Wood, P. M. (1982) Dev. Biol. 92, 449-460. [DOI] [PubMed] [Google Scholar]
- 3.Bunge, R. P. (1993) Curr. Opin. Neurobiol. 3, 805-809. [DOI] [PubMed] [Google Scholar]
- 4.Fawcett, J. W. & Keynes, R. J. (1990) Annu. Rev. Neurosci. 13, 43-60. [DOI] [PubMed] [Google Scholar]
- 5.Levi-Montalcini, R. (1987) Science 237, 1154-1162. [DOI] [PubMed] [Google Scholar]
- 6.Shooter, E. M. (2001) Annu. Rev. Neurosci. 24, 601-629. [DOI] [PubMed] [Google Scholar]
- 7.Huang, E. J. & Reichardt, L. F. (2003) Annu. Rev. Biochem. 72, 609-642. [DOI] [PubMed] [Google Scholar]
- 8.Kaibuchi, K., Kuroda, S. & Amano, M. (1999) Annu. Rev. Biochem. 68, 459-486. [DOI] [PubMed] [Google Scholar]
- 9.Bar-Sagi, D. & Hall, A. (2000) Cell 103, 227-238. [DOI] [PubMed] [Google Scholar]
- 10.Davis, R. J. (2000) Cell 103, 239-252. [DOI] [PubMed] [Google Scholar]
- 11.Chan, J. R., Cosgaya, J. M., Wu, Y. J. & Shooter, E. M. (2001) Proc. Natl. Acad. Sci. USA 95, 10459-10464. [Google Scholar]
- 12.Cosgaya, J. M., Chan, J. R. & Shooter, E. M. (2002) Science 298, 1245-1248. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi, J., Tsujimoto, G., Kaziro, Y. & Itoh, H. (2001) J. Biol. Chem. 276, 23362-23372. [DOI] [PubMed] [Google Scholar]
- 14.Miyamoto, Y., Yamauchi, J. & Itoh, H. (2003) J. Biol. Chem. 278, 29890-29900. [DOI] [PubMed] [Google Scholar]
- 15.Aghazadeh, B., Lowry, W. E., Huang, X.-Y. & Rosen, M. K. (2000) Cell 102, 625-633. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, K., Tsuji, T., Takada, Y., Miki, T. & Narumiya, S. (2000) J. Biol. Chem. 275, 17233-17236. [DOI] [PubMed] [Google Scholar]
- 17.Sander, E. E., van Delft, S., ten Klooster, J. P., Reid, T., van der Kammen, R. A., Michiels, F. & Collard, J. G. (1998) J. Cell Biol. 143, 1385-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottenstein, J. E. & Sato, G. H. (1980) Exp. Cell Res. 129, 361-366. [DOI] [PubMed] [Google Scholar]
- 19.Ritch, P. A., Carroll, S. L. & Sontheimer, H. (2003) J. Biol. Chem. 278, 20971-20978. [DOI] [PubMed] [Google Scholar]
- 20.McDonald, P. H., Chow, C. W., Miller, W. E., Laporte, S. A., Field, M. E., Lin, F. T., Davis, R. J. & Lefkowitz, R. J. (2000) Science 290, 1574-1577. [DOI] [PubMed] [Google Scholar]
- 21.Bennett, B. L., Sasaki, D. T., Murray, B. W., O'Leary, E. C., Sakata, S. T., Xu, W., Leisten, J. C., Motiwala, A., Pierce, S., Satoh, Y., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coso, O. A., Chiariello, M., Yu, J.-C., Teramoto, H., Crespo, P., Xu, N., Miki, T. & Gutkind, J. S. (1995) Cell 81, 1137-1146. [DOI] [PubMed] [Google Scholar]
- 23.Minden, A., Lin, A., Claret, F.-X., Abo, A. & Karin, M. (1995) Cell 81, 1147-1157. [DOI] [PubMed] [Google Scholar]
- 24.Just, I., Selzer, J., Wilm, M., von Eichel-Streiber, C., Mann, M. & Aktories, K. (1995) Nature 375, 500-503. [DOI] [PubMed] [Google Scholar]
- 25.Burbelo, P. D., Drechsel, D. & Hall, A. (1995) J. Biol. Chem. 270, 29071-29074. [DOI] [PubMed] [Google Scholar]
- 26.Mahanthappa, N. K., Anton, E. S. & Matthew, W. D. (1996) J. Neurosci. 16, 4673-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanazzi, G., Einheber, S., Westreich, R., Hannocks, M.-J., Bedell-Hogen, D., Marchionni, M. A. & Salzer, J. L. (2001) J. Cell Biol. 152, 1289-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinoda, J., Nakano, J., Iizuka, Y., Toba, T., Yazaki, T., Kawase, T. & Uyemura, K. (1997) J. Neurosci. Res. 50, 673-683. [DOI] [PubMed] [Google Scholar]
- 29.Cohen, J. A., Yachnis, A. T., Arai, M., Davis, J. G. & Scherer, S. S. (1992) J. Neurosci. Res. 31, 622-634. [DOI] [PubMed] [Google Scholar]
- 30.Grinspan, J. B., Marchionni, M. A., Reeves, M., Coulaloglou, M. & Scherer, S. S. (1996) J. Neurosci. 16, 6107-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garratt, A. N., Britsch, S. & Birchmeier, C. (2000) BioEssays 22, 987-996. [DOI] [PubMed] [Google Scholar]
- 32.Zhou, X.-F., Deng, Y.-S., Chie, E., Xue, Q., Zhong, J.-H., McLachian, E. M., Rush, R. A. & Xian, C. J. (1999) Eur. J. Neurosci. 11, 1711-1722. [DOI] [PubMed] [Google Scholar]
- 33.Huber, A. B., Kolodkin, A. L., Ginty, D. D. & Cloutier, J.-F. (2003) Annu. Rev. Neurosci. 26, 509-563. [DOI] [PubMed] [Google Scholar]
- 34.Huang, C., Rajfur, Z., Borchers, C., Schaller, M. D. & Jacobson, K. (2003) Nature 424, 219-223. [DOI] [PubMed] [Google Scholar]