Abstract
ATP-dependent chromatin remodeling complexes rearrange nucleosomes by altering the position of DNA around the histone octamer. Although chromatin remodelers and the histone variant H2A.Z colocalize on transcriptional control regions, whether H2A.Z directly affects remodeler association or activity is unclear. We determined the relative association of remodelers with H2A.Z chromatin and tested whether replacement of H2A.Z in a nucleosome altered the activity of remodeling enzymes. Many families of remodelers showed increased association with H2A.Z chromatin, but only the ISWI family of chromatin remodelers showed stimulated activity in vitro. An acidic patch on the nucleosome surface, extended by inclusion of H2A.Z in nucleosomes and essential for viability, is required for ISWI stimulation. We conclude that H2A.Z incorporation increases nucleosome remodeling activity of the largest class of mammalian remodelers (ISWI) and that it correlates with increased association of other remodelers to chromatin. This reveals two possible modes for regulation of a remodeler by a histone variant.
Keywords: Chromatin/Regulation, Chromatin/Remodeling, Chromatin/Structure, Gene/Promoters, Gene/Regulation, Gene/Transcription
Introduction
Histone H2A.Z substitutes for the canonical H2A histone in the nucleosome at nonrandom locations throughout the genome and has therefore been proposed to play a key role in chromatin regulation. Because H2A.Z is expressed throughout the cell cycle, unlike H2A, which is synthesized predominantly during the S phase, H2A.Z is a likely candidate for modulating the many functions of chromatin in addition to the packaging of DNA (1). High conservation of H2A.Z across all metazoans (>90%) relative to lower conservation with canonical H2A (∼64%) indicates that the H2A.Z variant might play a specific biological function (2). The importance of H2A.Z is further demonstrated both in knock-out mice and depletion from embryonic stem cells, showing that H2A.Z is essential for cell differentiation and organism development (3, 4).
H2A.Z compromises 5–10% of the total H2A in a human cell and localizes to ∼50% of annotated promoters regions, often at transcription start sites, supporting a role in transcriptional regulation (5, 6). In addition to its localization at promoters, H2A.Z is also detected at many loci staged for reorganization in response to stimulation such as at binding sites for the transcription factor glucocorticoid receptor (7), p53 (8), and estrogen receptor α (ERα)2 (9). These studies suggest that H2A.Z might play a regulatory role, either by causing a change in chromatin structure or by recruiting chromatin-modifying proteins.
Chromatin can be modified by numerous proteins, one class of which is ATP-dependent remodeling enzymes. Remodelers catalyze the hydrolysis of ATP and use the energy generated to move DNA relative to the histone octamer. Chromatin remodeling complexes are involved in numerous processes in the nucleus that, like H2A.Z, include roles in gene regulation and subsequent changes in cell identity. Because remodeling complexes bind directly to nucleosomes, replacing H2A.Z for H2A might alter either the recruitment or the function of remodelers. Families of remodeling complexes have been characterized and are categorized by the identity of the core ATPase motor protein (10). One class of remodelers, the “split ATPases” typified by SRCAP and p400 in humans, have been shown to specifically exchange canonical dimers in chromatin for H2A.Z-containing dimers (11). Other prominent classes of remodelers in human cells are the Swi/Snf family with BRG1 as the core ATPase, the ISWI family with either SNF2H or SNF2L as the core ATPase, and the CHD family with Mi-2 as the core ATPase. Although not shown to be involved in H2A.Z deposition, the functional and mechanistic differences between these three families of complexes may be influenced by the presence of H2A.Z in their chromatin substrate.
Several studies describe a connection between H2A.Z and chromatin remodelers not involved in deposition. In Saccharomyces cerevisiae, the H2A.Z variant Htz1 shows synthetic lethality with both remodeling motor enzymes Snf2 and Isw1. Parallel but redundant roles in specific gene expression were shown between H2A.Z and both Snf2 and Isw1 remodelers (12, 13). On Drosophila melanogaster salivary gland chromosomes, H2A.Z colocalizes with the ISWI remodeling complex RSF (14). In humans, H2A.Z is detected at sites of transcriptionally regulated DNase I hypersensitivity; accessibility of DNase I to some of these sites is mediated by the Swi/Snf motor protein BRG1 (7). Furthermore, recruitment of BRG1 to ERα-induced promoter TTF1 is dependent on H2A.Z deposition (15). Thus, chromatin remodeler associations with H2A.Z have been widely reported; however, direct evidence for H2A.Z specifying chromatin remodeling activity remains to be demonstrated.
In this report we address the association of human ATP-dependent remodelers with H2A.Z chromatin. We determined the physical interaction of remodelers on chromatin containing H2A.Z using cultured cells. We also measured the activity of remodelers in vitro to ascertain whether remodelers distinguish H2A.Z nucleosomes from H2A nucleosomes. We find that distinct classes of remodelers can be differentially associated with H2A.Z nucleosomes. Furthermore, the ISWI class of remodelers has greater activity on H2A.Z nucleosomes, thus suggesting an alternative mechanism for increased remodeling on variant chromatin.
MATERIALS AND METHODS
Chromatin Immunoprecipitation
Native nuclei from HeLa S3 cell lines stably expressing H2A or H2A.Z with a C-terminal FLAG affinity tag were purified using 10 strokes with a tight pestle Dounce homogenizer in chromosome isolation buffer (3.75 mm Tris-HCl, pH 7.7, 20 mm KCl, 0.5 mm EDTA, 1 mm dithiothreitol, 0.05 mm spermidine, 0.125 mm spermine, 1 mm phenylmethylsulfonyl fluoride, 0.1% digitonin) (16). Purified nuclei were digested in buffer A (20 mm HEPES, pH 7.7, 20 mm KCl, 0.5 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride) with 200 units/ml micrococcal nuclease (Roche Applied Science) and 3 mm CaCl2 at room temperature for 1 h. The reactions were stopped with 5 mm EGTA and centrifuged for 15 min at 10,000 × g. DNA fragments from the supernatant were predominantly mononucleosomal with a ladder extending up to tetranucleosomes. For purification the chromatinized nuclear extract was bound to M2-agarose beads (Sigma) for 3 h; washed in batch successively with 30 mm KCl, 300 mm KCl, and 300 mm KCl with 0.1% Tween 20; and eluted for 1 h at 4 °C with 1 mg/ml FLAG peptide. H2A and H2A.Z purifications were subjected to Western blot using the FLAG antibody as a loading control. The enrichment ratios were estimated by titrating the sample with the more prominent band until the band intensity was similar between the samples. For proteins where the differences were minor, for example ACF, quantitation was performed using a Typhoon optical scanner on membranes probed with Cy5-labeled secondary antibody (Amersham Biosciences). Western blots were performed on at least three biological replicates for each immunoprecipitation.
Antibodies
All of the antibodies used are commercially available: from Abcam, Snf2h (ab3749), p400 (ab5201), and RSF (ab43632); from Bethyl, Brg1 (A300–813A-1), Mi-2α (A301-219A-1), Mi-2β (A301–082A-1), ACF1 (A301–318A-1), and BAF155 (A301–021A-1); from Novus, BPTF (NB100–41418); from Diagenode, Tip5 (CS-090–100); and from Sigma, WSTF (W3766).
Nucleosome Assembly
Individual histone proteins were purified recombinantly in Escherichia coli after expression using recombinant vectors and assembled into octamers by the standard techniques described previously (17). Mononucleosomes were assembled on a 200-bp PCR fragment containing the TPT nucleosome positioning sequence by salt dialysis and further purified by glycerol gradient (18). Cy3- and Cy5-labeled mononucleosomes were assembled analogously using PCR fragments generated with 5′ end-labeled fluorophores. Mononucleosome with either a 5S or 601 positioning sequence were prepared likewise. Twelve nucleosome arrays were assembled on a 2.5-kb G5E4 fragment containing ten 5S repeats with a central 400 bp containing a unique HhaI restriction site. EcoRI digestion between the 5S repeats followed by native agarose electrophoresis monitored the degree of assembly. At least 90% of the 5S repeats appeared nucleosomal for both H2A and H2A.Z arrays.
Nucleosome Remodeling Assay
All of the remodeling enzymes were expressed using the baculovirus protein expression in Sf9 cells except for the Swi/Snf complex, which was purified from stable Ini1-FLAG expressing HeLa cells (19). In the restriction enzyme accessibility (REA) assay, remodeling is inferred from the cutting of a specific restriction enzyme site exposed upon the addition of ATP (20). An REA assay was performed on differentially labeled recombinant nucleosomes (30 nm each nucleosome, 3 nm remodeling enzyme, 4 mm MgCl2, 2 mm ATP-Mg, 4 units of PstI) (21). Subsequent kobs was calculated using the same REA assay but under single turnover conditions (1 nm nucleosome, 20–80 nm remodeling enzyme). Calculations were repeated for Brg1 and Snf2h on at least seven different nucleosome assemblies to ensure that the measured differences were not a result of the preparation or variability in quantitation (22). The reactions without ATP were performed in parallel to determine enzyme-independent background cutting, which was typically ∼10% for canonical nucleosomes and ∼15% for H2A.Z nucleosomes. Cleavage in the presence of ATP resulted in ∼50% cutting in the first 15 min of the reaction.
Glycerol Gradient
The nucleosome assembly reactions were pipetted onto a 10–30% gradient (50 mm Tris, pH 8.0, 100 mm KCl, 1 mm EDTA, 0.1% Nonidet P-40) and spun at 35,000 × g for 18 h using an Sw55Ti ultracentrifuge rotor. Eluted fractions were assessed by native gel electrophoresis.
RESULTS
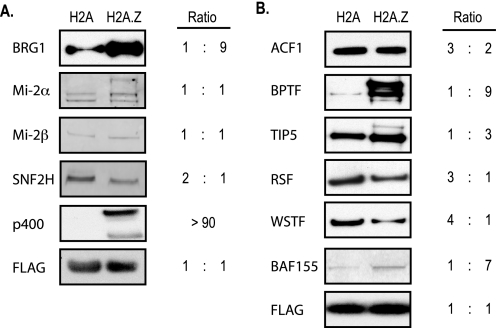
To determine which ATP-dependent remodeling enzymes are associated with chromatin containing H2A.Z, we purified nucleosome sized fragments of chromatin from cells expressing an affinity-tagged copy of either H2A or H2A.Z and looked for enrichment of motor proteins. The ATP-dependent remodeler responsible for H2A.Z deposition, p400 (8), was found solely at chromatin purified using H2A.Z-FLAG, indicating that these preparations are of high quality and are enriched for expected factors. Because H2A.Z has widespread localization at gene regulatory elements, we expected that many other remodelers might be enriched on H2A.Z chromatin. As expected, BRG1, the motor subunit for the human Swi/Snf complex, was considerably enriched with H2A.Z purified chromatin (Fig. 1A). CHD family members Mi-2α and Mi-2β were similarly present on H2A.Z chromatin. We conclude that several remodeling complexes are preferentially associated with H2A.Z chromatin relative to the proportion of H2A.Z in the genome. These associations might reflect associations in the intact cell or might reflect redistribution of proteins following cell lysis.
FIGURE 1.
All major families of ATP-dependent chromatin remodelers are bound to H2A.Z-containing chromatin. Western blots on purified H2A and H2A.Z chromatin were performed for ATP-dependent core motor proteins (A) and other remodeling complex subunits (B). The primary antibody used in each blot is indicated on the left. A Western blot against the affinity tag on the histone (FLAG) was used as a loading control. Changes in amount of protein present in the input of the purifications were not evident between HeLa, H2A-FLAG, or H2A.Z-FLAG nuclear extracts as determined by Western blot (not shown). The two lower bands observed for Mi-2α migrate at the expected molecular weight for the protein and likely reflect alternatively modified forms of the protein. The top slower migrating species observed only with H2A.Z is >100 kDa larger than expected and is of unknown identity.
The only major remodeling protein tested that was not enriched on H2A.Z chromatin was SNF2H, which is among the most abundant remodeling proteins in the nucleus. Although analysis of the motor protein alone revealed modest depletion of SNF2H in H2A.Z chromatin, there are several complexes containing SNF2H involved in various processes in addition to gene regulation; each of these complexes may be recruited at different levels to H2A.Z chromatin. To address whether depletion of Snf2h on H2A.Z was indicative of general depletion of hISWI-containing complexes, we probed for other subunits of hISWI complexes (Fig. 1B). As a control, we found that the Swi/Snf core subunit BAF155 is enriched on H2A.Z chromatin in agreement with BRG1 enrichment. ISWI complex subunit enrichment levels depended on the complex. Two subunits from separate ISWI complexes involved in transcription were enriched appreciably with H2A.Z, BPTF from hNURF, and Tip5 from NoRC. Therefore, ATP-dependent remodeling complexes important in gene regulation are more prevalent on H2A.Z chromatin. Interestingly, we note that both of these subunits also showed an additional slower migrating species that is potentially indicative of H2A.Z specific modifications.
Analysis of purified chromatin from HeLa cells revealed that certain remodeling enzymes are more prevalent on H2A.Z-containing chromatin. Enrichment of a remodeler with H2A.Z might provide one mechanism for increasing remodeling as has been indicated previously (7, 15). Additionally, inclusion of H2A.Z in chromatin could effect the proficiency of the remodeling reaction itself. Both mechanisms could act in concert or alternatively for H2A.Z to specify increased remodeling. To address whether incorporation of H2A.Z was sufficient to alter the remodeling reaction, we compared the activity of purified remodelers on the nucleosomes assembled using recombinant H2A or H2A.Z (Fig. 2).
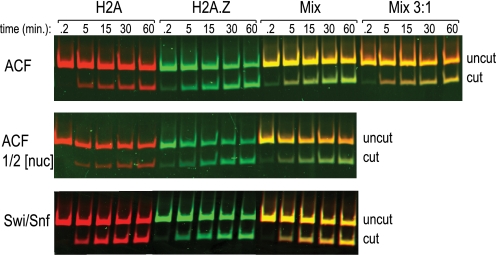
FIGURE 2.
Human ACF complex remodels H2A.Z-containing nucleosomes more rapidly than H2A nucleosomes. Cy5 end-labeled recombinant canonical nucleosomes (red) were mixed in equal proportion (30 nm) to Cy3 end-labeled recombinant H2A.Z nucleosomes (green) and remodeled in the presence of 2 mm ATP by the enzyme complex indicated on the left. Remodeling is inferred by digestion with Pst1. The reactions were stopped at the indicated time points, proteinase-digested, and resolved on a 6% acrylamide gel before scanning on a Typhoon imager. Time points from independent reactions with H2A-Cy5 and H2A.Z-Cy3 nucleosomes alone are shown first and indicate that both preparations of nucleosomes are competent for remodeling. The bottom panel is a replicate of ACF remodeling shown in the top panel but done with half the total concentration of each nucleosome (15 nm).
Remodeling reactions were performed with equal amounts of both H2A and H2A.Z nucleosomes in the same reaction but labeled with different fluorophores to distinguish between the products. Remodeling of nucleosomes was measured by an increased access to restriction enzyme cleavage at a site normally occluded in the nucleosome three or four helical turns from the entry point. Remodeling by purified Swi/Snf complex, which is elevated on H2A.Z chromatin in vivo, generated cleaved products from both H2A and H2A.Z with similar proportion (both the uncut and cut maintain a yellow color in the mixture; Fig. 2). Under these conditions, this implies a similar rate of remodeling of these two substrates by Swi/Snf and verifies that each of the substrates in the mixture can be remodeled. Unexpectedly, the human ISWI complex ACF, which is equally distributed on H2A.Z and canonical chromatin, remodeled H2A.Z nucleosomes more readily than it remodeled H2A nucleosomes (cut template is green). To offer a rough quantification of this difference, we mixed substrates at varying concentrations and found that the reaction favors the H2A.Z nucleosomes by an approximately 3:1 margin, as when nucleosomes were mixed in that ratio the color of the “cut” band indicated a roughly equal amount of cleavage of the two substrates. Similar results were seen at lower nucleosome concentrations, reaffirming the preference of ACF for H2A.Z-containing nucleosomes (Fig. 2).
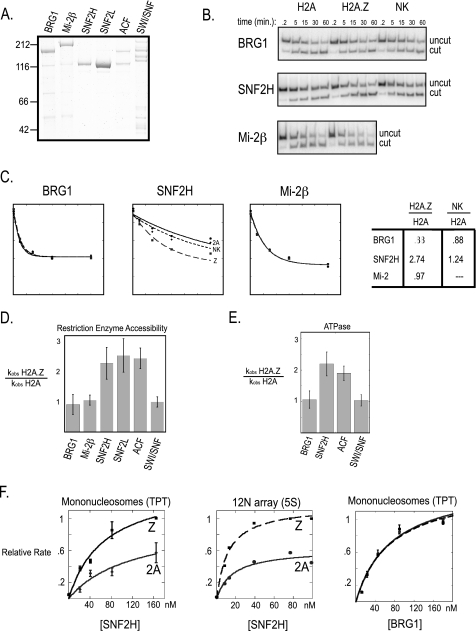
To confirm that H2A.Z specifies increased remodeling by ACF, we decided to more thoroughly quantify the differences. We determined the ratio of remodeling activity (kobsZ/kobs2A) on each substrate separately using the same restriction enzyme accessibility assay but under single turnover conditions where a rate can be calculated from fitting time points to an exponential decay function (22). Confirming the previous experiment, H2A.Z nucleosomes were remodeled more rapidly by the ACF complex than H2A-containing nucleosomes, whereas the Swi/Snf complex remodeled both at the same rate (Fig. 3, and C). To determine whether H2A.Z stimulation is a unique property of the ACF complex or a common property of core hISWI motor proteins, we tested recombinant SNF2H and SNF2L and found a similar H2A.Z-dependent increase in activity. Remodeling by core motor proteins BRG1 (Swi/Snf family member) and Mi-2β (CHD family member) were similar on the two substrates (Fig. 3D). We conclude that stimulation of remodeling on H2A.Z nucleosomes is a general feature of the ISWI family remodelers that is not seen with other prominent mammalian remodeling complexes (see also below).
FIGURE 3.
Increased efficiency of remodeling H2A.Z-containing nucleosomes is a general feature of ISWI family remodeling enzymes. A, colloidal blue staining of purified remodeling enzymes. B, sample time courses of restriction enzyme accessibility remodeling reactions for core motor proteins on P32 end-labeled nucleosome substrates. C, graphs of phosphorimaging quantitation of uncut substrate versus time from reactions shown in B. kobs for remodeling reactions is determined for the enzyme indicated by fitting the reaction to the exponential decay curve [S] = [S]ie−kt where [S]i is the initial substrate concentration and k is kobs. The ratios of kobs are shown on the right. D, summary of ratio kobs for reactions performed with recombinant H2A.Z-containing nucleosomes to recombinant canonical nucleosomes. The error bars represent standard deviations from the average of 7–26 different remodeling reactions from multiple enzyme preparations and nucleosome assemblies. E, kobs was measured by the rate of appearance of P32 labeled phosphate and is displayed as the ratio between H2A.Z nucleosomes to canonical nucleosomes. Free phosphate was resolved from hydrolyzed ATP by thin layer chromatography in 1 m formic acid, 0.5 m LiCl. F, rates were calculated and graphed as the percentage of the maximal rate within each titration to compare between replicates. The error bars represent the standard deviation between three replicates.
The increased activity of the SNF2H complexes on H2A.Z nucleosomes might reflect an increased efficiency in transferring the energy of ATP hydrolysis to remodeling or might reflect increased rates of ATP hydrolysis upon stimulation by this substrate. We therefore tested the rates of ATP hydrolysis by SNF2H on H2A and H2A.Z nucleosomes. The ability of SNF2H to hydrolyze ATP was stimulated by nucleosomes containing H2A.Z, as compared with H2A, in a manner similar to stimulation of remodeling. As a control, the rate of ATP hydrolysis was not different on the two substrates when BRG1 was used (Fig. 3E). Therefore ISWI family remodelers are stimulated by H2A.Z nucleosomes to similar levels using different assays.
Stimulation of SNF2H activity could be a result of either increased affinity for H2A.Z or improved proficiency of the remodeling reaction triggered by inclusion of H2A.Z in nucleosomes. We measured remodeling rates on H2A and H2A.Z nucleosomes over a broad range of concentrations of SNF2H that spanned the previously measured Km, nucleosome for ATPase activity (23) and found similar increases in remodeling of H2A.Z nucleosomes at all concentrations (Fig. 3F). The shape and magnitude of these plots are consistent with an increase in kcat for SNF2H remodeling of H2A.Z nucleosomes. Importantly, increased activity of SNF2H was also observed on a 12-nucleosome array over a broad range of concentrations (Fig. 3F). In conclusion, H2A.Z nucleosomes are more readily remodeled for a given unit of SNF2H.
One reason for the differences in activity of SNF2H on H2A.Z nucleosomes could be that these nucleosomes might display a different conformation of DNA than H2A nucleosomes. Increased affinity for particular translational positions on a nucleosome phasing sequence was reported previously for H2A.Z (24). Translational positions that increase the size of the DNA overhang are expected to increase remodeling activity more dramatically for ISWI family members (25, 26). To address this, mononucleosomes at different translational positions, which therefore contain different lengths of DNA overhang on each side of the nucleosome, were resolved by glycerol gradient and were remodeled by SNF2H (Fig. 4A). Similarly oriented H2A and H2A.Z fractions each recapitulated the increased SNF2H activity seen on H2A.Z nucleosomes. Moreover, the H2A.Z-dependent stimulation is maintained on different nucleosome phasing sequences as well as on a nucleosome array where the restriction site is located internally on DNA lacking a positioning sequence. We conclude that alternative lengths of DNA overhangs on mononucleosomes cannot explain the stimulation of ISWI family remodeling by H2A.Z. Therefore, hISWI family stimulation by H2A.Z is likely mediated by residues specific to the H2A.Z octamer and not through DNA.
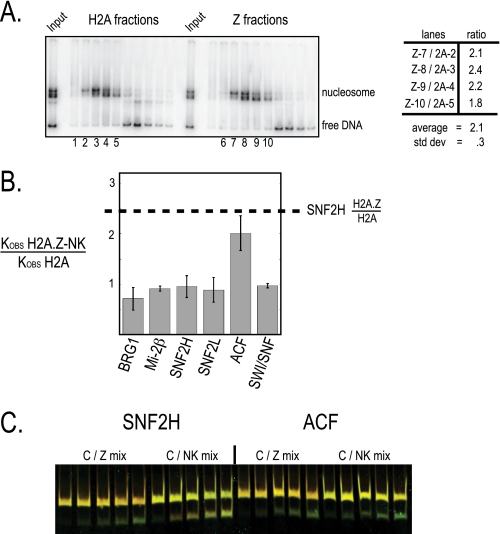
FIGURE 4.
ISWI family stimulation is independent of nucleosome position and dependent on the extended acidic patch on H2A.Z-containing nucleosomes. A, native PAGE of eluted glycerol gradient fractions of mononucleosomes recombinantly assembled on the TPT positioning sequence with a 45-bp overhang. Fractions of similar mobility within the native gel were compared as similarly positioned nucleosomes in REA assays with SNF2H. A summary of ratio of kobs between the indicated fractions is shown in the graph on the right. B, kobs of remodeling determined by REA assay are compared between the mutant H2A.Z mononucleosomes (NK) and canonical mononucleosomes for the enzyme indicated. The calculations were done as in Fig. 3D. The dashed line is from the data shown in Fig. 3D and is displayed as a reference point to compare H2A.Z-NK remodeling to wild type H2A.Z remodeling by SNF2H. C, REA assay with Cy5 end-labeled recombinant mononucleosomes (red) mixed with either Cy3 end-labeled H2A.Z nucleosomes or mutant H2A.Z (NK) mononucleosomes (green). Separate SNF2H and ACF remodeling reactions are indicated. The Cy3 label on the H2A.Z nucleosomes appears relatively less bright than the Cy3 label on the NK nucleosomes most likely because of decay of the Cy3 on the H2A.Z nucleosomes that were assembled earlier.
To verify that the H2A.Z protein is itself responsible for the increase in hISWI activity, we measured the remodeling activity of SNF2H on nucleosomes with mutations in biologically important H2A.Z residues. Dependence of SNF2H stimulation on such residues would suggest that this activity plays an important role in vivo. In Drosophila, the essential region of H2A.Z homolog H2Av mapped to a 16-amino acid region encompassing an extended acidic patch along the surface of the nucleosome (27). Within this patch on H2A.Z, replacement of acidic residues Asp94 and Ser95 to the analogous residues found in H2A (Asn94 and Lys95) reduces the acidic patch to canonical size and mimics developmental defects upon H2A.Z depletion in Xenopus embryos (28).
We assembled recombinant H2A.Z nucleosomes containing D94N,S95K mutations (NK) and measured the ability of SNF2H to remodel these nucleosomes. Unlike H2A.Z nucleosomes, the NK mutant nucleosomes were not more active as a substrate when SNF2H or SNF2L remodeling activity was tested (Fig. 4B). These mutations did not impact Swi/Snf remodeling or Mi-2β remodeling in control reactions. The reciprocal replacement of residues on H2A with the two acidic residues present in H2A.Z did not increase SNF2H activity (data not shown). In conclusion, the same residues required for H2A.Z function in vivo are necessary but not sufficient for the H2A.Z-specific increase in ISWI motor protein activity.
We were surprised to find, however, that remodeling by the ACF complex occurred at similar rates on both H2A.Z-WT and mutant H2A.Z-NK nucleosomes. To verify this result, we used the qualitative color-based protocol (described above; Fig. 2) to directly compare activity on these two substrates. Remodeling reactions were performed in the presence of both canonical nucleosomes (labeled with red) and either H2A.Z nucleosomes or NK nucleosomes (labeled with green; Fig. 4C). As shown above, remodeling of the canonical/H2A.Z mixture by Snf2h and ACF resulted in the more rapid appearance of the H2A.Z product (bottom green band). Remodeling of the canonical/NK mixture by the Snf2h motor protein resulted in concurrent appearance of both products (bottom yellow band), confirming that ISWI stimulation by H2A.Z depends on the acidic patch residues. However, remodeling with the ACF complex resulted in the more rapid appearance of the NK product (bottom green band). We conclude that the ACF complex displays less of a dependence upon the NK residues than the catalytic subunit, indicating that the extra hACF1 subunit either interacts with H2A.Z itself or alters the ability of the catalytic subunit to interact with H2A.Z.
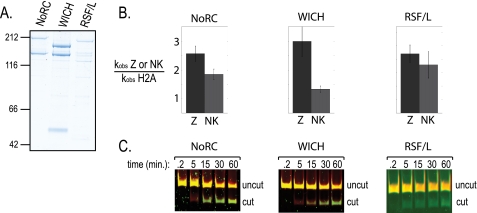
The stimulation of remodeling activity by H2A.Z might be more pronounced with hISWI complexes found to be more prevalent on H2A.Z chromatin in vivo. The activity of SNF2H-containing complexes can be altered depending on the particular associating subunit. Therefore, we compared the activity of the hISWI complex NoRC (TIP5/SNF2H), which is enriched on H2A.Z chromatin, to both WICH (WSTF/SNF2H) and RSF (RSF/SNF2L) complexes, which are enriched on H2A chromatin (Fig. 1). Each of these hISWI-containing complexes behaved similarly to ACF, in that each was more active on H2A.Z nucleosomes than on H2A nucleosomes (Fig. 5). As was seen with ACF, activity of the RSF complex was not significantly impacted by the NK mutation, in contrast to the individual motor subunits. We conclude that the stimulation of remodeling by H2A.Z is a general property of ISWI complexes that is shared by complexes whether or not they are enriched on chromatin that contains H2A.Z.
FIGURE 5.
H2A.Z stimulation of remodeling is shared with multiple ISWI complexes regardless of degree of in vivo enrichment. A, colloidal blue staining of recombinant ISWI complexes. B, summary of REA remodeling reactions for H2A.Z enriched SNF2H complex (NoRC) and both H2A.Z depleted complexes containing SNF2H (WICH) and SNF2L (RSF/L). The ratio of kobs of either H2A.Z or H2A.Z-NK nucleosomes to kobs for canonical nucleosomes is graphed for each enzyme complex. The error bars reflect standard deviation from the average of at least seven reactions on multiple nucleosome assemblies. C, REA assay with Cy5 end-labeled recombinant canonical nucleosomes (red) and Cy3 end-labeled recombinant H2A.Z nucleosomes (green) were performed as in Fig. 2.
DISCUSSION
Loci with inducible chromatin accessibility changes dependent on remodeling enzymes are also enriched for H2A.Z (7). Consistent with a relationship between the presence H2A.Z and chromatin remodeling function, we show that human ISWI complexes have higher activity when presented with H2A.Z-containing nucleosomes as compared with H2A-containing nucleosomes. Furthermore, enrichment of Swi/Snf complex members BRG1 and BAF155 with H2A.Z chromatin suggests that different families of remodelers use different strategies to strengthen the activity of remodelers in regions that contain H2A.Z. Association between Swi/Snf and H2A.Z may be a result of separate pathways that colocalize but offer no further functional significance, although we consider this possibility as less likely considering the importance of both proteins in the process of cell differentiation. The distinct properties of H2A.Z-containing nucleosomes result in increased enzymatic activity with both the isolated ATPase subunits of the human ISWI family and four different ISWI remodeling complexes, whereas similar effects are not seen with BRG1 or Swi/Snf. We infer that H2A.Z loci are sites that direct increased function of the ISWI family remodelers. Combined with our observations (Fig. 1) and those of others (7, 12–15) that other remodelers are enriched on H2A.Z chromatin, this suggests a general role for H2A.Z in specifying increased remodeling.
Why are two distinct strategies employed to enhance H2A.Z remodeling that differ between families of remodelers? ISWI-containing complexes are far more abundant in the nucleus than Swi/Snf with an estimate of 1 remodeling complex/∼80 nucleosomes, making increased association specific to H2A.Z difficult to regulate (29). However, ISWI family ATPase activity, unlike Swi/Snf family activity, is much more stimulated by nucleosomes than free DNA, indicating that activity of ISWI remodelers rely more heavily on amino acid residues on the histone octamer (30). Therefore, altered activity as a result of changes in solvent-accessible amino acids by replacement of histone H2A with histone H2A.Z may be an effective method of targeting particular ISWI functions. Consistent with the idea that ISWI proteins interact directly with H2A.Z, the S. cerevisiae remodeler Isw2 was found to bind atop the surface of the nucleosome lying directly across the position where the H2A.Z histone would reside (31). Therefore, changes in biophysical properties of remodeling on H2A.Z chromatin may be a more effective means of increasing ISWI activity with this variant than increased recruitment.
A second model for how replacement with H2A.Z enhances ISWI activity could be through changes in location of the N-terminal tail of histone H4. Residues 16–19 of histone H4 are required for ISWI remodeling and also interact with the acidic patch of histone H2A to mediate chromatin fiber folding (32, 33). Extension of the H2A acidic patch by inclusion of H2A.Z alters chromatin fiber folding presumably by altering interactions with H4 N-terminal tails (34). Therefore changes in H4 tail position or accessibility mediated by the extended acidic patch on H2A.Z could alter ISWI activity. In support of this model, the same residues of H2A.Z that extend the acidic patch are required for ISWI motor protein stimulation by H2A.Z (Fig. 4). However, the implications of the acidic patch are expected to be more apparent in chromatin fibers than on mononucleosomes, because they are expected to involve internucleosomal interactions. We observe similar effects of H2AZ on remodeling in arrays as in mononucleosomes (Fig. 3F), an observation not easily reconciled with a key role for H4-H2A interactions. Also of note is that ISWI complexes ACF and RSF/L, unlike ISWI complexes NoRC and WICH, are equally stimulated by the acidic patch H2A.Z mutant, suggesting that additional H2A.Z contacts with ISWI remodelers with residues outside of the acidic patch might be mediated by noncatalytic subunits.
One important biological role of chromatin remodeling that may be important to regions that contain H2A.Z is the positioning of nucleosomes at promoters. In yeast Isw2 is required for positioning of nucleosomes at transcription start sites to facilitate the proper direction of transcription (35). Htz1, the yeast H2A.Z homolog, is shown to flank transcription start sites and is therefore correlated with the same nucleosomes positioned by Isw2 (36–39). Consistent with H2A.Z nucleosomes also being important in nucleosome positioning in humans, promoter nucleosomes positioned by transcriptional stimulation by ERα, remain diffuse in the TTF1 promoter upon depletion of H2A.Z (15). Generally, positioning of H2A.Z is observed at transcription start sites in humans mainly at the +1 nucleosome downstream. It is possible that hISWI may be instrumental in positioning H2A.Z at human promoters to modulate either the direction or other aspects of transcription.
Positioning of nucleosomes by chromatin remodelers has other important roles in gene regulation such as allowing the transcription machinery to bind. Preferential loss of H2A.Z from the −1 nucleosome mainly at active genes directly overlaps RNA polymerase II-binding sites (5, 40). Therefore, the apparent loss or delocalization of H2A.Z mediated either by hISWI complexes or by hSwi/Snf may be important for proper RNA polymerase II binding or orientation. In summary, regulation of either of the H2A.Z nucleosomes flanking transcription start sites in humans could be an important function of remodeling enzymes to regulate gene expression.
Previously it has been reported that H2A.Z replacement in a nucleosome interferes with in vitro activity of yeast chromatin remodeling proteins Chd1, Isw1, Swi/Snf, and RSC (37). Remodeling activity was assayed by ATP-dependent mobilization of nucleosomes in a polyacrylamide gel. We have also seen an apparent decrease in activity on H2A.Z nucleosomes by this assay (supplemental Fig. S1). Presumably, increased affinity of H2A.Z for particular nucleosome positions may explain the lack of changes in mobility and therefore the apparent lack of remodeling activity when measured by a protocol such as this that examines the end point of the reaction. Other reports have described competent remodeling by a SNF2H-containing complex, ACF, despite an apparent lack of mobilization in a gel shift (26). The restriction enzyme protocol used here measures all types of nucleosome movement, not just events that lead to an altered final position, so detection of a broader set of remodeling events is expected. Increased access for restriction enzymes observed in vitro may reflect the ability of the remodelers to allow increased access of DNA-binding factors required for a biological function in vivo.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ERα
- estrogen receptor α
- REA
- restriction enzyme accessibility
- NK
- D94N,S95K mutation.
REFERENCES
- 1.Wu R. S., Bonner W. M. (1981) Cell 27, 321–330 [DOI] [PubMed] [Google Scholar]
- 2.Thatcher T. H., Gorovsky M. A. (1994) Nucleic Acids Res. 22, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faast R., Thonglairoam V., Schulz T. C., Beall J., Wells J. R., Taylor H., Matthaei K., Rathjen P. D., Tremethick D. J., Lyons I. (2001) Curr. Biol. 11, 1183–1187 [DOI] [PubMed] [Google Scholar]
- 4.Creyghton M. P., Markoulaki S., Levine S. S., Hanna J., Lodato M. A., Sha K., Young R., Jaenisch R., Boyer L. A. (2008) Cell 135, 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schones D. E., Cui K., Cuddapah S., Roh T. Y., Barski A., Wang Z., Wei G., Zhao K. (2008) Cell 132, 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tolstorukov M., Kharchenko P., Goldman J., Kingston R., Park P. (2009) Genome Res. 19, 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John S., Sabo P. J., Johnson T. A., Sung M. H., Biddie S. C., Lightman S. L., Voss T. C., Davis S. R., Meltzer P. S., Stamatoyannopoulos J. A., Hager G. L. (2008) Mol. Cell 29, 611–624 [DOI] [PubMed] [Google Scholar]
- 8.Gévry N., Chan H. M., Laflamme L., Livingston D. M., Gaudreau L. (2007) Genes Dev. 21, 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gévry N., Hardy S., Jacques P. E., Laflamme L., Svotelis A., Robert F., Gaudreau L. (2009) Genes Dev. 23, 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narlikar G. J., Fan H. Y., Kingston R. E. (2002) Cell 108, 475–487 [DOI] [PubMed] [Google Scholar]
- 11.Altaf M., Auger A., Covic M., Côté J. (2009) Biochem. Cell Biol. 87, 35–50 [DOI] [PubMed] [Google Scholar]
- 12.Santisteban M. S., Kalashnikova T., Smith M. M. (2000) Cell 103, 411–422 [DOI] [PubMed] [Google Scholar]
- 13.Lindstrom K. C., Vary J. C., Jr., Parthun M. R., Delrow J., Tsukiyama T. (2006) Mol. Cell. Biol. 26, 6117–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanai K., Furuhashi H., Yamamoto T., Akasaka K., Hirose S., Akhtar A. (2008) PLoS Genet. 4, e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gévry N., Hardy S., Jacques P. E., Laflamme L., Svotelis A., Robert F., Gaudreau L. (2009) Genes Dev. 23, 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) Nat. Cell Biol. 8, 458–469 [DOI] [PubMed] [Google Scholar]
- 17.Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Mol. Biol. 119, 1–16 [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F. M. (1987) Current Protocols in Molecular Biology, Greene Publishing Associates, Brooklyn, NY [Google Scholar]
- 19.Sif S., Stukenberg P. T., Kirschner M. W., Kingston R. E. (1998) Genes Dev. 12, 2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logie C., Peterson C. L. (1997) EMBO J. 16, 6772–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang E. Y., Ferreira H., Somers J., Nusinow D. A., Owen-Hughes T., Narlikar G. J. (2008) Biochemistry 47, 13726–13732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narlikar G. J., Phelan M. L., Kingston R. E. (2001) Mol. Cell 8, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 23.Aalfs J. D., Narlikar G. J., Kingston R. E. (2001) J. Biol. Chem. 276, 34270–34278 [DOI] [PubMed] [Google Scholar]
- 24.Fan J. Y., Gordon F., Luger K., Hansen J. C., Tremethick D. J. (2002) Nat. Struct. Biol. 9, 172–176 [DOI] [PubMed] [Google Scholar]
- 25.He X., Fan H. Y., Narlikar G. J., Kingston R. E. (2006) J. Biol. Chem. 281, 28636–28647 [DOI] [PubMed] [Google Scholar]
- 26.Yang J. G., Madrid T. S., Sevastopoulos E., Narlikar G. J. (2006) Nat. Struct. Mol. Biol. 13, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 27.Clarkson M. J., Wells J. R., Gibson F., Saint R., Tremethick D. J. (1999) Nature 399, 694–697 [DOI] [PubMed] [Google Scholar]
- 28.Ridgway P., Brown K. D., Rangasamy D., Svensson U., Tremethick D. J. (2004) J. Biol. Chem. 279, 43815–43820 [DOI] [PubMed] [Google Scholar]
- 29.Tsukiyama T., Daniel C., Tamkun J., Wu C. (1995) Cell 83, 1021–1026 [DOI] [PubMed] [Google Scholar]
- 30.Boyer L. A., Logie C., Bonte E., Becker P. B., Wade P. A., Wolffe A. P., Wu C., Imbalzano A. N., Peterson C. L. (2000) J. Biol. Chem. 275, 18864–18870 [DOI] [PubMed] [Google Scholar]
- 31.Kagalwala M. N., Glaus B. J., Dang W., Zofall M., Bartholomew B. (2004) EMBO J. 23, 2092–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorigo B., Schalch T., Bystricky K., Richmond T. J. (2003) J. Mol. Biol. 327, 85–96 [DOI] [PubMed] [Google Scholar]
- 33.Clapier C. R., Längst G., Corona D. F., Becker P. B., Nightingale K. P. (2001) Mol. Cell. Biol. 21, 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. (2004) Mol. Cell 16, 655–661 [DOI] [PubMed] [Google Scholar]
- 35.Whitehouse I., Rando O. J., Delrow J., Tsukiyama T. (2007) Nature 450, 1031–1035 [DOI] [PubMed] [Google Scholar]
- 36.Guillemette B., Bataille A. R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L. (2005) Plos. Biol. 3, e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B., Pattenden S. G., Lee D., Gutiérrez J., Chen J., Seidel C., Gerton J., Workman J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raisner R. M., Hartley P. D., Meneghini M. D., Bao M. Z., Liu C. L., Schreiber S. L., Rando O. J., Madhani H. D. (2005) Cell 123, 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H., Roberts D. N., Cairns B. R. (2005) Cell 123, 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. (2009) Nat. Genet. 41, 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.