Abstract
In this study we have profiled the free sterol content of cerebrospinal fluid by a combination of charge tagging and liquid chromatography-tandem mass spectrometry. Surprisingly, the most abundant cholesterol metabolites were found to be C27 and C24 intermediates of the bile acid biosynthetic pathways with structures corresponding to 7α-hydroxy-3-oxocholest-4-en-26-oic acid (7.170 ± 2.826 ng/ml, mean ± S.D., six subjects), 3β-hydroxycholest-5-en-26-oic acid (0.416 ± 0.193 ng/ml), 7α,x-dihydroxy-3-oxocholest-4-en-26-oic acid (1.330 ± 0.543 ng/ml), and 7α-hydroxy-3-oxochol-4-en-24-oic acid (0.172 ± 0.085 ng/ml), and the C26 sterol 7α-hydroxy-26-norcholest-4-ene-3,x-dione (0.204 ± 0.083 ng/ml), where x is an oxygen atom either on the CD rings or more likely on the C-17 side chain. The ability of intermediates of the bile acid biosynthetic pathways to activate the liver X receptors (LXRs) and the farnesoid X receptor was also evaluated. The acidic cholesterol metabolites 3β-hydroxycholest-5-en-26-oic acid and 3β,7α-dihydroxycholest-5-en-26-oic acid were found to activate LXR in a luciferase assay, but the major metabolite identified in this study, i.e. 7α-hydroxy-3-oxocholest-4-en-26-oic acid, was not an LXR ligand. 7α-Hydroxy-3-oxocholest-4-en-26-oic acid is formed from 3β,7α-dihydroxycholest-5-en-26-oic acid in a reaction catalyzed by 3β-hydroxy-Δ5-C27-steroid dehydrogenase (HSD3B7), which may thus represent a deactivation pathway of LXR ligands in brain. Significantly, LXR activation has been found to reduce the symptoms of Alzheimer disease (Fan, J., Donkin, J., and Wellington C. (2009) Biofactors 35, 239–248); thus, cholesterol metabolites may play an important role in the etiology of Alzheimer disease.
Keywords: Bile Acid, Brain, Lipid, Mass Spectrometry (MS), Steroid, LC-MS/MS, Cerebrospinal Fluid, Liver X Receptor, Nuclear Receptor, Oxysterol
Introduction
The steroid profile of the central nervous system is of considerable interest with respect to neurodegenerative disease (1–3). This is partly because of the high levels of cholesterol (cholest-5-en-3β-ol) in the central nervous system (4, 5), the potential neuroprotective role of neurosteroids (6), and the ability of some cholesterol metabolites to act as ligands to nuclear receptors, which are themselves implicated in neurodegenerative disease, e.g. the liver X receptors (LXRs)6 in Alzheimer disease (7). Furthermore, brain-derived cholesterol metabolites represent biomarkers for cerebral cholesterol homeostasis, which is deranged in certain neurodegenerative disease, and thus their measurement in body fluids offers a marker for the progression of such disorders (8). In this regard, urine and blood represent the most accessible body fluids (9, 10), but their composition is highly dependent on the activity of other organs. Alternatively, cerebrospinal fluid (CSF), although being less readily accessible, bathes the central nervous system, and its content is more likely to reflect cholesterol metabolism in brain itself. With this in mind, we have set to profile the sterol content of human CSF.
The levels of sterols in CSF are comparatively low; even cholesterol is present at a level of only 4–5 μg/ml (cf. 2 mg/ml in plasma) (11, 12), whereas the brain-derived oxysterol 24(S)-hydroxycholesterol (cholest-5-ene-3β,24(S)-diol, C5-3β,24(S)-diol) is present at a level of only about 1.5 ng/ml (cf. 40–60 ng/ml in plasma) (supplemental Table S1) (11–14). These values were determined by gas chromatography-mass spectrometry following hydrolysis of sterol fatty acid esters and derivatization, and thus represent the free plus fatty acid ester sterols as opposed to the levels of free unconjugated sterols, which are likely to be about an order of magnitude lower and are the likely bioactive forms. It should be noted that sulfated and glucuronidase oxysterols have also been found in plasma (15, 16) and that there is the possibility that sulfated sterols also have biological activity (17). 24(S)-Hydroxycholesterol is a net export product from the central nervous system (18) and is ultimately transported to the liver where it is metabolized into bile acids (19, 20). On the other hand, 27-hydroxycholesterol (cholest-5-ene-3β,26-diol, C5-3β,26-diol) is a net import product to the central nervous system (21), and recent data suggest that it is metabolized in brain to 7α-hydroxy-3-oxocholest-4-en-26-oic acid (CA4-7α-ol-3-one) (22). This acid is an intermediate in the acidic bile acid biosynthetic pathway (19) and can be formed extrahepatically (23), and its formation in brain arouses interest in other bile acid precursors and bile acids formed in brain. In 1997, Zhang et al. (24) showed that rat brain cells could metabolize 27-hydroxycholesterol to 7α-hydroxy-3-oxocholest-4-en-26-oic acid via 3β-hydroxycholest-5-en-26-oic acid (CA5-3β-ol) and 3β,7α-dihydroxycholest-5-en-26-oic acid (CA5-3β,7α-diol), and Mano et al. (25) demonstrated the conversion of 3β-hydroxychol-5-en-24-oic acid (BA5-3β-ol) to chenodeoxycholic acid (3α,7α-dihydroxy-5β-cholan-24-oic acid, 5β-BA-3α,7α-diol) via 3β,7α-dihydroxychol-5-en-24- oic acid (BA5-3β,7α-diol) and 7α-hydroxy-3-oxochol-4-en-24-oic acid (BA4-7α-ol-3-one) in rat brain tissue. Mano et al. (26) also demonstrated the presence of chenodeoxycholic acid, deoxycholic acid (3α,12α-dihydroxy-5β-cholan-24-oic acid, 5β-BA-3α,12α-diol), and cholic acid (3α,7α,12α-trihydroxy-5β-cholan-24-oic acid, 5β-BA-3α,7α,12α-triol) in rat brain, the chenodeoxycholic acid level being about 30 times greater than in serum. Furthermore, both C24 and C27 bile acids have been identified in human brain (27). Despite their presence in brain and blood (28), there are few reports of the presence of bile acids and their precursors in CSF of healthy individuals, although 7α-hydroxy-3-oxocholest-4-en-26-oic acid has been found in the CSF of individuals who underwent surgery for aneurysmal subarachnoid hemorrhage (29). The same group also identified high concentrations of this acid in chronic subdural hematoma (30).
Sterols and bile acids have traditionally been analyzed by gas chromatography-mass spectrometry; however, liquid chromatography (LC)-MS and LC-tandem mass spectrometry (tandem mass spectrometry or MSn) offers an attractive alternative (31). In this study, we have chosen to focus our attention on cholesterol metabolites, which are intermediates in the bile acid biosynthetic pathways, and to pay particular interest to those that possess either a 3β-hydroxy-5-ene or 3-oxo-4-ene structure in the AB rings (the ultimate primary bile acids have a 3α,7α-dihydroxy-5β(H) structure). To improve the response of such metabolites in LC-MS analysis when utilizing electrospray ionization, we have utilized a charge-tagging approach where analyte molecules are specifically tagged with a charged group to enhance their mass spectrometric detection (10, 32–38). We have selected the Girard P derivative that effectively tags a positively charged quaternary nitrogen group to the steroid skeleton.
The finding of bile acids and their precursors in human brain raises the question as to their biological function. They could simply be transport forms of cholesterol, or alternatively, they may be biologically active molecules. For instance, 3-oxocholest-4-en-26-oic acid (rechristened Δ4-dafachronic acid) is a ligand for an orphan nuclear receptor (DAF12) in the nematode Caenorhabditis elegans (39), and 3β-hydroxycholest-5-en-26-oic acid has been shown to activate LXRα in human embryonic kidney 293 cells (40). To clarify the question of biological function, we have tested the biological activity of a number of bile acids identified here in CSF as ligands for the LXRs α and β, both of which are expressed in brain (41, 42), Nur-related factor 1 (NURR1), an orphan nuclear receptor expressed in brain (43, 44), and the farnesoid X receptor (FXR), a nuclear receptor known to be activated by bile acids, e.g. chenodeoxycholic acid and deoxycholic acid (45). Each of these nuclear receptors form obligate heterodimers with the retinoid X receptor (RXR) and regulate gene expression through binding to response elements in the promoter regions of target genes.
EXPERIMENTAL PROCEDURES
Materials
HPLC water and HPLC grade solvents were from Fisher or Sigma. Authentic sterols, bile acids, and their precursors were from Avanti Polar Lipids (Alabaster, AL), Steraloids Inc. (London, UK), Sigma, or from previous studies in our laboratories (10, 32, 34). GP reagent (1-(carboxymethyl)pyridinium chloride hydrazide) was from TCI Europe (Oxford, UK), and cholesterol oxidase from Streptomyces sp. was from Sigma. Sep-Pak tC18 200-mg cartridges were from Waters. Luer-lock syringes were from BD Biosciences. CSF samples from nine subjects were part of a GlaxoSmithKline study and were provided with institutional review board and ethical approval.
Methods
Extraction of CSF for Analysis of Sterols
CSF (0.5 ml) was added dropwise to 2.1 ml of 99.9% ethanol, containing 10 μl of 24(RS)-[26,26,26,27,27,27-2H6]hydroxycholesterol (Avanti Polar Lipids) in propan-2-ol (4 ng/μl), in an ultrasonic bath. This solution was diluted to 70% ethanol by the addition of 0.4 ml of water, ultrasonicated for 2 min, and centrifuged at 14,000 × g at 4 °C for 30 min.
A 200-mg Sep-Pak tC18 cartridge (SPE1) was rinsed with 4 ml of 99.9% ethanol followed by 6 ml of 70% ethanol. CSF in 70% ethanol (3 ml) was applied to the cartridge and allowed to flow at a rate of ∼0.25 ml/min, and flow was aided by application of a slight pressure from a Luer-lock syringe. The flow-through and a column wash of 4 ml of 70% ethanol were collected (SPE1-Fr-1, Scheme 1). By testing the method with a solution of cholesterol and 25-hydroxycholesterol (cholest-5-ene-3β,25-diol, C5-3β,25-diol) in 70% ethanol, cholesterol was found to be retained on the column even after a 4-ml column wash, whereas 25-hydroxycholesterol elutes in the flow-through and column wash. Following a further wash with 4 ml of 70% ethanol (SPE1-Fr-2), cholesterol was eluted from the Sep-Pak column with 2 ml of 99.9% ethanol (SPE1-Fr-3). The column can be further stripped with an additional 2-ml aliquot of 99.9% ethanol to elute more hydrophobic sterols (SPE1-Fr-4). Each fraction was dried under reduced pressure using a vacuum concentrator (ScanLaf, Denmark).
SCHEME 1.
Sample preparation for analysis of sterols and bile acids by LC-MSn. By analyzing samples in parallel by routes A (without cholesterol oxidase) and B (with cholesterol oxidase), analytes possessing a 3-oxo group are differentiated from those oxidized to contain one (i.e. 3β-hydroxy-5-ene or 3β-hydroxy-5α-hydrogen sterols/bile acids).
Charge Tagging of Sterols
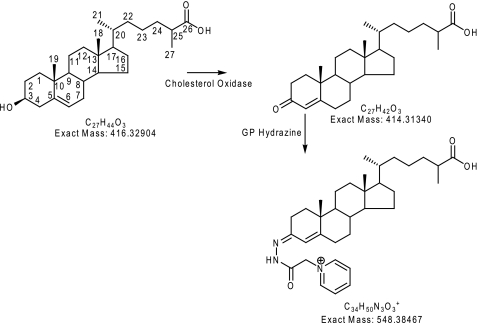
The sterol fractions from above were reconstituted in 100 μl of propan-2-ol, and a solution of 1 ml of 50 mm phosphate buffer (KH2PO4, pH 7) containing 3.0 μl of cholesterol oxidase (2 mg/ml in H2O, 44 units/mg of protein) was added to each. The mixture was incubated at 37 °C for 1 h and then quenched with 2 ml of methanol (Scheme 1, route B, and Scheme 2).
SCHEME 2.
Charge tagging of sterols and bile acids as exemplified by 3β-hydroxycholest-5-en-26-oic acid.
Glacial acetic acid (150 μl) was added to the reaction mixture above (now in ∼70% methanol), followed by 150 mg of GP reagent. The mixture was thoroughly vortexed and incubated at room temperature overnight in the dark.
SPE Extraction of Charge-tagged Sterols
Even when derivatized with GP reagent, sterols may be difficult to solubilize (or retain in solution) when using a highly aqueous mixture of methanol and water. This can make their extraction using reversed phase-solid phase extraction (SPE) challenging. To circumvent this problem a recycling procedure is used (10, 32).
A 200-mg Sep-Pak tC18 cartridge (SPE2) was washed with 6 ml of 100% methanol, 6 ml of 10% methanol and conditioned with 4 ml of 70% methanol. The derivatization mixture from above (∼3 ml of 70% methanol, 5% acetic acid, 3% propanol-2-ol, containing 150 mg of GP reagent and 6 μg of cholesterol oxidase) was applied to the column followed by 1 ml of 70% methanol and 1 ml of 35% methanol. The combined effluent (5 ml) was diluted with water (4 ml) to give 9 ml of ∼35% methanol. The resulting solution was again applied to the column followed by a wash of 1 ml of 17% methanol. To the combined effluent, 9 ml of water was added to give 19 ml of ∼17.5% methanol. This solution was again applied to the column followed by a wash with 6 ml of 10% methanol. At this point, all the derivatized sterols were extracted by the column, and excess derivatization reagent was in the flow-through and wash. Derivatized sterols were then eluted in three 1-ml portions of 100% methanol (SPE2-Fr-1, Fr-2, and Fr-3) followed by 1 ml of 99.9% ethanol (SPE2-Fr-4). LC-MSn analysis revealed that the derivatized sterols were present almost exclusively in the first 2 ml of methanol eluent (SPE2-Fr-1 and SPE2-Fr-2). The recovery of 24(RS)-[26,26,26,27,27,27-2H6]hydroxycholesterol was estimated to be in excess of 85%.
Cholesterol oxidase converts sterols with a 3β-hydroxy-5-ene function to 3-oxo-4-ene analogues (Scheme 2) and a 3β-hydroxy-5α-hydrogen function to a 3-oxo function. To identify sterols that naturally possess a 3-oxo function from those oxidized to contain one, CSF samples were analyzed in parallel in the presence (Scheme 1, route B) and absence (Scheme 1, route A) of cholesterol oxidase.
LC-MSn on the LTQ-Orbitrap XL
Chromatographic separation of GP-tagged sterols was performed on an Ultimate 3000 HPLC system (Dionex, Surrey, UK) utilizing a Hypersil GOLD reversed phase column (1.9 μm particles, 50 × 2.1 mm, Thermo Fisher, San Jose, CA). Mobile phase A consisted of 33.3% methanol, 16.7% acetonitrile containing 0.1% formic acid, and mobile phase B consisted of 63.3% methanol 31.7% acetonitrile containing 0.1% formic acid. After 1 min at 20% B, the proportion of B was raised to 80% B over the next 7 min and maintained at 80% B for a further 5 min, before returning to 20% B in 6 s and re-equilibration for a further 3 min, 54 s, giving a total run time of 17 min. The flow rate was maintained at 200 μl/min and eluent directed to the atmospheric pressure ionization source of an LTQ-Orbitrap XL (Thermo Fisher, San Jose, CA) mass spectrometer. This instrument is a hybrid linear ion-trap (LIT)-Orbitrap analyzer. The Orbitrap is a Fourier transform mass analyzer capable of high resolution (up to 100,000 full width at half-maximum height) and exact mass measurement.
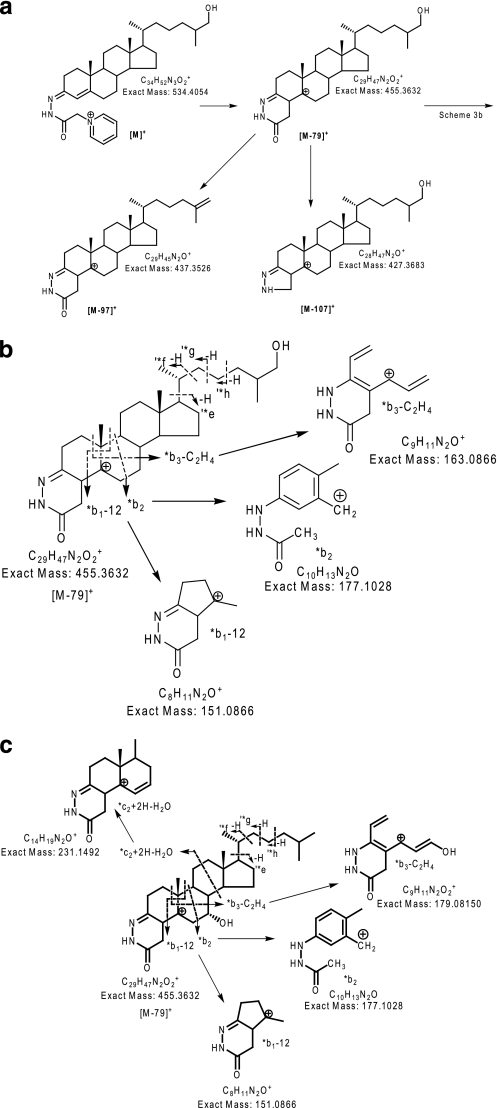
The Orbitrap was calibrated externally prior to each analytical session. Mass accuracy was better than 5 ppm. In any given chromatographic run, and in the mass range of GP-tagged sterols, measured mass values were found to be offset from the theoretical mass by a constant value ranging from +1 to +2 millimass units, for example. For LC-MS and LC-MSn analysis of reference compounds, the sample (1 pg/μl in 60% methanol, 0.1% formic acid) was injected (20 μl) onto the reversed phase column and eluted into the LTQ-Orbitrap at a flow rate of 200 μl/min. Two experimental methods were utilized. In the first experimental method, three scan events were performed as follows: a Fourier transform-MS scan in the Orbitrap analyzer over the m/z range 400–650 (or 300–800) at 30,000 resolution (full width at half-maximum height) with a maximum ion fill time of 500 ms, followed by data-dependent MS2 and MS3 events performed in the LIT with maximum ion fill times of 200 ms. For the MS2 and MS3 scans, three microscans were performed, the precursor ion isolation width was set at 2 (to select the monoisotopic ion) and the normalized collision energy at 30 and 35 (instrument settings), respectively. A precursor ion inclusion list was defined according to the m/z of the [M]+ ions of expected sterols (see supplemental Table S2) so that MS2 was preferentially performed on these ions in the LIT if their intensity exceeded a pre-set minimum (500 counts). If a fragment ion corresponding to a neutral loss of 79 Da from the precursor ion was observed in the MS2 event and was above a minimal signal setting (200 counts), MS3 was performed on this fragment. To maximize efficiency, the MS2 and MS3 event was performed at the same time as the high resolution mass spectrum was being recorded in the Orbitrap. The second experimental method involved a targeted multiple reaction monitoring approach (MRM). In event 1, the Orbitrap analyzer was scanned as above, and in event 2, the transition 534.4→455.4→ was monitored using collision energies of 30 and 35 for the MS2 and MS3 events, respectively (Scheme 3a). In event 3, the transition 540.4→461.4→ was monitored in a similar manner (to accommodate the 24(RS)-[2H6]hydroxycholesterol internal standard).
SCHEME 3.
Fragmentation of GP-tagged sterols and bile acids. a, major MS2 fragmentation routes for GP-tagged sterols and bile acids exemplified by 27-hydroxycholesterol. A 3-oxo-4-ene functionality was generated by oxidation of the native 3β-hydroxy-5-ene function by cholesterol oxidase prior to treatment with GP reagent. b and c, structurally informative fragment ions observed in MS3 ([M]+→[M − 79]+→) spectra of GP-tagged sterols exemplified by 27-hydroxycholesterol following cholesterol oxidase treatment (b) and 7α-hydroxycholesterol after similar treatment (c).
For the analysis of GP-tagged sterols from CSF, 12 μl of the first methanol fraction (1 ml) from the second SepPak C18 cartridge (SPE2-Fr-1) (equivalent to 6 μl of CSF assuming all the sterols elute in this methanol fraction) was diluted with 8 μl of 0.1% formic acid and 20 μl injected onto the LC column. MS, MS2, and MS3 spectra were recorded as described above. Other fractions from the SPE columns were analyzed in an identical fashion.
Quantification and Isotope Dilution Mass Spectrometry
The quantities of identified sterols and bile acids in CSF were determined by isotope dilution mass spectrometry against a known amount of added 24(SR)-[2H6]hydroxycholesterol (100% [2H6]) internal standard (IS). For monohydroxycholesterols (C5-3β,x-diol), which are present in CSF in their free form in low amounts (<1 ng/ml), quantification was performed using the MRM transitions 534.4→455.4→ for the GP-tagged sterols and 540.4→461.4→ for GP-tagged 24(SR)-[2H6]hydroxycholesterol IS. Peak areas were used for calculation of concentration (see Equation 1). Bile acids were present in greater abundance than monohydroxycholesterols, and this allowed their quantification from reconstructed ion chromatograms (RICs) recorded on the Orbitrap. RICs were generated from spectra recorded at 30,000 resolution, with an m/z tolerance of 10 ppm. Again peak areas were determined, and analyte levels were calculated by applying Equation 1.
 |
Equation 1 assumes that all analytes have an identical response factor to the internal standard, which is true for 3β-hydroxy-5-ene and 3-oxo-4-ene sterols without additional substituents in the A-ring (33). In some experiments, the IS was not included, in which case relative abundances (%RA) were determined against 7α-hydroxy-3-oxocholest-4-en-26-oic acid, the most abundant sterol/bile acid component found in CSF, by applying Equation 2.
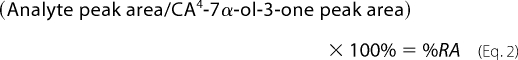 |
Luciferase Reporter Assay
The ability of the acidic cholesterol metabolites 3β-hydroxycholest-5-en-26-oic, 3β,7α-dihydroxycholest-5-en-26-oic, 7α-hydroxy-3-oxocholest-4-en-26-oic, 7α,12α-dihydroxy-3-oxo-5β-cholan-24-oic (5β-BA-7α,12α- diol-3-one), and 3-oxo-5β-cholan-24-oic (5β-BA-3-one) acids to activate several nuclear receptors was tested in luciferase assays. Transient transfection studies were performed in the mouse substantia nigra-like cell line SN4741. As the object of our study was CSF, we decided to use the SN4741 cell line, which is of neural origin. Cells were plated in 24-well plates (5 × 105 cells/well) 24 h before transfection and transfected with 1 μg of plasmid DNA/well complexed with 2 μl of Lipofectamine 2000 (Invitrogen). Cells were transfected with 400 ng of an LXR- or FXR- or NURR1-responsive luciferase reporter construct and 200 ng of LXRα, FXR, or NURR1. A reporter gene expressing the Renilla luciferase (pRL-TK, Promega) was co-transfected in all experiments as an internal control for normalization of transfection efficiency. After a 12-h incubation, the lipid/DNA mixture was replaced with fresh 2.5% serum medium containing vehicle or appropriate ligand (10 μm), as specified in each experiment. Luciferase activities were assayed 24 h later using Dual-Luciferase reporter assay system (Promega), following the manufacturer's protocol.
RESULTS
Extraction of Sterols from CSF
The extraction procedure was modified from our previous protocol for oxysterol analysis of plasma (10). However, the lesser availability of CSF necessitated the method to be scaled down to accommodate volumes of less than 1 ml. In the extraction procedure, it was important to separate cholesterol from its oxidized metabolites, i.e. oxysterols and bile acids, at an initial stage, as cholesterol can be oxidized in air to give autoxidation products that can be confused with those of biological origin (46, 47). The scaled down method was optimized using 25-hydroxycholesterol and cholesterol, and 25-hydroxycholesterol was found to elute in fraction 1 from the first SPE column (SPE1-Fr-1) and, in its GP-tagged form, in fractions 1 and 2 from the second SPE column (SPE2-Fr-1 and SPE2-Fr-2). Cholesterol eluted in fraction 3 from the first SPE column (SPE1-Fr-3) and, in its GP-tagged form, in fractions 1 and 2 from the second column (SPE2-Fr-1 and SPE2-Fr-2). These results were confirmed in tests in which added 24(SR)[2H6]hydroxycholesterol was extracted from CSF. 24(SR)-[2H6]Hydroxycholesterol was found to elute in fraction SPE1-Fr-1 and in its GP-tagged form in SPE2-Fr-1 and SPE2-Fr-2. Recoveries were estimated to be in excess of 85%. Only a very minor amount of cholesterol (<1 ng/ml) was found to leak into fraction SPE1-Fr-1.
Quantification and Isotope Dilution Mass Spectrometry
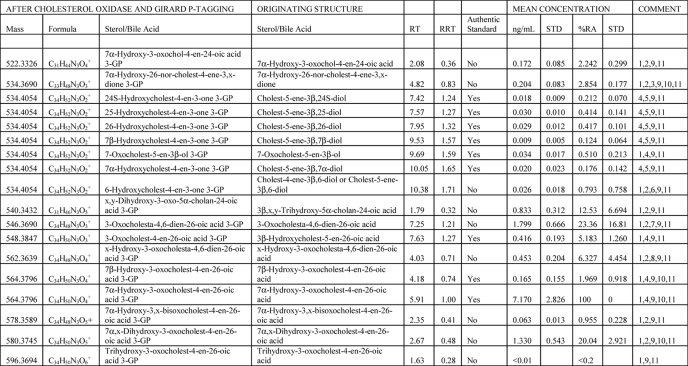
The “gold standard” method for analyte quantification by mass spectrometry is isotope dilution mass spectrometry. Here, an isotope-labeled version of the target analyte, added during the sample extraction step, is used as an internal standard. The isotope-labeled internal standard will behave in an (almost) identical fashion to the natural analyte during extraction, derivatization, and chromatography steps and give a similar response during mass spectrometry analysis. Thus, from the known amount of internal standard added and the ratio of peak areas measured for the analyte and internal standard during mass spectrometry analysis, the absolute amount of target analyte is calculated (Equation 1). In this study, 24(SR)-[2H6]hydroxycholesterol was used as the internal standard. Once GP-tagged, sterols and bile acids (with a 3-oxo-4-ene or 3β-hydroxy-5-ene structure) give a similar mass spectrometric response (33), and thus 24(SR)-[2H6]hydroxycholesterol was used as the internal standard for all detected analytes. The levels of sterols and bile acids identified in CSF are listed in Table 1. In some experiments, the IS was not included, in which case relative abundance (%RA, means ± S.D., nine subjects) was determined against 7α-hydroxy-3-oxocholest-4-en-26-oic acid (see Equation 2), the most abundant sterol/bile acid component found in CSF. %RA values are also given in Table 1. It should be noted that the values determined for bile acids are only quantitative estimates (i.e. approximations); this is a consequence of the possibility of differing extraction and purification efficiencies from the internal standard. Ideally, isotope-labeled internal standards should be used for each acid. This is, however, impractical in a discovery mode study, but it could be incorporated in a more targeted study.
TABLE 1.
Oxysterols and bile acids in CSF
The following abbreviations were used: RT, retention time/min; RRT, retention time relative to 7α-hydroxy-3-oxocholest-4-en-26-oic acid; STD, S.D.; %RA, % relative abundance (7α-hydroxy-3-oxocholest-4-en-26-oic acid = 100%, n = 9).
1 Quantitative estimate was based on 24(RS)-[26,26,26,27,27,27-2H6]hydroxycholesterol internal standard. Mean concentration was ± S.D. (n = 6).
2 Identification was based on exact mass and MSn spectra.
3 26-Norsterol is a likely decomposition product of a 24-oxo-26-acid. Alternatives to the oxo group x are an enol or epoxy group; all add 14 Da to the sterol structure.
4 Identification was based on comparison with authentic standards.
5 Quantification was based on 24(RS)-[26,26,26,27,27,27-2H6]hydroxycholesterol internal standard. Mean concentration was ± S.D. (n = 6).
6 Cholest-4-ene-3β,6-diol and/or cholest-5-ene-3β,6-diol are decomposition products of 5,6-epoxycholestan-3β-ol and cholestane-3β,5α,6β-triol.
7 3-Oxocholesta-4,6-dien-26-oic acid is a dehydration product of 7α-hydroxy-3-oxocholest-4-en-26-oic acid.
8 x-Hydroxy-3-oxocholesta-4,6-dien-26-oic acid is a likely dehydration product of 7α-x-dihydroxy-3-oxocholest-4-en-26-oic acid.
9 Retention time/min (RT) for compounds eluting in chromatograms shown in Figs. 1–4. Relative retention time (RRT, mean) was to 7α-hydroxy-3-oxocholest-4-en-26-oic acid.
10 Resolved syn and anti conformers.
11 %RA against 7α-hydroxy-3-oxocholest-4-en-26-oic acid (n = 9).
Identification of Sterols and Bile Acids in CSF
Although it is not uncommon to base the identification of a metabolite solely on the measurement of molecular weight, particularly in the era of metabolomics and systems biology, our identification strategy was based on a combination of functional group-specific derivatization (charge tagging), exact mass measurement (<5 ppm), MS2 and MS3 analysis, and chromatographic retention time. Where possible, experimental data for presumptively identified metabolites are compared with that obtained for authentic standards.
Oxysterols
Oxysterols can be defined as oxygenated forms of cholesterol, and this definition includes hydroxycholesterols, e.g. 24(S)-hydroxycholesterol, and oxocholesterols, e.g. 7- oxocholesterol (7-oxocholest-5-en-3β-ol, C5-3β-ol-7-one). Hydroxycholesterols may also be called bile alcohols, as many are intermediates in the biosynthesis of bile acids from cholesterol (31). Table 1 lists the oxysterols identified in this study.
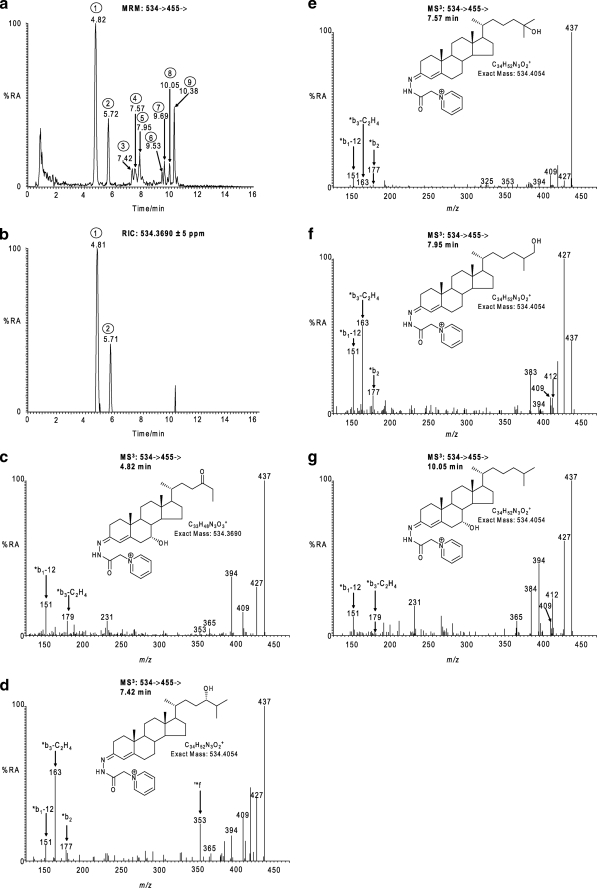
The initial focus of this work was on the identification and relative quantification of oxysterols in CSF. Once charge-tagged by the GP reagent, oxysterols can be analyzed by LC-electrospray ionization-MSn with high sensitivity (< pg injected), and the oxysterol content of CSF was initially probed by generation of exact mass (5 ppm) RICs for all potential oxysterols with one to five additional oxygen functions on the cholesterol skeleton (see supplemental Table S2). Oxysterols are present in their unconjugated form in CSF at low levels (<1 ng/ml); therefore, to further maximize the sensitivity of analysis, an MRM-like experiment was performed in the LIT in parallel to the acquisition of high resolution mass scans in the Orbitrap (see the second experimental method under “Experimental Procedures”). GP-tagged oxysterols offer three advantages for this type of analysis as follows: (i) charge tagging improves parent molecule ionization by 2–3 orders of magnitude (33); (ii) GP-tagged oxysterols give an abundant [M − 79]+ ion upon MS2 ([M]+→); and (iii) MS3 spectra generated by the [M]+→[M − 79]+→ transition are structurally informative (Scheme 3, b and c). Thus, by using this protocol, it was possible to identify and quantify 24(S)-hydroxycholesterol (peak 3 in Fig. 1a), 25-hydroxycholesterol (peak 4 in Fig. 1a), 27-hydroxycholesterol (peak 5 in Fig. 1a), 7β-hydroxycholesterol (peak 6 in Fig. 1a), 7α-hydroxycholesterol (peak 8 in Fig. 1a), and 6-hydroxycholesterol (peak 9 in Fig. 1a) and also 7-oxocholesterol (peak 7 in Fig. 1a). Identifications were based on exact mass and comparison of retention time and MS2 and MS3 spectra with those of authentic standards (Fig. 1 and supplemental Fig. S1). The levels of these free oxysterols in CSF were very low ranging from 0.009 ± 0.005 ng/ml for 7β-hydroxycholesterol (cholest-5-ene-3β,7β-diol, C5-3β,7β-diol) to 0.026 ± 0.018 ng/ml for 6-hydroxycholesterol (cholest-4-ene-3β,6-diol, C4-3β,6-diol, or cholest-5-ene-3β,6-diol, C5-3β,6-diol) and 0.034 ± 0.017 ng/ml for 7-oxocholesterol (Table 1). The ratio of 24(S)- to 27- to 7α-hydroxycholesterol (cholest-5-ene-3β,7α-diol, C5-3β,7α-diol) in CSF was found to be 1:1.6:1.1 (six subjects). These are three enzymatically formed oxysterols, 24(S)-hydroxycholesterol being formed from cholesterol in a reaction catalyzed by neuron-specific cytochrome (CYP) P450 46A1 (48), 27-hydroxycholesterol being formed in many tissues in a reaction catalyzed by CYP27A1 (49), and 7α-hydroxycholesterol being formed hepatically in a CYP7A1-catalyzed reaction (50). The ratio of these three oxysterols in CSF thus gives an indication of cholesterol metabolism in brain with respect to oxysterol transport from the periphery into CSF. Of the other oxysterols, 25-hydroxycholesterol can be formed enzymatically by a cholesterol 25-hydroxylase (51) and also by CYP46A1 (48), but it is also an autoxidation product of cholesterol, as are 7β-hydroxycholesterol and 7-oxocholesterol (46), whereas 6-hydroxycholesterol is a decomposition product of the autoxidation products 5,6-epoxycholesterol (5,6-epoxycholestan-3β-ol, C-3β-ol-5,6-epoxide) and cholestane-3β,5α,6β-triol (C-3β,5α,6β-triol) (33).
FIGURE 1.
Identification of monohydroxycholesterols and hydroxynorcholestenedione in CSF. a, MRM chromatogram for the transition 534→455→. b, RIC for the exact m/z 534.3690 ± 5 ppm. c–g, MS3 [534→455→] spectra of the peaks eluting as follows: 4.82 min, peak 1 (c); 7.42 min, peak 3 (d); 7.57 min, peak 4 (e); 7.95 min, peak 5 (f); and 10.05 min, peak 8 (g); in chromatogram a. The MRM chromatogram (a) and MS3 spectra (c–g) were generated in the LIT, the RIC (b) was generated in parallel in the Orbitrap analyzer. The MS3 spectra correspond to GP-tagged 7α-hydroxy-26-norcholest-4-ene-3,x-dione (c); 24(S)-hydroxycholesterol (d); 25-hydroxycholesterol (e); 27-hydroxycholesterol (f); and 7α-hydroxycholesterol (g). Proposed structures of the GP-tagged molecules are shown as insets to the appropriate spectra. GP-tagged 7α-hydroxy-26-norcholest-4-ene-3,x-dione appears in chromatograms (a and b) as syn and anti conformers (peaks 1 and 2). Chromatograms and spectra are from sterols isolated from CSF. Some confusion may arise concerning the nomenclature of 27-hydroxycholesterol and related compounds. According to rules of priority of numbering the correct description of 27-hydroxycholesterol is 25(R),26-hydroxycholesterol. However, the common name is 27-hydroxycholesterol on account of its formation via the mitochondrial CYP27A1-catalyzed hydroxylation of cholesterol, and this will be the name used here, although we will use the abbreviation C5-3β,26-diol in accord with the systematic name cholest-5-ene-3β,26-diol recommended by Fahy et al. (74). In other cases, we adopt the nomenclature recommended by Fahy et al. (74) and the Lipid Maps consortium. %RA, % relative abundance.
C27 Bile Acids
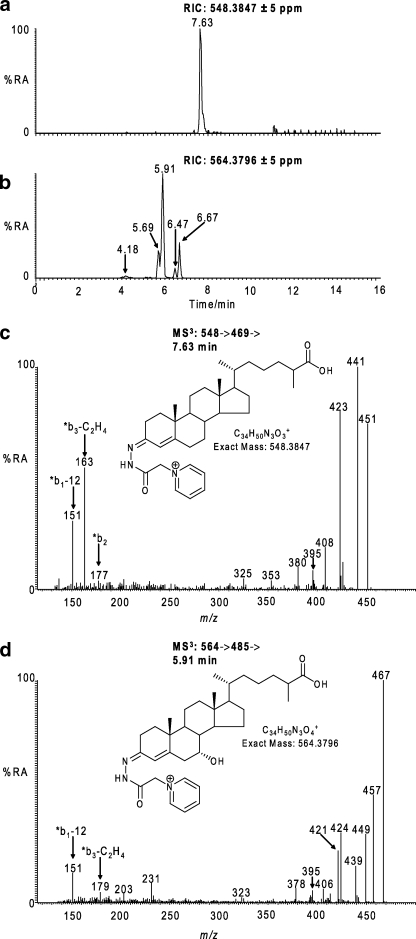
C27 bile acids, including 3β-hydroxycholest-5-en-26-oic acid, 3β,7α-dihydroxycholest-5-en-26-oic, and 7α-hydroxy-3-oxocholest-4-en-26-oic acid, are prevalent in blood (22, 28) (see supplemental Table S1) and have been identified in cells of the rodent central nervous system following incubation with 27-hydroxycholesterol (24), whereas 7α-hydroxy-3-oxocholest-4-en-26-oic acid has been identified as an export product from human brain into plasma (22). We thus probed for the presence of C27 bile acids in human CSF. Again, using a combination of exact mass, retention time, MS2 and MS3 spectra, and comparison with authentic standards, the C27 bile acids 3β-hydroxycholest-5-en-26-oic acid and 7α-hydroxy-3-oxocholest-4-en-26-oic acid were identified (Fig. 2). It was possible to differentiate endogenous 7α-hydroxy-3-oxocholest-4-en-26-oic acid from that generated by treatment with bacterial cholesterol oxidase, as the abundance of the GP-tagged compound did not change when derivatization was performed in the presence or absence of the bacterial cholesterol oxidase. Similarly, GP-tagged 3-oxocholest-4-en-26-oic acid (CA4-3-one) was only identified following treatment with bacterial cholesterol oxidase and thus must originate from endogenous 3β-hydroxycholest-5-en-26-oic acid. It is of interest to note that there is no endogenous 3-oxocholest-4-en-26-oic acid in CSF in light of its ability to activate the DAF-12 orphan nuclear receptor in C. elegans (39). However, HSD3B7, the necessary 3β-hydroxysteroid dehydrogenase, requires the presence of a 7α-hydroxyl group and a side chain longer than that in C21 steroids (52). The levels of 7α-hydroxy-3-oxocholest-4-en-26-oic and 3β-hydroxycholest-5-en-26-oic acid in CSF were found to be 7.170 ± 2.826 and 0.416 ± 0.193 ng/ml, respectively. These C27 bile acids could originate in plasma and be transferred to CSF or alternatively be formed in brain from 27-hydroxycholesterol (22, 24), which is itself suggested to be imported into brain from plasma (21). The possibility also exists that C27 bile acids could be formed in brain from cholesterol itself.
FIGURE 2.
Identification of hydroxycholestenoic and hydroxyoxocholestenoic acids in CSF. a and b, RICs for the exact m/z 548.3847 (a) and 564.3796 (b) ±5 ppm. c, MS3 [548→469→] spectrum of the peak eluting at 7.63 min in RIC (a). d, MS3 [564→485→] spectrum of the peak eluting at 5.91 min in RIC (b). The MS3 spectra correspond to GP-tagged 3β-hydroxycholest-5-en-26-oic (c) and 7α-hydroxy-3-oxocholest-4-en-26-oic acids (d). Structures of the GP-tagged molecules are shown as insets to the appropriate spectra. GP-tagged 7α-hydroxy-3-oxocholest-4-en-26-oic acid appears as syn and anti conformers in RIC (b). The syn and anti confirmers appear to give split peaks in RIC (b) (i.e. 5.69 and 5.91 min and 6.47 and 6.67 min). The MS3 spectra for these peaks are indistinguishable; however, only the latter eluting peaks (5.91 min, 6.67 min) appear in the RIC of the authentic reference compound. The origin of the early eluting peaks is unknown but may be related to the stereochemistry of the C-17 side chain. 7β-Hydroxy-3-oxocholest-4-en-26-oic is the minor component eluting at 4.18 min in RIC (b). Chromatograms and spectra are from sterols isolated from CSF, and recorded as indicated in Fig. 1. %RA, % relative abundance.
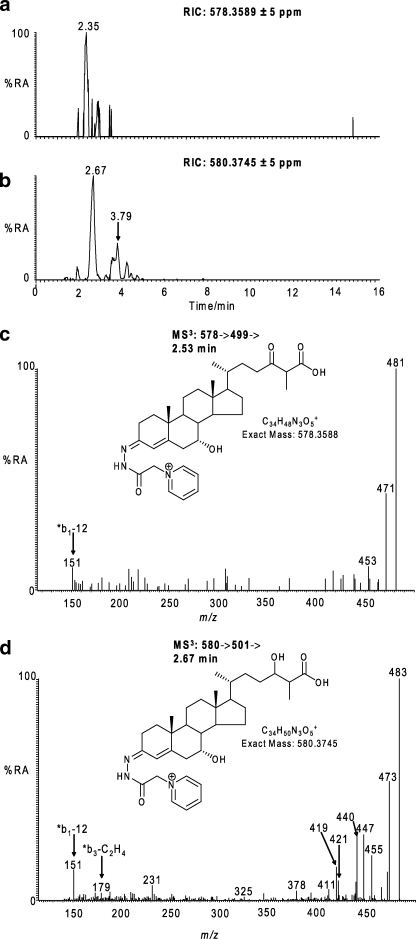
In our search for other C27 bile acids in CSF, a candidate [M]+ ion was observed at m/z 580.3745 with a retention time compatible with a GP-tagged dihydroxy-3-oxocholest-4-en-26-oic acid (Table 1). The [M]+ ion gave MS2 and MS3 spectra consistent with one hydroxyl group at position 7α with the second on the C or D rings or on the side chain (i.e. CA4-7α,x-diol-3-one) (Fig. 3). The abundance of this bile acid was 1.330 ± 0.543 ng/ml. There was also evidence for a trihydroxy-3-oxocholest-4-en-26-oic acid with the appropriate mass and appearing very early in the chromatogram, but its level was too low (<0.01 ng/ml) to allow the recording of a structurally informative MS3 spectrum.
FIGURE 3.
Identification of hydroxybisoxocholestenoic and dihydroxyoxocholestenoic acids. a and b, RICs for the exact m/z 578.3589 (a) and 580.3745 (b) ± 5 ppm. c, MS3 [578→499→] spectrum of the peak eluting at 2.35 min in RIC (a). d, MS3 [580→501→] spectrum of the peak eluting at 2.67 min in RIC (b). The MS3 spectra correspond to GP-tagged 7α-hydroxy-3,x-bisoxocholest-4-en-26-oic acid (c), and 7α,x-dihydroxy-3-oxocholest-4-en-26-oic acid (d). Proposed structures of the GP-tagged molecules are shown as insets to the appropriate spectra. GP-tagged 7α,x-dihydroxy-3-oxocholest-4-en-26-oic acid appears as syn and anti conformers appearing at 2.67 and 3.79 min in RIC (b). Chromatograms and spectra are from sterols isolated from CSF and recorded as indicated in Fig. 1. %RA, % relative abundance.
Bile acids with both a 7-hydroxy-4-ene and 7-hydroxy-5-ene structure are known to be labile and to dehydrate to give conjugated dienes (31). There is in fact evidence for this by the appearance of peaks at m/z 562.3639 (580.3745 − H2O) and 546.3690 (564.3796 − H2O) assigned to hydroxy-3-oxocholesta-4,6-dien-26-oic acid (CA4,6-x-ol-3-one, 0.453 ± 0.204 ng/ml) and 3-oxocholesta-4,6-dien-26-oic acid (CA4,6-3-one, 1.799 ± 0.666 ng/ml), respectively, on the basis of their retention time and MS3 spectra (Table 1 and supplemental Fig. S1). There is also molecular weight evidence for the presence of a dihydroxy-3-oxocholesta-4,6-dien-26-oic acid (CA4,6-x,y-diol-3-one); however, the MS3 spectrum, although weak, favors the structural isomer 7α-hydroxy-3,x-bisoxocholest-4-en-26-oic acid (CA4-7α-ol-3,x-dione, 0.063 ± 0.013 ng/ml) (Fig. 3). It is intriguing to note that 7α-hydroxy-3,24-bisoxocholest-4-en-26-oic acid (CA4-7α-ol-3,24-dione) and 7α,24-dihydroxy-3-oxocholest-4-en-26-oic acid (CA4-7α,24-diol-3-one) are precursors of C24 bile acids in the biosynthetic pathway from 24(S)-hydroxycholesterol (19, 27), and there is evidence for the presence of the necessary enzymes in brain, i.e. CYP46A1 (48), CYP39A1 (53), CYP27A1 (24, 54, 55), HSD3B7 (24), very long chain acyl-CoA synthetase (56), and l-bifunctional protein (57) to account for their formation (Scheme 4).
SCHEME 4.
Suggested pathways for biosynthesis of bile acids found in CSF. The acidic pathway starts with 26-hydroxylation of cholesterol by CYP27A1, and the 24(S)-hydroxycholesterol pathway by 24(S)-hydroxylation of cholesterol by CYP46A1. Compounds drawn in black are identified in CSF, those in blue are presumptively identified, and those in red were not detected or were present in trace quantities only. With the exception of α-methylacyl-CoA racemase (necessary for the acidic pathway), all of the enzymes (or mRNA transcripts) required for the formation of 7α-hydroxy-3-oxochol-4-en-24-oic acid are present in brain. Abbreviations, Swiss-Prot accession number, and reference to the enzyme, where applicable (or mRNA transcript), in brain are as follows: CYP27A1, cytochrome P450 27A1, Q02318 (54); CYP46A1, cytochrome P450 46A1, Q9Y6A2 (48); CYP7B1, cytochrome P450 7B1, O75881 (75); CYP39A1, cytochrome P450 39A1, Q9NYL5 (53); HSD3B7, 3β-hydroxysteroid dehydrogenase type 7, Q9H2F3 (24); VLCS, very long chain acyl-CoA synthetase, O35488 (56); AMACR, α-methylacyl-CoA racemase, Q9UHK6; BCOX, branched chain acyl-CoA oxidase, Q99424 (76); DBP, d-bifunctional protein, P51659 (57); LBP, l-bifunctional protein, Q08426 (57); SCPx, sterol carrier protein x, P22307 (77). Metabolites identified as LXRα ligands in SN4741 cells are indicated by LXR+.
During our profiling studies for oxysterols, we monitored the MRM transition 534→455→ ([M]+→[M − 79]+→) in the LIT in parallel with recording high resolution exact mass spectra in the Orbitrap analyzer (Fig. 1a). A pair of early eluting peaks was observed in the MRM chromatogram (4.82 and 5.72 min in Fig. 1a), but unexpectedly, did not appear in the RIC for m/z 534.4054 corresponding to the m/z of the [M]+ ion of a GP-tagged monohydroxycholesterol. By searching the mass spectra recorded when these early peaks elute for ions of nominal mass 534, the dominant peak was found at m/z 534.3690. This mass corresponds to a [M]+ ion with elemental composition C33H48N3O3+, which prior to GP tagging fits a sterol of elemental composition C26H40O3. The MS2 and MS3 spectra of these [M]+ ions indicate a 7α-hydroxy-26-norcholest-4-en-3-one sterol structure (0.204 ± 0.083 ng/ml) with an additional oxo-, epoxy-, or hydroxyalkene function in the CD-rings or side chain (Fig. 1). It is interesting to note that 24-oxo-26-norcholestanes are known decarboxylation products of 24-oxocholestan-26-oic acids (58, 59), which can themselves be formed from 24(S)-hydroxycholesterol. Also, C26 bile alcohols have been described previously in the glucuronide fraction from human urine (60, 61).
C24 Bile Acids
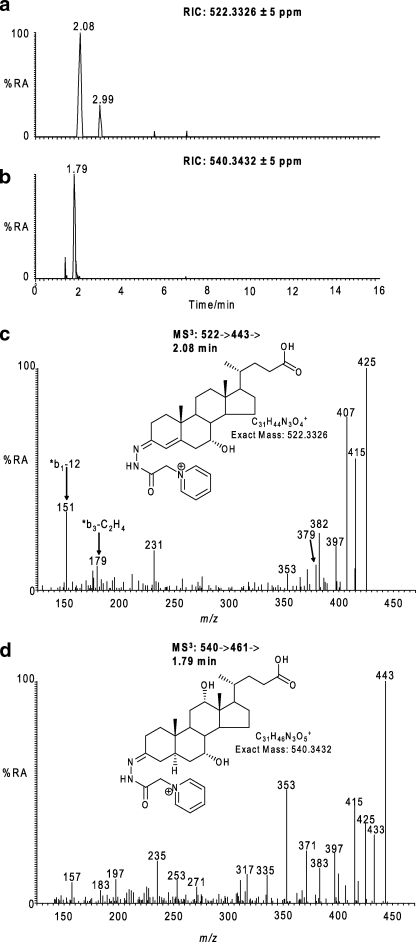
C24 bile acids are formed from C27 bile acids by the process of β-oxidation in the peroxisomes (19). This involves oxidation and thiolation to give a 24-oxo-26-oyl-CoA structure followed by β-oxidation to the C24 bile acid. The thiolation and β-oxidation reactions are carried out by bile acyl-CoA synthetase (liver-specific) or very long chain acyl-CoA synthetase and sterol carrier protein x, respectively, the latter two of which are expressed in rodent brain (Swiss-Prot O35488, P11915).7 We thus searched for the presence of C24 precursors of primary bile acids in CSF. Two potential C24 bile acids were identified, the latter eluting a peak of which has a mass and gave MS2 and MS3 spectra compatible with 7α-hydroxy-3-oxochol-4-en-24-oic acid (Fig. 4), which is a precursor of chenodeoxycholic acid. Its level was estimated at 0.172 ± 0.085 ng/ml. The earlier eluting peak had a mass and gave MS2 and MS3 spectra compatible with a trihydroxycholanoic acid (BA-triol) possibly with a 3β-hydroxy-5α-hydrogen functionality (i.e. 3β,x,y-trihydroxy-5α-cholan-24-oic acid, 5α-BA-3β,x,y-triol). Its level was estimated at 0.833 ± 0.312 ng/ml.
FIGURE 4.
Identification of C24 bile acids in CSF. RICs for the exact m/z 522.3326 (a) and 540.3432 (b) ± 5 ppm are shown. c, MS3 [522→443→] spectrum of the peak eluting at 2.08 min in RIC (a). d, MS3 [540→461→] spectrum of the peak eluting at 1.79 min in RIC (b). The MS3 spectra correspond to GP-tagged 7α-hydroxy-3-oxochol-4-en-24-oic (c) and a trihydroxycholanoic acid, possibly 3β,x,y-trihydroxy-5α-cholan-24-oic acid (d). Proposed structures of the GP-tagged molecules are shown as insets to the appropriate spectra. Chromatograms and spectra are from sterols isolated from CSF and recorded as indicated in Fig. 1. %RA, % relative abundance.
Sterols and Bile Acids as Ligands to LXRs, NURR1, and FXR
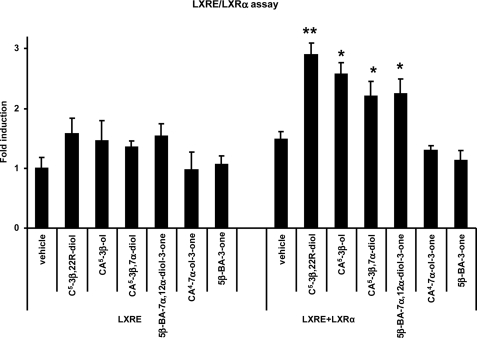
Earlier studies have shown 24(S)-, 25, and 27-hydroxycholesterols to be ligands to the LXRs (62–64). Here, we utilize an LXR-response element/LXRα luciferase assay to examine the activational capacity of intermediates in the acidic pathway of bile acid biosynthesis downstream of 27-hydroxycholesterol. Our data show that 3β-hydroxycholest-5-en-26-oic and 3β,7α-dihydroxycholest-5-en-26-oic acids, but not 7α-hydroxy-3-oxocholest-4-en-26-oic acid, have the ability to activate LXRα and therefore act as LXR ligands. Additional experiments were also performed on other intermediates in bile acid biosynthesis. Although 3-oxo-5β-cholan-24-oic acid could not activate LXRα, 7α,12α-dihydroxy-3-oxo-5β-cholan-24-oic acid was found to be an LXR ligand (Fig. 5).
FIGURE 5.
Analysis of the nuclear receptor activational capacity of acidic intermediates of bile acid biosynthesis. Analysis of luciferase activity in SN4741 cells transfected with an LXR-responsive luciferase reporter construct (LXRE) and LXRα, as indicated, and stimulated for 24 h with 22(R)-hydroxycholesterol (cholest-5-ene-3β,22(R)-diol, C5-3β,22R-diol; 10 μm) a known LXRα ligand (62, 63), or the acidic compounds indicated, i.e. 3β-hydroxycholest-5-en-26-oic acid, CA5-3β-ol; 3β,7α-dihydroxycholest-5-en-26-oic acid, CA5-3β,7α-diol; 7α,12α-dihydroxy-3-oxo-5β-cholan-24-oic acid, 5β-BA-7α,12α-diol-3-one; 7α-hydroxy-3-oxocholest-4-en-26-oic acid, CA4-7α-ol-3-one; and 3-oxo-5β-cholan-24-oic acid, 5β-BA-3-one. The firefly luciferase activity was normalized to Renilla luciferase activity, and the values are expressed as fold activation over the normalized basal LXR response element-luciferase activity set to 1. Data are means ± S.E. (n = 3), *, p < 0.05; **, p < 0.01 compared with vehicle treatment. %RA, % relative abundance.
To test the specificity of the activation observed by the acidic compounds described above, luciferase assays were performed using a luciferase reporter gene under the control of the farnesoid X receptor-response element. In SN4741 cells transfected with FXR, chenodeoxycholic acid activated FXR. On the contrary, none of the C27 or C24 acids tested showed a significant effect on FXR activation (supplemental Fig. S2). Similar experiments were performed using a luciferase reporter gene under the control of a DR5 element. NURR1/RXR heterodimers bind to DR5 elements on the promoter of target genes. In SN4741 cells transfected with NURR1, 9-cis-retinoic acid activated the RXR/NURR1 heterodimer. On the contrary, none of the C27 or C24 acids tested showed a significant effect on RXR/NURR1 activation (supplemental Fig. S2).
Thus our results indicate that 3β- hydroxycholest-5-en-26-oic, 3β,7α- dihydroxycholest-5-en-26-oic, and 7α,12α-dihydroxy-3-oxo-5β-cholan-24-oic acids are specific LXRα ligands.
DISCUSSION
Oxysterols are biologically active molecules. They can suppress the synthesis of cholesterol via interaction with the molecular machinery for transcription of the enzymes of the mevalonate pathway (65) and also encourage cholesterol export by acting as ligands to the LXRs and activating transcription of genes for cholesterol transport (62, 63). Importantly, it is only specific isomers that are biologically active, e.g. 24(S)- and 25-hydroxycholesterol are both suppressors of cholesterol synthesis and enhancers of cholesterol export, whereas 7α-hydroxycholesterol and 7-oxocholesterol only weakly suppress cholesterol synthesis and are not ligands to the LXRs (63, 65, 66). Thus, the exact chemical structure of this class of molecule is critically important for biological activity. Recent evidence suggests that the initial steps of the acidic pathway of bile acid biosynthesis operate in brain (22, 24), with the conversion of 27-hydroxycholesterol to 7α-hydroxy-3-oxocholest-4-en-26-oic acid via either 3β-hydroxycholest-5-en-26-oic acid and 3β,7α-dihydroxycholest-5-en-26-oic acid or via cholest-5-ene-3β,7α,26-triol (C5-3β,7α,26-triol) and its 7α-hydroxy-3-oxo-4-ene analogue (7α,26-dihydroxycholest-4-en-3-one, C4-7α,26-diol-3-one) (22, 24) (Scheme 4). In CSF we observe only very minor quantities (0.029 ± 0.012 ng/ml) of the biologically active oxysterol 27-hydroxycholesterol (64) but much higher levels of 7α-hydroxy-3-oxocholest-4-en-26-oic acid (7.170 ± 2.826 ng/ml). We thus tested the activity of 7α-hydroxy-3-oxocholest-4-en-26-oic acid as an LXR ligand in a luciferase assay. It did not activate the LXRα receptor, but interestingly its precursors 3β-hydroxycholest-5-en-26-oic acid and 3β,7α-dihydroxycholest-5-en-26-oic acid did activate LXR in the luciferase assay. This indicates that intermediates of the acidic pathway of bile acid biosynthesis present in brain and possessing biological activity are efficiently deactivated before secretion into CSF. None of the compounds tested for LXR activity here activated either NURR1 or FXR. There is, however, precedence for bile acid involvement with the nervous system. In sea lamprey, Li et al. (67) found allocholic acid (3α,7α,12α-trihydroxy-5α-cholan-24-oic acid, 5α-BA-3α,7α,12α-triol) and petromyzonol sulfate (5α-cholan-3α,7α,12α,24-tetraol 24-sulfate) to be potent stimulants of the adult olfactory system interacting with specific olfactory receptor sites and functioning as migratory pheromones. Furthermore, the brain is affected in severe liver disease with high plasma bile acid levels, in so-called hepatic encephalopathy (68).
Bile acids are biosynthesized extrahepatically via the acidic pathway starting with 27-hydroxycholesterol and also via the 24(S)-hydroxycholesterol pathway (19). The latter pathway is initiated in brain by 24(S)-hydroxylation of cholesterol by neuron-specific CYP46A1 (48), followed by 7α-hydroxylation by the presumed liver-specific enzyme CYP39A1 (69). However, recent data indicate that CYP39A1 protein is also expressed in eye and its mRNA in brain (53, 70), opening a route for extrahepatic bile acid biosynthesis via this pathway. In fact, all of the enzymes necessary for the biosynthesis of the primary bile acid chenodeoxycholic acid by the 24(S)-hydroxycholesterol pathway are expressed in brain (Scheme 4). This hypothesis is supported here by the identification in CSF of 7α-hydroxy-3-oxochol-4-en-24-oic acid, an immediate precursor of chenodeoxycholic acid, and the partial identification of 7α,x-dihydroxy-3-oxocholest-4-en-26-oic and 7α-hydroxy-3,x-bisoxocholest-4-en-26-oic acids, and 7α-hydroxy-26-norcholest-4-ene-3,x-dione (26-nor-C4-7α-ol-3,x-dione), where x may be position 24, i.e. giving 7α,24-dihydroxy-3-oxocholest-4-en-26-oic acid, 7α-hydroxy-3,24-bisoxocholest-4-en-26-oic acid, and 7α-hydroxy-26-norcholest-4-ene-3,24-dione. If this is the case, then 7α-hydroxy-26-norcholest-4-ene-3,24-dione is probably a decomposition product of 7α-hydroxy-3,24-bisoxocholest-4-en-26-oic acid. Similar decarboxylation reactions have been observed with other 24-oxo-C27 acids (58, 59). As 24(S)-hydroxycholesterol is biologically active, its conversion to bile acids in brain could represent a method for in situ deactivation. Here, we have identified 7α-hydroxy-3-oxochol-4-en-24-oic acid in CSF at a level of 0.172 ± 0.085 ng/ml. Mano et al. (25) demonstrated that rat brain cytosolic preparation contains an enzyme(s) capable of converting 7α-hydroxy-3-oxochol-4-en-24-oic acid to chenodeoxycholic acid; however, this activity was low, and the Δ4-3-oxosteroid 5β-reductase and 3α-hydroxysteroid dehydrogenase were not identified. Interestingly, epiallopregnanolone (3β-hydroxy-5α-pregnan-20-one, 5α-P-3β-ol-20-one) is present in rat brain (71, 72), presumably formed from progesterone (pregn-4-ene-3,20-dione, P4-3,20-dione) via activities of a 5α-reductase and 3β-hydroxysteroid dehydrogenase, whereas progesterone and testosterone have also been found to produce 5α-saturated derivatives in pre-viable human fetus (73). This raises the intriguing possibility that the same enzyme system may be responsible for the formation of the partially identified trihydroxycholanoic acid found here in CSF, i.e. 3β,x,y-trihydroxy-5α-cholan-24-oic acid (5α-BA-3β,x,y-triol). Although chenodeoxycholic acid has been identified in rat brain (26), it was only found after extensive protein denaturation, leading to the conclusion that it is noncovalently bound to protein in brain. The methodology used in this study was designed for the analysis of sterols and intermediates in the bile acid biosynthetic pathways but not primary bile acids that possess a 3α-hydroxy group rather than a 3β-hydroxy or oxo group. We are therefore currently developing a methodology that will allow the extraction and high sensitivity analysis of primary bile acids from brain and CSF, results from which will be reported in future publications.
In this study, we have identified the major cholesterol metabolite found in human CSF to be 7α-hydroxy-3-oxocholest-4-en-26-oic acid (Table 1). This is an intermediate in the acidic (and neutral/classical) pathway(s) of bile acid biosynthesis. Its immediate precursors in the acidic pathway, 3β-hydroxycholest-5-en-26-oic acid and 3β,7α-dihydroxycholest-5-en-26-oic acid, were each shown to activate the LXRα in a luciferase assay (Scheme 4 and Fig. 5), whereas 7α-hydroxy-3-oxocholest-4-en-26-oic acid was inactive. The data indicate that LXR ligands are efficiently deactivated in brain before passage into CSF. This is true whether the ligands are supplied via the bloodstream (21) or produced in brain (22, 24).
Supplementary Material
This work was supported by the United Kingdom Research Councils, Biotechnology and Biological Sciences Research Council Grants BBC5157712 and BBC5113561, and Engineering and Physical Sciences Research Council Grant EP/F014341/1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
W. J. Griffiths, S. Heidelberger, J. Turton, and Y. Wang, unpublished observations.
- LXR
- liver X receptor
- CSF
- cerebrospinal fluid
- CYP
- cytochrome P450
- FXR
- farnesoid X receptor
- GP
- Girard P
- HSD
- hydroxysteroid dehydrogenase
- IS
- internal standard
- LC-MS
- liquid chromatography-mass spectrometry
- LIT
- linear ion trap
- MRM
- multiple reaction monitoring
- MSn
- mass spectrometry with multiple fragmentation
- NURR1
- nuclear receptor related 1
- RIC
- reconstructed ion chromatogram
- RXR
- retinoid X receptor
- SPE
- solid phase extraction
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1.Björkhem I., Cedazo-Minguez A., Leoni V., Meaney S. (2009) Mol. Aspects Med. 30, 171–179 [DOI] [PubMed] [Google Scholar]
- 2.Griffiths W. J., Wang Y. (2009) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 2778–2805 [DOI] [PubMed] [Google Scholar]
- 3.Leoni V. (2009) Scand. J. Clin. Lab. Invest. 69, 22–25 [DOI] [PubMed] [Google Scholar]
- 4.Lütjohann D. (2006) Acta Neurol. Scand. Suppl.185, 33–42 [DOI] [PubMed] [Google Scholar]
- 5.Dietschy J. M., Turley S. D. (2004) J. Lipid Res. 45, 1375–1397 [DOI] [PubMed] [Google Scholar]
- 6.Weill-Engerer S., David J. P., Sazdovitch V., Liere P., Eychenne B., Pianos A., Schumacher M., Delacourte A., Baulieu E. E., Akwa Y. (2002) J. Clin. Endocrinol. Metab. 87, 5138–5143 [DOI] [PubMed] [Google Scholar]
- 7.Fan J., Donkin J., Wellington C. (2009) Biofactors 35, 239–248 [DOI] [PubMed] [Google Scholar]
- 8.Björkhem I. (2006) J. Intern. Med. 260, 493–508 [DOI] [PubMed] [Google Scholar]
- 9.Alvelius G., Hjalmarson O., Griffiths W. J., Björkhem I., Sjövall J. (2001) J. Lipid Res. 42, 1571–1577 [PubMed] [Google Scholar]
- 10.Griffiths W. J., Hornshaw M., Woffendin G., Baker S. F., Lockhart A., Heidelberger S., Gustafsson M., Sjövall J., Wang Y. (2008) J. Proteome Res. 7, 3602–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leoni V., Lütjohann D., Masterman T. (2005) J. Lipid Res. 46, 191–195 [DOI] [PubMed] [Google Scholar]
- 12.Schönknecht P., Lütjohann D., Pantel J., Bardenheuer H., Hartmann T., von Bergmann K., Beyreuther K., Schröder J. (2002) Neurosci. Lett. 324, 83–85 [DOI] [PubMed] [Google Scholar]
- 13.Papassotiropoulos A., Lütjohann D., Bagli M., Locatelli S., Jessen F., Buschfort R., Ptok U., Björkhem I., von Bergmann K., Heun R. (2002) J. Psychiatr. Res. 36, 27–32 [DOI] [PubMed] [Google Scholar]
- 14.Leoni V., Shafaati M., Salomon A., Kivipelto M., Björkhem I., Wahlund L. O. (2006) Neurosci. Lett. 397, 83–87 [DOI] [PubMed] [Google Scholar]
- 15.Yang Y., Griffiths W. J., Nazer H., Sjövall J. (1997) Biomed. Chromatogr. 11, 240–255 [DOI] [PubMed] [Google Scholar]
- 16.Meng L. J., Griffiths W. J., Nazer H., Yang Y., Sjövall J. (1997) J. Lipid Res. 38, 926–934 [PubMed] [Google Scholar]
- 17.Fine J. M., Sorensen P. W. (2008) J. Chem. Ecol. 34, 1259–1267 [DOI] [PubMed] [Google Scholar]
- 18.Lütjohann D., Breuer O., Ahlborg G., Nennesmo I., Sidén A., Diczfalusy U., Björkhem I. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9799–9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell D. W. (2003) Annu. Rev. Biochem. 72, 137–174 [DOI] [PubMed] [Google Scholar]
- 20.Björkhem I., Andersson U., Ellis E., Alvelius G., Ellegard L., Diczfalusy U., Sjövall J., Einarsson C. (2001) J. Biol. Chem. 276, 37004–37010 [DOI] [PubMed] [Google Scholar]
- 21.Heverin M., Meaney S., Lütjohann D., Diczfalusy U., Wahren J., Björkhem I. (2005) J. Lipid Res. 46, 1047–1052 [DOI] [PubMed] [Google Scholar]
- 22.Meaney S., Heverin M., Panzenboeck U., Ekström L., Axelsson M., Andersson U., Diczfalusy U., Pikuleva I., Wahren J., Sattler W., Björkhem I. (2007) J. Lipid Res. 48, 944–951 [DOI] [PubMed] [Google Scholar]
- 23.Lund E., Andersson O., Zhang J., Babiker A., Ahlborg G., Diczfalusy U., Einarsson K., Sjövall J., Björkhem I. (1996) Arterioscler. Thromb. Vasc. Biol. 16, 208–212 [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Akwa Y., el-Etr M., Baulieu E. E., Sjövall J. (1997) Biochem. J. 322, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mano N., Sato Y., Nagata M., Goto T., Goto J. (2004) J. Lipid Res. 45, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 26.Mano N., Goto T., Uchida M., Nishimura K., Ando M., Kobayashi N., Goto J. (2004) J. Lipid Res. 45, 295–300 [DOI] [PubMed] [Google Scholar]
- 27.Ferdinandusse S., Denis S., Faust P. L., Wanders R. J. (2009) J. Lipid Res. 50, 2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Axelson M., Mörk B., Sjövall J. (1988) J. Lipid Res. 29, 629–641 [PubMed] [Google Scholar]
- 29.Nagata K., Seyama Y., Shimizu T. (1995) Neurol. Med. Chir. 35, 294–297 [DOI] [PubMed] [Google Scholar]
- 30.Nagata K., Takakura K., Asano T., Seyama Y., Hirota H., Shigematsu N., Shima I., Kasama T., Shimizu T. (1992) Biochim. Biophys. Acta 1126, 229–236 [DOI] [PubMed] [Google Scholar]
- 31.Griffiths W. J., Sjövall J. (2010) J. Lipid Res. 51, 23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths W. J., Wang Y., Alvelius G., Liu S., Bodin K., Sjövall J. (2006) J. Am. Soc. Mass Spectrom. 17, 341–362 [DOI] [PubMed] [Google Scholar]
- 33.Karu K., Hornshaw M., Woffendin G., Bodin K., Hamberg M., Alvelius G., Sjövall J., Turton J., Wang Y., Griffiths W. J. (2007) J. Lipid Res. 48, 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., So K. M., Bodin K., Theofilopoulos S., Sacchetti P., Hornshaw M., Woffendin G., Karu K., Sjövall J., Arenas E., Griffiths W. J. (2009) Mol. Biosyst. 5, 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirk J. M., Tarbin J., Keely B. J. (2006) Rapid Commun. Mass Spectrom. 20, 1247–1252 [DOI] [PubMed] [Google Scholar]
- 36.Cowan D. A., Kicman A. T., Kubli-Garfias C., Welchman H. J. (2008) Steroids 73, 621–628 [DOI] [PubMed] [Google Scholar]
- 37.Higashi T., Nishio T., Hayashi N., Shimada K. (2007) Chem. Pharm. Bull. 55, 662–665 [DOI] [PubMed] [Google Scholar]
- 38.Woo H. K., Go E. P., Hoang L., Trauger S. A., Bowen B., Siuzdak G., Northen T. R. (2009) Rapid Commun. Mass Spectrom. 23, 1849–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motola D. L., Cummins C. L., Rottiers V., Sharma K. K., Li T., Li Y., Suino-Powell K., Xu H. E., Auchus R. J., Antebi A., Mangelsdorf D. J. (2006) Cell 124, 1209–1223 [DOI] [PubMed] [Google Scholar]
- 40.Song C., Liao S. (2000) Endocrinology 141, 4180–4184 [DOI] [PubMed] [Google Scholar]
- 41.Sacchetti P., So K. M., Hall A. C., Liste I., Steffensen K. R., Theofilopoulos S., Parish C. L., Hazenberg C., Richter L. A., Hovatta O., Gustafsson J. A., Arenas E. (2009) Cell Stem Cell 5, 409–419 [DOI] [PubMed] [Google Scholar]
- 42.Fan X., Kim H. J., Bouton D., Warner M., Gustafsson J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13445–13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law S. W., Conneely O. M., DeMayo F. J., O'Malley B. W. (1992) Mol. Endocrinol. 6, 2129–2135 [DOI] [PubMed] [Google Scholar]
- 44.Jankovic J., Chen S., Le W. D. (2005) Prog. Neurobiol. 77, 128–138 [DOI] [PubMed] [Google Scholar]
- 45.Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., Dent P. (2009) J. Lipid Res. 50, 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schroepfer G. J., Jr. (2000) Physiol. Rev. 80, 361–554 [DOI] [PubMed] [Google Scholar]
- 47.Ruan B., Wilson W. K., Pang J., Gerst N., Pinkerton F. D., Tsai J., Kelley R. I., Whitby F. G., Milewicz D. M., Garbern J., Schroepfer G. J., Jr. (2001) J. Lipid Res. 42, 799–812 [PubMed] [Google Scholar]
- 48.Lund E. G., Guileyardo J. M., Russell D. W. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7238–7243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersson S., Davis D. L., Dahlbäck H., Jörnvall H., Russell D. W. (1989) J. Biol. Chem. 264, 8222–8229 [PubMed] [Google Scholar]
- 50.Jelinek D. F., Andersson S., Slaughter C. A., Russell D. W. (1990) J. Biol. Chem. 265, 8190–8197 [PMC free article] [PubMed] [Google Scholar]
- 51.Lund E. G., Kerr T. A., Sakai J., Li W. P., Russell D. W. (1998) J. Biol. Chem. 273, 34316–34327 [DOI] [PubMed] [Google Scholar]
- 52.Furster C., Zhang J., Toll A. (1996) J. Biol. Chem. 271, 20903–20907 [DOI] [PubMed] [Google Scholar]
- 53.Shafaati M., O'Driscoll R., Björkhem I., Meaney S. (2009) Biochem. Biophys. Res. Commun. 378, 689–694 [DOI] [PubMed] [Google Scholar]
- 54.Heverin M., Bogdanovic N., Lütjohann D., Bayer T., Pikuleva I., Bretillon L., Diczfalusy U., Winblad B., Björkhem I. (2004) J. Lipid Res. 45, 186–193 [DOI] [PubMed] [Google Scholar]
- 55.Gilardi F., Viviani B., Galmozzi A., Boraso M., Bartesaghi S., Torri A., Caruso D., Crestani M., Marinovich M., de Fabiani E. (2009) Neuroscience 164, 530–540 [DOI] [PubMed] [Google Scholar]
- 56.Berger J., Truppe C., Neumann H., Forss-Petter S. (1998) FEBS Lett. 425, 305–309 [DOI] [PubMed] [Google Scholar]
- 57.Itoh M., Suzuki Y., Akaboshi S., Zhang Z., Miyabara S., Takashima S. (2000) Brain Res. 858, 40–47 [DOI] [PubMed] [Google Scholar]
- 58.Yuri M., Tokumoto M., Hara N., Fujimoto Y., Kobayashi N., Morisaki M. (1993) Chem. Pharm. Bull. 41, 1327–1329 [DOI] [PubMed] [Google Scholar]
- 59.Bun-ya M., Maebuchi M., Kamiryo T., Kurosawa T., Sato M., Tohma M., Jiang L. L., Hashimoto T. (1998) J. Biochem. 123, 347–352 [DOI] [PubMed] [Google Scholar]
- 60.Karlaganis G., Almé B., Karlaganis V., Sjövall J. (1981) J. Steroid Biochem. 14, 341–345 [DOI] [PubMed] [Google Scholar]
- 61.Karlaganis G., Karlaganis V., Sjövall J. (1984) J. Lipid Res. 25, 693–702 [PubMed] [Google Scholar]
- 62.Lehmann J. M., Kliewer S. A., Moore L. B., Smith-Oliver T. A., Oliver B. B., Su J. L., Sundseth S. S., Winegar D. A., Blanchard D. E., Spencer T. A., Willson T. M. (1997) J. Biol. Chem. 272, 3137–3140 [DOI] [PubMed] [Google Scholar]
- 63.Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu X., Menke J. G., Chen Y., Zhou G., MacNaul K. L., Wright S. D., Sparrow C. P., Lund E. G. (2001) J. Biol. Chem. 276, 38378–38387 [DOI] [PubMed] [Google Scholar]
- 65.Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Muneton S., Sjövall J., Jovanovic J. N., Griffiths W. J. (2008) J. Proteome Res. 7, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W., Sorensen P. W., Gallaher D. D. (1995) J. Gen. Physiol. 105, 569–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siciliano M., Milani A., Marra L., Arringoli D., Rossi L. (1986) Quad. Sclavo. Diagn. 22, 355–361 [PubMed] [Google Scholar]
- 69.Li-Hawkins J., Lund E. G., Bronson A. D., Russell D. W. (2000) J. Biol. Chem. 275, 16543–16549 [DOI] [PubMed] [Google Scholar]
- 70.Ikeda H., Ueda M., Ikeda M., Kobayashi H., Honda Y. (2003) Lab. Invest. 83, 349–355 [DOI] [PubMed] [Google Scholar]
- 71.Liu S., Sjövall J., Griffiths W. J. (2003) Anal. Chem. 75, 5835–5846 [DOI] [PubMed] [Google Scholar]
- 72.Ebner M. J., Corol D. I., Havlíková H., Honour J. W., Fry J. P. (2006) Endocrinology 147, 179–190 [DOI] [PubMed] [Google Scholar]
- 73.Mickan H. (1972) Steroids 19, 659–668 [DOI] [PubMed] [Google Scholar]
- 74.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Jr., Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., Shimizu T., Spener F., van Meer G., VanNieuwenhze M. S., White S. H., Witztum J. L., Dennis E. A. (2005) J. Lipid Res. 46, 839–861 [DOI] [PubMed] [Google Scholar]
- 75.Stapleton G., Steel M., Richardson M., Mason J. O., Rose K. A., Morris R. G., Lathe R. (1995) J. Biol. Chem. 270, 29739–29745 [DOI] [PubMed] [Google Scholar]
- 76.Baumgart E., Vanhooren J. C., Fransen M., Marynen P., Puype M., Vandekerckhove J., Leunissen J. A., Fahimi H. D., Mannaerts G. P., van Veldhoven P. P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13748–13753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seedorf U., Assmann G. (1991) J. Biol. Chem. 266, 630–636 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.