Abstract
The GTF2IRD1 gene is of principal interest to the study of Williams-Beuren syndrome (WBS). This neurodevelopmental disorder results from the hemizygous deletion of a region of chromosome 7q11.23 containing 28 genes including GTF2IRD1. WBS is thought to be caused by haploinsufficiency of certain dosage-sensitive genes within the deleted region, and the feature of supravalvular aortic stenosis (SVAS) has been attributed to reduced elastin caused by deletion of ELN. Human genetic mapping data have implicated two related genes GTF2IRD1 and GTF2I in the cause of some the key features of WBS, including craniofacial dysmorphology, hypersociability, and visuospatial deficits. Mice with mutations of the Gtf2ird1 allele show evidence of craniofacial abnormalities and behavioral changes. Here we show the existence of a negative autoregulatory mechanism that controls the level of GTF2IRD1 transcription via direct binding of the GTF2IRD1 protein to a highly conserved region of the GTF2IRD1 promoter containing an array of three binding sites. The affinity for this protein-DNA interaction is critically dependent upon multiple interactions between separate domains of the protein and at least two of the DNA binding sites. This autoregulatory mechanism leads to dosage compensation of GTF2IRD1 transcription in WBS patients. The GTF2IRD1 promoter represents the first established in vivo gene target of the GTF2IRD1 protein, and we use it to model its DNA interaction capabilities.
Keywords: DNA-binding Protein, Evolution, Gene Regulation, Genetic Diseases, Transcription Regulation, Gtf2ird1, Williams Beuren Syndrome
Introduction
Williams-Beuren syndrome (WBS)4 is a relatively rare disorder with an estimated incidence of 1:7,500–1:20,000 (1). In 95% of cases, this deletion is 1.5 Mb long and results from illegitimate recombination during meiosis between two blocks of low copy repeats (LCRs) that flank the deletion domain (2, 3). To explain the causes of WBS in molecular terms, it is first necessary to identify the genes that underpin each of the disorders. The only symptom that fulfills this to date is SVAS. Haploinsufficiency of elastin due to loss of the ELN gene leads to narrowing of the large elastic aorta and may also affect the pulmonary, coronary, and carotid arteries (4). Accumulating evidence from patients with atypical hemizygous deletions within the critical region indicate that the many of the remaining symptoms, in particular the craniofacial abnormalities, the visuospatial construction deficit and the hypersociability, can be attributed to two genes at the telomeric end of the deletion region, GTF2IRD1 and GTF2I (5–7). These genes share sequence homology and are adjacent, indicating that they have arisen by duplication and divergence from a common ancestor. Functional evidence suggests that these genes encode nuclear proteins with DNA binding capabilities and are widely considered to be transcription factors with specific gene targets (8, 9).
The first reported gene product of GTF2IRD1 was MusTRD1, which was isolated in a yeast one-hybrid screen for proteins that could bind to a DNA enhancer element present in the TNNI1 gene (10). Human, mouse, and Xenopus orthologs of the gene were subsequently isolated in three independent yeast one-hybrid screens as GTF3 (11), BEN (12), and XWBSCR11 (13). Herein, we will refer to protein and gene by the approved symbol GTF2IRD1. A comparison of the bait sequences used in each of the four yeast one-hybrid assays revealed a common core binding sequence of GGATTA and subsequent DNA binding studies confirmed this as the core recognition motif (14–16). In the yeast one-hybrid studies, GTF2IRD1 protein was implicated in the regulation of the genes from which each of the baits were derived; TNNI1, Hoxc8, and GSC.
In this report, we explore an autoregulatory feedback circuit that regulates the levels of GTF2IRD1 transcript. This mechanism is responsible for an observed increase in the levels of transcript produced from the targeted Gtf2ird1 allele in knock-out mice and leads to dosage compensation of GTF2IRD1 transcript in cell lines derived from WBS patients. This calls into question whether the GTF2IRD1 protein is haploinsufficient in WBS. We demonstrate that this mechanism is controlled directly by GTF2IRD1 binding to a highly conserved upstream region of its own gene and show that the binding affinity is critically dependent upon multiple interactions of the repeat domains with at least two binding sites. These data constitute the first definitive example of an interaction between GTF2IRD1 and a target gene, supportable by in vivo data, and therefore serves as a valuable model system for the study of GTF2IRD1 DNA binding.
EXPERIMENTAL PROCEDURES
Knock-out
The mouse Gtf2ird1 allele was targeted in 129R1 ES cells using homologous arms flanking exon 2 inserted into the pPGKneobpALox2DTA plasmid. The neomycin cassette was subsequently removed using cre/lox excision by mating to C57BL/6JArc mice carrying the Tg(CMV-Cre)1Cgn transgene (17). The mutant allele was backcrossed onto C57BL/6JArc, and these experiments were conducted on the N5 generation.
Protein Expression Analysis
C2C12 cells were washed with ice-cold phosphate-buffered saline, sonicated, and lysed in RIPA buffer supplemented with protease inhibitor mixture (Roche) for 30 min at 4 °C. Cell lysates were centrifuged at 13,000 × g for 20 min to remove cell debris and precleared by incubation with protein G-Sepharose beads (Roche) for 1 h at 4 °C. The anti-GTF2IRD1 antibody (WBSCR11 (M-19), cat. no. sc-14714, Santa Cruz Biotechnology) was coupled to protein G beads for 1 h at 4 °C. Pre-cleared lysates were incubated with the antibody-bound beads at 4 °C overnight. For the peptide block experiment, WBSCR11 (M-19) antigenic peptides were added during antibody precoupling and during incubation with precleared lysates. Beads were washed in RIPA buffer three times and proteins were eluted by boiling in 2× SDS sample buffer. One-fifth of immunoprecipitated eluent was separated by 6% SDS-polyacrylamide gels and analyzed by immunoblotting with the anti-GTF2IRD1 antibody, WBSCR11 (M-19).
Wild-type and mutant cDNA fragments were amplified from brown adipose tissue-derived cDNA samples using mIRD1ex1F and mIRD1ex7/8R (see supplemental Table S2) and inserted into a pre-existing pCDNA3.1 (Invitrogen) expression plasmid containing mouse Gtf2ird1 isoform 3α7 to recreate the full-length wild-type and mutant transcripts present in the mice. These plasmids were transiently transfected into COS-7 cells using Lipofectamine (Invitrogen). Protein extracts were made with RIPA buffer using standard methods, electrophoresed on 7.5% SDS-polyacrylamide gels and analyzed by immunoblotting with the anti-GTF2IRD1 antibodies, WBSCR11 (M-19), cat. no. sc-14714 (Santa Cruz Biotechnology) and G21, raised in sheep against the peptide CNNAKVPAKDNIPKRK.
Cell Lines and Immunofluorescence
Lymphoblastoid cell lines derived from six WBS patients and six relatives were chosen (Coriell Cell Repository; see supplemental Table S1). Cell lines were maintained in RPMI 1640 (GIBCO) supplemented with glutamine and 15% fetal calf serum according to Coriell protocols.
Inducible C2C12 Myc-GTF2IRD1 (mouse isoform 3α7) (18) clones were produced using the RheoSwitch system (New England Biolabs). Parental C2C12 cells grown in low glucose GlutaMAX DMEM (GIBCO) with 20% fetal calf serum were transfected with pNEBR-R1 and neomycin-resistant clones were tested for high expression of the RheoActivator and RheoReceptor proteins. A high expressing clone was transfected with the pNEBRX1-Hygro plasmid containing a cDNA encoding N-terminal Myc-tagged mouse GTF2IRD1 and hygromycin-resistant clones were screened for inducible Myc-GTF2IRD1 expression.
Myc-GTF2IRD1 was identified by immunofluorescence using cells grown on glass coverslips, fixed in 4% paraformaldehyde, 0.125% Triton, incubated with the anti-Myc 9E10 monoclonal antibody (Sigma), followed by goat anti-mouse Alexa Fluor 488 (Molecular Probes Invitrogen) secondary antibody and mounted in Vectashield (Vector Laboratories) containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI).
Six inducible cell clones were identified, and some mosaicism of inducible expression was apparent in all. Silencing of expression was observed to increase rapidly with passage number. The line expressing the most in the greatest number of cells was used for subsequent transfection assays.
RNA Expression Analysis
Mouse tissues were dissected and stored in RNAlater (Ambion). Cell lines were washed with phosphate-buffered saline and homogenized in TriReagent (Sigma) by trituration. RNA was isolated according to the TriReagent instructions. First-strand cDNA synthesis was performed using 1 μg of total RNA, M-MLV reverse transcriptase (Promega), and oligo (dT)15 primers (Promega).
RNA was electrophoresed in MOPS/formaldehyde gels and blotted according to standard methods (Hybond, GE Healthcare). Membranes were hybridized with a 32P-labeled probe derived from the full-length mouse Gtf2ird1 cDNA (isoform mouse 3α7) at 65 °C according to standard protocols (19).
Quantitative PCR was performed in a Corbett Rotor-Gene 2000 using the QuantiTect SYBR green PCR kit (Qiagen). Each measurement was made in duplicate using 5 μl of diluted cDNA template. Primer sequences are shown in supplemental Table S2. For the human lymphoblastoid cell lines, measurements were made in duplicate from RNA collected on 3 separate occasions and normalized to GAPDH measurements made in the same way. This was done to minimize errors associated with cell culture conditions.
Electrophoretic Mobility Shift Assay (EMSA)
Human GTF2IRD1, GTF2IRD1ΔLZ (lacking the first 81 amino acids at the N terminus, see supplemental Table S2), mouse GTF2IRD1 isoforms 3α5 and 3α7 and the mouse GTF2I (TFII-I) β isoform were produced by in vitro coupled transcription-translation using T7-primed TnT rabbit reticulocyte lysate (Promega) and 1 μg of pCDNA3.1 plasmid (Invitrogen) containing the cDNA of interest. Labeled probes were made in two ways. For the B1 combinations, complementary oligonucleotides were synthesized with an AGCT 5′-overhang at both ends, which were filled in using the DECAprimeII 5x-dCTP reaction buffer and Exo- Klenow enzyme (Ambion) in the presence of [32P]dCTP. For the wild-type mouse GUR fragment and the various mutant forms, double-stranded DNA fragments were generated by PCR using GUR primers (see supplemental Table S2). The template was derived from C57BL/6 mouse genomic DNA and the template for fragments containing the middle mutation was synthesized as a single stranded oligonucleotide (GUR MID MUT, mouse). All fragments were labeled by incubation of the denatured DNA with DECAprimeII 5×-dCTP reaction buffer, Exo- Klenow enzyme (Ambion) and [32P]dCTP in the presence of the same primers used to amplify the fragment initially. Unincorporated nucleotides were removed using G-25 spin column (GE Healthcare). DNA binding reactions and gel running conditions were as described previously (10).
Bioinformatics
genome sequences of human, mouse, Xenopus tropicalis, and Takifugu rubripes were obtained from the Ensembl genome server. EST sequences for mouse and human were derived from the NCBI data base and the X. tropicalis EST from the X. tropicalis project at the Sanger Centre UK. ESTs for fugu or any other fish species do not include entries that show exon 1 of the GTF2IRD1 gene. Therefore, exon 1 and the associated GUR were manually curated using a combination of homology searches between T. rubripes and Tetraodon nigroviridis genomic sequences derived from the Ensembl data base and the use of the GENSCAN server at MIT. BLAST alignments were performed using the bl2seq alignment tool available at NCBI with genomic sequence from the various species spanning the whole gene plus 10-kb upstream.
RESULTS
To determine the function of Gtf2ird1 and to examine its potential role in WBS, we created a mouse knock-out (KO) by targeted deletion of exon 2 (Fig. 1A), which contains the start codon. Homozygous Gtf2ird1tm1Hrd mice have no obvious developmental defects, but show specific behavioral changes, a motor coordination deficit and evidence of altered GABAergic neuronal function.5
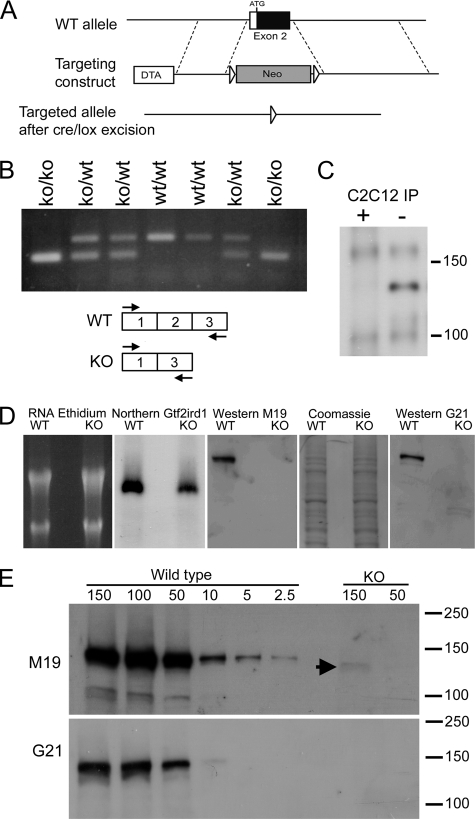
FIGURE 1.
Generation of the Gtf2ird1tm1Hrd mutant allele and the consequences for transcription and translation. A, homologous recombination of the targeting construct with the wild-type (WT) allele using positive neomycin (Neo) selection and negative diphtheria toxin (DTA) selection. The Neo cassette was subsequently removed using the flanking LoxP sites (arrowheads). B, RTPCR analysis of Gtf2ird1 transcripts in brown adipose tissue from a litter of mice segregating the mutant allele using primers in exon 1 and exon 3 (shown in scheme). A novel exon 1–3 splice transcript is produced with equal efficiency to the wild-type transcript. C, Western blot of proteins immunoprecipitated (IP) from C2C12 cell extracts using the M19 antibody for IP and as probe. The immunoprecipitation was conducted in the presence (+) or absence (−) of a blocking peptide antigen to demonstrate specificity. D, analysis of transcription and translation from a wild-type Gtf2ird1 cDNA and the equivalent mutant cDNA lacking exon 2. Extracts from transfected COS-7 cells show that both transcripts are abundantly produced (Northern Gtf2ird1) but the GTF2IRD1 peptide is below detectable levels in cells expressing the mutant transcript (KO) using two antibodies, M19 and G21. Equal loading is shown by ethidium bromide staining of RNA and Coomassie Blue staining of protein in the M19 Western blot. E, Western blot analysis using sequential dilutions of protein extract from cells transfected with the wild-type construct compared with high concentration loadings of mutant extract (KO), probed with M19 and G21 antibodies. A faint band is visible at the highest concentration in the M19 analysis (arrowhead). Numbers represent μg of total protein.
Analysis of Transcription and Translation in Gtf2ird1tm1Hrd Mice
An initial examination of Gtf2ird1 expression in tissues from KO mice suggested that a mutant transcript lacking exon 2 is produced from the targeted allele. Because exon 2 and its splice junctions were removed entirely, we predicted that exon 1 was splicing directly to exon 3. To test the efficiency of this process, we examined the activity of Gtf2ird1 in brown adipose tissue, which we have established previously as one of the highest Gtf2ird1-expressing tissues in the adult mouse (20). RNA was extracted from adult littermates segregating the Gtf2ird1tm1Hrd allele, and RTPCR was conducted using primers that bind to sequences in exon 1 and exon 3 to determine the relative proportions of wild-type and mutant transcript (Fig. 1B). In heterozygous KO mice, the level of mutant transcript was deemed to be approximately equal to the level of transcript produced from the wild-type allele.
The first downstream AUG in the mutant transcript resides in exon 3 and is out of frame. The AUG in this reading frame is followed 5 codons later by a premature termination codon. The mutant transcript is clearly not subject to nonsense-mediated decay (NMD) as shown above (Fig. 1B), which is probably because of the close proximity of the start codon to the premature termination codon. Short nonsense open reading frames can escape NMD as seen in mutants of the β-globin gene (21).
Translation of an in-frame-truncated GTF2IRD1 peptide could result from translational re-initiation beginning at the second AUG at residue Met-65, also located in exon 3. Detection of endogenous GTF2IRD1 protein is extremely difficult, probably because of a very low general abundance, but is possible by immunoprecipitation in C2C12 cells, which are known to express high levels of Gtf2ird1 transcript (Fig. 1C). However, measuring reduced endogenous levels of any potential translational re-initiation products is harder still and well below the possible limits of detection. Therefore, a mutant cDNA representative of a transcript from the KO allele (corresponding to the full-length mouse 3α7 isoform (18) but lacking exon 2), was cloned into an expression vector and transfected into COS-7 cells. Transcription from this mutant cDNA was comparable to the wild-type 3α7 control; although ∼2-fold lower (Fig. 1D). By loading a large amount of protein extract (150 μg) from cells transfected with the mutant construct, a faint band corresponding to a truncated GTF2IRD1 peptide could be detected using the anti-GTF2IRD1, WBSCR11 (M-19) antibody (Fig. 1E). A similar analysis using the G21 antibody was unable to detect any mutant protein, even at the highest concentrations (Fig. 1E). By comparison with dilutions of the extracts derived from the wild-type transfections and taking into account the relative levels of Gtf2ird1 and Gtf2ird1tm1Hrd transcript (measured by quantitative RTPCR); it was possible to estimate that the efficiency of production of the mutant peptide is ∼3% of normal levels.
Negative Autoregulation Controls GTF2IRD1 Transcription in Mice and Humans
Initial examination of the mutant transcript in the Gtf2ird1tm1Hrd KO mice suggested that levels were higher than normal. To examine this in detail, we used RNA extracted from a number of tissues and performed Northern blotting (Fig. 2A) and quantitative RTPCR (Fig. 2B) using primers that amplify mutant and wild-type transcript indiscriminately (mIRD1ex6F- mIRD1ex7/8R). Total levels of Gtf2ird1 transcript were significantly higher in heterozygous and homozygous Gtf2ird1tm1Hrd KOs, with levels in the homozygous nulls approximately double the normal level of transcript (Fig. 2B). This result concurred with the band intensities seen in Northern blots of RNA extracted from a number of representative mice (Fig. 2A). The most plausible explanation for these findings was the existence of a negative feedback mechanism that increased Gtf2ird1 transcription in response to the depletion of GTF2IRD1 protein.
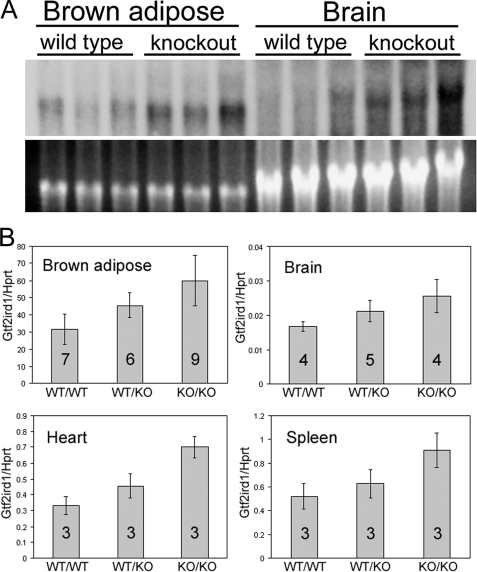
FIGURE 2.
Gtf2ird1 transcript production approximately doubles in the tissues of Gtf2ird1tm1Hrd knock-out mice. A, Northern blot of total RNA extracted from brown adipose and brain tissue of three wild-type and three homozygous knock-out mice showing that the 3.5-kb Gtf2ird1 band is more intense in all three knock-out mice. Close scrutiny also shows an expected slight reduction in transcript length because of the loss of exon 2 (129 bp) in the knock-out samples. B, quantitative RT-PCR analysis of Gtf2ird1 levels. Error bars show S.D., and the numbers indicate the number of mice used. Unpaired two-tailed Student's t tests show a difference between wild-type (WT/WT) and homozygous null (KO/KO) genotypes with a probability of: p = 0.0011 for brown adipose, p = 0.012 for brain, p = 0.0019 for heart, and p = 0.022 for spleen.
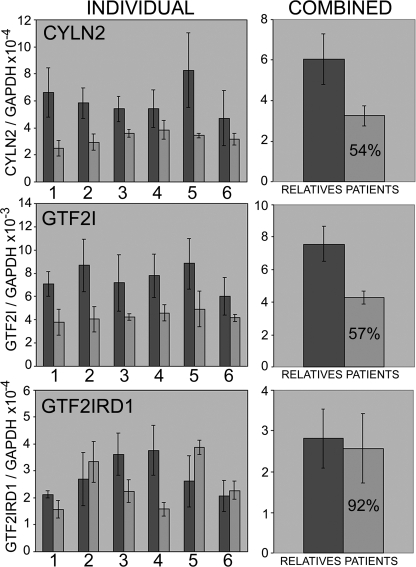
We reasoned that if such a negative autoregulatory mechanism were conserved in humans, WBS patients, who only have one allele of GTF2IRD1, might undergo a form of dosage compensation and restore GTF2IRD1 transcript levels to normal. We examined the expression of GTF2IRD1 and its flanking genes in lymphoblastoid cell lines derived from six WBS patients and six relatives using quantitative RTPCR (Fig. 3) with primers designed within the coding regions. The mean expression levels of the flanking genes CYLN2 and GTF2I were just above the expected 50% of normal. However, mean levels of GTF2IRD1, although slightly lower than normal (92%), were not significantly different from levels in their unaffected relatives (Fig. 3). This result was confirmed using a separate RTPCR assay (see supplemental Fig. S1) using primers designed to amplify a different portion of the GTF2IRD1 cDNA. GTF2IRD1 transcript levels were highly variable between individuals of all genotypes, suggesting that this is typical behavior, at least in this cell type, and is not associated with the WBS condition.
FIGURE 3.
Quantitative RT-PCR analysis of CYLN2, GTF2I, and GTF2IRD1 expression in lymphoblastoid cell lines derived from Williams-Beuren syndrome patients and their relatives. Error bars show S.D., and the figure in the patient bar indicates the estimated mean expression level as a percentage of the relatives' mean. Paired two-tailed Student's t tests show differences between the means of genotype groups with a probability of: CYLN2 p = 0.004575, GTF2I p = 0.000342, and GTF2IRD1 p = 0.559668.
The GTF2IRD1 Gene Contains a Conserved GTF2IRD1 DNA Binding Domain
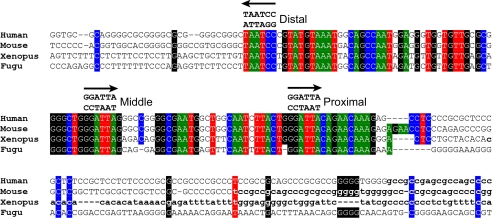
Because GTF2IRD1 protein binds DNA in a sequence-specific manner (14–16), we reasoned that the negative autoregulatory mechanism might involve direct binding of GTF2IRD1 protein to an enhancer element within the GTF2IRD1 allele. On the assumption that such a binding region would be conserved, we conducted phylogenetic footprinting analysis using the genome sequence of GTF2IRD1 genes from human, mouse, chicken, zebrafish, X. tropicalis, and T. rubripes. Within non-transcribed domains, a region immediately adjacent to the transcription start site in mouse and human emerged as a clear candidate for this hypothesis. The GTF2IRD1 upstream region (GUR), which we have defined as a 104-bp region of high homology, shows 82% sequence identity between human and fish species. Significantly, the GUR contains three canonical GTF2IRD1 recognition sequences (Fig. 4). Assuming equal and random base distributions, the incidence of the sequence GGATTA or the inverted TAATCC, should occur by chance every 512 bp. Within the 150 kb of DNA containing the human GTF2IRD1 gene, the observed mean frequency is 549 bp. It is unlikely, therefore, that these three sites are present within 104 bp by chance.
FIGURE 4.
ClustalW alignment of the GUR in four vertebrate species. Colored boxes indicate identical base residues, and the arrows indicate the position and orientation of three highly conserved GTF2IRD1 recognition sequences. The core motifs are perfectly conserved, but conservation quickly diminishes either side of the flanking sites. The transition to bold lowercase indicates the start of the most 5′ ESTs found in data base searches. Human EST BM544769, Mouse EST CJ178886, X. tropicalis EST CN092749. No informative Fugu ESTs were found.
It is interesting to note that the GGATTA sites themselves show 100% identity between all of the species, and the sequence homology ends abruptly either side of the proximal and distal GGATTA sites. Apart from the conjunction of these 3 sites, the region is relatively unremarkable. It does not contain a conventional TATA box but does have a well-conserved CCAAT box and potential elements that match a number of known transcription factor recognition sequences, but their significance is, so far, unknown.
GTF2IRD1 Binding to the GUR Requires a Tandem Interaction
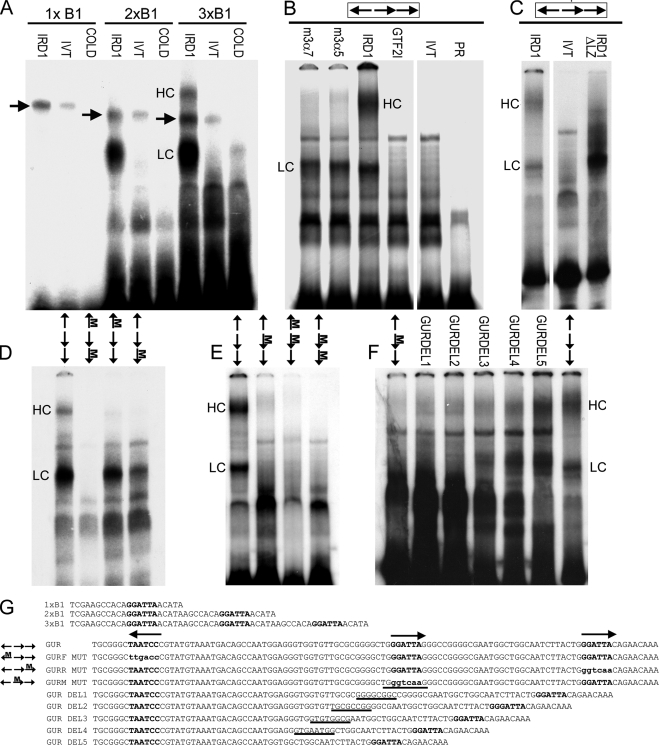
The cluster of well-conserved GGATTA binding sites indicates a strong potential for interaction with the GTF2IRD1 protein. Furthermore, the presence of three sites might indicate that the protein has a preference for multiple binding sites. Therefore, we tested the relative affinity of GTF2IRD1 for multiple GGATTA binding sites in an EMSA using artificial trimeric and dimeric DNA probes based on the sequence used in the TNNI1 yeast one-hybrid screen originally used to isolate GTF2IRD1 (10). The affinity of in vitro translated GTF2IRD1 for a single GGATTA was below the level of detection (Fig. 5A). However, with two sites the interaction was strong, and with three sites band intensity increased further and a second higher shift complex (HC) became visible.
FIGURE 5.
EMSA of GTF2IRD1 proteins showing affinity for the GUR and the tandem nature of the binding. A, binding of human GTF2IRD1 to synthetic multimers of the TNNI1-derived probe, B1, originally used as a yeast one-hybrid bait (10). GTF2IRD1 (IRD1) does not bind detectably to single copies, but binds well to B1 dimers (2xB1) and trimers (3xB1). A lower shift complex (LC) is present in both, but a higher shift complex (HC) appears with the trimer. Probes combined with unprogrammed in vitro translation mix (IVT) or with GTF2IRD1 and excess cold competitor oligonucleotide (COLD) showed no shift. An unknown endogenous shift complex (arrows) present in the in vitro translation mix shows only a minor affinity increase as a result of multimerization. B, GUR fragment is bound efficiently by the two most abundant mouse isoforms of GTF2IRD1, 3α7, and 3α5 and by human GTF2IRD1, but not by the mouse β isoform of TFII-I (GTF2I). No shift occurs in the IVT control or probe-only lane (PR). C, deletion of the leucine zipper domain of GTF2IRD1 (IRD1ΔLZ) intensifies affinity of the LC, but ablates the HC, demonstrating that the HC contains a GTF2IRD1 dimer. D and E, probes containing mutations in single GGATTA sites or in combinations were tested with human GTF2IRD1 protein to determine changes in affinity. Position of the mutation (M) is indicated on the scheme of the GUR (arrows). F, mutation of the middle site alone leads to absence of binding, but the shifts are restored by reducing the distance between the flanking sites, as illustrated using a series of probes with successive deletions (GURDEL1–5). G, sequences of the double-stranded probes used in the EMSA: binding sites are indicated in bold type, and mutations are indicated in lowercase bold type. In the GUR DEL series, the residues deleted in each successive probe are indicated by underlines. Gaps between images indicate where irrelevant lanes have been removed.
The GUR of the mouse was amplified from genomic DNA by PCR and labeled for EMSA analysis. Mouse and human GTF2IRD1 isoforms bound with high affinity to the GUR probe, whereas the related GTF2I (TFII-I) peptide encoded by Gtf2i did not (Fig. 5B). Human GTF2IRD1 formed a lower shift complex (LC) and a higher shift complex (HC) with the GUR probe. On the assumption that the HC may be caused by dimerization of GTF2IRD1, which is reportedly mediated via the leucine zipper (LZ) (15), we made an N-terminal deletion of human GTF2IRD1 that lacks the LZ motif and found that this peptide fails to form the HC shift, whereas the intensity of the LC is significantly enhanced (Fig. 5C).
To determine whether binding is dependent on the presence of all three GGATTA recognition sequences, we prepared a series of DNA probes with 3-bp mutations within the core motif, changing GGATTA into GGTCAA, either singly or in combination (Fig. 5, D–F). Mutation of the distal site does not impact significantly on the LC, but the HC becomes very weak; whereas, mutation of the proximal site reduces binding of both. When both flanking sites are mutated, binding is lost entirely (Fig. 5D). Mutation of the middle site also results in ablation of binding (Fig. 5E). This could mean that the middle binding site is obligatory, or it may indicate that the distance between the two flanking sites is too great for the GTF2IRD1 protein to span. To discriminate between these possibilities, a series of probes were made with progressive 8-bp deletions within the middle region (Fig. 5G), which showed that binding is restored once the distance between the flanking sites is reduced to 57 bp and becomes progressively stronger as the distance narrows (Fig. 5F).
The GUR Is an Effective Promoter/Enhancer That Can Be Repressed by GTF2IRD1
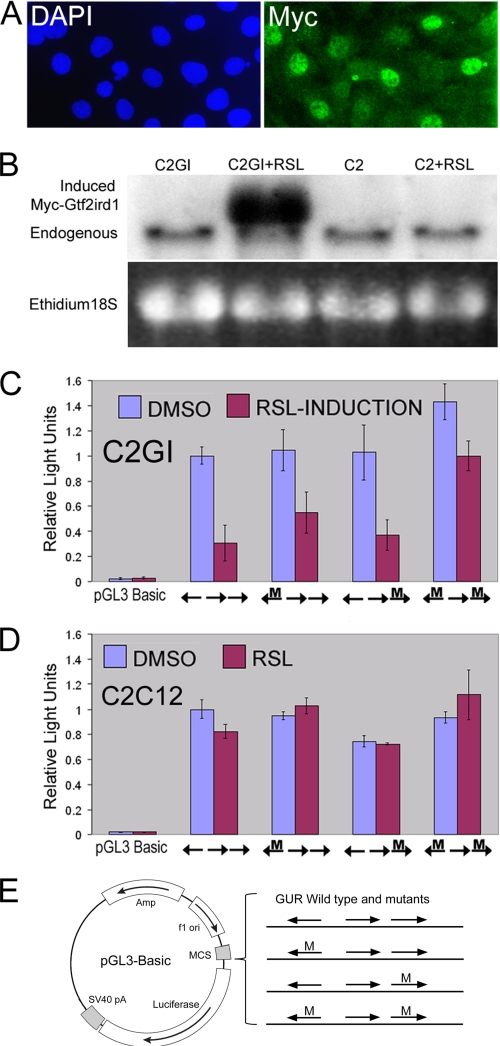
The next step was to examine whether the GUR has promoter/enhancer capabilities and to determine whether GTF2IRD1 binding can negatively regulate this function. We amplified a 193-bp fragment containing the GUR, which was inserted into a pGL3-luciferase reporter construct. A fragment longer than the conserved 104-bp GUR was used to take advantage of convenient internal restriction sites. Constructs containing 3-bp mutations in the flanking GGATTA binding sites were also made to determine the consequences of reduced DNA binding capability, as shown in the EMSA studies. These reporter constructs were transiently transfected into a C2C12 cell clone (C2GI) that contained an inducible Myc-GTF2IRD1 construct (Fig. 6, A and B). Inducible expression of the tagged protein allowed low level expression in a high proportion of cells (∼50%), which could be monitored using immunofluorescence (Fig. 6A). The mouse C2C12 myoblast cell line was chosen because endogenous Gtf2ird1 transcript, and protein is abundant in these cells (Figs. 6B and 1C), so it was assumed that these cells would have the appropriate factors necessary for GUR activation.
FIGURE 6.
The GUR acts as an effective enhancer/promoter in cell culture assays and is negatively regulated by exogenously derived inducible GTF2IRD1. A, inducible Myc-GTF2IRD1 clonal cell line C2GI, with nuclei stained by DAPI and Myc-tagged GTF2IRD1 detected by anti-Myc immunofluorescence (Myc). B, Northern blot of C2GI and parental C2C12 (C2) cell RNA with or without the inducer RSL. C, C2GI inducible GTF2IRD1 cells with pGL3-Basic and pGL3-GUR constructs containing the wild-type GUR or with mutations (M) in the flanking recognition sites. Induction of GTF2IRD1 leads to a repression of the reporter containing the wild-type GUR. Mutations in the GTF2IRD1 binding sites result in attenuation of repression. Luciferase activity was set at 1 for the wild-type GUR, and measurements were normalized relative to this to combine experiments. Error bars indicate the S.D. of duplicate measurements in two separate experiments. D, C2C12 parental cell line control experiment showing that repression is not caused by the induction ligand RSL. A single experiment conducted in duplicate. Error bars indicate range. E, diagram of reporter constructs used in C and D. The wild-type GUR and permutations containing 3-bp mutations (M) in the GGATTA site were introduced into pGL3-Basic in the appropriate orientation.
Transfection of pGL3-GUR, containing the wild-type 193-bp GUR fragment, led to levels of luciferase activity 50-fold greater than the parental plasmid (Fig. 6, C and D), which contains no promoter or enhancer. However, luciferase levels were ∼10-fold lower than those produced by the transfection of the pGL3-Control plasmid, which contains a strong SV40 viral promoter and enhancer (data not shown). Induction of the exogenous Myc-GTF2IRD1 by the addition of the RheoSwitch ligand (RSL) resulted in a 70% reduction in luciferase activity in cells transfected with pGL3-GUR (Fig. 6C) but had no effect on the expression of the control SV40-luciferase reporter (data not shown). The induction ligand RSL had no independent effect on luciferase levels in parental C2C12 cells transfected with the GUR-luciferase reporters (Fig. 6D). Reporter constructs with mutant GGATTA binding sites showed reduced efficiency of GTF2IRD1 repression. The construct with both flanking sites mutated showed a repression of only 30% (Fig. 6C), thus supporting the findings of the DNA binding studies in the link between the interaction of the protein and its target sequence.
DISCUSSION
In this study, we have shown that the gene GTF2IRD1 is subject to negative autoregulation using a mechanism that involves direct binding of the GTF2IRD1 protein to a highly conserved region (GUR) within the GTF2IRD1 locus. The nature of the binding interaction is unusual, in that it involves the simultaneous binding of at least two separate DNA binding domains to a minimum of two identical GGATTA recognition sequences.
Consequences of Gtf2ird1 Mutation in the Mouse
A number of mutations of the mouse Gtf2ird1 locus now exist and differences in phenotypes are emerging. The KO we have generated involves the targeted deletion of exon 2, which results in the efficient production of a 1–3 spliced mutant transcript that escapes NMD. Translation of an N-terminal truncated form of GTF2IRD1 from this mutant transcript occurs with an estimated efficiency of 3%. Because levels of transcript in KO mice are approximately double wild-type levels, we estimate that the truncated peptide is present at 6% of normal GTF2IRD1 protein levels. The phenotypic consequences of this mutation appear to be very similar to another KO line, in which exons 2–5 are deleted (22), and the Gtf2ird1tm2(LacZ)Hrd mutant line, in which LacZ has been inserted into exon 2 (20). Homozygous null mice from these lines do not have major developmental abnormalities, but show defects in brain function and behavioral alterations (22).5
The random insertion of a c-myc transgene led to the production of another Gtf2ird1 mutant mouse line (Tg(Alb1-Myc)166.8), due to a 40-kb deletion removing exon 1 and the Gtf2ird1 promoter (23). Homozygous null mice of this line have a mild craniofacial abnormality (24) and increased brain ventricle volume (25) but exhaustive behavioral testing was not done. Craniofacial abnormalities have not been observed in the targeted gene KOs, and it is possible that the phenotypic differences are because of genetic background or additional effects on genes adjacent to Gtf2ird1 in the (Tg(Alb1-Myc)166.8) transgenic line due to a disruption of gene regulation resulting from the large 40-kb deletion.
A gene trap mutant was recently reported in which a LacZ-neomycin fusion cassette had inserted into intron 22 of the Gtf2ird1 locus (26). In contrast to the other studies, homozygosity for this mutation was embryonic lethal. The authors argue that alternative splicing and alternative promoter usage ensures the survival of some GTF2IRD1 protein in other Gtf2ird1 mutant lines, thus explaining the less severe phenotype. This explanation is unlikely as transcription of Gtf2ird1 in the (Tg(Alb1-Myc)166.8) mutants was undetectable (24), and analysis of the mutant transcript produced in the Gtf2ird1tm1Hrd mice reported here indicate very low levels of protein synthesis. All of the reported splice isoforms and (putative) alternative promoter-driven transcripts (18, 27) include exons 2 and 3, and would, therefore, be subject to the same translation constraints as the 3α7 isoform used in the above analysis. Therefore, our estimate of 6% total protein synthesis in homozygous Gtf2ird1tm1Hrd mice applies, regardless of alternative splicing possibilities. It is hard to imagine how such a small amount of residual protein is sufficient to explain these significant phenotypic differences. Furthermore, the exon 1 alternatives 1a and 1b (27), and the much more frequently used exon 1, all cluster together within the same 1-kb region of the genome, which would suggest that all transcripts fall under the same regulatory constraints, even if there is some wobble around the transcription start site.
An alternative explanation for the phenotypic difference arises from the fact that the gene-trap strategy does not prevent GTF2IRD1 protein synthesis, but creates a peptide fusion between most of the N-terminal region of GTF2IRD1 (72%) and β-GEO. This fusion protein might retain sufficient GTF2IRD1 peptide to permit interaction with its usual protein partners or DNA targets but the presence of the additional peptide attachment may severely disrupt normal function. This dominant-negative explanation predicts that heterozygous mutants would have more severe consequences than a hemizygous Gtf2ird1 deletion. This does appear to be the case, as some of the gene-trap mutants show more severe abnormalities than another recently reported mouse line in which 7 contiguous genes including Gtf2ird1 are hemizygously deleted (28).
Analysis of GTF2IRD1 Autoregulation in WBS Patients
Analysis of GTF2IRD1 transcript levels in WBS patients is obviously hampered by access to suitable tissue samples. Therefore, most analyses have focused on lymphoblastoid cell lines derived from blood. The results presented here are supported by several other studies (29–31) showing that GTF2IRD1 transcript levels are not significantly different from controls, whereas transcription levels from the flanking genes CYLN2 and GTF2I were 50% of normal, as expected.
In all studies, GTF2IRD1 transcript levels were highly variable between WBS patients and between the controls, suggesting that this is typical behavior and not related to the WBS deletion. However, the evidence for autoregulation in lymphoblastoid cells is, nevertheless, compelling. The degree to which the autoregulation is effective in restoring normal expression levels of GTF2IRD1 protein in the developing affected tissues (e.g. brain) of WBS patients is unknown and would be ethically impossible to ascertain. Therefore, a role for GTF2IRD1 in the cause of WBS due to a slight reduction in expression cannot be ruled out. It might be argued that the autoregulatory mechanism may not operate in all cell types and lymphoblastoid cells are unrepresentative of tissues affected by WBS. In this regard, one analysis shows GTF2IRD1 levels to be significantly reduced in fibroblasts from WBS patients (30). These differences may arise due to the very low levels of GTF2IRD1 expression that would be expected in these cell types, by extrapolation from expression analysis in the mouse (20). All of the tissues tested in the mouse, which were chosen on the basis of moderate to high levels of Gtf2ird1 expression (20), showed similar levels of autoregulation, including the brain. It seems unlikely that such a mechanism, with strong evidence of conservation across all vertebrates, would operate in all of the tissues of one organism but selectively in another species.
Binding of GTF2IRD1 to the GUR
EMSA analysis using in vitro translated human and mouse GTF2IRD1 proteins revealed that all isoforms tested have the ability to bind to the GUR. Furthermore, high affinity binding is dependent on simultaneous interactions of the protein with at least two separate GGATTA recognition sequences. Reduction to a single recognition site leads to loss of binding and separation of the recognition sequences beyond 57 bp also ablates binding. These data are consistent with a model in which the GTF2IRD1 protein interacts in a tandem fashion with dual recognition sequences, and this can be further enhanced by dimeric interactions when three binding sites are present (Fig. 7). However, this model may be oversimplistic as it must be noted that the HC also appears when only two sites are present in the GUR DEL series of probes. This may indicate that DNA sequence outside of the GGATTA core can also influence the conformation of the interaction.
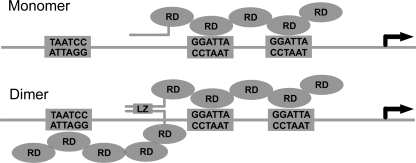
FIGURE 7.
Model of the proposed mechanism for GTF2IRD1 binding to the GUR. EMSA experiments suggest that the middle and proximal GGATTA sites, which are both in the same orientation, are bound in tandem by two RDs of GTF2IRD1. Binding to the middle and distal sites is also possible, but has a lower affinity. Formation of the dimer is dependent on the presence of the leucine zipper domain. The orientation of the protein on the DNA and identification of which binding site the RDs contact is speculative.
Deletion studies and purification of GST fusion peptides has shown that the DNA binding functions of GTF2IRD1 are localized within the repeat domains (RDs). These studies indicate that RD2, 3, 4, and 5 all have DNA binding properties although RD3 lacks sequence specificity (14–16). RD4 has the greatest individual affinity (16) so it is likely that the tandem interaction involves RD4 and one other RD. The evidence presented shows that the predominant human isoform of GTF2IRD1 (1α1) readily forms homodimeric and monomeric interactions with the GUR, whereas homodimeric interactions of the mouse isoforms (3α5 and 3α7) are virtually undetectable. The ability to form dimers could be influenced by sequence outside of the well-conserved leucine zipper domain or RDs. Alternatively, the addition of a sixth RD in rodent species has altered the protein conformation and reduced this capacity. Extending the comparison of binding to GTF2IRD1 from Xenopus and fish species, or examination of mouse splice isoforms that contain 5 RDs would address this question.
GTF2IRD1 has been isolated in four separate yeast one-hybrid studies (10–13) and the bait DNA sequence, containing a single GGATTA, was triplicated in all of them, which is a common strategy to maximize the chance of trapping potential prey proteins. On the basis of work presented here, it is likely that these baits created an artificial selection bias for GTF2IRD1, which shows vastly increased affinity for multiple binding sites. Whereas most studies have concluded that GTF2IRD1 acts as a transcriptional regulator of specific gene targets, future studies should now address whether the affinity of GTF2IRD1 for the single GGATTA site present in the proposed targets, TNNI1 (10, 11), GSC (13), or Hoxc8 (12) is sufficient to be of biological importance.
Evolutionary Considerations
It is worth considering why the GTF2IRD1 feedback mechanism exists and why it has been so faithfully conserved in vertebrates. It cannot have evolved as a dosage compensation mechanism as there would be no requirement under normal circumstances. It might act as a homeostatic mechanism that maintains GTF2IRD1 protein at a set level. However, this would have to be context dependent as transcript levels vary considerably between cell types (20). Furthermore, modeling studies suggest that strong negative autoregulatory circuits increase noise rather than reducing it (32), and this might explain why transcript levels of GTF2IRD1 show high variability.
Alternatively, negative autoregulation circuits can dramatically enhance protein synthesis rise-time to steady state levels (33), and this could provide cells with much greater control over temporal expression. The kinetic behavior of genes involved in transcriptional regulation is an important consideration for the understanding of regulatory gene networks and this may prove to be an essential part of GTF2IRD1 function.
Supplementary Material
Acknowledgments
We thank Richard Harvey and Christine Biben for supplying the Tg(CMV-Cre)1Cgn mice.
This work was supported by a Project Grant 423401 from the Australian National Health and Medical Research Council (NHMRC).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. S1.
M. Howard, E. Tay, S. Palmer, J. Widagdo, J. Hook, F. Lemckert, K. Popovic, P. Gunning, M. Spitzer, M. Murphy, A. Hannan, and E. Hardeman, submitted for publication.
- WBS
- Williams-Beuren syndrome
- WSCP
- Williams syndrome cognitive profile
- SVAS
- supravalvular aortic stenosis
- LCR
- low copy repeat
- EMSA
- electrophoretic mobility shift assay
- GUR
- GTF2IRD1 upstream region
- HC
- higher shift complex
- LC
- lower shift complex
- LZ
- leucine zipper
- RD
- repeat domain
- NMD
- nonsense-mediated decay
- RIPA
- radioimmune precipitation assay buffer
- MOPS
- 4-morpholinepropanesulfonic acid
- EST
- expressed sequence tag
- RSL
- RheoSwitch ligand
- KO
- knock-out.
REFERENCES
- 1.Strømme P., Bjørnstad P. G., Ramstad K. (2002) J. Child Neurol. 17, 269–271 [DOI] [PubMed] [Google Scholar]
- 2.Osborne L. R., Mervis C. B. (2007) Expert. Rev. Mol. Med. 9, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valero M. C., de Luis O., Cruces J., Pérez Jurado L. A. (2000) Genomics 69, 1–13 [DOI] [PubMed] [Google Scholar]
- 4.Curran M. E., Atkinson D. L., Ewart A. K., Morris C. A., Leppert M. F., Keating M. T. (1993) Cell 73, 159–168 [DOI] [PubMed] [Google Scholar]
- 5.Hirota H., Matsuoka R., Chen X. N., Salandanan L. S., Lincoln A., Rose F. E., Sunahara M., Osawa M., Bellugi U., Korenberg J. R. (2003) Genet. Med. 5, 311–321 [DOI] [PubMed] [Google Scholar]
- 6.Karmiloff-Smith A., Grant J., Ewing S., Carette M. J., Metcalfe K., Donnai D., Read A. P., Tassabehji M. (2003) J. Med. Genet. 40, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tassabehji M. (2003) Hum. Mol. Genet. 12, R229–R237 [DOI] [PubMed] [Google Scholar]
- 8.Roy A. L. (2007) Biochim. Biophys. Acta. 1769, 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson P. D., Webb M., Beckett W., Hinsley T., Jowitt T., Sharrocks A. D., Tassabehji M. (2007) FEBS Lett. 581, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 10.O'Mahoney J. V., Guven K. L., Lin J., Joya J. E., Robinson C. S., Wade R. P., Hardeman E. C. (1998) Mol. Cell. Biol. 18, 6641–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo S., Vullhorst D., Venepally P., Cheng J., Karavanova I., Buonanno A. (2001) Mol. Cell. Biol. 21, 8490–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayarsaihan D., Ruddle F. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7342–7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ring C., Ogata S., Meek L., Song J., Ohta T., Miyazono K., Cho K. W. (2002) Genes Dev. 16, 820–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polly P., Haddadi L. M., Issa L. L., Subramaniam N., Palmer S. J., Tay E. S., Hardeman E. C. (2003) J. Biol. Chem. 278, 36603–36610 [DOI] [PubMed] [Google Scholar]
- 15.Vullhorst D., Buonanno A. (2003) J. Biol. Chem. 278, 8370–8379 [DOI] [PubMed] [Google Scholar]
- 16.Vullhorst D., Buonanno A. (2005) J. Biol. Chem. 280, 31722–31731 [DOI] [PubMed] [Google Scholar]
- 17.Schwenk F., Baron U., Rajewsky K. (1995) Nucleic Acids Res. 23, 5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tay E. S., Guven K. L., Subramaniam N., Polly P., Issa L. L., Gunning P. W., Hardeman E. C. (2003) Biochem. J. 374, 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Church G. M., Gilbert W. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer S. J., Tay E. S., Santucci N., Cuc Bach T. T., Hook J., Lemckert F. A., Jamieson R. V., Gunnning P. W., Hardeman E. C. (2007) Gene Expr. Patterns 7, 396–404 [DOI] [PubMed] [Google Scholar]
- 21.Inácio A., Silva A. L., Pinto J., Ji X., Morgado A., Almeida F., Faustino P., Lavinha J., Liebhaber S. A., Romão L. (2004) J. Biol. Chem. 279, 32170–32180 [DOI] [PubMed] [Google Scholar]
- 22.Young E. J., Lipina T., Tam E., Mandel A., Clapcote S. J., Bechard A. R., Chambers J., Mount H. T., Fletcher P. J., Roder J. C., Osborne L. R. (2008) Genes Brain Behav. 7, 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durkin M. E., Keck-Waggoner C. L., Popescu N. C., Thorgeirsson S. S. (2001) Genomics 73, 20–27 [DOI] [PubMed] [Google Scholar]
- 24.Tassabehji M., Hammond P., Karmiloff-Smith A., Thompson P., Thorgeirsson S. S., Durkin M. E., Popescu N. C., Hutton T., Metcalfe K., Rucka A., Stewart H., Read A. P., Maconochie M., Donnai D. (2005) Science 310, 1184–1187 [DOI] [PubMed] [Google Scholar]
- 25.van Hagen J. M., van der Geest J. N., van der Giessen R. S., Lagers-van Haselen G. C., Eussen H. J., Gille J. J., Govaerts L. C., Wouters C. H., de Coo I. F., Hoogenraad C. C., Koekkoek S. K., Frens M. A., van Camp N., van der Linden A., Jansweijer M. C., Thorgeirsson S. S., De Zeeuw C. I. (2007) Neurobiol. Dis. 26, 112–124 [DOI] [PubMed] [Google Scholar]
- 26.Enkhmandakh B., Makeyev A. V., Erdenechimeg L., Ruddle F. H., Chimge N. O., Tussie-Luna M. I., Roy A. L., Bayarsaihan D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makeyev A. V., Bayarsaihan D. (2009) Gene 433, 16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H. H., Roy M., Kuscuoglu U., Spencer C. M., Halm B., Harrison K. C., Bayle J. H., Splendore A., Ding F., Meltzer L. A., Wright E., Paylor R., Deisseroth K., Francke U. (2009) EMBO Mol. Med. 1, 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collette J. C., Chen X. N., Mills D. L., Galaburda A. M., Reiss A. L., Bellugi U., Korenberg J. R. (2009) J. Hum. Genet. 54, 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merla G., Howald C., Henrichsen C. N., Lyle R., Wyss C., Zabot M. T., Antonarakis S. E., Reymond A. (2006) Am. J. Hum. Genet. 79, 332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonell A., Del Campo M., Magano L. F., Kaufmann L., Martinez de la Iglesia J., Gallastegui F., Flores R., Schweigmann U., Fauth C., Kotzot D., Perez-Jurado L. A. (2009) J. Med. Genet., Epub ahead of print [DOI] [PubMed] [Google Scholar]
- 32.Stekel D. J., Jenkins D. J. (2008) BMC Syst. Biol. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenfeld N., Elowitz M. B., Alon U. (2002) J. Mol. Biol. 323, 785–793 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.