Abstract
The archaeal ribosomal stalk complex has been shown to have an apparently conserved functional structure with eukaryotic pentameric stalk complex; it provides access to eukaryotic elongation factors at levels comparable to that of the eukaryotic stalk. The crystal structure of the archaeal heptameric (P0(P1)2(P1)2(P1)2) stalk complex shows that the rRNA anchor protein P0 consists of an N-terminal rRNA-anchoring domain followed by three separated spine helices on which three P1 dimers bind. Based on the structure, we have generated P0 mutants depleted of any binding site(s) for P1 dimer(s). Factor-dependent GTPase assay of such mutants suggested that the first P1 dimer has higher activity than the others. Furthermore, we constructed a model of the archaeal 50 S with stalk complex by superposing the rRNA-anchoring domain of P0 on the archaeal 50 S. This model indicates that the C termini of P1 dimers where translation factors bind are all localized to the region between the stalk base of the 50 S and P0 spine helices. Together with the mutational experiments we infer that the functional significance of multiple copies of P1 is in creating a factor pool within a limited space near the stalk base of the ribosome.
Keywords: Methods/X-ray Crystallography, Protein/Protein-Protein Interactions, Protein/Structure, RNA/Ribosome Function, RNA/Ribosome Structure, Translation/Control
Introduction
Ribosomes from all domains of life contain a highly flexible protuberance, the so-called stalk, at the GTPase-associated center in the large subunits, that participates in the interaction of the ribosomes with GTP-bound translation factors (1, 2). The stalk structure is formed by a set of strongly acidic proteins, together with an anchor protein. Compared with detailed information on the fine structure and function of the complex composed of multicopy stalk protein L12 and the anchor protein L10 in eubacterial ribosomes (3–11), less is known about ribosomal stalk in archaea or in eukaryotes.
Eukaryotic ribosomes contain two types of acidic stalk proteins P1 and P2 (12–14), the amino acid sequences of which show almost no similarity to eubacterial L12 (15). In contrast, the eukaryotic anchor protein P0 shows some amino acid sequence identity with eubacterial L10 (see Fig. 1A (16)). The amino acid sequences of eukaryotic P0 and P1/P2 are much more closely related to their archaeal counterparts (Fig. 1, A and B), although there is a single P1/P2 ortholog in archaea (we refer to these proteins as archaeal P0 and P1, respectively, throughout this report). Evidence from binding experiments indicates that eukaryotic P1 and P2 form a stable heterodimer by the interaction of each N-terminal region (17–22). Two P1–P2 dimers bind through their N-terminal halves to limited regions within the C-terminal half of P0 (23–25). The eukaryotic pentameric complex P0(P1–P2)(P1–P2) binds to the 1070 region of 23 S/28 S rRNA (Escherichia coli numbering is used throughout) with much higher affinity than the E. coli stalk complex (26). On the other hand, archaeal P1 forms a homodimer and three such dimers bind to the C-terminal region of archaeal P0, which includes a sequence homologous to the P1/P2 heterodimer-binding site within eukaryotic P0 (27). The archaeal heptameric stalk complex P0(P1)2(P1)2(P1)2 has also been confirmed by mass spectrometric analyses (27).
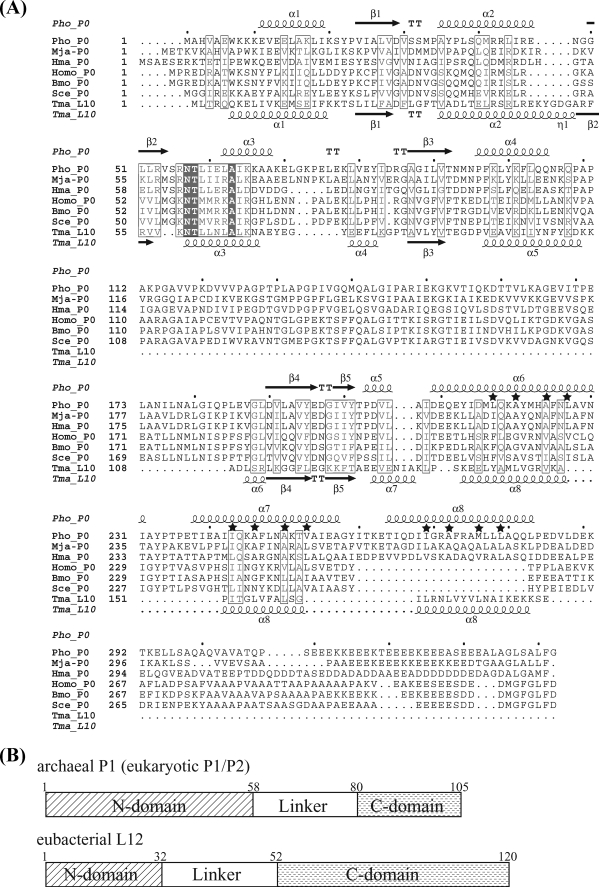
FIGURE 1.
Sequence comparison of the stalk complexes. A, sequence alignment of archaeal/eukaryotic P0 and eubacterial L10. This alignment was prepared using Web service of ClustalW and modified based on the structural comparison of Pho_P0 and Tma_L10. Pho, P. horikoshii OT3; Mja, Methanocaldococcus jannaschii DSM 2661; Hma, H. marismortui; Homo, Homo sapiens; Bmo, Bombyx mori; Sce, S. cerevisiae; and Tma, Thermotoga maritima. The secondary structures of archaeal P0 and eubacterial L10 are indicated above and below the sequences, respectively. The α-helices and β-strands are represented as helices and arrows, respectively, and β-turns are marked TT. Hydrophobic residues important for P1 binding are marked by stars. The figure was prepared using ESPript (available on-line). B, schematic representation of domain organization of archaeal P1, eukaryotic P1/P2, and eubacterial L12 (E. coli).
Functional aspects of the eukaryotic and archaeal stalk complexes in protein synthesis have been successfully investigated by replacements with the L10(L12)2(L12)2 complex of E. coli 50 S subunits and in vitro functional assays (21, 24, 26–28). The experimental evidence indicates that eukaryotic P0(P1–P2)(P1–P2) complex is responsible for ribosomal specificity to eukaryotic translation factors and 10-fold slower GTPase turnover in eukaryotic 80 S ribosome/eEF2 system than that in the eubacterial 70 S ribosome/EF-G system (26). Interestingly, archaeal stalk complex P0(P1)2(P1)2(P1)2 provides access to eukaryotic elongation factors at levels comparable to the eukaryotic stalk complex (28). Therefore, the archaeal heptameric stalk complex and the eukaryotic pentameric complex apparently share conserved functional structures, despite the striking differences between archaeal and eukaryotic stalk dimers, i.e. there are three archaeal homodimers versus two eukaryotic heterodimers. The knowledge of higher resolution structure of the archaeal stalk complex seems to be able to provide a useful clue for elucidation of the molecular details of the eukaryotic as well as archaeal ribosomal GTPase-associated center.
Here, we present the crystal structure of the stalk complex of the thermophilic archaea Pyrococcus horikoshii. The solved heptameric P0(P1)2(P1)2(P1)2 structure shows characteristics distinct from the stalk complex L10(L12)2(L12)2(L12)2, of a thermophilic eubacterial species, reported by Diaconu et al. (9). Based on the crystal structure, we successfully generated amino acid substitution mutants of P0, which disrupted P1 binding to each of the three binding sites. Furthermore, we constructed a model of 50 S ribosome with stalk complex by superposing the N-terminal domain of P0 of P0(P1)2(P1)2(P1)2 on the 50 S subunit. Combining the superposed model with functional analysis of these mutants, we discuss how three P1 dimers work together in protein synthesis.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The coding regions for Pyrococcus horikoshii ribosomal protein P0, P1, and L11 were inserted into the E. coli expression vectors pET28c or pET3a (Novagen) and cloned, as described previously (28). The plasmids for P0ΔC58 (lacking the C-terminal region encoding residue 285–342) of P0 and P1ΔC50 (encoding residue 1–58) of P1 were constructed by introducing a stop codon after codon 284 in the P0 expression plasmid and a stop codon after codon 58 in the P1 expression plasmid, respectively, using a QuikChangeTM site-directed mutagenesis kit according to the manufacturer's protocol. The DNA fragment for P0(Δ111–180), in which the region for residues 111–180 of P0 was truncated, was amplified by nested PCR method (29) and inserted into pET28c (Novagen). The resultant plasmid was used to make the plasmid for P0(Δ111–180)ΔC58 (encoding residues 1–110 fused with residues 181–284 of P0) by introducing a stop codon after codon 58 as described above. All plasmids for point mutants of P0 were also constructed by using the QuikChangeTM method as described above using the plasmid encoding full-length P1 as the template.
Protein Preparation
Individual plasmids were transformed into E. coli BL21 (DE3) codon plus, and the P. horikoshii ribosomal proteins were expressed in E. coli cells. After heat treatment of the cell extracts at 70 °C for 30 min to remove the endogenous E. coli proteins, the expressed proteins were purified by anion- or cation-exchange chromatography under denaturing conditions with 6 m urea, as described previously (28). The P0–P1 complex was usually reconstituted by mixing of isolated P0 and P1 samples in the presence of 6 m urea at a molar ratio of 1:8 and then by renaturation, as described previously (28). The complex sample for crystallization was prepared as follows. E. coli cells expressing P0ΔC58 and P1ΔC50 were mixed, suspended in phosphate-buffered saline containing 1% Triton X-100, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride, and then lysed by sonication. The soluble fraction of the lysate was heated at 70 °C for 30 min to remove the endogenous E. coli proteins and to reconstitute the P0ΔC58-P1ΔC50 complex. The protein complex sample formed was dialyzed overnight against buffer B (1 mm dithiothreitol, 20 mm Tris-HCl, pH 8.0) and applied to a HiTrap QXL column (Amersham Biosciences) pre-equilibrated with buffer B. The P0ΔC58-P1ΔC50 complex was eluted with a linear gradient of 0–1 m NaCl. The complex fraction was further purified with a HiTrap Heparin HP column (Amersham Biosciences) using the same linear gradient. The isolated complex was finally purified with a HiLoad 26/60 Superdex 200-pg column (Amersham Biosciences) in buffer C (100 mm KCl, 1 mm dithiothreitol, 20 mm Tris-HCl, pH 7.6). Expression of the selenomethioning-containing P0ΔC58 was performed in E. coli B834(DE3) codon plus. Purification of P0ΔC58-P1ΔC50 complex containing selenomethionine derivative was carried out as described above, except that all buffer used contained 10 mm dithiothreitol.
Crystallization and Data Collection
Initial crystallization trials were performed by using the sitting drop, vapor-diffusion method using a series of crystallization kits produced by Hampton Research (Laguna Niguel, CA) and Emerald BioSystems (Bainbridge Island, WA). Each drop was prepared by mixing 1.0 μl of protein solution with the same volume of reservoir solution. The crystals of native and selenomethionine derivative appeared under condition number 11 of MembFac (100 mm magnesium chloride hexahydrate, 0.1 m sodium acetate trihydrate, pH 4.6, 12% (w/v) polyethylene glycol 6000). After optimization of the crystallization conditions (pH, precipitant content and concentration, and protein concentration), the best crystals were obtained against 0.1 m MgCl2, 0.1 m sodium acetate trihydrate, pH 4.2–4.4, 6–10% polyethylene glycol 6000 at 20 °C within a few days.
The Se-SAD data of the present complex were collected at 2.35-Å resolution at BL38B1 using wavelength of 0.98914 Å based on the fluorescence spectrum of the selenium atom, while the 2.13-Å resolution native data were collected at BL41XU at SPring-8 using a wavelength of 1.0 Å. All data sets were collected under 100 K after crystals were soaked in reservoir solution containing an additional 15% glycerol and flash-cooled under a stream of liquid nitrogen. The crystal of P0ΔC58-P1ΔC50 complex belongs to the space group P212121 with one P0ΔC58-P1ΔC50 molecule in the asymmetric unit corresponding to the VM value of 2.2 Å3Da−1. All data sets were indexed, integrated, scaled, and merged using the HKL2000 program (30). Statistics are shown in Table 1.
TABLE 1.
Statistics of data collection and refinement
Values in parentheses refer to the highest resolution shell.
| Native | Selenomethionine | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | |
| Unit cell (Å) | ||
| a, b, c (Å) | 54.9, 104.4, 136.2 | 55.1, 104.3, 135.7 |
| Resolution (Å) | 50.0-2.13 (2.21-2.13) | 50.0-2.35 (2.43-2.35) |
| Completeness (%) | 99.1 (93.0) | 99.2 (93.7) |
| Average redundancy | 6.8 (4.7) | 4.4 (3.4) |
| I/σ(I) | 12.4 (2.0) | 10.0 (0.97) |
| Rsym (%) | 10.6 (40.8) | 6.2 (47.1) |
| Refinement | ||
| Resolution range (Å) | 35–2.13 | |
| Number of used reflections | 44,207 | |
| Completeness (%) | 99.04 | |
| R-factor (%) | 22.0 | |
| Rfree-factor (%)a | 26.0 | |
| Total number of nonhydrogen atoms | ||
| Protein | 4,298 (1,676 + 437 × 6 molecules) | |
| Water | 440 | |
| Averaged B factors (Å2) | ||
| Protein | 44.3 | |
| Water | 55.5 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.012 | |
| Bond angles (°) | 1.552 | |
a Rfree-factor value was calculated for R-factor, using a subset (7.1%) of reflections that were not used for refinement.
Structure Determination and Refinement
Using Se-SAD data, six of the eight selenium sites were located using the program SHELXD (31), and the phases were re-calculated with the program SOLVE (32). After phase improvement, the initial model was built to ∼73% by the RESOLVE (33, 34). The additional model building and refinement were carried out semi-automatically using LAFIRE (35) with CNS (36) on the native data. After several iterations of LAFIRE, the model was refined to an R factor of 31.6% at the resolution range 20.0–2.13 Å. After manual model check and fit with COOT (37), water molecules were located, and the model was finally refined to an R/Rfree factor of 22.0/26.0% using LAFIRE with REFMAC5 (38), and TLS refinement (39) was used at the final step. The stereochemical quality was analyzed using the program PROCHECK (40). The final model has 92.2%, 6.6%, and 1.0% of the residues (non-Gly and non-Pro) in the most favored, allowed, and generously allowed regions, respectively, and only one residue fell in a disallowed region. The refinement statistics for the structure are listed in Table 1. The final atomic coordinates are deposited in the Protein Data Bank under the accession numbers 3A1Y.
Functional Assays
Ribosomal 50 S subunits were prepared from the L11-deficient E. coli strain AM68 (41), as described previously (42). The 50 S core particles deficient in both L10–L12 complex and L11 were prepared by extraction of the L11-deficient 50 S subunits in a solution containing 50% ethanol and 0.5 m NH4Cl at 0 °C, as described previously (42). Eukaryotic elongation factors eEF1α and eEF2 were isolated from pig liver as described before (43). The P0–P1 complex variants composed of truncation mutants of P0 and P1 were mixed with 50 S cores, together with P. horikoshii L11, to construct the hybrid 50 S particles, as described previously (28). The eEF2-dependent GTPase activity and eEF1α/eEF2-dependent polyphenylalanine synthesis were assayed as described in our previous reports (28).
eEF2 Binding Assay
A mixture (50 ml) containing 30 pmol of 50 S subunit cores, 90 pmol of 30 S subunits, 90 pmol of L11, 120 pmol of P0–P1 complex variants, 90 pmol of pig eEF2, 500 mm GMPPNP,4 10 mm MgCl2, 50 mm NH4Cl, 5 mm 2-mercaptoethanol, and 20 mm Tris-HCl (pH 7.6) was incubated at 37 °C for 10 min. This reaction mixture was loaded onto a 20% sucrose cushion in buffer containing 10 mm MgCl2, 50 mm NH4Cl, 5 mm 2-mercaptoethanol, and 20 mm Tris-HCl (pH 7.6). The ribosome·eEF2·GMPPNP complex was pelleted by ultracentrifugation at 60,000 rpm and 4 °C for 3 h using an Hitachi S100-AT4 rotor. The pellet was dissolved in a small volume of buffer consisting of 10 mm MgCl2, 50 mm NH4Cl, 5 mm 2-mercaptoethanol, and 20 mm Tris-HCl (pH 7.6). The eEF2 factor cosedimented with ribosomes (0.5 A260 unit) was analyzed by SDS-PAGE, followed by staining with Coomassie Brilliant Blue.
RESULTS
Overall Structure of Archaeal Stalk Complex
We expressed P0 and P1 from the hyperthermophilic archaeon Pyrococcus horikoshii separately and reconstructed the stalk complex. Diffraction-quality crystals were obtained after truncation of the C-terminal 58 amino acids of P0 (P0ΔC58, residues 1–284) and 50 amino acids of P1 (P1ΔC50, residues 1–58). We refer to these truncation variants as P0 and P1 hereafter where no conflict is expected. The structure was solved by a single-wavelength diffraction method using selenium atoms. The asymmetric unit contained one stalk complex composed of one P0 and three dimers of P1: P0(P1)2(P1)2(P1)2. All except the insertion region (residues 111–180) specific for archaeal and eukaryotic P0 (described later) were built based on the electron density map. Fig. 2A shows the overall structure of the archaeal stalk complex determined at 2.1-Å resolution. The general features of the complex are similar to the eubacterial stalk complex (9), in that it is a banana-shaped molecule consisting of the P0 N-terminal rRNA-anchoring domain followed by a long helical spine on which three P1 dimers are bound. However, the two complexes also differ significantly in many respects.
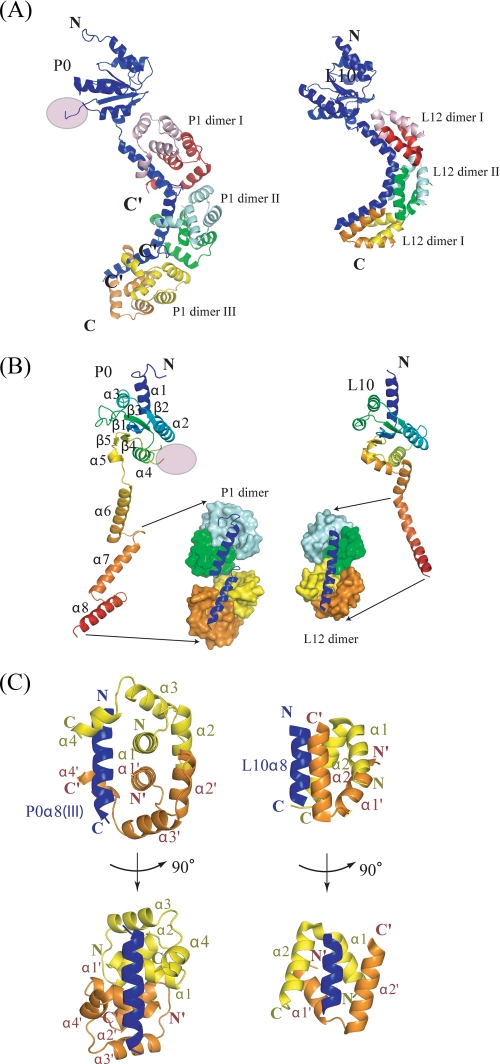
FIGURE 2.
Structural comparison between archaeal (left) and eubacterial (right) stalk complex. A, ribbon representation of the overall structure of the archaeal (left) and eubacterial (right) stalk complex showing the N-domain at the top. The disordered insertion region is shown as a pink circle. P0 (or L10) is colored blue, while three P1 (or L12) dimers bound to the spine helices are colored pink/red, cyan/green, and yellow/orange, respectively. The truncated C termini of all P1 dimers are indicated by C′. B, rainbow representation of P0 (left) and L10 (right) showing their N-terminal domains have a similar fold except for the disordered insertion region. P1 dimers (or L12 dimers) bound to the second and third spine helices (the first are not depicted for clarity) are shown as a space-filling model. C, orthogonal view of the interactions between P0 and P1 dimer (left), and L10 and L12 dimer (right). The secondary structures are labeled in the same color as in the structure, where the prime symbol refers to the second monomer. All structure figures were prepared with the program PyMOL (DeLano Scientific LLC, San Carlos, CA).
P0 Structure
P0ΔC58 is composed of an N-terminal rRNA-anchoring domain (residues 1–206), a short linker (residues 207–208), and a helical spine (residues 209–282). The N-terminal domain is an α/β structure with a five-stranded anti-parallel β-sheet surrounded by five helices. Based on the tertiary structure obtained, we aligned the primary structures of eukaryotic/archaeal P0 and eubacterial L10 as shown in Fig. 1A, which clearly identifies a 70-residue insertion (residues 111–180) specific for archaea and eukarya within the N-terminal domain. This insertion region corresponds to the disordered region in the present crystal. With the exception of this insertion, the N-terminal domains of P0 (or L10) from all three domains of life are highly similar (Figs. 1A and 2B). This structural similarity of the N-terminal domain is consistent with the results of previous studies indicating that the archaeal or eukaryotic stalk complex binds to the E. coli 50 S core, which lacks the L10(L12)2(L12)2 complex (26, 28).
In contrast to the structural similarity of the N-terminal domain, the helical spine of archaeal P0 has different structural characteristics from that of eubacterial L10; unlike eubacterial L10, the spine of which consists of one curved helix with two kinks, the archaeal spine is composed of three independent α-helices (α6: Glu209–Ile231, α7: Thr238–Ala257, α8: Thr264–Gln282) connected with short (6 residues) linkers (Fig. 2B). Hereafter, we refer to these as helices I, II, and III, respectively. The first helix is three residues longer than the others, the latter of which consists of 20 residues (30 Å). Thus, the overall length of the archaeal spine (∼75 residues) is approximately twice as large as that of eubacteria (∼38 residues). Furthermore, archaeal/eukaryote P0 has a 50-residue extension at the C terminus (Fig. 1A), which was truncated for crystallization in the present study.
Each of these α-helices of the P0 spine is associated with one P1 dimer (Figs. 2, A–C). As three binding helices of the P0 spine are spaced by the 6-residue linkers, the contact between bound P1 dimers is loose (only 1.3% of the accessible surface area on each monomer is buried), in comparison with the packing of L12 dimers on the eubacterial spine (4.6% of the accessible surface per dimer is buried) (Fig. 2B).
P1 Structure
The N-terminal domain of archaeal P1 (P1ΔC50) solved in the present study is composed of four α-helices: α1 (Met1–Val13), α2 (Glu19–Ala29), α3 (Glu35–Glu46), and α4 (Asn49–Lys56) (Fig. 2C). The hydrophobic interactions within α1 and α2 pairs of archaeal P1 are responsible for the P1–P1 dimerization, which is formed by two stacked pairs of anti-parallel helices α1–α1′ and α2–α2′ (Fig. 2C). Upon dimerization, ∼17% of accessible surface area on each monomer is buried. These structural features are very different from the eubacterial counterpart L12 in which much smaller domains each consisting of two α-helices form dimers (Figs. 1B and 2C). The lack of a structural relationship is consistent with the observation that there is no sequence homology between archaeal P1 and eubacterial L12. A DALI search (44) using both monomers and dimers of P1 showed that the Z-scores of the largest structural similarity were less than 4.5, indicating that the N-terminal domain of archaeal P1 is unique in both protein fold and in dimerization mode.
Interaction between P0 and P1
Consistent with the results of previous biochemical analyses, three P1 dimers are bound to the three helices of the spine to form a stalk complex. Although the eubacterial helical spine is accommodated by the hydrophobic cavity created by a pair of parallel oriented helices (α2–α2′) of L12 dimers, the archaeal P0 spine helices are placed on the strands composed of helices α1 and α1′ of P1 dimers and are clasped by the helical arms of α4 and α4′ (Fig. 2C).
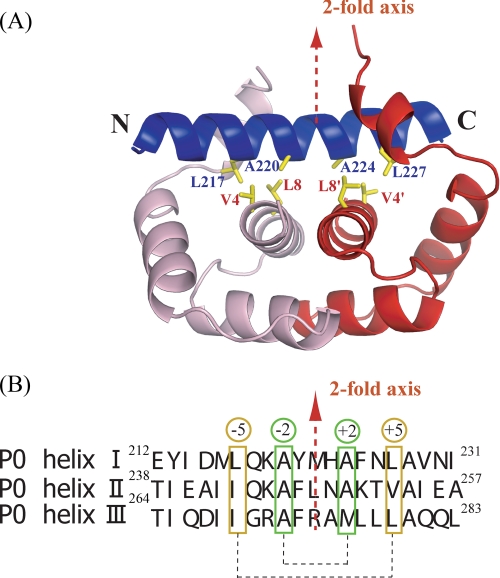
Fig. 3A shows a P1 dimer bound to a P0 helix viewed from the direction perpendicular to both P0 helix and P1 dyad axes. As the 2-fold symmetry axis of the P1 dimer runs almost perpendicularly to each helical axis of the P0 spine, we expected that the amino acid residues of P0 important for P1 binding are also distributed with this symmetry on the α-helices despite the intrinsic polar nature of the α-helix. Fig. 3B shows sequence alignment of the three α-helices of the P0 spine based on the structure in complex with P1. Although the degree of sequence similarity is not high, consensus residues consistent with the 2-fold symmetry are readily detectable. Most important are alanyl residues positioned at ±2 from the center of each helix (Fig. 3). Hydrophobic residues at ±5 from the center of each helix are also important. These residues are Leu217, Ala220, Ala224, and Leu227 on helix I, Ile243, Ala246, Ala250, and Val253 on helix II, and Ile269, Ala272, Met276, and Leu279 on helix III. They make hydrophobic interactions with Val4 and Leu8 of P1. Such 2-fold distribution of the hydrophobic residues important for the interactions is also seen, albeit less obvious at ±1,2 positions in eubacterial L10 (Fig. 1A).
FIGURE 3.
Interactions between P0 spine helix and P1 dimer. A, close-up view of P0 spine helix I (blue) and first P1 dimer (pink/red). The hydrophobic residues important for binding are shown as sticks. The 2-fold axis of the P1 dimer is shown as a red broken line. B, the sequences of three spine helices. The hydrophobic residues important for the binding of P1 dimers are marked in green and orange boxes. They are positioned at ±2 and ±5 from the 2-fold dimer axis.
Construction of P0 Mutants Depleted of Any Binding Site(s) for P1 Dimers
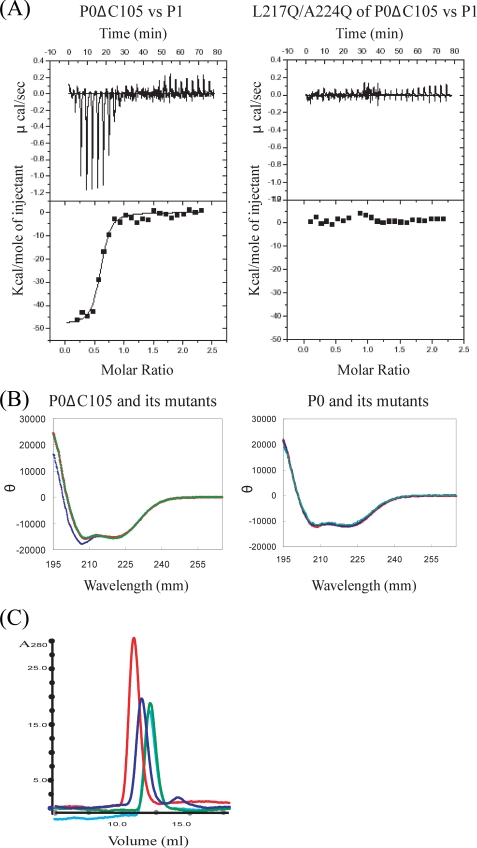
Based on these structural features, we constructed P0 mutants each of the P1-binding sites of which was disrupted individually. For this purpose, we first prepared the deletion mutant P0ΔC105 of which only the first P1-binding helix (helix I) remained intact (27). Then, we generated a double mutation variant L217Q/A224Q and quadruple mutation variant L217Q/A220Q/A224Q/L227Q in which the important hydrophobic residues for P1 binding were substituted by glutamine. The isothermal titration calorimetry measurement showed both variants had lost the P1-binding ability (Fig. 4A). Subsequent CD measurements of these variants confirmed the structural integrity for the former but not for the latter (Fig. 4B). Therefore, for the further experiments we used the double mutation variants. The gel-filtration experiment and native electrophoresis showed that each of the following double mutation variants actually lost P1 binding site of spine helices, I (L217Q/A224Q), II (I243Q/A250Q), and III (A272Q/L279Q), respectively (Fig. 4C). Therefore, we have succeeded in generating P0 mutants depleted of any binding site(s) on any helices. These mutants allow us to study the individual role of each P1 dimer in factor binding within the ribosome.
FIGURE 4.
The isothermal titration calorimetry measurement, CD spectra, and gel-filtration experiment. A, representative plots from isothermal titration calorimetry experiments of interaction between P0 mutants and P1 at 70 °C are illustrated with raw data in the upper panel and fitting curves (continuous lines) in the lower panel. The concentrations of P0 mutants and P1 dimer were 0.0085 and 0.1 mm, respectively. B, CD spectrum of P0DC105, double mutation variant L217Q/A224Q of P0ΔC105 and quadruple mutation variant L217Q/A220Q/A224Q/L227Q of P0ΔC105 are shown as red, green, and blue lines, respectively (left). CD spectrum of P0, mutation variant P0Mt(I, II) and P0Mt(I, III) are shown as red, green, and blue lines, respectively (right). C, the result of the gel-filtration experiment for stalk complexes. The plots of P0(P1)2(P1)2(P1)2, P0Mt(I)(P1)2II(P1)2IIII, P0Mt(I, III)(P1)2II, and P0Mt(I, II)(P1)2III are shown as red, blue, light blue, and green lines, respectively.
Functional Contributions of Individual P1 Dimers on the Spine
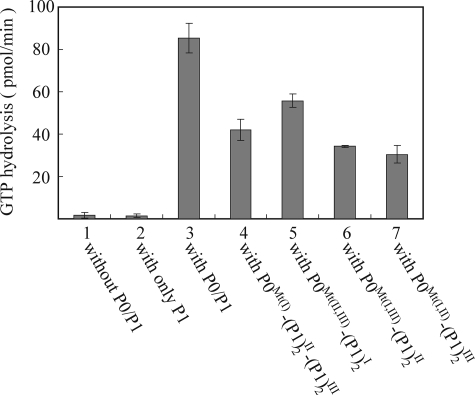
Functionality of the archaeal stalk complexes used in the present study was examined by replacement with E. coli L10·L12 stalk complex on the 50 S subunits in vitro, as described previously (28). Accessibility of the resultant hybrid ribosomes to elongation factor eEF2 was evaluated by eEF2-dependent GTPase activity (Fig. 5). As a positive control, the hybrid ribosome carrying intact P0 (P0WT) and P1 (column 3 in Fig. 5) showed activity at levels comparable to those reported previously (27, 28). The activity with P0Mt(II, III) (column 5 in Fig. 5), which binds P1 only at helix I, was 65% that of the control, whereas that with P0Mt(I, III) (column 6 in Fig. 5) and P0Mt(I, II) (column 7 in Fig. 5), which bind P1 only at helix II and III, respectively, was 35–40% of the control. The activity with P0Mt(I), which binds P1 at both helices II and III, was 49% of the control. These results indicated that each P1 dimer contributes to the eEF2-dependent GTPase activity at least partially. It is interesting that P1 dimer at helix I showed obviously higher activity than that at helices II and III.
FIGURE 5.
Contribution of individual P1 stalk dimers to eEF2-dependent GTPase activity. E. coli 50 S core (2.5 pmol) was preincubated without P0/P1 (column 1) or with only P1 (80 pmol) (column 2). The same core was also incubated with 10 pmol of the following complexes: column 3, intact P0(P1)2I(P1)2II(P1)2III complex reconstituted from purified P0 and P1; column 4, the complex formed with the P0 mutant (L217Q/A224Q), P0Wt(I)(P1)2II(P1)2III; column 5, the complex formed with the P0 mutant (I243Q/A250Q/A272Q/L279Q), P0Wt(II,III)(P1)2I; column 6, the complex with the P0 mutant (L217Q/A224Q/A272Q/L279Q), P0Wt(I,III)(P1)2II; column 7, the complex formed with the P0 mutant (L217Q/A224Q/I243Q/A250Q), P0Wt(I,II)(P1)2III. These hybrid samples were then assayed for eukaryotic eEF2-dependent GTPase activity in the presence of PhL11 and E. coli 30 S subunits (7.5 pmol). The superscripts of P1 indicate the bound position on spine of P0.
Structure of Stalk Complex on 50 S Ribosome
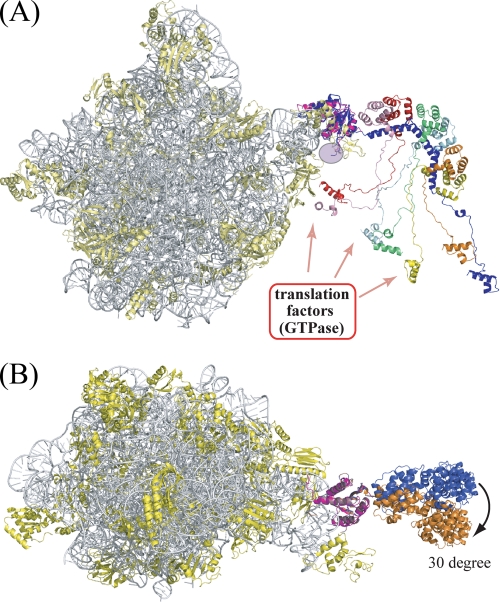
The placement of stalk complex on the 50 S ribosomal subunit is the key to address how the stalk complex exerts its function of recruiting/releasing translation factors to/from the Sarcin/Ricin loop of the ribosome during protein synthesis. The most recent structure of the 50 S ribosome subunit from Haloarcula marismortui (hereafter, Hm50S) showed part of the N-terminal rRNA-anchoring domain of P0 (hereafter HmP0) (PDB ID: 2QA4) (45). The P0 from Pyrococcus horikoshii (hereafter, PhP0) showed a high degree of similarity to HmP0 in both sequence and structure with 34% identical residues and root mean square difference of 0.44 Å for 102 residues (PhP0: residues 3–105, HmP0: residues 5–107) of the N-terminal domain, respectively. Therefore, we were able to place the present complex structure on the Hm50S subunit model by superposing the N-terminal domain of PhP0 onto that of HmP0 as shown in Fig. 6A. The model of 50 S with stalk complex showed that the spine is curved toward the stalk base area, including the Sarcin/Ricin loop region, which constitutes the factor binding site in the ribosome. The N-terminal domains of P1 dimers mounted on the spine helices from the outer side of the spine curve with all C termini oriented toward the inner side of the spine curve (Fig. 6A). Moreover, 2Fo − Fc and Fo − Fc maps of Hm50S subunit calculated using published structure factor data and coordinates showed the density blobs not occupied by either model of Hm50S subunit or superposed P0(P1)2(P1)2(P1)2 at around the regions close to the location of the first P1 dimer. Subsequently, we fitted the spine with P1 dimer into map blobs by changing only the conformation of the short linker bridging N-terminal rRNA-anchoring domain and the spine helices, without disrupting hydrophobic contacts between the N-terminal domain of P0 and the P1 dimer at spine helix I. The spine of the resulting P0 model was rotated about 30° along the anteroposterior axis (Fig. 6B), suggesting that the spine of P0 carrying P1 is flexibly connected to the N-terminal domain with limited movement in a restricted direction.
FIGURE 6.
Stalk complex model superposed on the H. marismortui 50 S subunit. The 23 S rRNA and ribosomal proteins of the H. marismortui 50 S subunit (PDB ID: 2QA4) are indicated as gray and yellow ribbons, respectively, while the N-terminal domain of HmP0 is colored purple. A, a side view of the 50 S ribosome carrying the stalk complex. The stalk complex is colored as in Fig. 2A. This figure includes all the truncated C-terminal parts of P0 and P1 to emphasize that they are all in a limited space near the stalk base of the ribosome (see “Discussion”). B, top view of the 50 S subunit carrying stalk complex in two positions. The stalk complex colored blue is the same model as in A, whereas the stalk complex colored orange is a model that fits into the residual electron density of the H. marismortui 50 S subunit.
Roles of the Domains Specific for Archaeal/Eukaryote P0 and P1
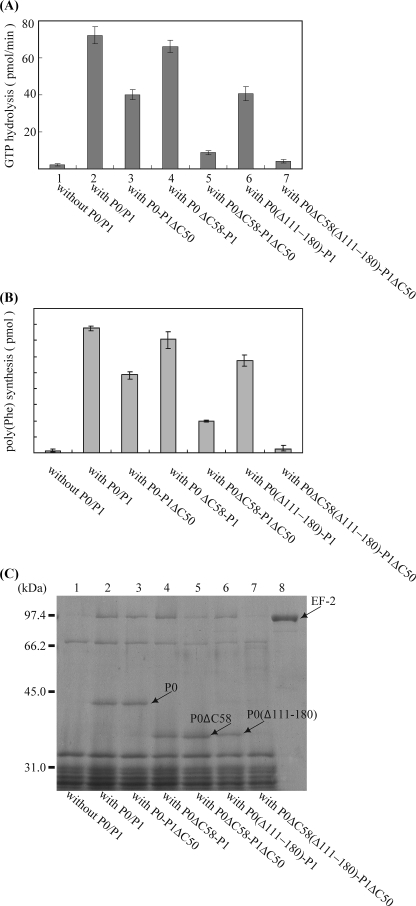
The results of amino acid sequence analysis showed that the C-terminal region of P0 is specific for archaea and eukarya and has some similarity to the C-terminal region of P1 (16). Interestingly, we also discovered that the disordered region (residues 111–180) of P0 is another specific domain for archaeal/eukaryotic P0. To address how these regions affect the function of the stalk complex, we constructed the following deletion mutants of P0: P0ΔC58 lacking the archaea/eukarya-specific C-terminal region, P0(Δ111–180), which lacks the internal disordered region, and P0(Δ111–180)ΔC58, which lacks both the C-terminal and internal regions. The stalk complexes were reconstituted with these P0 samples, together with P1 or P1ΔC50, and tested for their functional activity by using the hybrid ribosome system, as shown in Fig. 5. The activity was evaluated by assays for eEF-2-dependent GTPase (Fig. 7A), eEF1α/eEF2-dependent polyphenylalanine synthesis (Fig. 7B) (27, 28), and eEF2 binding to the hybrid ribosomes (Fig. 7C). Deletion of the C-terminal region of P0 showed only slight effects (column 4 in Fig. 7 (A and B) and lane 4 of Fig. 7C), whereas deletion of the C-terminal region of six copies of P1 had larger effects on GTPase activity (53% of WT), polyphenylalanine synthesis (60% of WT), and eEF2 binding (column 3 in Fig. 7 (A and B) and lane 3 of Fig. 7C). Deletion of the internal disorder region also gave a partial effect on GTPase activity (55% of WT), polyphenylalanine synthesis (75% of WT), and also in eEF2 binding (column 6 in Fig. 7 (A and B) and lane 6 of Fig. 7C). By deletions of the C-terminal regions of both P0 and P1, the activity was considerably reduced not only in GTPase (12% of WT) and polyphenylalanine synthesis (25% of WT), but also in eEF2 binding (column 5 in Fig. 7 (A and B) and lane 5 of Fig. 7C). The stalk complex with the triple truncation P0(Δ111–180)ΔC58-P1ΔC50 showed almost no activity in factor binding, GTPase, or polyphenylalanine synthesis (column 7 in Fig. 7 (A and B) and lane 7 of Fig. 7C).
FIGURE 7.
Functional contributions of the C-terminal regions of P1/P0 and of the insertion domain of P0. A, effects of the truncation mutations of the stalk complex on eEF2-dependent GTPase. E. coli 50 S core (2.5 pmol) was preincubated without stalk complex (column 1) or with 20 pmol of the following complexes: column 2, P0–P1 (P0: wild type; P1: wild-type); column 3, P0–P1ΔC50; column 4, P0ΔC58-P1; column 5, P0ΔC58-P1ΔC50; column 6, P0(Δ111–180)-P1; and column 7, P0(Δ111–180)ΔC58-P1ΔC50. The resultant particles were then assayed for eukaryotic eEF2-dependent GTPase activity in the presence of PhL11 (7.5 pmol) and E. coli 30 S subunits (7.5 pmol). B, the same samples (10 pmol of core) as in A were assayed for eukaryotic eEF1α- and eEF2-dependent poly(U)-directed polyphenylalanine synthesis in the hybrid ribosome system (42). C, effects of the truncation mutations of the stalk complex on eEF2 binding. E. coli 50 S core (30 pmol) was preincubated without any stalk complex (lane 1) or with 120 pmol of the following complexes: lane 2, P0–P1; lane 3, P0–P1ΔC50; lane 4, P0ΔC58-P1; lane 5, P0ΔC58-P1ΔC50; lane 6, P0(Δ111–180)-P1; and lane 7, P0(Δ111–180)ΔC58-P1ΔC50. The resultant particles were then incubated with eukaryotic eEF2 (lane 8, eEF2 alone) and GMPPNP, together with PhL11 (90 pmol) and E. coli 30 S subunits (90 pmol), and the eEF2·ribosome complexes were recovered by ultracentrifugation. A given amount of each complex was analyzed by SDS-PAGE. The gels were stained with Coomassie Brilliant Blue.
DISCUSSION
Archaeal P0 and Eubacterial L10
The ribosome stalk complex is conserved in all three domains of life and plays a crucial role in the ribosome-mediated stimulation of translation factor-dependent GTP hydrolysis. In the present study, we determined the first structure of archaeal stalk complex. The structure was compared with the eubacterial stalk complex determined previously (9). The archaeal stalk complex exhibits features in common with eubacterial stalk complex in terms of the N-terminal rRNA-anchoring domain followed by the C-terminal spine helices to which P1 (or L12 in eubacteria) dimers bind. However, archaeal P0 spine is approximately twice as large as that of eubacterial L10 to allow binding of much larger P1 dimers in a mode different from eubacterial L12. Archaea and eubacteria have presumably evolved their P0 (L10) spines to recruit P1 or L12 dimers of similar function with different structures.
Archaeal P1 and Eubacterial L12
Archaeal P1 (or eukaryotic P1–P2) shows no sequence similarity with the eubacterial counterpart, L12. The results of the present study indicated that the tertiary structures of these two proteins are also unrelated. Despite lack of an evolutionary relationship between these two proteins, they have general features in common probably as a result of convergent evolution. Both P1 and L12 have an N-terminal domain that binds to α-helical spine by the hydrophobic interactions after dimerization. They also have a C-terminal functional domain in common, and these two domains are connected through a long hydrophobic linker. In the present study, we truncated the C-terminal 50 residues of P1. Secondary structural prediction suggested that the C-terminal region (∼20–25 residues) of archaeal P1 forms a two-helix bundle, and the rest (∼25–30 residues) is a conserved hydrophobic linker. On the other hand, a previous experiment showed that the C-terminal domain of eubacterial L12 is much larger (∼70 residues). Thus, P1 and L12 have a similar domain organization (base, arm, and hand domains) but with very different domain sizes (Fig. 1B).
Implementation of the Eukaryotic Stalk
Sequence alignment showed that eukaryotic P0 has similar features to archaeal P0, including the N-terminal insertion domain and C-terminal extension domain. Most of the structural features obtained in the present study of the archaeal stalk should be applicable to that of eukaryotic complex except that the latter spine is composed of two helices rather than three (27) (Fig. 1A). Similarly, the eukaryotic P1–P2 dimer is structurally homologous to the archaeal P1 dimer even though P1–P2 forms a heterodimer. It is likely that the eukaryotic P0 and P1–P2 dimers interact with each other in a similar mode as that of archaeal P0–P1 interaction. Based on the 2-fold symmetry of the P1 homodimer on archaeal P0 spine, we identified the pseudosymmetry arrangement of hydrophobic residues important for P1 binding. This pseudo dyad axis is not obvious in the eukaryotic P0 sequence. However, the hydrophobic residues important for P1/P2 binding are detectable (Fig. 1A). It seems that compensatory changes of amino residues occurred in the P0 sequence to interact with asymmetrical P1–P2 heterodimer.
Role of Insertion Domain
The results of the present study indicated that archaeal and eukaryotic P0 have an insertion (residues 111–180) at the N-terminal rRNA-anchoring domain, which is not conserved in eubacterial L10. Although this domain was not constructed in the present structure, the model of stalk complex on the 50 S ribosome suggests that this domain is located between the N-terminal domain of L11 and H42/H43/H44 of 23 S rRNA, suggesting that this domain may function by interacting with these regions. The present results on eEF2-dependent GTPase assay and eEF2 recruitment using the mutant P0(Δ111–180) suggest that this archaeal/eukaryotic-specific domain is a helper for eEF2-dependent GTPase turnover. This is supported by the observation in the cryoelectron microscopy structure of 80S·eEF2 complex that eEF2 interacts with this stalk base area (46). This is also consistent with the fact that the insertion domain carries the mutations in Saccharomyces cerevisiae mutants resistant to the antifungal compound sordarin, which acts on eEF2 (47, 48).
Multiple Copies of P1 Dimers and Factor Recruitment
What is the biological significance of the multiple copies of P1 dimers on the P0 spine? Are there any distinct roles for each dimer? To answer these questions, a system that allows measurement of the individual activity of each P1 dimer is necessary. Based on the structure, we have successfully developed such a system and measured the activities of individual P1 dimers bound at three positions. These studies showed that P1 dimers at any position can function in the absence of the others. The experiment also showed that the first P1 on helix I is more active than the others. There are two possible explanations for these observations. First, position 1 may be more favored than the others for the function. Second, the P1 dimer bound at the first position may have some additional role other than simply recruiting/releasing the factors. Inspection of the structure suggested that the P1 dimers bound at the first position seem to keep the P0 spine at the optimum orientation by direct interaction with N-terminal rRNA-anchoring domain.
The rate of factor binding to the ribosome is much faster than expected from simple diffusion. In eubacteria, it has been speculated that the unusually fast rate is due to the flexibility at the hinge region connecting L10 base and L10 spine as well as at the L12 linkers, which allows L12 to recruit factors from a wide region (9). The present structure superposed on the Hm50S structure suggested that some type of flexibility is expected at the hinge region of P0, but the movement seems to be within a limited range. In addition, such a complex structure suggests that C-terminal domains (factor binding sites) of P1 are oriented toward a limited spatial area between P0 spine and the stalk base of the 50 S subunit. The C-terminal domain of P1 is probably constrained over a narrow range, rather than being mobile over a wide area. We infer that the functional significance of multiple copies of P1 is not only in their role in picking up factors from a wide area, but also in creating a factor pool within a limited space near the stalk base of the ribosome.
Acknowledgments
We thank Dr. Y. Tanaka and Dr. S. Takeda for their help during measurements of CD and isothermal titration calorimetry, and J. Yu for structural refinement. We also thank the staff of beamline BL38B2 and BL41XU, SPring-8, Japan, for their help with data collection.
This work was supported by the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan and the Naito Foundation (to T. U.).
The atomic coordinates and structure factors (code 3A1Y) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GMPPNP
- guanylyl imidodiphosphate
- Hm50S
- 50 S ribosome subunit from H. marismortui
- PhP0
- P0 from P. horikoshii
- rRNA
- ribosomal RNA
- WT
- wild type
- Se-SAD
- single-wavelength anomalous diffraction method using selenomethionine as scatteror.
REFERENCES
- 1.Möller W., Maassen J. A. (1986) in Structure, Function, and Genetics of Ribosomes (Hardesty B., Kramer G. eds) pp. 309–325, Springer-Verlag New York Inc., New York [Google Scholar]
- 2.Chandra Sanyal S., Liljas A. (2000) Curr. Opin. Struct. Biol. 10, 633–636 [DOI] [PubMed] [Google Scholar]
- 3.Traut R. R., Dey D., Bochkariov D. E., Oleinikov A. V., Jokhadze G. G., Hamman B., Jameson D. (1995) Biochem. Cell Biol. 73, 949–958 [DOI] [PubMed] [Google Scholar]
- 4.Gudkov A. T. (1997) FEBS Lett. 407, 253–256 [DOI] [PubMed] [Google Scholar]
- 5.Bocharov E. V., Sobol A. G., Pavlov K. V., Korzhnev D. M., Jaravine V. A., Gudkov A. T., Arseniev A. S. (2004) J. Biol. Chem. 279, 17697–17706 [DOI] [PubMed] [Google Scholar]
- 6.Christodoulou J., Larsson G., Fucini P., Connell S. R., Pertinhez T. A., Hanson C. L., Redfield C., Nierhaus K. H., Robinson C. V., Schleucher J., Dobson C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10949–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder F. A., Bouakaz L., Lundell A., Venkataramana M., Liljas A., Akke M., Sanyal S. (2004) Biochemistry 43, 5930–5936 [DOI] [PubMed] [Google Scholar]
- 8.Datta P. P., Sharma M. R., Qi L., Frank J., Agrawal R. K. (2005) Mol. Cell 20, 723–731 [DOI] [PubMed] [Google Scholar]
- 9.Diaconu M., Kothe U., Schlünzen F., Fischer N., Harms J. M., Tonevitsky A. G., Stark H., Rodnina M. V., Wahl M. C. (2005) Cell 121, 991–1004 [DOI] [PubMed] [Google Scholar]
- 10.Savelsbergh A., Mohr D., Kothe U., Wintermeyer W., Rodnina M. V. (2005) EMBO J. 24, 4316–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgstrand M., Mandava C. S., Mulder F. A., Liljas A., Sanyal S., Akke M. (2007) J. Mol. Biol. 365, 468–479 [DOI] [PubMed] [Google Scholar]
- 12.Maassen J. A., Schop E. N., Brands J. H., van Hemert F. J., Lenstra J. A., Möller W. (1985) Eur. J. Biochem. 149, 609–616 [DOI] [PubMed] [Google Scholar]
- 13.Rich B. E., Steitz J. A. (1987) Mol. Cell Biol. 7, 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wool I. G., Chan Y. L., Glück A. (1995) Biochem. Cell Biol. 73, 933–947 [DOI] [PubMed] [Google Scholar]
- 15.Liao D., Dennis P. P. (1994) J. Mol. Evol. 38, 405–419 [DOI] [PubMed] [Google Scholar]
- 16.Shimmin L. C., Ramirez C., Matheson A. T., Dennis P. P. (1989) J. Mol. Evol. 29, 448–462 [DOI] [PubMed] [Google Scholar]
- 17.Tchórzewski M., Boldyreff B., Issinger O. G., Grankowski N. (2000) Int. J. Biochem. Cell Biol. 32, 737–746 [DOI] [PubMed] [Google Scholar]
- 18.Guarinos E., Remacha M., Ballesta J. P. (2001) J. Biol. Chem. 276, 32474–32479 [DOI] [PubMed] [Google Scholar]
- 19.Gonzalo P., Lavergne J. P., Reboud J. P. (2001) J. Biol. Chem. 276, 19762–19769 [DOI] [PubMed] [Google Scholar]
- 20.Shimizu T., Nakagaki M., Nishi Y., Kobayashi Y., Hachimori A., Uchiumi T. (2002) Nucleic Acids Res. 30, 2620–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naganuma T., Shiogama K., Uchiumi T. (2007) Genes Cells 12, 501–510 [DOI] [PubMed] [Google Scholar]
- 22.Briceño V., Camargo H., Remacha M., Santos C., Ballesta J. P. (2009) Int. J. Biochem. Cell Biol. 41, 1315–1322 [DOI] [PubMed] [Google Scholar]
- 23.Lalioti V. S., Pérez-Fernández J., Remacha M., Ballesta J. P. (2002) Mol. Microbiol. 46, 719–729 [DOI] [PubMed] [Google Scholar]
- 24.Hagiya A., Naganuma T., Maki Y., Ohta J., Tohkairin Y., Shimizu T., Nomura T., Hachimori A., Uchiumi T. (2005) J. Biol. Chem. 280, 39193–39199 [DOI] [PubMed] [Google Scholar]
- 25.Krokowski D., Boguszewska A., Abramczyk D., Liljas A., Tchórzewski M., Grankowski N. (2006) Mol. Microbiol. 60, 386–400 [DOI] [PubMed] [Google Scholar]
- 26.Uchiumi T., Hori K., Nomura T., Hachimori A. (1999) J. Biol. Chem. 274, 27578–27582 [DOI] [PubMed] [Google Scholar]
- 27.Maki Y., Hashimoto T., Zhou M., Naganuma T., Ohta J., Nomura T., Robinnson C. V., Uchiumi T. (2007) J. Biol. Chem. 282, 32827–32833 [DOI] [PubMed] [Google Scholar]
- 28.Nomura T., Nakano K., Maki Y., Naganuma T., Nakashima T., Tanaka I., Kimura M., Hachimori A., Uchiumi T. (2006) Biochem. J. 396, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong D., Bajaj S. P. (1993) BioTechniques 15, 874–878 [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao M., Zhou Y., Tanaka I. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 189–196 [DOI] [PubMed] [Google Scholar]
- 36.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W, Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 38.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 39.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 41.Dabbs E. R. (1979) J. Bacteriol. 140, 734–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchiumi T., Honma S., Nomura T., Dabbs E. R., Hachimori A. (2002) J. Biol. Chem. 277, 3857–3862 [DOI] [PubMed] [Google Scholar]
- 43.Iwasaki K., Kaziro Y. (1979) Methods Enzymol. 60, 657–676 [DOI] [PubMed] [Google Scholar]
- 44.Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 45.Kavran J. M., Steitz T. A. (2007) J. Mol. Biol. 371, 1047–1059 [DOI] [PubMed] [Google Scholar]
- 46.Spahn C. M., Gomez-Lorenzo M. G., Grassucci R. A., Jørgensen R., Andersen G. R., Beckmann R., Penczek P. A., Ballesta J. P., Frank J. (2004) EMBO J. 23, 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gómez-Lorenzo M. G., García-Bustos J. F. (1998) J. Biol. Chem. 273, 25041–25044 [DOI] [PubMed] [Google Scholar]
- 48.Santos C., Rodríguez-Gabriel M. A., Remacha M., Ballesta J. P. (2004) Antimicrob. Agents Chemother. 48, 2930–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]