Abstract
Lysine 48-linked polyubiquitin chains usually target proteins for 26 S proteasomal degradation; however, this modification is not a warrant for destruction. Here, we found that efficient degradation of a physiological substrate UbcH10 requires not only an exogenous polyubiquitin chain modification but also its unstructured N-terminal region. Interestingly, the unstructured N-terminal region of UbcH10 directly binds the 19 S regulatory complex of the 26 S proteasome, and it mediates the initiation of substrate translocation. To promote ubiquitin- dependent degradation of the folded domains of UbcH10, its N-terminal region can be displaced by exogenous proteasomal binding elements. Moreover, the unstructured N-terminal region can initiate substrate translocation even when UbcH10 is artificially cyclized without a free terminus. Polyubiquitinated circular UbcH10 is completely degraded by the 26 S proteasome. Accordingly, we propose that degradation of some polyubiquitinated proteins requires two binding interactions: a polyubiquitin chain and an intrinsic proteasomal binding element in the substrates (likely an unstructured region); moreover, the intrinsic proteasomal binding element initiates substrate translocation regardless of its location in the substrates.
Keywords: Protease/ATP-dependent, Proteases/Proteasomes, Proteases/Ubiqiuitin, Proteases/Ubiquitination, Protein/Degradation, Protein/Processing
Introduction
The 26 S proteasome-mediated protein degradation is highly regulated and plays pivotal roles in many cellular processes including cell cycle progression, gene transcription, and signal transduction (1). The 26 S proteasome is a 2.4-MDa complex consisting of the four ring-stacked, barrel-shaped 20 S proteasome and the 20-subunit regulatory complex, PA700 (19 S regulatory particle) (1). PA700 is essential for processing polyubiquitinated proteins (2), it possesses activities of binding a polyubiquitin (polyUb)4 chain, which engages the substrates to the 26 S proteasome; deubiquitinating the substrates, which could clear the steric restriction of a polyUb chain to facilitate substrate translocation; hydrolyzing ATP that could be used for unfolding the substrates to feed them into the narrow substrate translocation channel consisting of the axial pore of the ATPase ring in PA700 and the α-subunit ring chamber of the 20 S proteasome. Eventually, unfolded substrates are translocated into the central β chamber of the 20 S proteasome for proteolysis. There are three distinct peptidase activities located in the central degradation chamber: the β1-subunit has a caspase-like activity, the β2-subunit has a trypsin-like activity, and the β5-subunit has a chymotrypsin-like activity (2).
Binding a substrate to the 26 S proteasome is the initial step for proteolysis. In ubiquitin (Ub)-dependent degradation, three endogenous subunits of the 26 S proteasome have been shown to bind Ub or polyUb chains: the S5a subunit binds a polyUb chain through its two Ub interacting motifs (3); the Adrm1 subunit binds mono or diUb through a pleckstrin-homology domain interaction (4, 5); and the S6′ ATPase subunit binds a polyUb chain as identified by a chemical cross-linking assay (6). Usually, a tetraUb is the minimal chain length for providing a high binding affinity to the 26 S proteasome (7). Besides the endogenous proteasomal subunits, several studies have shown that shuttle proteins, such as Rad23, Dsk2, and free Rpn10 (S5a), can help delivery of polyubiquitinated proteins to the yeast 26 S proteasome and promote degradation (8). However, delivering a polyubiquitinated protein to the 26 S proteasome is not a warrant for proteasomal destruction because efficient degradation requires coordinated actions of substrate recognition, deubiquitination, unfolding, translocation, polypeptide hydrolysis, and ATP hydrolysis.
Some substrates including both unstructured and folded proteins are degraded by the 26 S proteasome without a polyubiquitin chain modification. Interestingly, degradation of unstructured proteins by the 26 S proteasome is ATP hydrolysis-independent, indicating that substrate translocation does not consume energy (9, 10). Presumably, unstructured substrates can enter into the degradation chamber by passive diffusion as proposed by the Brownian ratchet model (11). However, we recently found that the 26 S proteasome degrades unstructured proteins at obviously different rates (12), suggesting that the 26 S proteasome discriminates unstructured proteins for proteolysis. Using protein fusion techniques, several studies have shown that an unstructured region could promote degradation (13–15). At least one function of an unstructured region is to initiate substrate translocation and thus, promote degradation (16). Furthermore, depending on the location of the unstructured region, a substrate could translocate into the degradation chamber from either a free terminus (processive degradation) or an internal site (endoproteolysis). Unequivocal evidence for initiating degradation from an internal site has been demonstrated by using cyclized unstructured proteins, which lack free termini as model substrates (15). Whereas unambiguous biochemical evidence for endoproteolysis of polyubiquitinated proteins is currently lacking. Physiologically, endoproteolysis might be the mechanism for regulating partial protein proteolysis in processing some transcription factors based on studies in intact cells or using crude cell lysates (17, 18). For example, releasing of the transcriptionally active N-terminal p90 domain from Spt23, a process stimulated by mono or oligoubiquitination and subsequent proteasome degradation (19, 20), can still proceed when the C terminus is blocked from degradation by the methotrexate-stabilized dihydrofolate reductase domain in yeast cells (18). Thus, degradation has to initiate from an internal site of Spt23 to produce the intact N-terminal p90 domain.
Proteasomal degradation of ornithine decarboxylase (ODC) is a well-characterized example for Ub-independent proteolysis of folded proteins (21). Upon binding to antizyme 1, the C-terminal 37-amino acid PEST domain of ODC is exposed and mediates Ub-independent degradation of ODC by initiating translocation (22). By fusion with other proteins, the PEST domain of ODC has been shown to facilitate protein degradation in cells (23). Interestingly, in addition to its role for initiating translocation, the PEST domain of ODC needs to bind the 26 S proteasome to mediate Ub-independent degradation (24), reminiscent of a role of polyUb chains that tether polyubiquitinated proteins to the 26 S proteasome. Moreover, the binding function of the PEST domain can be bypassed by providing an alternative proteasomal binding element such as fusing ODC to the Rpn10 (S5a) subunit of the proteasome (24), which delivers ODC to the 26 S proteasome by integrating it into the 26 S proteasome complex (24, 25).
In this report, we reconstituted Ub-dependent degradation of the human mitotic-specific E2 enzyme UbcH10 to evaluate substrate recognition and translocation. We found that, in addition to a polyUb chain, a direct interaction between the unstructured N-terminal region of UbcH10 and the 26 S proteasome is mandatory for UbcH10 degradation. These results reveal that the 26 S proteasome recognizes some polyubiquitinated substrates via binding both a polyUb chain and an intrinsic proteasomal binding element in the substrates. Furthermore, we found that the N-terminal region of UbcH10 initiates translocation even when UbcH10 is cyclized. Circular UbcH10 undergoes complete degradation in the presence of a polyUb chain. Thus, our results provide unequivocal in vitro evidence that the 26 S proteasome can catalyze endoproteolysis of polyubiquitinated, folded proteins.
EXPERIMENTAL PROCEDURES
Reagents
Ubiquitin aldehyde, epoxomicin, MG132, and the thrombin cleavage capture kit were purchased from Calbiochem. Succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcourmarin (Suc-LLVY-AMC) was purchased from Bachem. Antibodies were purchased against the following proteins: UbcH10 (Boston Biochemical), Cdc27 (Santa Cruz) and polyhistidine (Sigma). Anti-proteasome antibodies were gifts from Dr. G. N. DeMartino (University of Texas Southwestern Medical Center).
Plasmids, Recombinant Protein Expression, and Purification
The coding sequence of full-length UbcH10 or N-terminally truncated UbcH10 mutant (ΔN10-, ΔN20-, or ΔN28-UbcH10) was subcloned into the pET28(a) vector (Novagen) using the NheI and XhoI sites. Thus, a His6 tag and a thrombin cleavage site were appended at the N terminus of UbcH10 or UbcH10 mutants, followed by a C-terminal His6 tag. To obtain circular UbcH10 or ΔN10-UbcH10, DNA coding sequence of UbcH10 or ΔN10-UbcH10 was subcloned into the pTWIN1 vector (New England Biolabs) using the SapI site. Certain linkers were introduced to facilitate protein cyclization as indicated in the figure legends. To generate PEST-ΔN28-UbcH10 expression plasmid, the coding sequences of the 37-amino acid PEST domain in ODC and ΔN28-UbcH10 were inserted into the pET28(a) vector by two step ligations using the NdeI, EcoRI, and XhoI sites. To generate Syn1–28-ΔN28-UbcH10, the coding sequences of the N-terminal 28 amino acids of α-synuclein and ΔN28-UbcH10 were sequentially inserted into the PstI, EcoRI, and XhoI sites of the pET28(a) vector in which the original NcoI site was replaced with the PstI site. All plasmids were verified by DNA sequencing analysis. All proteins were expressed in Escherichia coli BL21 (DE3) and induced by adding 0.2–0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h at 37 °C or 6 h at room temperature. His6-tagged or GST-tagged proteins were purified by using Nickel-nitrilotriacetic acid resin (Invitrogen) or glutathione-Sepharose 4B resin (GE Healthcare) according to the manufacturers' instructions. Circular UbcH10 or ΔN10-UbcH10 was produced and purified according to the protocol provided by New England Biolabs. K48-linked Ub4 chain was synthesized according to an established protocol (26).
Circular Dichroism (CD) Spectroscopy
CD measurements were carried out with a J-810 Jasco spectropolarimeter using a 1.0-mm pathlength quartz cuvette. Spectra were recorded from 250–200 nm with 8 average scans at room temperature. 7.0 μm UbcH10 or UbcH10 mutants were kept in 20 mm Tris, pH 7.2, 20 mm NaCl, 0.5 mm β-mercaptoethanol. Buffer alone was recorded and subtracted as the blank. The α-helix content was calculated based on the absorption at 222 nm (27).
In Vitro Polyubiquitination of UbcH10
Ubiquitination of UbcH10 or UbcH10 mutants was performed according to our previously published method (12) using the anaphase-promoting complex/cyclosome (APC/C) complex E3 ligase immunoprecipitated from Xenopus egg extracts.
Determination of Ubiquitination Sites by Mass Spectroscopy
A sample from in-gel trypsin digestion of Ub4-UbcH10 or Ub4-ΔN28-UbcH10 was analyzed on a LTQ-FT Ultra hybrid mass spectrometer (Thermo Fisher). Peptide desalting and separation were achieved using a dual capillary/nano pump HPLC system (Agilent 1200, Palo Alto, CA). The nano pump run was 60 min at a flow rate of 350 nl/min. A 70-min gradient from 12 to 35% acetonitrile was used to separate the peptides. The HPLC run was monitored by sequentially recording MS scans in the ion cyclotron resonance (ICR) cell, whereas three MS/MS were obtained in the ion trap via collision-induced dissociation (CID). A raw distiller (UCSF) was used to create de-isotoped centroided peak lists from the raw spectra. These peak lists were searched against all human entries in the SwissProt protein data base using MascotTM server (Version 2.2, Matrix Science). For searches, mass tolerances were ± 10 ppm for MS peaks, and ± 0.6 Da for MS/MS fragment ions. Trypsin specificity was used allowing for 1 missed cleavage. The variable modifications of Met oxidation, protein N-terminal acetylation, peptide N-terminal Q pyroglutamic acid formation, glycine-glycine of lysine were allowed for the search. An expected value (p value) of <0.05 was considered significant.
In Vitro Proteasomal Degradation Assay
The 26 S proteasome was purified from bovine red blood cells according to a previous report (10). The purified 26 S proteasomes usually contain 50% doubly capped and 50% singly capped proteasomes as determined by a native-PAGE assay. We preincubated a 1:2 molar ratio of purified 26 S proteasomes with purified PA700 to produce predominantly doubly capped 26 S proteasome. Degradation was performed according to our published method (10, 28).
Size Exclusion Spin Column Assay and Gel Filtration Assay
50 nm PA700 or doubly capped 26 S proteasome in the degradation buffer was preinhibited by 100 μm epoxomicin prior to the supplementation of 250 nm UbcH10 or UbcH10 mutants. The mixtures were incubated for 5 min at room temperature, then loaded into Micro Bio-Spin 30 chromatography columns (Bio-Rad), which were pre-equilibrated in the degradation buffer and centrifuged according to the manufacturer's instruction. The flow-through was immediately mixed with 5× SDS sample buffer. Proteins in the flow-through were analyzed by immunoblotting. Homemade Sephadex G-100 spin columns were used to assay the interaction between Ub4 conjugates and the 26 S proteasome. The binding assay contained 18 nm of doubly capped 26 S proteasome and 100 nm Ub4 conjugates. The 26 S proteasome was preincubated with 2 μm ubiquitin aldehyde and 5 mm 1,10-phenanthroline to block its deubiquitinating activities and 100 μm epoxomicin to block its proteolytic activities.
To assay the interaction between UbcH10 and the 26 S proteasome by gel filtration, 200 nm of 26 S proteasome was preincubated with 200 μm MG132 at room temperature for 10 min before adding 2 μm of UbcH10 or ΔN28 UbcH10. After 10 min of incubation, 300-μl mixtures were injected into a Superdex 200 (10/30) gel filtration column on a fast performance liquid chromatography system. Fractions were immunoblotted with an anti-S7 (a subunit of PA700) antibody or an anti-UbcH10 antibody. The 26 S proteasome, UbcH10, or ΔN28 UbcH10 alone was run as a control.
RESULTS
The Unstructured N-terminal Region of UbcH10 Can Be Cleaved by the 26 S Proteasome
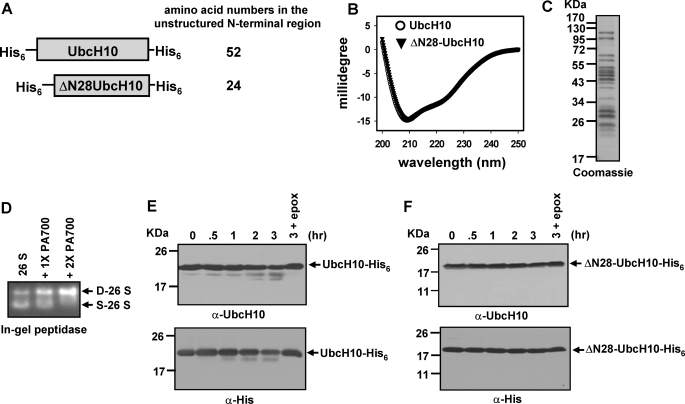
Ub-dependent degradation of the E2 enzyme UbcH10 is critical to maintain the levels of several proteins that are substrates of the APC/C E3 Ub ligase during the G1-S phase transition (29). Structurally, UbcH10 has four α-helices and four-stranded antiparallel β-sheets with an unstructured N-terminal region containing the first 29 amino acids (30). To determine whether the unstructured N-terminal region of UbcH10 could function similarly as the C-terminal PEST domain of ODC to mediate Ub-independent degradation, we generated bacterial expression constructs for purification of full-length UbcH10 or N-terminally truncated UbcH10 in which the first 28 amino acids were deleted (Fig. 1A). Of note, all UbcH10 and its N-terminally truncated mutants used in this study contained a His6 tag on both N and C termini except where otherwise specified. ΔN28-UbcH10 had indistinguishable secondary structures in comparison with full-length UbcH10 as determined by CD spectrometry (Fig. 1B), indicating that deletion of the unstructured N-terminal region has no effect on the UbcH10 folded structure. Based on absorptions at 222 nm in CD spectra, UbcH10 and ΔN28-UbcH10 contained 21.2% α-helix content, consistent with the value of 26.1% calculated from the crystal structure (30). Next, we determined whether UbcH10 and ΔN28-UbcH10 can be degraded by purified 26 S proteasomes (Fig. 1, C and D). To exclude any potential effect of the unstructured His6 tag and the thrombin site at the N terminus on degradation, the N-terminal His6/thrombin tag in UbcH10 or ΔN28-UbcH10 was removed by thrombin cleavage for this assay (data not shown), whereas the C-terminal His6 tag was intact. UbcH10-His6 was very slowly cleaved by purified 26 S proteasomes with the accumulation of a large fragment that was recognized by an anti-UbcH10 and an anti-polyhistidine antibody (Fig. 1E), indicating that only the N-terminal region of UbcH10 is accessible for cleavage. Consistent with this finding, deletion of the unstructured N-terminal region caused ΔN28-UbcH10-His6 to be resistant to cleavage by the 26 S proteasome (Fig. 1F). Together, these results demonstrate that the unstructured N-terminal region of UbcH10 can initiate proteasomal degradation; however, it cannot drive complete proteolysis.
FIGURE 1.
The unstructured N-terminal region in UbcH10 initiates translocation for partial cleavage by the 26 S proteasome. A, schematic representations of UbcH10 and N-terminally truncated ΔN28-UbcH10 constructs. Both constructs have an N-terminal His6 tag followed by a thrombin cleavage site and a C-terminal His6 tag. The amino acid numbers in the unstructured N-terminal region are shown on the right side. B, far-UV CD spectra of 7 μm UbcH10 or ΔN28-UbcH10. C, Coomassie Blue-stained SDS-PAGE of 5 μg of purified 26 S proteasomes. D, assembly of purified 26 S proteasome with PA700 to form the doubly capped 26 S proteasome. The assembly reactions contained 3 μg of purified 26 S proteasomes and 1 or 2 μg of PA700. Formation of the doubly capped 26 S proteasome was assayed by the in-gel peptidase assay using Suc-LLVY-AMC as the fluorogenic substrate. D-26 S and S-26 S represent doubly capped and singly capped 26 S proteasomes, respectively. E and F, unstructured N-terminal region of UbcH10, but not the folded domains (ΔN28-UbcH10), can be cleaved by the 26 S proteasome. The N-terminal His6 tags in both UbcH10 and ΔN28-UbcH10 were removed by thrombin cleavage in these assays. The lane denoted 3 + epox indicates that the 26 S proteasome was preincubated with 100 μm epoxomicin to inhibit its proteolytic activities prior to the supplementation of substrates, and the reactions were stopped at the 3-h time point.
A K48 Ub4 Chain Efficiently Targets ΔN28-UbcH10 to the 26 S Proteasome, but Does Not Support Its Proteolysis
We have previously reported that K48-linked Ub4-UbcH10 is efficiently degraded by purified 26 S proteasomes (10). To determine whether a K48 Ub4 chain can target N-terminally truncated UbcH10 for proteasomal degradation, we generated bacterial expression constructs for purification of UbcH10 deletion mutants in which the first 10, 20, or 28 amino acids were deleted (Fig. 1A and data not shown). A previous study showed that the N-terminal truncation impairs autoubiquitination of UbcH10 catalyzed by Cdh1-activated APC/C, but allows ubiquitination catalyzed by Cdc20-activated APC/C (29). The role of the N-terminal region of UbcH10 to enhance UbcH10 autoubiquitination by Cdh1 is unclear. We immunoprecipitated Xenopus APC/C E3 Ub ligase from metaphase egg extracts that lack Cdh1. We found that all UbcH10 mutants were able to be conjugated with a K48-linked Ub4 chain as efficiently as full-length UbcH10 in the presence of E1 and immunoprecipitated APC/C E3 Ub ligase. Furthermore, the conjugated Ub4 chain was resistant to 10 mm dithiothreitol treatment (data not shown), indicating the conjugate is not the ubiquitination intermediate where Ub is linked to the catalytic cysteine of E2s by a thioester bond.
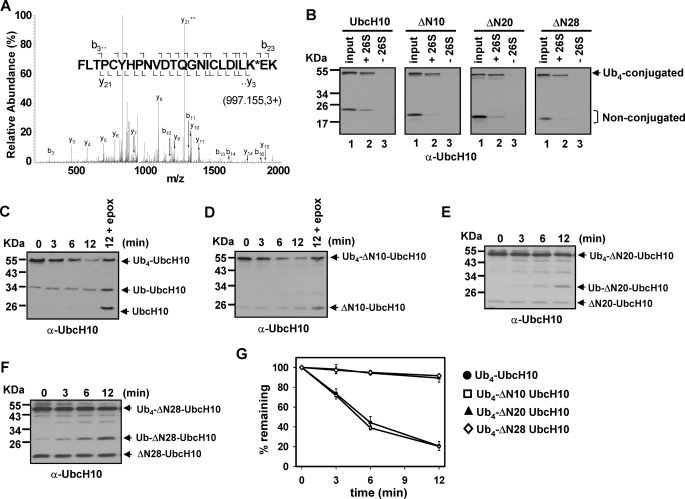
We first attempted to identify the K48 Ub4 conjugation site in UbcH10 and ΔN28 UbcH10. Trypsin digestion followed by mass spectrometric determination found that K48 Ub4 was conjugated on the lysine 119 in both UbcH10 and ΔN28 UbcH10 (Fig. 2A and data not shown), demonstrating that they use the same lysine residue for Ub4 conjugation. We next evaluated whether Ub4-conjugated UbcH10 or UbcH10 mutants bind efficiently to the 26 S proteasome using a size exclusion spin column assay. The Sephadex G-100 (exclusion limit of 100 kDa) trapped Ub4-conjugated UbcH10 or UbcH10 mutants after centrifugation (lane 3 in Fig. 2B). In contrast, Ub4 conjugates were coeluted with the 2.4-MDa 26 S proteasome after centrifugation when they were mixed together (lane 2 in Fig. 2B), indicating a direct interaction between them. Furthermore, we examined whether Ub4-conjugated UbcH10 or UbcH10 mutants are able to efficiently bind the S5a subunit of the 26 S proteasome, one of the primary polyUb chain receptors of mammalian 26 S proteasomes (3). Not surprisingly, we found that Ub4-conjugatged UbcH10 and its N-terminally truncated mutants all efficiently bound to GST-S5a, but not GST (supplemental Fig. S1). Therefore, we conclude that a K48-linked Ub4 chain can target UbcH10 and its N-terminal deletion mutants to the 26 S proteasome.
FIGURE 2.
A K48-linked tetraubiquitin chain targets N-terminally truncated UbcH10 to the 26 S proteasome, but cannot promote degradation. A, K48 Ub4 is conjugated on lysine 119 of UbcH10 and ΔN28-UbcH10. The [MH]3+ precursor ion 997.15 resulted in the identification of UbCH10 peptide 98–121 with Lys-119 modified by 114.04 Da, corresponding to two glycine residues left on lysine after tryptic digestion. Assigned fragment ions are shown on the peptide sequence. The identification of fragment ions y3, b22, and b23 supports Lys-119 as the site of ubiquitination. The Ub4 conjugation site for ΔN28-UbcH10 is the same as UbcH10 (data not shown). B, K48 Ub4-conjugated UbcH10 or N-terminally truncated UbcH10 mutants (ΔN10, ΔN20, and ΔN28) efficiently bind the 26 S proteasome. The size-exclusion spin column assay was performed according to the method described under “Experimental Procedures.” C and D, K48 Ub4-UbcH10 and Ub4-ΔN10-UbcH10 are efficiently degraded by purified 26 S proteasomes. The reactions contained 13.5 nm purified 26 S proteasome and 100 nm ubiquitinated substrates. E and F, K48 Ub4-ΔN20-UbcH10 and Ub4-ΔN28-UbcH10 are slowly deubiquitinated by the 26 S proteasome without degradation. Degradation assays were analogous to C and D. G, densitometric quantitation of degradation/deubiquitination of Ub4-conjugated UbcH10 and N-terminal UbcH10 deletions. Data represent the mean values of three independent degradations ± S.D.
Next, we examined whether Ub4-conjugated UbcH10 mutants are proteolytic substrates of purified 26 S proteasomes. Compared with the 12-min degradation, inhibition of the 26 S proteasome by epoxomicin caused the accumulation of more non-ubiquitinated UbcH10 or ΔN10-UbcH10 concomitantly with more Ub4-conjugated forms left (Fig. 2, C, D, and G), indicating degradation. In contrast, Ub4-ΔN20-UbcH10 and Ub4-ΔN28-UbcH10 were only slowly deubiquitinated without degradation as evidenced by the accumulation of mono-, di-, and tri-ubiquitinated UbcH10 mutants (Fig. 2, E, F, and G). Similar results were obtained when only C-terminally His6-tagged UbcH10 or ΔN28 UbcH10 was used for ubiquitination, and then proteasomal degradation (supplemental Fig. S2), indicating that the degradation-resistance of ΔN28-UbcH10 is not caused by the N-terminal His6 tag. Also, an N-terminally truncated UbcH10 mutant (ΔN27-UbcH10) was found to be stable in HeLa cell lysates, although it is ubiquitinated (29). Thus, the unstructured N-terminal region of UbcH10 is necessary for mediating its Ub-dependent degradation.
The Length of the Unstructured Region per se Does Not Account for the Degradation-Resistance of Ub4-ΔN28-UbcH10
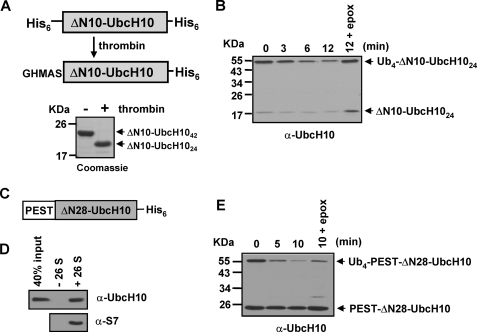
Next, we sought to understand why the N-terminal region of UbcH10 is required to mediate Ub-dependent degradation. One possible reason for the degradation-resistance of Ub4-ΔN20- and Ub4-ΔN28-UbcH10 is that both of them have shorter unstructured N-terminal regions compared with that in full-length or ΔN10-UbcH10 (Fig. 1A and data not shown). To assess this possibility, we cleaved the N-terminal His6/thrombin tag in ΔN10-UbcH10 by thrombin digestion (Fig. 3A). The resulting shorter ΔN10-UbcH10 has an unstructured N-terminal region containing a total of 24 amino acids in which 19 amino acids are from the original UbcH10 N terminus (termed ΔN10-UbcH1024). The length of the unstructured N-terminal region in ΔN10-UbcH1024 is equal to that in ΔN28-UbcH10, which mostly consists of a His6 tag and the thrombin cleavage sequence. CD spectroscopic determination revealed that deletion of the N-terminal His6 tag had no effect on the secondary structures of ΔN10-UbcH10 (data not shown). After conjugation with a K48-linked Ub4 chain, Ub4-ΔN10-UbcH1024 was efficiently degraded by purified 26 S proteasomes (Fig. 3B), demonstrating that the length of the unstructured N terminus is not responsible for the degradation-resistance of Ub4-ΔN28-UbcH10.
FIGURE 3.
The length of the unstructured N-terminal region per se does not determine Ub-dependent UbcH10 degradation. A, schematic representation and Coomassie Blue-stained SDS-PAGE for removal of the N-terminal His6 tag from His6-ΔN10-UbcH10-His6 by thrombin digestion. After cleavage, the length of the unstructured N-terminal region of ΔN10-UbcH10 was reduced to 24 amino acids. B, K48 Ub4-ΔN10-UbcH1024 is efficiently degraded by purified 26 S proteasomes. Degradation assays were analogous to Fig. 2C. C, schematic representation of the construct with the PEST domain of ODC fused at the N terminus of ΔN28-UbcH10. D, PEST-ΔN28-UbcH10 binds the 26 S proteasome. The binding assay was performed using the Bio-Spin 30 spin column as described under “Experimental Procedures.” E, fused PEST region promotes Ub-dependent degradation of ΔN28-UbcH10. Degradation assays were analogous to Fig. 2C.
A Translocation Initiation Site Is Not Sufficient to Promote Ub-dependent UbcH10 Degradation
Another possible reason for the degradation-resistance of Ub4-ΔN20- and Ub4-ΔN28-UbcH10 is that they might lack a translocation initiation site. However, His6-ΔN28-UbcH10-His6 was slowly cleaved by purified 26 S proteasomes with the accumulation of a large degradation fragment recognized by an anti-UbcH10 antibody (supplemental Fig. S3A), indicating that the unstructured N-terminal His6/thrombin tag region in His6-ΔN28-UbcH10-His6 can initiate substrate translocation for partial cleavage. Similarly, the 24-amino acid unstructured N terminus in ΔN10-UbcH1024 was slowly cleaved by the 26 S proteasome as well (supplemental Fig. S3B). Thus, degradation of Ub4-conjugated ΔN10-UbcH1024 but His6-ΔN28-UbcH10-His6 suggests that the unstructured N-terminal region of UbcH10 has an additional function besides initiating substrate translocation.
Exogenous Proteasomal Binding Elements Promote Degradation of ΔN28-UbcH10
The C-terminal PEST domain of ODC was shown to promote degradation of its fused proteins (22). We, therefore, examined whether the unstructured PEST domain can replace the N-terminal region of UbcH10 to mediate degradation of ΔN28-UbcH10. To test this, we fused the PEST domain of ODC at the N terminus of ΔN28-UbcH10 (Fig. 3C). The size exclusion spin column assay showed that PEST-ΔN28-UbcH10 bound the 26 S proteasome (Fig. 3D), whereas ΔN28-UbcH10 did not (Fig. 4A), indicating that the PEST domain mediates the substrate binding of the 26 S proteasome. Interestingly, although PEST-ΔN28-UbcH10 was only slowly cleaved by the 26 S proteasome (supplemental Fig. S3C) at a similar rate to that of His6-ΔN28-UbcH10-His6 (supplemental Fig. S3A) or UbcH10 (Fig. 1E), K48 Ub4-PEST-ΔN28-UbcH10 was rapidly degraded by purified 26 S proteasomes (Fig. 3E). Similarly, we also found that the unstructured N-terminal region of α-synuclein can mediate binding of ΔN28-UbcH10 to the 26 S proteasome, and promote Ub-dependent degradation of ΔN28-UbcH10 (supplemental Fig. S4). Thus, exogenous proteasomal binding elements can promote Ub-dependent degradation of ΔN28-UbcH10.
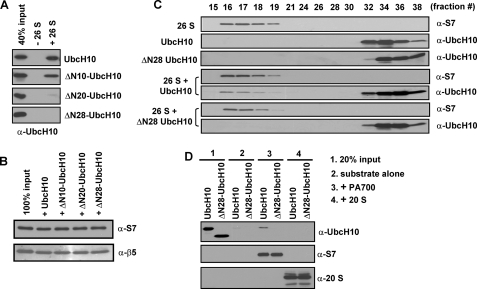
FIGURE 4.
The N-terminal region of UbcH10 directly binds the 26 S proteasome. A and B, the N-terminal region of UbcH10 binds the 26 S proteasome. The binding assay was performed using the Bio-Spin 30 spin column as described under “Experimental Procedures.” The elutions were immunoblotted with an anti-UbcH10 antibody (A), an anti-S7 (a PA700 subunit) or an anti-β5 (a 20 S proteasome subunit) antibody (B). C, UbcH10, but not ΔN28-UbcH10, interacts with the 26 S proteasome as confirmed by gel filtration. Proteins in different fractions were immunoblotted with appropriate antibodies. D, UbcH10, but not ΔN28-UbcH10, interacts with PA700, the regulatory complex of the 26 S proteasome. The binding assays were similar to those in A.
The Unstructured N-terminal Region of UbcH10 Directly Binds the 26 S Proteasome
The above studies suggest that the N-terminal region of UbcH10 might directly bind the 26 S proteasome and thus, promote efficient degradation. To test this, we performed the size exclusion spin column assay to determine whether UbcH10 directly binds the 26 S proteasome. UbcH10 and its N-terminal deletions were trapped inside the spin column (exclusion limit of 40 kDa, lane 2 in Fig. 4A). However, only UbcH10 and ΔN10-UbcH10 were found in the flow-through together with the 26 S proteasome when they were mixed with the 26 S proteasome prior to being applied to the spin columns (lane 3 in Fig. 4A), indicating a direct interaction between UbcH10 or ΔN10-UbcH10 and the 26 S proteasome. ΔN20-UbcH10 or ΔN28-UbcH10 did not bind the 26 S proteasome although equal amounts of the 26 S proteasome were recovered in the flow-through for all four reactions (Fig. 4, A and B). Based on densitometric analysis, one 26 S proteasome molecule can bind approximately two UbcH10 molecules as judged by the fact that the mixture in the binding assay contained 50 nm of doubly capped 26 S and 250 nm of UbcH10 and the bound fraction was equal to that of 40% UbcH10 input (100 nm) (Fig. 4A). Next, we further confirmed the interaction between UbcH10 and the 26 S proteasome by gel filtration. We detected that UbcH10 co-eluted with the 26 S proteasome, whereas ΔN28-UbcH10 was absent in the fractions containing the 26 S proteasome (Fig. 4C). Moreover, we found that PA700 bound a small amount of UbcH10, but not ΔN28-UbcH10 (Fig. 4D). In contrast, the 20 S proteasome did not bind either of the substrates (Fig. 4D). These results indicate that binding of UbcH10 is possibly mediated by PA700, the regulatory complex of the 26 S proteasome. In our assay, PA700 had much less affinity than the 26 S proteasome to bind UbcH10 (Fig. 4, A and D), presumably the conformational change of PA700 upon 26 S proteasomal assembly enhances substrate binding. Combining these data with those in Fig. 2, our results indicate that the unstructured N-terminal region of UbcH10 directly binds the 26 S proteasome, and this binding is necessary for Ub-dependent UbcH10 degradation.
The 26 S Proteasome Degrades Ub4-conjugated Circular UbcH10
The above data indicate that the unstructured N-terminal region of UbcH10 binds the 26 S proteasome and initiates substrate translocation for Ub-dependent degradation. Next, we used UbcH10 as a model substrate to investigate if degradation can initiate from an internal site of the substrate for Ub-dependent degradation. To examine this, we inserted the coding region of UbcH10 into the pTWIN1 vector for production of circular UbcH10 (31) (Fig. 5A). SDS-PAGE showed that purified UbcH10 contained about 20% linear and 80% circular forms judged by thrombin cleavage (Fig. 5B). Structurally, UbcH10 cyclization did not affect its secondary structures as determined with CD spectrometry (data not shown). Both forms of UbcH10 were able to be conjugated with a K48-linked Ub4 chain when the APC/C E3 Ub ligase was presented (lower panel in Fig. 5B). Non-ubiquitinated circular and linear forms of UbcH10 were not obviously degraded by purified 26 S proteasomes in a time course of 12-min incubation (Fig. 5C). In striking contrast, Ub4-conjugated linear and circular UbcH10 were efficiently degraded by purified 26 S proteasomes (Fig. 5D). Degradation of circular UbcH10 was judged by a time-dependent disappearance of Ub4-conjugated circular UbcH10 with no concomitant accumulation of the non-ubiquitinated form. Accumulation of non-ubiquitinated circular UbcH10 is usually observed when only deubiquitination occurs. Such a scenario did occur when the proteasome-specific inhibitor, epoxomicin, was used to block the proteolytic activities (Fig. 5D). Furthermore, the rates of degradation of Ub4-linear UbcH10 and Ub4-circular UbcH10 were comparable (Figs. 2G and 5D), indicating that the 26 S proteasome does not have a preference for substrates with free termini compared with those where degradation has to initiate from an internal site.
FIGURE 5.
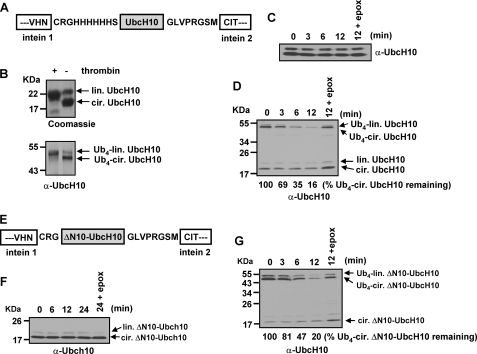
The 26 S proteasome catalyzes Ub-dependent endoproteolysis. A, schematic representation of the pTWIN 1 construct for purification of circular UbcH10. Cyclization of UbcH10 was initiated after cleavage of the intein 1 and intein 2 domains. The original N terminus-containing unstructured region has 54 amino acids: a 10-residue His6 tag region, the endogenous unstructured N-terminal region of UbcH10 (29 amino acids), the unstructured C terminus of UbcH10 (7 residues) followed by an 8-residue thrombin cleavage sequence (GLVPRGSM). B, verification of circular UbcH10 and K48 Ub4-circular UbcH10 by thrombin digestion. 5 μg of purified circular UbcH10 (upper gel) or 100 ng of Ub4-circular UbcH10 (lower blot) was mixed with 0.2 units of thrombin. Production of linear form of proteins was visualized by Coomassie Blue staining or immunoblotting. C, 26 S proteasome cannot degrade circular UbcH10. Degradation assays were analogous to Fig. 1E. D, K48-linked Ub4-circular UbcH10 is efficiently degraded by the 26 S proteasome. Degradation assays were analogous to Fig. 2C. E, schematic representation of the pTWIN 1 construct for purification of circular ΔN10-UbcH10. After cyclization, the original N terminus-containing unstructured region has 37 amino acids: an N-terminal linker (3 residues), the N-terminal region of ΔN10-UbcH10 (19 amino acids), the 7-residue unstructured C terminus of UbcH10 followed by an 8-amino acid thrombin cleavage sequence (GLVPRGSM). F, 26 S proteasome cannot degrade circular ΔN10-UbcH10. G, Ub4-circular ΔN10-UbcH10 is efficiently degraded by the 26 S proteasome. Degradation assays were analogous to Fig. 2C.
Next, we examined whether a shorter unstructured region in circular ΔN10-UbcH10 can induce endoproteolysis. After cyclization, the original N terminus-containing unstructured region in circular ΔN10-UbcH10 contained a total of 37 amino acids (Fig. 5E). Production of circular ΔN10-UbcH10 was confirmed by thrombin digestion (data not shown). Non-ubiquitinated circular ΔN10-UbcH10 was not degraded by purified 26 S proteasomes during a time course of 24 min of incubation (Fig. 5F). In contrast, Ub4 circular ΔN10-UbcH10 was efficiently degraded (Fig. 5G). Taken together, these results provide unequivocal in vitro evidence that polyubiquitinated, folded proteins can be endoproteolytically degraded by the 26 S proteasome.
DISCUSSION
Binding Interactions via Both a PolyUb Chain and an Intrinsic Proteasomal Binding Element in the Substrates Drive Ub- dependent Degradation
Our results indicate that proteasomal degradation of some proteins requires two binding interactions: an exogenous polyUb chain and an intrinsic proteasomal binding element in the substrates. We have thus far not succeeded in identifying the proteasomal binding site(s) of the unstructured N-terminal region of UbcH10 using UV-induced chemical cross-linking strategies. However, UbcH10 interacts with PA700 albeit with a weaker affinity compared with the 26 S proteasome (Fig. 4D). Presumably, it interacts with the ATPase subunit(s) because the ATPase ring of the 26 S proteasome has a chaperone-like activity that binds loosely folded proteins (32, 33). The axial pore of the ATPase ring is the entrance of the substrate translocation channel leading to the degradation chamber. Therefore, by binding on the ATPase ring substrates are proximally localized at the entrance of the substrate translocation channel (step 1 in Fig. 6). However this substrate localization is not sufficient to drive UbcH10 unfolding for complete degradation (Fig. 1, E and F) unless a polyUb chain provides an additional binding force (Fig. 2, C and D). How these two binding interactions coordinate to drive substrate degradation is not clear. It is speculative that the high proteasomal binding affinity of K48-linked polyUb chains and the time-consuming deubiquitination process allow a time window for the ATPase ring-engaged substrates (through an intrinsic proteasomal binding element in the substrates) to be unfolded by mechanical forces generated by ATP hydrolysis. Thus, these two binding interactions could coordinate substrate unfolding and translocation, which are likely coupled processes in proteasomal degradation (34, 35).
FIGURE 6.
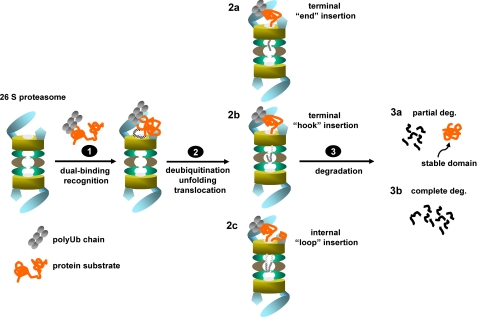
A model for recognition and translocation of polyubiquitinated proteins for proteasomal degradation. The 26 S proteasome recognizes polyubiquitinated proteins by two binding factors via both the attached polyUb chain and an intrinsic proteasome binding element in the substrates that likely binds on the ATPase ring of the 26 S proteasome (step 1). The dual-binding substrate recognition promotes subsequent deubiquitination, unfolding, and translocation (step 2). The intrinsic proteasomal binding element in the substrates can initiate translocation by insertion of a terminal “end” (model 2a), a terminal “hook” (model 2b) into the degradation chamber if it locates proximally to either the N or C terminus; or an internal “loop” (model 2c) if it is away from either terminus. Accompanying substrate translocation, substrates undergo deubiquitination-coupled degradation (step 3), where the substrates are either partially processed (3a) or completely degraded into small peptides (3b).
Degradation of unstructured proteins does not require ATP hydrolysis (9, 10), suggesting that unstructured elements can passively diffuse into the proteolytic chamber of the proteasome. This could explain why all of the tested unstructured elements, including the N-terminal region of UbcH10, the PEST sequence of ODC, the His/thrombin element and the N-terminal region of α-synuclein, were very slowly cleaved by the 26 S proteasome. These results indicate that all these unstructured elements are able to initiate substrate translocation. However, only unstructured sequences that bind the 26 S proteasome support Ub-dependent degradation of ΔN28-UbcH10, suggesting that not any unstructured elements in a substrate can support Ub-dependent degradation. Future studies are necessary to validate this model using various unstructured elements.
The dual binding interactions for substrate recognition in Ub-dependent degradation is distinct from targeting Ub-independent degradation carried out by the 26 S proteasome or its kin in bacteria, in which a single targeting signal was found to be sufficient to promote degradation. For example, in bacteria, the ClpA or ClpX AAA ATPase complex associates with the ClpP protease and the resulting ClpAP or ClpXP complex degrades proteins bearing degron peptides at either the N or C terminus of a protein (36). The most characterized one of these peptide signals is the 11-residue ssrA peptide that is added to the C termini of nascent polypeptides on stalled ribosomes (37). The ssrA peptide directly interacts with the ATPase ring of ClpA or ClpX, which facilitates substrate unfolding and translocation into the degradation chamber of the ClpP starting with the ssrA tag (38). In Ub-independent degradation of ODC, the proteasomal binding function of the C-terminal PEST domain of ODC can be separated from its translocation initiation function because the binding function can be bypassed by providing an alternative proteasomal binding site (24).
Dual Substrate Binding Interactions Trigger Coordinated Degradation Actions
One intriguing observation is that deubiquitination of the non-degradable Ub4-ΔN20- or Ub4-ΔN28-UbcH10 occurred at a much slower rate than that of the degradation-coupled deubiquitination of Ub4-UbcH10 or Ub4-ΔN10-UbcH10 (Fig. 2, C–G). It is likely that dual binding interactions via a polyUb chain and an intrinsic proteasomal binding element in the substrate dock the substrate into the 26 S proteasome substrate accommodation pocket, which triggers coordinated actions of substrate unfolding, deubiquitination, translocation, and ATP hydrolysis. Therefore, all degradation actions could be geared up and thus, accelerated. However if the substrate binds the 26 S proteasome only through a polyUb chain while itself is not engaged, the 26 S proteasome can catalyze some of its activities including deubiquitination independent of degradation. Interestingly, degradation-coupled deubiquitination is ATP hydrolysis-dependent (39, 40), while deubiquitination of non-degradable substrates is ATP hydrolysis-independent (10). Thus, whether a substrate has an intrinsic proteasomal binding element might define a layer of substrate selectivity in Ub-dependent proteolysis.
Endoproteolysis of Polyubiquitinated Proteins
Based on the crystal structure of the 20 S proteasome (41) and the EM image of the 26 S proteasome (42), the substrate translocation channel is ∼130–170 Å in length. Biochemical studies, using model substrates for examining Ub-independent degradation, have demonstrated that both the processive model and the endoproteolytic model are applicable for proteasomal degradation (15, 43). Physiologically, endoproteolysis might be the mechanism for mediating proteasome-catalyzed partial processing, which is critical for releasing some transcription activators/repressors from their precursors (17, 18). Using artificially cyclized UbcH10 as a model substrate, we showed that K48-linked Ub4-circular UbcH10 was efficiently degraded by purified 26 S proteasomes (Fig. 5D). This result provides the first unambiguous biochemical evidence that degradation of a polyubiquitinated, folded protein can initiate from an internal site with concurrent translocation of two polypeptides through the substrate translocation channel. Furthermore, we found that the 37-amino acid unstructured region in circular ΔN10-UbcH10 was sufficient to mediate endoproteolysis (Fig. 5G). It is likely that, as a loop insertion, the 37-amino acid unstructured region is too short to cover the entire 130–170 Å long translocation channel taking the fact that each residue in a polypeptide chain is 3.8 Å in length. Therefore, this result suggests that the loop formed by the proteasomal binding element is not necessary to be long enough to directly reach the catalytic sites of the proteasome. Accordingly, we propose three models that initiate translocation of a polyubiquitinated protein for degradation (step 2 in Fig. 6): “the terminal end” (model 2a) and “the terminal hook” models (model 2b) illustrate the canonical processive model for translocation of proteins for proteasomal degradation, where the proteasomal binding element in the substrate is very close to a terminus. Either model 2a or model 2b could contribute to the degradation of UbcH10 in which the N-terminal region is the proteasomal binding element. “The internal loop” model (model 2c) accounts for cases in which the proteasomal binding element in a substrate is away from either of the termini. To mediate proteolysis, a polypeptide loop can be formed by the intrinsic proteasomal binding region, and then the loop is elongated and pushed into the substrate translocation channel accompanying substrate unfolding. Usually, proteins are completely degraded by the 26 S proteasome, while partial degradation is forced when an unfolding-resistant domain or a translocation halting signal is encountered (step 3 in Fig. 6) (17, 20, 22). Although only a few proteins have been found to be partially processed by the proteasome in vivo, endoproteolysis and partial degradation could be common themes in proteasome-mediated proteolysis as many proteins have internal unstructured loops that could serve as both the intrinsic proteasomal binding element and the translocation initiation site.
Supplementary Material
Acknowledgments
We thank J. Maller and G. N. DeMartino for providing valuable reagents.
This work was supported by grants from the American Heart Association, the Basil O'Conner Starter Scholar Research Award, and the American Cancer Society (to C.-W. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- polyUb
- polyubiquitin
- APC/C
- anaphase-promoting complex/cyclosome
- CD
- circular dichroism
- ODC
- ornithine decarboxylase
- Ub
- ubiquitin
- Suc-LLVY-AMC
- succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcourmarin
- GST
- glutathione S-transferase
- CID
- collision-induced dissociation
- MS
- mass spectroscopy.
REFERENCES
- 1.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 2.DeMartino G. N., Slaughter C. A. (1999) J. Biol. Chem. 274, 22123–22126 [DOI] [PubMed] [Google Scholar]
- 3.Young P., Deveraux Q., Beal R. E., Pickart C. M., Rechsteiner M. (1998) J. Biol. Chem. 273, 5461–5467 [DOI] [PubMed] [Google Scholar]
- 4.Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., Groll M. (2008) Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam Y. A., Lawson T. G., Velayutham M., Zweier J. L., Pickart C. M. (2002) Nature 416, 763–767 [DOI] [PubMed] [Google Scholar]
- 7.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsasser S., Finley D. (2005) Nat. Cell Biol. 7, 742–749 [DOI] [PubMed] [Google Scholar]
- 9.Smith D. M., Kafri G., Cheng Y., Ng D., Walz T., Goldberg A. L. (2005) Mol. Cell 20, 687–698 [DOI] [PubMed] [Google Scholar]
- 10.Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) Mol. Cell 24, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith D. M., Benaroudj N., Goldberg A. (2006) J. Struct. Biol. 156, 72–83 [DOI] [PubMed] [Google Scholar]
- 12.Zhang N. Y., Tang Z., Liu C. W. (2008) J. Biol. Chem. 283, 20288–20298 [DOI] [PubMed] [Google Scholar]
- 13.Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 14.Zhang M., Pickart C. M., Coffino P. (2003) EMBO J. 22, 1488–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. (2003) Science 299, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrader E. K., Harstad K. G., Matouschek A. (2009) Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian L., Holmgren R. A., Matouschek A. (2005) Nat. Struct. Mol. Biol. 12, 1045–1053 [DOI] [PubMed] [Google Scholar]
- 18.Piwko W., Jentsch S. (2006) Nat. Struct. Mol. Biol. 13, 691–697 [DOI] [PubMed] [Google Scholar]
- 19.Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 20.Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., Jentsch S. (2000) Cell 102, 577–586 [DOI] [PubMed] [Google Scholar]
- 21.Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 22.Zhang M., MacDonald A. I., Hoyt M. A., Coffino P. (2004) J. Biol. Chem. 279, 20959–20965 [DOI] [PubMed] [Google Scholar]
- 23.Loetscher P., Pratt G., Rechsteiner M. (1991) J. Biol. Chem. 266, 11213–11220 [PubMed] [Google Scholar]
- 24.Takeuchi J., Chen H., Coffino P. (2007) EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janse D. M., Crosas B., Finley D., Church G. M. (2004) J. Biol. Chem. 279, 21415–21420 [DOI] [PubMed] [Google Scholar]
- 26.Pickart C. M., Raasi S. (2005) Methods Enzymol. 399, 21–36 [DOI] [PubMed] [Google Scholar]
- 27.Kwok S. C., Hodges R. S. (2003) J. Biol. Chem. 278, 35248–35254 [DOI] [PubMed] [Google Scholar]
- 28.Jacobson A. D., Zhang N. Y., Xu P., Han K. J., Noone S., Peng J., Liu C. W. (2009) J. Biol. Chem. 284, 35485–35494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rape M., Kirschner M. W. (2004) Nature 432, 588–595 [DOI] [PubMed] [Google Scholar]
- 30.Lin Y., Hwang W. C., Basavappa R. (2002) J. Biol. Chem. 277, 21913–21921 [DOI] [PubMed] [Google Scholar]
- 31.Evans T. C., Jr., Benner J., Xu M. Q. (1999) J. Biol. Chem. 274, 18359–18363 [DOI] [PubMed] [Google Scholar]
- 32.Braun B. C., Glickman M., Kraft R., Dahlmann B., Kloetzel P. M., Finley D., Schmidt M. (1999) Nat. Cell Biol. 1, 221–226 [DOI] [PubMed] [Google Scholar]
- 33.Strickland E., Hakala K., Thomas P. J., DeMartino G. N. (2000) J. Biol. Chem. 275, 5565–5572 [DOI] [PubMed] [Google Scholar]
- 34.Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Mol. Cell 34, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang F., Wu Z., Zhang P., Tian G., Finley D., Shi Y. (2009) Mol. Cell 34, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoskins J. R., Sharma S., Sathyanarayana B. K., Wickner S. (2001) Adv. Protein Chem. 59, 413–429 [DOI] [PubMed] [Google Scholar]
- 37.Keiler K. C., Waller P. R., Sauer R. T. (1996) Science 271, 990–993 [DOI] [PubMed] [Google Scholar]
- 38.Martin A., Baker T. A., Sauer R. T. (2008) Mol. Cell 29, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 40.Yao T., Cohen R. E. (2002) Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 41.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 42.Walz J., Erdmann A., Kania M., Typke D., Koster A. J., Baumeister W. (1998) J. Struct. Biol. 121, 19–29 [DOI] [PubMed] [Google Scholar]
- 43.Navon A., Goldberg A. L. (2001) Mol. Cell 8, 1339–1349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.