Abstract
Many bacterial pathogens, including Pseudomonas aeruginosa, have a nonhomologous end joining (NHEJ) system of DNA double strand break (DSB) repair driven by Ku and DNA ligase D (LigD). LigD is a multifunctional enzyme composed of a ligase domain fused to an autonomous polymerase module (POL) that adds ribonucleotides or deoxyribonucleotides to DSB ends and primer-templates. LigD POL and the eukaryal NHEJ polymerase λ are thought to bridge broken DNA ends via contacts with a duplex DNA segment downstream of the primer terminus, a scenario analogous to gap repair. Here, we characterized the gap repair activity of Pseudomonas LigD POL, which is more efficient than simple templated primer extension and relies on a 5′-phosphate group on the distal gap strand end to confer apparent processivity in filling gaps of 3 or 4 nucleotides. Mutations of the His-553, Arg-556, and Lys-566 side chains implicated in DNA 5′-phosphate binding eliminate the preferential filling of 5′-phosphate gaps. Mutating Phe-603, which is imputed to stack on the nucleobase of the template strand that includes the 1st bp of the downstream gap duplex segment, selectively affects incorporation of the final gap-closing nucleotide. We find that Pseudomonas Ku stimulates POL-catalyzed ribonucleotide addition to a plasmid DSB end and promotes plasmid end joining by full-length Pseudomonas LigD. A series of incremental truncations from the C terminus of the 293-amino acid Ku polypeptide identifies Ku-(1–229) as sufficient for homodimerization and LigD stimulation. The slightly longer Ku-(1–253) homodimer forms stable complexes at both ends of linear plasmid DNA that protect the DSBs from digestion by 5′- and 3′-exonucleases.
Keywords: DNA/Polymerase, DNA/Protein Interaction, DNA/Repair, Enzymes/Mechanisms, Nucleic Acid/Enzymology, Protein/Binding/DNA
Introduction
Ku and DNA ligase D (LigD)2 are the central agents of the bacterial nonhomologous end joining (NHEJ) pathway of DNA double strand break (DSB) repair (1, 2). The bacterial taxa that encode Ku and LigD include diverse pathogens of humans and plants, e.g. Mycobacterium, Pseudomonas, Bacillus, and Agrobacterium. The signature feature of NHEJ in mycobacteria is that half the repair events at blunt or complementary 5′-overhang DSBs are unfaithful, because the DSB ends are extended by polymerases or resected by nucleases prior to sealing by ligase (3, 4). Ku is a homodimeric protein (5, 6) that binds DSB ends and recruits LigD (5). LigD is a large multifunctional enzyme consisting of an ATP-dependent ligase domain, a polymerase domain (POL), and a 3′-phosphoesterase domain (3, 5–10).
Biochemical characterization of the POL and phosphoesterase components suggests that they provide a means of remodeling the 3′ ends of broken DNA strands prior to sealing by the ligase component (3, 5–17). The LigD POL domain catalyzes either nontemplated single nucleotide additions to a blunt-ended duplex DNA or fill-in synthesis at a 5′-tailed duplex DNA. These are the molecular signatures of mutagenic bacterial NHEJ in vivo at blunt-end and 5′-overhang DSBs, respectively (3, 4). A mutation of the LigD POL active site results in increased fidelity of blunt-end DSB repair in vivo by virtue of eliminating the nontemplated nucleotide insertions at the unfaithful recombination junctions (4). Thus, LigD POL is a direct catalyst of mutagenic NHEJ in vivo. LigD POL can also add nontemplated nucleotides to a single-stranded primer, which could be relevant to unfaithful NHEJ at a 3′-overhang DSB end (18).
The polymerase activities of LigD POL in vitro are optimal in the presence of manganese. rNTPs are preferred over dNTPs as substrates for both nontemplated blunt-end addition and templated fill-in synthesis. During templated synthesis in the presence of dNTPs on a DNA primer-template with an 18-nucleotide 5′ tail, the primer is elongated to the end of the template strand and then further extended with a single nontemplated nucleotide. LigD POL can also add templated ribonucleotides to a DNA primer, but extension is limited to about four cycles of rNMP incorporation because the primer-template is rendered progressively less active as ribonucleotides accumulate at the 3′ end (10). These properties suggest that the initial insertions preceding the strand sealing step of NHEJ might involve rNMP incorporation and that the ability of LigD to use rNTPs as substrates might be advantageous for the repair of chromosomal breaks that arise in quiescent cells, insofar as the dNTP pool might be limiting when bacteria are not actively replicating. Indeed, we reported previously that in vitro repair of a double strand break by Pseudomonas LigD requires the POL module and results in incorporation of an alkali-labile ribonucleotide at the repair junction (17). Such results illuminate an underlying logic for the domain organization of LigD, whereby the polymerase and phosphoesterase domains can heal the broken 3′ end to generate a ribonucleotide terminus favored by the bacterial NHEJ ligases (17).
Another notable property of LigD POL is that it is relatively “unfaithful” and can incorporate a mispaired nucleotide during templated primer extension when the correct nucleotide is not available. Incorrect nucleotides are added much more rapidly with rNTP substrates than with dNTPs, no matter what the mispair configuration (13). Furthermore, 3′ mispairs can be extended by LigD POL, again more rapidly with rNTP substrates than with dNTPs. These results fortify the suggestion that LigD POL might fill in short 5′-overhangs with ribonucleotides when repairing DSBs.
Abasic sites are one of the most common forms of DNA damage, and they can exert profound effects when polymerases encounter an abasic lesion in the template DNA strand. LigD POL can add a nucleotide opposite an abasic lesion, and here again ribonucleotides are inserted more rapidly than are deoxynucleotides (13). LigD POL displays feeble activity in extending a preformed primer terminus opposing an abasic site, but it can readily bypass the lesion by slippage of the primer 3′ di- or trinucleotide and realignment to the template sequence distal to the abasic site (13). Slippage and realignment might aid in bridging a DSB and thereby permit cross-break synthesis during certain NHEJ scenarios in vivo.
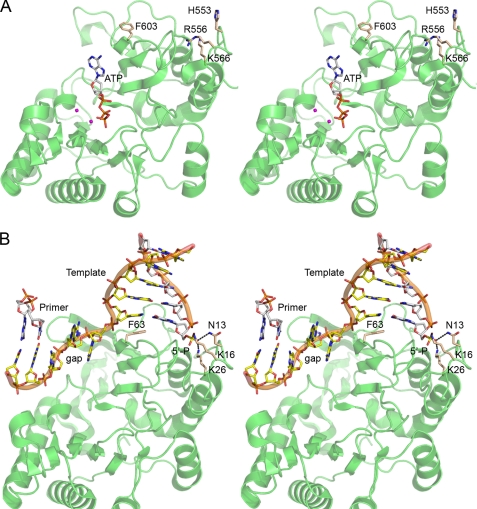
The 1.5 Å crystal structure of Pseudomonas LigD POL in complex with ATP and two manganese ions (Fig. 1 A, PDB 2faq) provided insights to the mechanism, catalytic repertoire, and origins of the enzyme (11). POL is a globular protein composed of 11 β-strands, organized mainly as two central sheets, flanked by seven α-helices. A six-strand antiparallel sheet, a three-strand antiparallel sheet, and the respective interstrand loops form a central cleft to which an NTP substrate binds in the absence of any instruction from a DNA primer-template. The crystal structure of M. tuberculosis LigD POL bound to GTP (PDB 2irx) (12) is very similar to that of Pseudomonas POL (DALI Z score 32.7). The fold of LigD POL most closely resembles the catalytic domain of archaeal DNA primases (PDB 1g71, 1v34, and 1zt2; Z scores 11.8 to 14.1) (19–21) and, to a lesser degree, an archaeal plasmid-encoded primase-polymerase (PDB 1ro0; Z score 6.6) (22). The LigD POL fold is unrelated to that of the pol X polymerase family, which includes human DNA pol λ, a low fidelity polymerase, and a central agent of mutagenic eukaryal NHEJ (23, 27, 28).
FIGURE 1.
Structures of bacterial LigD POL bound to NTP substrate or gap-like DNA. A, stereo view of P. aeruginosa LigD POL with ATP (stick model) and two manganese ions (magenta spheres) bound in the active site (PDB 2faq). B, stereo view of M. tuberculosis LigD POL bound to a gap-like DNA (PDB 2r9l), with the MtuPOL oriented so that it is superimposed on the PaePOL structure in A. The three amino acids (Asn-13, Lys-16, and Lys-26) that comprise the gap DNA 5′-phosphate binding pocket of MtuPOL are shown as stick models, as is the phenylalanine (Phe-63) that stacks on the template strand base of the 1st bp of the downstream gap duplex segment. The equivalent amino acids in PaePOL (His-553, Arg-556, Lys-566, and Phe-603) are shown as sticks models in A.
Important clues to the NHEJ activity of LigD POL and pol λ emerged from the crystal structures of the respective enzymes in binary complexes with gapped or gap-like DNAs in which the distal duplex boundary of the gap contained a DNA strand with a 5′-monophosphate terminus (24, 25). Although neither complex contained an NTP and in neither case was the 3′-OH primer terminus engaged productively in the active site, both structures revealed the following: (i) coordination of the distal 5′-phosphate anion by amino acid side chains of the enzyme; (ii) compression or distortion of the template single strand within the gap segment; and (iii) stacking of the first nucleobase of the distal gap duplex segment on an aromatic side of the polymerase (a tryptophan in pol λ or a phenylalanine in LigD POL). The structure of the complex of mycobacterial LigD POL with a gap-like DNA (PDB 2r9l) (25) is shown in Fig. 1B, with the primer strand pared back (to eliminate the unpaired bases) to the point where it is fully base-paired with the template strand; this DNA complex now mimics a 3-nucleotide gap.
The LigD POL and pol λ structures focus attention on the distal 5′-phosphate as a principal anchor point for polymerase on DNA and on gap filling as a preferred mode of nucleotide addition (12, 24, 25). Moreover, the structures suggest a scenario for cross-gap synthesis during NHEJ in which the polymerase binds to the 5′-phosphate terminus of one DSB end but adds nucleotides to the 3′-OH terminus of the other DSB end and continues incorporating nucleotides to the point that it fills in whatever gap separates the DSBs. This mechanism is quite distinct from the standard model of polymerization in which the primer end of the primer-template is the dominant factor in polymerase recruitment. Subsequent structures of pol λ bound to gapped DNA in a catalytically active conformation with the next templated dNTP (26) have underscored the importance of the distal DNA 5′-phosphate binding pocket and the flexibility of the template strand in the gap.
Our previous studies of Pseudomonas aeruginosa LigD POL (PaePOL) were performed using conventional primer-templates in which the primer strand was 5′-32P-labeled. Whereas PaePOL could extend virtually all the input primer strands, and was very effective at scavenging limiting (submicromolar) amounts of nucleoside triphosphates (10), the templated extension reactions generally required a large molar excess of PaePOL over primer-template (13), suggesting that a simple 5′-tailed duplex might not be the optimal substrate. Here, we take a cue from the mycobacterial POL and human pol λ DNA complexes and examine the effects of a downstream duplex, with and without a 5′-phosphate, to probe the gap filling capacity of PaePOL. We find that gap filling is indeed more efficient than simple templated primer extension and that the 5′-phosphate is a key mediator of this effect, by conferring processivity in filling gaps of 3 or 4 nucleotides. We report mechanistically informative effects of PaePOL mutations in the 5′-phosphate-binding and DNA gap-binding sites on gap filling activity.
We extend the present analysis of NHEJ structure-function relations to the Pseudomonas Ku protein, which has a distinctive C-terminal domain of unknown function. A series of incremental C-terminal truncations were evaluated for their effects on Ku heterodimerization, DNA end binding, and functional cooperation with LigD.
EXPERIMENTAL PROCEDURES
Recombinant LigD, POL, and POL Mutants
Full-length PaeLigD and the autonomous POL domain (amino acids 533–840) were produced in Escherichia coli as N-terminal His10 fusions and purified from soluble bacterial extracts by nickel-agarose chromatography as described previously (10). Missense mutations were introduced into the POL open reading frame by two-stage PCR overlap extension. The PCR products were digested with NdeI and BamHI and then inserted into pET16b. The inserts were sequenced completely to confirm the desired mutations and exclude the acquisition of unwanted changes during amplification and cloning. The POL mutants were produced in E. coli as His10 fusions and then purified from soluble bacterial lysates in parallel with wild-type POL. Protein concentrations were determined with the Bio-Rad dye-binding reagent using bovine serum albumin as the standard.
Polymerase Assays
The primer-template and gapped DNAs used as substrates for PaePOL were formed by annealing gel-purified 5′-32P-labeled 11-, 12-, 13-, or 14-mer oligodeoxyribonucleotides to a 4-fold molar excess of an unlabeled complementary 25-mer single strand, a 3′-tailed hairpin strand with a 5′-OH terminus, or a 3′-tailed hairpin strand with a 5′-phosphate terminus. The DNA mixtures in 0.2 m NaCl were heated at 95 °C for 7 min, followed by slow cooling to room temperature. Polymerase reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 5 mm MnCl2, 100 μm rNTPs or dNTPs, 1 pmol (50 nm) of 5′-32P-labeled DNA substrate, and enzyme as specified were incubated at 37 °C for 20 min. The reactions were quenched by adjusting the mixtures to 10 mm EDTA, 48% formamide. The products were resolved by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 m urea in TBE (90 mm Tris borate, 2.5 mm EDTA).
Recombinant Ku and Ku Truncation Mutants
The C-terminal deletion mutants of P. aeruginosa Ku were constructed by PCR amplification of the Ku gene with antisense strand primers that introduced stop codons immediately following the codons for Phe-215, Arg-229, Glu-240, Asp-253, Ala-263, Glu-272, and Lys-283 plus a BamHI site 3′ of the new stop codons. The truncated Ku open reading frames were digested with NdeI and BamHI and then inserted into pET16b. The inserts of the pET-His10KuΔ plasmids were sequenced completely to exclude the acquisition of unwanted change during amplification and cloning. The pET plasmids encoding full-length Ku and the KuΔ mutants were transformed into E. coli BL21(DE3). Cultures (1 liter) of E. coli BL21(DE3)/pET-His10Ku were grown at 37 °C in Luria-Bertani medium containing 0.1 mg/ml ampicillin until the A600 reached 0.6. The cultures were adjusted to 0.5 mm isopropyl-β-d-thiogalactopyranoside and then incubated at 17 °C for 15 h. The cells were harvested by centrifugation, and the pellets were stored at −80 °C. All subsequent steps were performed at 4 °C. Thawed bacteria were resuspended in 50 ml of lysis buffer (50 mm Tris-HCl (pH 7.5), 0.5 m NaCl, 200 mm Li2SO4, 10% glycerol, 15 mm imidazole). Lysozyme and Triton X-100 were added to final concentrations of 50 μg/ml and 0.1%, respectively. The lysates were sonicated to reduce viscosity, and insoluble material was removed by centrifugation. The supernatants were applied to 2-ml columns of Ni2+-nitrilotriacetic acid-agarose (Qiagen) that had been equilibrated with lysis buffer. The columns were washed with 20 ml of lysis buffer and then eluted stepwise with 4-ml aliquots of buffers containing 50, 100, 200, and 500 mm imidazole in 50 mm Tris-HCl (pH 7.5), 0.4 m NaCl, 10% glycerol. The polypeptide compositions of the fractions were monitored by SDS-PAGE. Ku was recovered predominantly in the 500 mm imidazole eluate fraction. Protein concentrations were determined with the Bio-Rad dye-binding reagent, using bovine serum albumin as the standard.
Glycerol Gradient Sedimentation
Aliquots of the nickel-agarose preparations of the KuΔ proteins were mixed with catalase (40 μg), bovine serum albumin (40 μg), and cytochrome c (40 μg) in 200 μl of buffer containing 50 mm Tris-HCl (pH 7.5), 200 mm NaCl, 10% glycerol. The mixtures were applied to 4.8-ml of 15–30% glycerol gradients containing 50 mm Tris-HCl (pH 8.0), 0.2 m NaCl, 1 mm EDTA, 2.5 mm DTT, 0.1% Triton X-100. The gradients were centrifuged in a Beckman SW 50 rotor at 50,000 rpm for 18 h at 4 °C. Fractions (∼0.2 ml) were collected from the bottoms of the tubes. The polypeptide compositions of the odd-numbered gradient fractions were analyzed by SDS-PAGE.
RESULTS
Filling a 4-Nucleotide Gap with Deoxynucleotides Is Stimulated by a 5′-Phosphate on the Distal Duplex
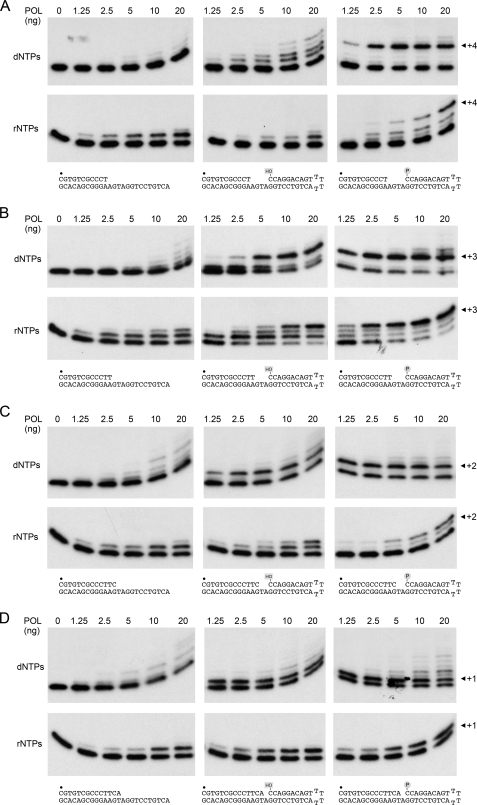
Gapped DNA substrates for PaePOL were prepared by annealing a 5′-32P-labeled primer strand (either an 11-, 12-, 13-, or 14-mer) to the 3′ tail of a 39-mer 3′-tailed hairpin duplex oligonucleotide to form the series of substrates depicted in Fig. 2, which have 4-, 3-, 2-, or 1-nucleotide gaps, respectively. Two otherwise identical hairpin duplexes were employed that had either a 5′-OH or a 5′-phosphate terminus at the distal boundary of the gap (Fig. 2). The gapped substrates were tested for primer extension activity with PaePOL, in parallel with standard primer-templates formed by annealing the same radiolabeled primers to a complementary single strand DNA oligonucleotide identical in sequence to the template strand of the gapped DNAs. The primer extension and gap filling reactions were performed in the presence of 100 μm dNTPs with increasing amounts of input PaePOL, albeit always with the DNA (50 nm) in excess over the enzyme.
FIGURE 2.
Gap filling versus template primer extension. Reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 5 mm MnCl2, 100 μm rNTPs or dNTPs, 1 pmol (50 nm) of 32P-labeled DNAs (shown at the bottom of the figure), and 0, 1.25, 2.5, 5, 10, or 20 ng of PaePOL (corresponding to 0, 1.7, 3.4, 6.8, 13.5, and 27 nm enzyme) were incubated for 20 min at 37 °C. The products were resolved by PAGE and visualized by autoradiography. The 5′-32P label is indicate by ●.
The salient findings are illustrated in Fig. 2A, where we see that there is scant dNMP addition to the standard primer-template at substoichiometric PaePOL levels. The few primers that have reacted are extended by just one, two, or three nucleotides, rather than to the end of the template strand, suggesting that the enzyme is not acting processively. The creation of a 4-nucleotide gap by the distal hairpin duplex with a 5′-OH end clearly stimulated primer utilization, although most of the reactive primers were extended by just one or two dNMPs. By contrast, the 4-nucleotide gap with a 5′-phosphate on the distal duplex was filled in completely, at even the lowest levels of input POL, to yield a +4 extension product, and there was scant accumulation of +1, +2, or +3 elongation intermediates (Fig. 2A). Thus, the 5′-phosphate group appeared to render PaePOL processive when filling in a 4-nucleotide gap with dNMPs. From the fraction of primers extended by 5 ng of PaePOL with the 5′-phosphate gap (61%), 5′-OH gap (24%), and primer-template (6.5%) DNAs, we estimate that the gap duplex per se increases primer utilization ∼4-fold and the 5′-phosphate exerts an additional 2.5-fold stimulation. Our inferences about substrate preference and processivity were underscored by the kinetic profile of gap filling versus templated primer extension, performed at a PaePOL/primer ratio of ∼0.7 (supplemental Fig. S1A). Whereas little (<3%) of the primer-template was extended by 4 nucleotides after 5 min, a substantial fraction (55%) of the 5′-phosphate gap DNAs was filled in completely. The 5′-phosphate group enhanced the rate of gap filling compared with the 5′-OH gap, such that +4 products were detected by 20 s and increased in abundance thereafter, with little accumulation of elongation intermediates.
Effects of Gap Size on dNMP Addition
Similar positive effects of a downstream duplex and 5′-phosphate were noted when PaePOL was presented with a 3-nucleotide gapped substrate versus a primer-template (Fig. 2B). Whereas only 2% of primer-templates were elongated by 3 or more nucleotides by 5 ng of POL, 34% of the 5′-OH gaps and 67% of the 5′-phosphate gaps were closed completely to yield +3 products (Fig. 2B). Full gap closure occurred at lower POL levels on the 5′-phosphate DNA than the 5′-OH DNA. Whereas almost no intermediate species were seen with the 5′-phosphate gapped DNA, there was a clear +1 intermediate accumulating at lower POL levels on the 5′-OH gapped substrate. The kinetic profile of the 3-nucleotide gap filling with dNMPs highlighted the same themes, especially faster gap closure on the 5′-phosphate DNA (supplemental Fig. S1B). To wit, 1% of primer-templates were elongated by 3 or more nucleotides after 1 min, compared with 7% of the 5′-OH gaps and 43% of the 5′-phosphate gaps.
The advantage of the 5′-phosphate versus 5′-OH began to erode as the gap was reduced to 2 nucleotides, although the gap duplex still increased the yield of ≥+2 products compared with the primer-template, whether as a function of enzyme (Fig. 2C) or reaction time (supplemental Fig. S1C). We discerned no preference for the 5′-phosphate versus 5′-OH in the context of a 1-nucleotide gap, although again the gap duplex had a positive effect on primer utilization compared with the standard primed template (Fig. 2D and supplemental Fig. S1D). It was noteworthy that PaePOL could extend a few nucleotides into the duplex segment after closing the 1-nucleotide gap; minor extension products past the closed gap were also detected on the longer gapped DNAs. We presume this reflects fraying of the duplex end. (Note that the POL domain has no 5′-exonuclease activity.)
Gap Filling with Ribonucleotides
Here, as previously (13), we noted that rNTPs were superior to dNTPs as substrates for the first nucleotide addition step to a standard primer-template, whether as a function of enzyme (Fig. 2A) or reaction time (supplemental Fig. S1A). The predominant outcome at substoichiometric enzyme levels was addition of a singe rNMP. Ribonucleotide addition to a 4-nucleotide gapped DNA with a downstream 5′-OH duplex terminus was limited to one or two steps (Fig. 2A and supplemental Fig. S1A). By contrast, when a 5′-phosphate was present at the distal margin of the gap, PaePOL was able to complete the fill-in and yield a +4 product (Fig. 2A and supplemental Fig. S1A). Processive closure of the gap appeared to be less efficient with rNTPs than dNTPs, but this effect (seen as the persistence of elongation intermediates) reflects the previously documented decline in primer utilization as each ribonucleotide is added to the DNA primer end (14). Thus, the 5′-phosphate of the gap overcomes, to some extent, the inhibitory effect of an RNA:DNA hybrid structure at the primer terminus.
The salutary effects of the gap duplex on rNMP incorporation were more easily appreciated with the 3-nucleotide gapped DNA. The 5′-OH gap sufficed to allow complete gap closure and generation of a +3 product, which was virtually undetectable when PaePOL was reacted in parallel with a simple primer-template (Fig. 2B and supplemental Fig. S1B). In both cases, the +1 species accumulated to substantial levels before the second rNMP was added. By contrast, complete gap filling with a +3 product was the predominant outcome when 2.5–20 ng of PaePOL reacted with the 5′-phosphate gapped DNA (Fig. 2B), and there was comparatively less accumulation of the +1 and +2 intermediates during the gap filling reaction (Fig. 2B and supplemental Fig. S1B). The percentages of the input primers elongated by 3 or more rNMPs by 10 ng of POL in Fig. 2B were as follows: 67% for the 5′-phosphate gap; 30% for the 5′-OH gap; and 2% for the primer-template. Thus, the 5′-phosphate moiety appeared to enhance the processivity of 3- and 4-nucleotide gap filling with both rNTP and dNTP substrates. The 5′-phosphate effect on ribonucleotide addition was ablated when the gap was shortened to 2-nucleotides or 1-nucleotide (Fig. 2, C and D, and supplemental Fig. S1, C and D).
Effects of Mutations in the Gap 5′-Phosphate Binding Pocket
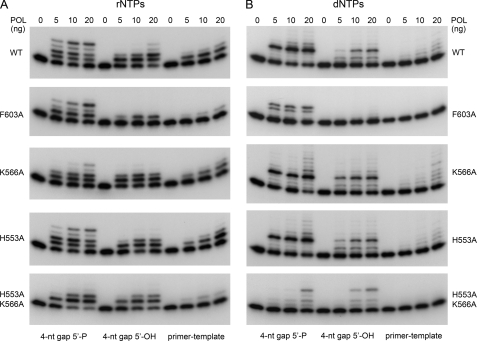
The structure of Mycobacterium tuberculosis LigD-POL (MtuPOL) in complex with a gap-like DNA highlighted a network of electrostatic and polar contacts to the gap 5′-phosphate from side chains Lys-16, Lys-26, and Asn-13 (Fig. 1B) (25). The equivalent residues in PaePOL are Arg-556, Lys-566, and His-553 (Fig. 1A). To gauge the role of these putative 5′-phosphate binding groups in the PaePOL gap repair activities, we changed them to alanine, individually and in combination. Mutants R556A, K566A, H553A, H553A/K566A, and H553A/R556A/K566A were produced in E. coli as His10-tagged fusions and purified from soluble bacterial extracts by nickel-agarose chromatography, in parallel with wild-type PaePOL (supplemental Fig. S2). The H553A change had no apparent effect on rNMP additions (Fig. 3A) or dNMP additions (Fig. 3B) to the 4-nucleotide gapped DNAs or to the equivalent primer-template DNA. The K566A mutation had little or no impact on dNMP additions (Fig. 3B), but it appeared to efface the difference in efficacy of rNMP addition to 5′-phosphate versus 5′-OH 4-nucleotide gapped DNAs (Fig. 3A). This trend matured fully with the H553A/K566A double mutation, which equalized PaePOL ribonucleotide addition activity on the 5′-phosphate and 5′-OH 4-nucleotide gapped DNAs, limited in either case to one or two steps (Fig. 3A). H553A/K566A ablated the preference of PaePOL for a 5′-phosphate gap with dNTPs and the processivity in gap closure (Fig. 3B). These results underscored the critical, albeit functionally redundant, roles of His-553 and Lys-566 in docking PaePOL at the gap 5′-phosphate downstream of the primer site. The functional overlap of these two side chains in PaePOL is consistent with the fact that their Asn-13 and Lys-26 counterparts in MtuPOL make atomic contacts to the same 5′-phosphate oxygen atom (25).
FIGURE 3.
Effects of POL mutations on gap filling. Reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 5 mm MnCl2, 100 μm rNTPs (A) or dNTPs (B), 50 nm 32P-labeled DNAs as specified, and 0, 1.25, 2.5, 5, 10, or 20 ng of wild type (WT) or mutant PaePOL as specified (corresponding to 0, 1.7, 3.4, 6.8, 13.5, and 27 nm enzyme) were incubated for 20 min at 37 °C. The products were resolved by PAGE and visualized by autoradiography.
The R556A change equalized PaePOL gap filling activity with rNTPs on the 5′-phosphate and 5′-OH 4-nucleotide gapped substrates, such that neither gap was closed completely (supplemental Fig. S3B). (The equivalent MtuPOL K16A mutation negated the stimulation by a gapped 5′-phosphate of rNMP addition to a 2-nucleotide gapped DNA (25).) By contrast, PaePOL R556A remained capable of processively closing the 4-nucleotide gap with dNTPs (supplemental Fig. S3A). We would attribute the distinctive effects of R556A versus H553A/K566A to the fact that the Arg-556 equivalent in MtuPOL (Lys-16) makes atomic contact with a different 5′-phosphate oxygen than do the other two residues that comprise the phosphate-binding pocket (Fig. 1B). Superposition of PaePOL on the MtuPOL·DNA structure suggests that Arg-556 would make a bidentate contact to two oxygen atoms of the gap 5′-phosphate. As might be expected, the H553A/R556A/K556A triple mutation ablated the 5′-phosphate preference for both rNMP and dNMP additions by PaePOL at a 4-nucleotide gap (supplemental Fig. S3).
Role of Phenylalanine Stacking on the Distal Gap Duplex 5′- Nucleobase
A shared feature of the structures of MtuPOL and pol λ bound to gapped DNAs is the stacking of an aromatic side chain (Trp-274 in pol λ and Phe-63 in MtuPOL) against the nucleobase of the template strand that includes the 1st bp of the downstream gap duplex segment (24, 25). In the MtuPOL·DNA structure, Phe-63 and its surrounding β-strand insinuate between adjacent nucleotides of the template strand and splay them apart, thereby kinking the gap single strand into a trajectory that is nearly orthogonal to the duplex DNA at the end of the gap (Fig. 1B). To interrogate the significance of this distortion, we changed the corresponding phenylalanine in PaePOL (Phe-603) to alanine, purified the F603A protein (supplemental Fig. S2), and assayed the mutant for gap filling activity. F603A suppressed dNMP addition to the 4-nucleotide gapped DNA with a distal 5′-OH end and to the standard primer-template (Fig. 3B), signifying that this residue and its imputed contacts to the template strand were of general importance to template-primed DNA synthesis by PaePOL. The F603A mutant retained activity in dNMP addition to the 4-nucleotide gapped DNA with a distal 5′-phosphate end but with the novel twist that a significant fraction of the extension events seemed to halt after incorporation of three dNMPs (Fig. 3B), a phenomenon not seen during processive gap filling by wild-type PaePOL. Because F603A did not accumulate +2 or +1 extension products on the 5′-phosphate gapped DNA, we surmise that the Phe-603 interaction with the template is specifically implicated in the final dNMP addition step of gap closure. This view would be consistent with the idea that maintaining the “cracked-open” conformation of the primer-template duplex and distal duplex segments assists the last gap closure reaction.
Similar inferences were drawn from the effects of F603A on rNMP additions (Fig. 3A). Compared with wild-type PaePOL, the F603A mutant displayed lower activity in single rNMP addition on the primer-template DNA and decreased primer utilization with the 5′-OH gapped DNA, which was limited to a single round of dNMP incorporation at 20 ng of input F603A, in contrast to the prevalence of +2 addition products generated by wild-type PaePOL under the same conditions (Fig. 3A). Loss of the Phe-603 side chain exerted a striking effect on rNMP addition to the 5′-phosphate gapped DNA, whereby +1 and +3 additions predominated and virtually no complete filling of the 4-nucleotide gap was observed. These results underscore the aforementioned importance of Phe-603 interaction with the template strand bases, especially in the final gap closure step when the 5′-phosphate of the downstream duplex is anchored in the phosphate-binding pocket.
Ku Stimulates POL Activity at Plasmid DSB Ends
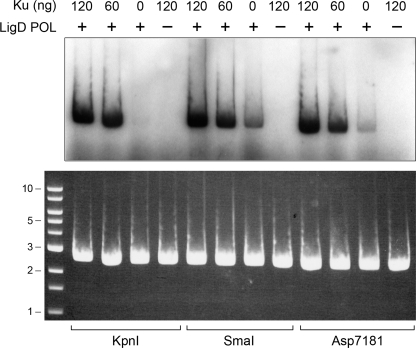
P. aeruginosa Ku (PaeKu) is a homodimer of a 293-amino acid polypeptide (6). The homologous M. tuberculosis Ku homodimer interacts physically and functionally with mycobacterial LigD (3, 5), especially with the LigD-POL domain (9). Here, we examined the properties of PaeKu and its impact on PaeLigD function. First, we queried whether PaeKu affects the ribonucleotide addition activity of PaePOL at a DSB end. Circular pUC19 plasmid DNA was digested with single-cutting restriction endonucleases KpnI, SmaI, or Asp-7181 to generate linear double strand DNAs with 3′-overhang, blunt, or 5′-overhang termini (Fig. 4). The linear plasmids served as primers for incorporation by PaePOL of [32P]AMP at the DSB 3′ ends. PaePOL per se displayed a basal polymerase activity, with blunt DNA being the best substrate for AMP addition. POL activity was stimulated by inclusion of Ku at a ∼3:1 molar ratio of Ku homodimer to the input DNA ends (Fig. 4). Ku itself had no polymerase activity.
FIGURE 4.
Ku stimulates POL activity. Reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 5 mm MnCl2, 1 μm [α-32P]ATP, 0.25 μg of linear pUC19 DNA (∼145 fmol of plasmid; 290 fmol of double strand breaks) with 3′-overhang (KpnI-cut), blunt (SmaI-cut), or 5′-overhang (Asp-7181-cut) ends, 0.2 μg (5.4 pmol) of PaePOL (where indicated by +) and 0, 60, or 120 ng of Ku (corresponding to 0, 0.85, or 1.7 pmol of Ku homodimer) were incubated at 37 °C for 20 min. The reactions were quenched by adjusting the mixtures to 0.2% SDS, 6 mm EDTA. The products were resolved by electrophoresis through a vertical 15-cm 1% agarose gel containing 0.05% ethidium bromide in TAE (40 mm Tris acetate, 1 mm EDTA). A photograph of the gel under UV illumination is shown in the bottom panel. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left. The gel was then dried on DEAE paper, and radiolabeled material was visualized by autoradiography (top panel).
Effects of C-terminal Truncations on Ku Activity and Quaternary Structure
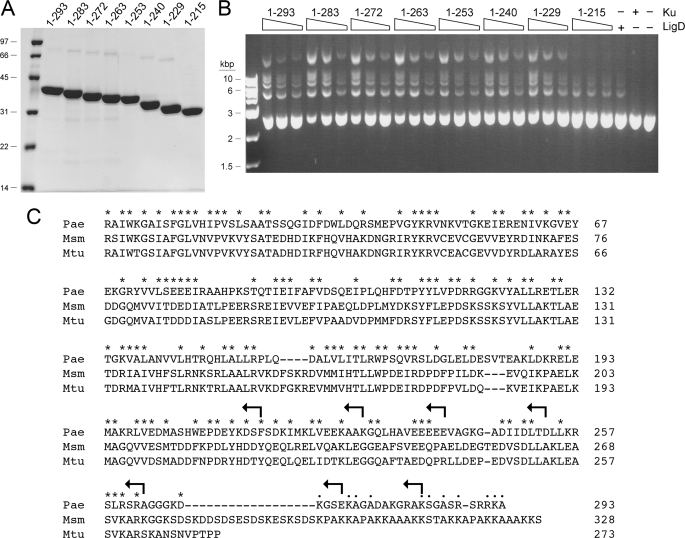
PaeKu displays extensive primary structure similarity to the Mycobacterium smegmatis (Msm) and M. tuberculosis (Mtu) Ku polypeptides over its N-terminal 262-amino acid segment (Fig. 5C). The three Ku proteins diverge with respect to the length and primary structures of their respective C-terminal peptides (Fig. 5C). PaeKu and MsmKu have extended C-terminal domains that have no counterpart in MtuKu. The 25-amino acid PaeKu C-domain is conspicuously rich in basic side chains (9 Lys/Arg residues) and in Ala/Gly/Ser (13 residues). The analogous segment of MsmKu is separated from the conserved Ku core domain by a linker rich in Lys, Asp, and Glu. To probe the function of the distinctive C-terminal peptide of PaeKu, and of the distal portion of the conserved Ku core domain, we created a series of seven incremental C-terminal truncations terminating at the positions indicated by arrows in Fig. 5C. Full-length PaeKu-(1–293) and the seven KuΔ mutants were produced in E. coli as His10 fusions and purified from soluble bacterial extracts by nickel-agarose chromatography. SDS-PAGE analysis revealed comparable purity and the expected incremental changes in electrophoretic mobility with each truncation (Fig. 5A).
FIGURE 5.
Ku stimulates LigD ligase activity. A, nickel-agarose preparations of full-length Ku-(1–293) and the indicated KuΔ mutants were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is show. The positions and sizes (kDa) of marker polypeptides are indicated on the left. B, reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 5 mm MnCl2, 0.25 mm ATP, 0.2 μg of KpnI-digested pUC19 DNA (∼115 fmol of plasmid; 230 fmol of double strand breaks), 0.1 μg (1 pmol) of PaeLigD, and 30, 60, or 120 ng (from right to left in each titration series) of full-length Ku or KuΔ mutants as specified were incubated for 30 min at 37 °C. Control reaction mixtures contained 0.1 μg of LigD alone, 120 ng of Ku alone, or neither protein. The reactions were quenched by adjusting the mixtures to 0.2% SDS, 6 mm EDTA. The products were resolved by electrophoresis through a horizontal 1% agarose gel containing TBE (90 mm Tris borate, 2.5 mm EDTA) and 0.05% ethidium bromide. A photograph of the gel under UV illumination is shown. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left. C, amino acid sequences of the Ku polypeptides of P. aeruginosa (Pae), M. smegmatis (Msm), and M. tuberculosis (Mtu) are aligned. Positions of side chain identity or similarity in all three proteins are indicated by an asterisk. Gaps in the alignment are indicated by a dash. The C termini of the KuΔ mutants are indicated by arrows.
We showed previously that PaeKu stimulated plasmid end joining by Agrobacterium NHEJ ligases (6). Here, we show that PaeKu also stimulates end joining by PaeLigD (Fig. 5B). The reaction mixtures contained ∼120 fmol of KpnI-digested pUC19 DNA, corresponding to 240 fmol of DSB ends. Whereas addition of a molar excess of PaeLigD (1 pmol) over the available DSB ends resulted in little end joining, the inclusion of full-length PaeKu-(1–293) stimulated the formation of a ladder of DNA concatemers, comprising dimer, trimer, and higher order products (Fig. 5B). Ku by itself had no end joining activity, as expected.
The salient findings were that six of the seven KuΔ mutants were as adept as wild-type PaeKu at stimulating plasmid end joining by PaeLigD. These were as follows: Ku-(1–283), Ku-(1–272), Ku-(1–263), Ku-(1–253), Ku-(1–240), and Ku-(1–229). We surmise that neither the C-terminal Lys/Arg-rich peptide nor the distal segment of the conserved Ku core (from amino acids 229 to 263) is critical for the functional interaction of PaeKu with PaeLigD, at least with respect to plasmid end joining in vitro. By contrast, the Ku-(1–215) protein did not stimulate LigD end joining activity (Fig. 5B), signifying that the segment from amino acids 216 to 229 includes structural elements important for Ku function.
The effects of the C-terminal truncations on Ku quaternary structure were gauged by zonal velocity sedimentation in glycerol gradients. Marker proteins catalase (native size 248 kDa; a homotetramer of a 62-kDa subunit), bovine serum albumin (66-kDa monomer), and cytochrome c (12-kDa monomer) were included as internal standards. The sedimentation profiles were assessed by SDS-PAGE of the gradient fractions collected from the bottoms of the tubes (supplemental Fig. S4). Ku-(1–272), Ku-(1–263), Ku-(1–253), Ku-(1–240), and Ku-(1–229) sedimented as single components at essentially the same position as bovine serum albumin (and well resolved from cytochrome c), consistent with a homodimeric quaternary structure for each. By contrast, Ku-(1–215) sedimented as a single lighter component, well separated from bovine serum albumin, that overlapped the heavy side of the cytochrome c peak (supplemental Fig. S4). We conclude that Ku-(1–215) is a monomer and infer that the segment from amino acids 216 to 229 is necessary for Ku dimerization. Taken together, the results of the end joining and sedimentation analyses suggest that Ku dimerization is required for Ku stimulation of end joining by LigD.
Binding of Ku to DSB Ends
Biochemical and genetic studies of mycobacterial Ku show that it binds linear DNAs in vitro (5) and protects DSBs from end resection in vivo (4). The DNA end-binding properties of Pseudomonas Ku have not been studied previously. Here, we incubated wild-type PaeKu and the various KuΔ mutants with linear pUC19 DNA and then analyzed the mixtures by native agarose gel electrophoresis, after which the gel was stained with ethidium bromide to visualize the DNA. Each Ku protein, with the exception of Ku-(1–215), elicited some degree of Ku·DNA complex formation, as evinced by the formation of either discretely or diffusely migrating DNAs of retarded electrophoretic mobility relative to the input linear plasmid (Fig. 6). We surmise that Ku dimerization is necessary for Ku binding to linear duplex DNA.
FIGURE 6.
Ku binding to linear plasmid DNA. Reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 2% glycerol, 0.25 μg of SmaI-digested pUC19 DNA (∼145 fmol of plasmid; 290 fmol of double strand breaks), and 0.25, 0.5, or 1 μg of full-length Ku or KuΔ mutants as specified were incubated at 22 °C for 20 min. Ku was omitted from the control reactions shown in lane −. The mixtures were analyzed by electrophoresis through a horizontal 1% agarose gel in TBE. The gel was then stained with 0.05% ethidium bromide and photographed under UV illumination. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left.
The pattern of the DNA mobility shift differed in informative ways between PaeKu proteins of different length. Full-length Ku-(1–293) yielded one or two discretely shifted DNA complexes visible within a background smear of DNA extending to the top of the agarose gel (Fig. 6). The smear effect was exacerbated by deletion of the C-terminal 10-amino acid peptide, such that interaction of Ku-(1–283) with the linear plasmid converted most of the input DNA into a diffusely migrating high molecular weight form (Fig. 6). This diffuse Ku·DNA complex was eliminated by the next incremental truncation to place the C terminus at residue 272, a maneuver that deleted the Lys/Arg-rich C-terminal domain. These findings engender a speculation that the C-terminal peptides of a PaeKu homodimer bound to linear DNA can make electrostatic contacts via their cluster of Lys and Arg side chains with the phosphodiester backbones of other DNA molecules, thereby forming a high molecular weight DNA network that we see as the “smear shift.”
The Ku-(1–272), Ku-(1–263), and Ku-(1–253) proteins, which include just the conserved Ku core domain, caused the sequential formation of discrete “singly shifted” and “doubly shifted” Ku·DNA complexes as the concentration of Ku was increased. A simple interpretation is that this pattern signifies stable binding of a Ku dimer, first to one end and then to both ends of the linear plasmid DNA. The sequential evolution of the two shifted complexes suggests that Ku end binding is not overtly cooperative. (If it were cooperative, the doubly shifted complex ought to predominate at limiting Ku concentrations.)
The affinity of Ku for the plasmid DNA ends appeared to vary depending on the C-terminal margin. Ku-(1–253) was the best binder, eliciting virtually complete conversion to a doubly shifted complex at 1 μg of input Ku (∼0.8 μm Ku homodimer). Further truncations of the C terminus resulted in sharply reduced yield of the two shifted Ku·DNA complexes; compare Ku-(1–240) and Ku-(1–229) to Ku-(1–253) (Fig. 6). Because DNA binding in the native gel assay relies on stability of the Ku·DNA complex during the electrophoretic separation, we suspect that the relatively superior binding of Ku-(1–253) reflects a slower off rate. (Note that the C-terminal truncation mutants did not vary significantly in stimulating end joining by LigD, suggesting that longevity of the Ku·DNA complex might not be a limiting factor for ligation.)
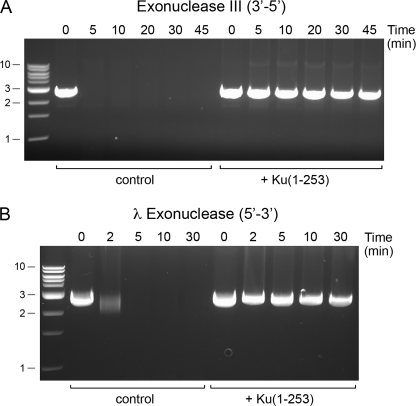
Ku Protects DSB Ends from Exonucleases
The native gel shift, revealing two discrete Ku·DNA complexes on linear DNA, suggests that Ku binds at each DSB end. Yet, the gel shift does not exclude alternative scenarios, e.g. that two Ku homodimers bind sequentially to the same DNA end or that Ku accesses the DNA end and then migrates internally. To probe how Ku is disposed on the linear plasmid, we incubated SmaI-cut pUC19 with Ku-(1–253) and then treated the Ku·DNA complexes with E. coli exonuclease III (a 3′–5′ double strand DNA exonuclease) or bacteriophage λ exonuclease (a 5′–3′ double strand DNA exonuclease). Whereas the “naked” linear plasmid DNA was rapidly digested by either exonuclease III or λ exonuclease, the inclusion of Ku afforded virtually complete protection from both exonucleases (Fig. 7). Because protein binding at only one DSB end, or at random internal sites, will not prevent end-resection by exonuclease III or λ exonuclease, we can confidently conclude that Ku-(1–253) is bound stably at, or closely adjacent to, both DSB ends of the exonuclease-resistant linear DNAs.
FIGURE 7.
Ku protects DSB ends. Reaction mixtures (140 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 2% glycerol, 1.75 μg of SmaI-digested pUC19, and 7 μg Ku-(1–253) were incubated at 22 °C for 20 min. Ku was omitted from the control reactions. An aliquot (20 μl) was withdrawn prior to exonuclease addition (time 0). Exonuclease digestions were performed by supplementing the remaining 120 μl of DNA-containing mixtures (±Ku) with 60 μl of a solution containing 2 mm MgCl2 and 150 units of exonuclease III (New England Biolabs) (A) or 60 μl of a solution containing 7.5 mm MgCl2 and 25 units of λ exonuclease (New England Biolabs) (B). The mixtures were then incubated at 37 °C. Aliquots (30 μl) were withdrawn at the times specified and quenched immediately with 0.2% SDS, 5 mm EDTA. The samples were analyzed by electrophoresis through a horizontal 1% agarose gel in TBE containing 0.05% ethidium bromide. Photographs of the gels under UV illumination are shown. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left.
DISCUSSION
This study of Pseudomonas LigD POL underscores the primacy of the gap filling functions of dedicated NHEJ polymerases and the influence of the gap 5′-phosphate end, a theme elaborated initially for human pol λ (24, 27) and subsequently for mycobacterial LigD POL (12, 25). The key findings here are as follows. (i) The DNA polymerase activity of PaePOL, which is distributive when adding dNMPs to a standard primer-template (10, 11, 13), is apparently processive when filling a 3- or 4-nucleotide gap that has a 5′-phosphate on the downstream duplex. (ii) The RNA polymerase activity of PaePOL, which is much faster than dNMP addition during the first catalytic cycle (13), but is self-inhibited with each further cycle of rNMP addition to a standard primer-template (14), is enhanced by a 5′-phosphate gap that stimulates gap closure with a 3- or 4-nucleotide RNA patch. (iii) The salutary effects of the gap 5′-phosphate on primer utilization, processivity, and resulting gap closure are mediated by a trio of conserved residues (His-553, Arg-556, and Lys-566) that directly coordinate the gap 5′-phosphate in the homologous MtuPOL·DNA complex (Fig. 1).
These results are consistent with the view enunciated previously (24) that the position of an NHEJ polymerase on its preferred gapped DNA substrate is dictated by the interactions of the enzyme with the gap 5′-phosphate rather than by its interactions (presumable weaker) with the primer 3′-OH terminus. Recent structural studies emphasize the distortion of the single strand segment that serves as the template for gap filling (Fig. 1) (25, 26). In the crystal structure of pol λ bound at the 2-nucleotide 5′-phosphate gap with the incoming templated dNTP in the active site and paired appropriately, the template single strand is “scrunched” so that the next template nucleobase is extrahelical (26). The net effect is that the primer terminus and the gap 5′-phosphate occupy the same positions when pol λ binds to the 2-nucleotide gap as when it is bound to the 1-nucleotide gap (26). In other words, the gap template strand is compressed to bring the primer end into the active site and then decompressed as each nucleotide is added. Although there is, as yet, no crystal structure of a bacterial LigD POL bound in a catalytically productive manner to a gapped DNA, it seems likely that the position of MtuPOL at the 5′-phosphate of the downstream duplex is preserved during gap filling.
Here, we provide evidence that the conserved Phe-603 side chain (that stacks on the template base that pairs with the 5′-phosphate nucleotide of the gap duplex) plays a role in maintaining the necessary kink between the primer-template segment and the distal duplex segment of the gapped DNA substrate. Changing Phe-603 to alanine selectively affected the final gap-closing nucleotide addition step on a 4-nucleotide 5′-phosphate gapped template. The F603A mutation also suppressed nucleotide additions to a standard primer-template, suggesting that insertion of Phe-603 between template strand bases (as seen in MtuPOL in Fig. 1B) is pertinent even when there is no downstream duplex. It is worth noting that Phe-603 and its conserved neighbor Phe-604 project in opposite directions from the main chain β-strand that splits apart adjacent bases in the template DNA (Fig. 1B). Whereas Phe-603 (equivalent to Phe-63 in MtuPOL) stacks on a template base on one side of the β-strand, Phe-604 (or Phe-64 in MtuPOL) stacks on the nucleobase of the incoming rNTP substrate in the polymerase active site on the opposite side of the β-strand (10, 12). Indeed, Phe-604 is the only side chain that contacts the ATP adenine in the PaePOL·ATP·(Mn2+)2 complex (10). As might be expected, Phe-604 is critical for templated rNMP and dNMP additions by PaePOL (10). When POL acts on standard primer-template, the position of the Phe-Phe “wedge” on the template strand is presumably determined by primer 3′-OH terminus, which implies that the Phe-603 interaction with the template strand would need to be reset after each polymerization step, either by POL dissociation and rebinding (consistent with the apparently distributive action on a primer-template) or else by a serial base flipping mechanism. However, when the POL is tethered to the 5′-phosphate of a downstream gap duplex, the position of the Phe-Phe wedge is fixed, and the enzyme can processively fill the short gap as the gap single strand “unscrunches,” and the primer end moves away from the NTP site after each cycle.
This study also provides new insights to structure-function relationships in Pseudomonas Ku and its interactions with Pseudomonas LigD. PaeKu stimulated ribonucleotide addition to DSB ends by PaePOL and DSB end sealing by LigD. We were especially interested in the distinctive C-terminal peptide domain of PaeKu (and MsmKu) that is apparently absent in MtuKu. We found that this basic peptide domain is dispensable for Ku homodimerization and for Ku stimulation of DSB sealing by LigD. The noteworthy effect of the basic C-terminal peptide was to promote the formation of higher order Ku-DNA networks that migrated diffusely during agarose gel electrophoresis, presumably via interactions of the cationic tails of the Ku homodimer with the anionic phosphodiester backbone of other DNAs. The diffuse Ku·DNA complexes were eliminated as soon as the C-terminal peptide was deleted, revealing the ability of the conserved Ku core domain to form two discrete shifted complexes on linear plasmid DNA. The added value, if any, of the C-domains in PaeKu (or MsmKu) remains to be determined. Conceivably, they might stabilize two approximated DSB ends by cross-break contacts to the DNA backbone. Alternatively, they might comprise a docking site for other DNA repair proteins.
By deleting further into the Ku core domain, we delineated a distal margin at position 229 for Ku homodimerization and LigD stimulation. Further truncation to position 215 eliminated both of those functions, as well as binding to linear plasmid DNA. A ψ-blast search with PaeKu yielded a primary structure alignment to the B chain (Ku80 subunit) of the human Ku70/Ku80 heterodimer bound to DNA (PDB 1jey) (29). The N terminus of PaeKu up to amino acid 236 (which embraces the Ku segment sufficient for homodimerization) aligned to the segment of Ku80 from residues 247 to 507. Inspection of the human Ku·DNA complex revealed that the deletion of PaeKu residues 216–229 (corresponding to Ku80 residues 487–500) would remove an entire α-helix that includes a major constituent of the dimer interface and the circumferential DNA interface (29). Thus, we can rationalize the results of the C-terminal deletion analysis in light of the human Ku-DNA structure. Our attempts to conduct a similar study of N-terminal deletions of PaeKu were stymied by insolubility of the recombinant Ku-NΔ proteins we produced in E. coli. In retrospect, this is perhaps not surprising, insofar as modeling the bacterial Ku homodimer based on the human Ku·DNA complex indicates that deleting 12 amino acids from the N terminus of PaeKu would eliminate the first β-strand of a seven-strand β-barrel that includes the core of the Ku tertiary structure (29).
The ability of the conserved PaeKu core homodimer to bind to DSB ends and protect them from digestion by two different DNA exonucleases resonates with previous genetic and biochemical studies of mycobacterial Ku. The majority of Ku-dependent NHEJ events at blunt and 5′-overhangs in M. smegmatis occur without nucleolytic resection (4). By contrast, when ku is ablated, nearly all of the residual NHEJ events at blunt and 5′-overhang DSBs involve deletions, and most of these have deletions at both DSB ends (4). Thus, Ku is implicated in protecting DSBs from resection in vivo. Furthermore, mycobacterial Ku protects linear plasmid DNA from digestion in vitro by the mycobacterial AdnAB complex (30), a DSB-resecting motor-nuclease machine implicated in mycobacterial DSB repair.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM63611.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- LigD
- ligase D
- NHEJ
- nonhomologous end joining
- DSB
- DNA double strand break
- POL
- polymerase module
- pol
- polymerase
- DTT
- dithiothreitol
- PDB
- Protein Data Bank.
REFERENCES
- 1.Shuman S., Glickman M. S. (2007) Nat. Rev. Microbiol. 5, 852–861 [DOI] [PubMed] [Google Scholar]
- 2.Pitcher R. S., Brissett N. C., Doherty A. J. (2007) Annu. Rev. Microbiol. 61, 259–282 [DOI] [PubMed] [Google Scholar]
- 3.Gong C., Bongiorno P., Martins A., Stephanou N. C., Zhu H., Shuman S., Glickman M. S. (2005) Nat. Struct. Mol. Biol. 12, 304–312 [DOI] [PubMed] [Google Scholar]
- 4.Aniukwu J., Glickman M. S., Shuman S. (2008) Genes Dev. 22, 512–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weller G. R., Kysela B., Roy R., Tonkin L. M., Scanlan E., Della M., Devine S. K., Day J. P., Wilkinson A., d'Adda di, Fagagna F., Devine K. M., Bowater R. P., Jeggo P. A., Jackson S. P., Doherty A. J. (2002) Science 297, 1686–1689 [DOI] [PubMed] [Google Scholar]
- 6.Zhu H., Shuman S. (2007) Nucleic Acids Res. 35, 3631–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akey D., Martins A., Aniukwu J., Glickman M. S., Shuman S., Berger J. M. (2006) J. Biol. Chem. 281, 13412–13423 [DOI] [PubMed] [Google Scholar]
- 8.Della M., Palmbos P. L., Tseng H. M., Tonkin L. M., Daley J. M., Topper L. M., Pitcher R. S., Tomkinson A. E., Wilson T. E., Doherty A. J. (2004) Science 306, 683–685 [DOI] [PubMed] [Google Scholar]
- 9.Pitcher R. S., Tonkin L. M., Green A. J., Doherty A. J. (2005) J. Mol. Biol. 351, 531–544 [DOI] [PubMed] [Google Scholar]
- 10.Zhu H., Shuman S. (2005) J. Biol. Chem. 280, 418–427 [DOI] [PubMed] [Google Scholar]
- 11.Zhu H., Nandakumar J., Aniukwu J., Wang L. K., Glickman M. S., Lima C. D., Shuman S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitcher R. S., Brissett N. C., Picher A. J., Andrade P., Juarez R., Thompson D., Fox G. C., Blanco L., Doherty A. J. (2007) J. Mol. Biol. 366, 391–405 [DOI] [PubMed] [Google Scholar]
- 13.Yakovleva L., Shuman S. (2006) J. Biol. Chem. 281, 25026–25040 [DOI] [PubMed] [Google Scholar]
- 14.Zhu H., Shuman S. (2005) J. Biol. Chem. 280, 25973–25981 [DOI] [PubMed] [Google Scholar]
- 15.Zhu H., Wang L. K., Shuman S. (2005) J. Biol. Chem. 280, 33707–33715 [DOI] [PubMed] [Google Scholar]
- 16.Zhu H., Shuman S. (2006) J. Biol. Chem. 281, 13873–13881 [DOI] [PubMed] [Google Scholar]
- 17.Zhu H., Shuman S. (2008) J. Biol. Chem. 283, 8331–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephanou N. C., Gao F., Bongiorno P., Ehrt S., Schnappinger D., Shuman S., Glickman M. S. (2007) J. Bacteriol. 189, 5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Augustin M. A., Huber R., Kaiser J. T. (2001) Nat. Struct. Biol. 8, 57–61 [DOI] [PubMed] [Google Scholar]
- 20.Ito N., Nureki O., Shirouzu M., Yokoyama S., Hanaoka F. (2003) Genes Cells 8, 913–923 [DOI] [PubMed] [Google Scholar]
- 21.Lao-Sirieix S. H., Nookala R. K., Roversi P., Bell S. D., Pellegrini L. (2005) Nat. Struct. Mol. Biol. 12, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 22.Lipps G., Weinzierl A. O., von Scheven G., Buchen C., Cramer P. (2004) Nat. Struct. Mol. Biol. 11, 157–162 [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Diaz M., Bebenek K., Krahn J. M., Pedersen L. C., Kunkel T. A. (2006) Cell 124, 331–342 [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Diaz M., Bebenek K., Krahn J. M., Blanco L., Kunkel T. A., Pedersen L. C. (2004) Mol. Cell 13, 561–572 [DOI] [PubMed] [Google Scholar]
- 25.Brissett N. C., Pitcher R. S., Juarez R., Picher A. J., Green A. J., Dafforn T. R., Fox G. C., Blanco L., Doherty A. J. (2007) Science 318, 456–459 [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Diaz M., Bebenek K., Larrea A. A., Havener J. M., Perera L., Krahn J. M., Pedersen L. C., Ramsden D. A., Kunkel T. A. (2009) Nat. Struct. Mol. Biol. 16, 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Díaz M., Bebenek K., Sabariegos R., Domínguez O., Rodríguez J., Kirchhoff T., García-Palomero E., Picher A. J., Juárez R., Ruiz J. F., Kunkel T. A., Blanco L. (2002) J. Biol. Chem. 277, 13184–13191 [DOI] [PubMed] [Google Scholar]
- 28.Lee J. W., Blanco L., Zhou T., Garcia-Diaz M., Bebenek K., Kunkel T. A., Wang Z., Povirk L. F. (2004) J. Biol. Chem. 279, 805–811 [DOI] [PubMed] [Google Scholar]
- 29.Walker J. R., Corpina R. A., Goldberg J. (2001) Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 30.Sinha K. M., Unciuleac M. C., Glickman M. S., Shuman S. (2009) Genes Dev. 23, 1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.