Abstract
The neonatal Fc receptor (FcRn) regulates the serum half-life of both IgG and albumin through a pH-dependent mechanism that involves salvage from intracellular degradation. Therapeutics and diagnostics built on IgG, Fc, and albumin fusions are frequently evaluated in rodents regarding biodistribution and pharmacokinetics. Thus, it is important to address cross-species ligand reactivity with FcRn, because in vivo testing of such molecules is done in the presence of competing murine ligands, both in wild type (WT) and human FcRn (hFcRn) transgenic mice. Here, binding studies were performed in vitro using enzyme-linked immunosorbent assay and surface plasmon resonance with recombinant soluble forms of human (shFcRnWT) and mouse (smFcRnWT) receptors. No binding of albumin from either species was observed at physiological pH to either receptor. At acidic pH, a 100-fold difference in binding affinity was observed. Specifically, smFcRnWT bound human serum albumin with a KD of ∼90 μm, whereas shFcRnWT bound mouse serum albumin with a KD of 0.8 μm. shFcRnWT ignored mouse IgG1, and smFcRnWT bound strongly to human IgG1. The latter pair also interacted at physiological pH with calculated affinity in the micromolar range. In all cases, binding of albumin and IgG from either species to both receptors were additive. Cross-species albumin binding differences could partly be explained by non-conserved amino acids found within the α2-domain of the receptor. Such distinct cross-species FcRn binding differences must be taken into consideration when IgG- and albumin-based therapeutics and diagnostics are evaluated in rodents for their pharmacokinetics.
Keywords: Antibodies, Protein/Ligand Binding, Receptors/Recycling, Blood, Protein Structure, Albumin, Biodistribution, Half-life, Immunoglobulin G, Neonatal Fc Receptor
Introduction
The major histocompatibility class I-related neonatal Fc receptor (FcRn)3 is a versatile receptor that regulates serum IgG half-life, transport of IgG across intestinal epithelia and placenta, as well as enhancement of neutrophil phagocytosis of immune complexes, as reviewed previously (1). Moreover, the receptor plays a role in antibody-mediated antigen presentation by dendritic cells (2). FcRn has also been found to salvage albumin from intracellular degradation (3), in a fashion similar to that described for IgG, which involves receptor ligand interactions in acidified endosomal compartments (1). Hence, FcRn affects diverse and important immunological and non-immunological processes.
FcRn is a heterodimeric receptor consisting of a transmembrane heavy chain (HC) that is non-covalently associated with β2-microglobulin (β2m). Consequently, the significance of FcRn has been extensively documented in knockout mouse models lacking β2m or the HC. Such deficient mice have IgG serum levels of 20–30% that of wild-type mice and a 60% reduced level of MSA (3, 4). A human example is the rare familial hypercatabolic hypoproteinemia syndrome that is characterized by reduced serum levels of both hIgG and HSA (5). An explanation was provided when deficient FcRn expression was demonstrated as a result of a point mutation in the β2m-encoding gene sequence that disrupts efficient secretion (6). Thus, FcRn is truly bifunctional and contributes to maintaining the high levels of IgG as well as albumin in serum, with levels amounting to ∼12 and ∼40 mg/ml, respectively, in mice and humans.
The FcRn HC consists of three ectodomains (α1, α2, and α3), a short transmembrane region, and a cytoplasmic tail (1). Mutagenesis and crystallographic studies have uncovered that the FcRn-IgG interaction is mediated by Fc-localized residues, especially Ile-253, His-310, and His-435, and acidic surface-exposed residues on the α2-domain of the HC (7–9). The interaction is strictly pH-dependent with binding at acidic pH and no or very weak binding at physiological pH. The histidines are mainly responsible for the pH dependence, because they are protonated under acidic conditions. Although the FcRn-albumin interaction is less well characterized, data indicate that domain III of albumin binds to the HC α2-domain at a site distant from the IgG binding site, because His-166 is crucial for the interaction (10, 11). Thus, both ligands may bind simultaneously in a pH-dependent manner.
Knowledge of FcRn-IgG biology explains the prolonged half-life of IgG Fc-fused therapeutics (1, 4, 12, 13). Understanding of the FcRn-IgG interaction at the atomic level has prompted the development of novel IgG-based therapeutics with point mutations in their Fc part that modulate serum half-life (14–19). Furthermore, improved half-life and efficiency of a number of small therapeutic molecules and proteins that are normally cleared rapidly from the circulation have been achieved by strategies such as chemical conjugation or genetic fusion to albumin itself (20–25) or any of several albumin binding molecules (26–29).
Mice are routinely used as convenient first line models for preclinical evaluation of such therapeutics. Thus, it is crucial to understand if and how mFcRn interacts with human ligands. Indeed, mFcRn has been shown to be rather promiscuous in its binding to IgG. It binds IgG from different species, including hIgG. On the other hand, hFcRn discriminates binding to mIgG (except for weak binding to mIgG2b) (30). This latter finding has greatly contributed to the understanding of the fast clearance and disappointing therapeutic effects obtained using monoclonal mIgGs in human trials. However, fast or intermediate clearance can also be favorable, as demonstrated for IgG immunoconjugates approved for cancer imaging and therapy (16, 31).
Mice have recently been constructed that lack the mFcRn HC and are transgenic for the human counterpart (4). Such mice express hFcRn that is exposed to the murine ligands, mIgG and MSA. Interestingly, they are found to be unable to protect mIgG from degradation. Nothing is known about the cross-species interaction between FcRn and albumin.
Herein, we report on important cross-species ligand-FcRn binding differences. Specifically, at acidic pH shFcRnWT binds MSA strongly while ignoring mIgG binding, whereas smFcRnWT binds hIgG1 strongly and HSA very weakly. The cross-species differences in albumin binding could partly be explained by non-conserved amino acid variations found in the vicinity of the conserved His-166 of the HC. In vivo, the consequences of weak binding of HSA to smFcRnWT may facilitate rapid clearance in the presence of high amounts of endogenous MSA. Such cross-species kinetic differences have great relevance for preclinical pharmacokinetics and biodistribution evaluations of engineered therapeutic and diagnostic IgGs, Fc, and albumin fusions in rodents.
EXPERIMENTAL PROCEDURES
PCR and Subcloning
The cDNA segments encoding truncated hFcRn HC and human β2m (hβ2m) were PCR-amplified from a U937 cell line (ATCC) cDNA library followed by subcloning of the fragments into the pcDNA3-GST vector, all as previously described (32). A mouse liver cDNA library (Zyagen) was used to PCR-amplify a cDNA encoding a truncated version of the mFcRn HC (encoding the endogenous native leader sequence, α1, α2, and α3 domains; 293 amino acids) using the primers mFcRnForw and mFcRnRev, listed in supplemental Table 1. Primers were designed to allow in-frame ligation of the fragment upstream of a cDNA encoding a glutathione S-transferase (GST) tag from Schistosoma japonicum into the pcDNA3-GST-hβ2m-oriP vector, which also contains a cDNA-encoding hβ2m and the Epstein-Barr virus origin of replication (oriP) (32). The final vector was sequenced and denoted pcDNA3-mFcRnWT-GST-hβ2m-oriP.
Construction of Mutant FcRn Variants
A single amino acid-substituted mFcRn variant was constructed by mutating His-168 to alanine by site-directed mutagenesis using the plasmid pcDNA3-mFcRnWT-GST-hβ2m-oriP and the primers mFcRnH168AForw and mFcRnH168ARev. Three double mutant FcRn variants, named hFcRnE117A/E118A, hFcRnR164L/E165G, and mFcRnL166R/G168E, were constructed using the templates pcDNA3-mFcRnWT-GST-hβ2m-oriP and pcDNA3-hFcRnWT- GST-hβ2m-oriP. The primer sequences used are all listed in supplemental Table 1.
Expression and Purification of Soluble FcRn Variants
For transient transfections, the hFcRn- and mFcRn-encoding plasmids were transfected into HEK 293E cells (ATCC) using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. HEK 293E cells were cultured in Dulbecco's modified Eagle's medium (BioWhittaker) using standard conditions. Pooled media were filtrated and applied on a GSTrap FF column (5 ml, Amersham Biosciences) connected to a semiautomatic workstation and recorder, and purifications were performed essentially as recommended in the manufacturer's manual. Eluted fractions were pooled, concentrated, and analyzed under non-reducing or reducing condition using β-mercaptoethanol (Sigma-Aldrich). Samples of 2 μg of each receptor were applied on a 12% SDS-PAGE (Bio-Rad). Protein concentrations were determined using a NanoDrop N-1000 spectrophotometer (NanoDrop Technologies).
Construction, Production, and Purification of IgG Variants
A mouse plasmacytoma cell line producing chimeric human IgG1 (hIgG1) anti-3-iodo-4-hydroxy-5-nitrophenacetyl (NIP) was a gift from Dr. M. Neuberger (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK). The construction of this antibody has been described before (33). Pure preparations of anti-NIP mIgG1 and mIgG2b were gifts from Dr. Gregory Winter (Centre for Protein Engineering, Medical Research Council Centre, UK). A single amino acid-substituted chimeric hIgG1 variant was constructed by mutating His-435 (numbering according to the EU index) to alanine by site-directed mutagenesis using the primers hIgG1H435Aforw and hIgG1H435Arev (listed in supplemental Table 1) and the template vector pLNOH2/Cγ1 (34), which contains the gene fragment encoding the constant HC of hIgG1. The mutant vector denoted pLNOH-hIgG1H435A was transiently expressed in HEK 293E cells by co-transfection with the pLNOKλ vector encoding the mouse lambda light chain as above. Chimeric hIgG1H435A was purified on NIP-coupled Sepharose as previously described (35). The integrity of expressed protein was verified by non-reducing SDS-PAGE analyses followed by Western blotting using a horseradish peroxidase-conjugated polyclonal rabbit anti-human Fc (Amersham Biosciences) and horseradish peroxidase-conjugated anti-murine lambda light chain (Southern Biotech) (data not shown).
Size-exclusion Chromatography Purification of Albumin Variants
Monomeric fractions of MSA (Calbiochem) and HSA (Sigma-Aldrich) were purified by size-exclusion chromatography on Superdex 200 (2.6 × 60 cm, Amersham Biosciences) operated on a gradient fraction collector (Pharmacia Biotech). The column was loaded with 1.5–5 ml of sample at a concentration of 75–100 mg/ml. As elution buffer, 0.05 m Tris, 0.2 m NaCl, 2 mm EDTA, 0.02% NaN3 was used, and the mixture was filtrated through a 0.22-μm filter prior to use. The purity of the collected fractions was tested by size-exclusion chromatography analysis on an analytical Superdex 200 (1 × 30 cm) operated on an LKB high-performance liquid chromatograph equipped with a Titan pump and eluted at 0.3 ml/min.
ELISA
Microtiter wells (Nunc) were coated with 100 μl of bovine serum albumin-NIP at 1 μg/ml, incubated overnight at 4 °C, and washed three times with PBS/0.005% Tween 20 (PBS/T), pH 7.4. They were then blocked with 4% skimmed milk (Acumedia) for 1 h at room temperature and washed as above. Serial dilutions (1 μg/ml to 0.0004 μg/ml) of anti-NIP hIgG1, hIgG1H435A, mIgG1, and mIgG2b were added for 1 h at room temperature and washed with PBS/T, pH 6.0 or pH 7.4. 1 μg/ml GST-tagged smFcRn or shFcRn variants preincubated with a horseradish peroxidase-conjugated goat anti-GST antibody (Amersham Biosciences) were added for 1 h at room temperature followed by washing with PBS/T, pH 6.0, or PBS/T, pH 7.4. Binding was visualized using tetramethylbenzidine substrate (Calbiochem). Binding to MSA or HSA was performed using serial dilutions of albumin (200 μg/ml to 0.010 μg/ml) coated in microtiter wells. The following steps were as described above.
SPR Analyses
SPR analyses were performed on a BIAcore 3000 instrument (Amersham Biosciences) using CM5 chips, and immobilization of smFcRn-GST and shFcRn-GST variants or smFcRn (kind gift from Dr. Sally Ward, University of Texas Southwestern Medical Center, Dallas, TX) was performed using the amine coupling kit (Amersham Biosciences). Protein samples (10 μg/ml) were injected in 10 mm sodium acetate at pH 4.5 (Amersham Biosciences), all as described by the manufacturer. Unreacted moieties on the surface were blocked with 1 m ethanolamine. For all experiments, phosphate buffer (67 mm phosphate buffer, 0.15 m NaCl, 0.005% Tween 20) at pH 6.0 or 7.4, or HBS-P buffer (0.01 m HEPES, 0.15 m NaCl, 0.005% surfactant P20) at pH 7.4 were used as running buffer or dilution buffer. Kinetic measurements were performed using a low density immobilized surface (100–200 resonance units (RU)). Serial dilutions of hIgG1 (2000.0–31.2 nm), mIgG1 (1000.0–15.6 nm), MSA (20.0–0.3 μm), and HSA (200.0–3.1 μm) were injected at pH 6.0 or 7.4, at a flow rate of 50 μl/min at 25 °C. Additive binding was recorded by injecting HSA (10 μm), MSA (5 μm), hIgG1 (100 nm), or mIgG1 (100 nm) alone or two at a time at 25 °C at 20 μl/min at pH 6.0 over immobilized shFcRn (∼600 RU) or smFcRn (∼600 RU). Competitive binding was measured by injecting shFcRn (50 nm) or smFcRn (100 nm) alone or together with different amounts of HSA or MSA (10.0–0.05 μm) over immobilized HSA (∼2600 RU) or MSA (∼2000 RU). In all cases, to correct for nonspecific binding and bulk buffer effects, responses obtained from the control surfaces and blank injections were subtracted from each interaction curve. Kinetic rate values were calculated using predefined models (Langmuir 1:1 ligand model, heterogeneous ligand model, and steady-state affinity model) provided by using BIAevaluation 4.1 software. The closeness of the fit, described by the statistical value χ2, which represents the mean square, was lower than 2.0 in all affinity estimations.
Sequence Analyses
ClustalW was used for amino acid sequence alignments. The NCBI accession numbers of the FcRn HC sequences: NM_004107 (human), NM_176657 (bovine), NM_033351 (rat), and NM_010189 (mouse). For the β2m sequences: AAA51811 (human), NP_776318 (bovine), NP_036644 (rat), and NP_033865 (mouse).
RESULTS
Preclinical evaluations of novel IgGs, Fc, and albumin fusions are frequently performed in rodents. Thus, in vitro interaction analyses of such constructs regarding cross-species FcRn binding may give information valuable when predicting in vivo biodistribution and efficacy.
Construction and Expression of a Chimeric smFcRn Variant
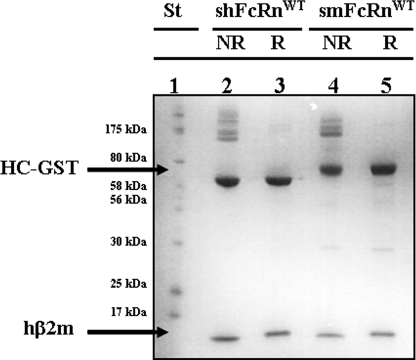
A cDNA segment encoding the three ectodomains (amino acids 1–269) of mFcRn HC was PCR-amplified from a mouse liver cDNA library and found to be identical with published sequences (data not shown). The HC was then expressed as fusion to GST after transient transfection of HEK 293E cells as described before (32). The vector used also carried the hβ2m cDNA. Harvested cell supernatants were pooled and applied to a GSTrap column for capture of chimeric smFcRnWT-GST molecules. SDS-PAGE analyses under non-reducing and reducing conditions showed the appearance of two main bands at ∼75 and ∼12 kDa that represent the GST-tagged mouse FcRn HC and hβ2m, respectively (Fig. 1). The shFcRn HC prepared in the same fashion migrated as a band of ∼65 kDa, which is in agreement with previous reports of heavier glycosylation of mFcRn than the human form (36, 37). Both receptor fractions contained bands of higher molecular weight, which represent covalent aggregates that resolve under reducing conditions. This is in agreement with previous reports for other GST fusion molecules (38, 39). The total amount of secreted chimeric smFcRnWT obtained was ∼0.4 mg/liter supernatant, slightly higher than that reported for production of shFcRnWT (32). Thus, mFcRn HC was shown to assemble with hβ2m in HEK 293E cells, and the heterodimer was secreted as a chimeric receptor.
FIGURE 1.
SDS-PAGE analyses of soluble receptor preparations. Secreted GST-tagged smFcRnWT and shFcRnWT molecules were purified from supernatants harvested from transiently transfected HEK 293E cells and analyses by 12% SDS-PAGE. Lane 1 shows protein standard. Lanes 2 and 3 show non-reduced (NR) and reduced (R) samples of shFcRnWT, respectively. Lanes 4 and 5 show NR and R samples of smFcRnWT. The bands corresponding to GST fused HCs and hβ2m are indicated by arrows.
Functional Integrity Determined by ELISA
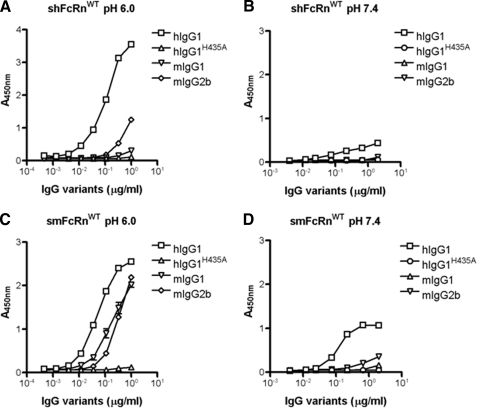
The functional integrity of the chimeric smFcRnWT was confirmed by testing its IgG-binding properties. Binding to mIgG and hIgG variants were investigated and compared side by side with the binding ability of the human counterpart using a pH-dependent ELISA. Dilutions of anti-NIP IgG variants were captured on NIP-conjugated bovine serum albumin-coated microtiter wells. GST-tagged receptors were then added at acidic or physiological pH, and binding was detected using a horseradish peroxidase-conjugated anti-GST antibody. Fig. 2 (A and B) shows pH-dependent binding of shFcRn to hIgG1, whereas a hIgG1 mutant, hIgG1H435A, did not interact at either pH. Moreover, mIgG1 did not bind and mIgG2b bound weakly, all in agreement with previous findings (30). Repeating the assays under the same conditions showed that the chimeric smFcRnWT variant interacted with mIgG1 and mIgG2b at acidic pH (Fig. 2C) and only very weakly at pH 7.4 (Fig. 2D). Binding to hIgG1 was considerably stronger than to the mIgG subclasses, and hIgG1 bound both at acidic and physiological pH (Fig. 2, C and D). Taken together, the results are as those previously reported for the murine receptor (40, 41), and thus, chimeric GST-tagged smFcRnWT has the same IgG-binding properties as the mouse receptor counterpart.
FIGURE 2.
pH-dependent binding of shFcRnWT and smFcRnWT to IgG variants in ELISA. Binding of shFcRnWT (A) and smFcRnWT (C) to hIgG1, hIgG1H435A, mIgG1, and mIgG2b at pH 6.0. Binding of shFcRnWT (B) and smFcRnWT (D) to hIgG1, hIgG1H435A, mIgG1, and mIgG2b at pH 7.4. The numbers given represent the mean of triplicates.
We next explored the interaction of the soluble receptor variants with albumin. Dilutions of monomeric size-exclusion chromatography isolated MSA and HSA (supplemental Fig. 1) were coated directly in ELISA wells, and pH-dependent binding studies were performed. Fig. 3 (A and B) shows binding of shFcRnWT to both HSA and MSA, respectively, but not at physiological pH. Thus, shFcRn does not discriminate against binding to MSA as it does to mIgGs. smFcRnWT bound both MSA and HSA, although lower binding responses were obtained compared with shFcRn binding (Fig. 3, C and D). No detectable binding was seen to either albumin variant at pH 7.4.
FIGURE 3.
pH-dependent binding of shFcRnWT and smFcRnWT to albumin variants in ELISA. Binding of shFcRnWT to HSA (A) and MSA (B) at pH 6.0 and 7.4. Binding of smFcRnWT to MSA (C) and HSA (D) at pH 6.0 and 7.4. Numbers given represent the mean of triplicates.
Determination of Binding Kinetics by SPR Analyses
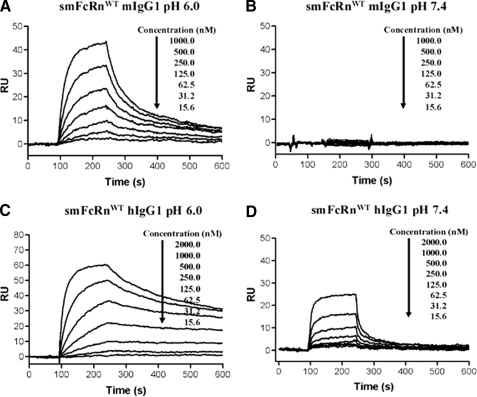
The expressed receptor domains are normally cell bound and exposed to circulating or pinocytosed soluble ligands. Thus, all measurements were run using covalently immobilized receptor and injection of IgG or albumin. Dilutions of mIgG1 were injected over CM5 surface of smFcRnWT, and reversible, concentration-dependent binding was observed at pH 6.0 (Fig. 4A), in contrast to almost negligible binding responses at pH 7.4 (Fig. 4B). The SPR data were fitted to the heterogeneous ligand binding model. This model has been used extensively to evaluate the FcRn-IgG interaction when FcRn is immobilized (12, 37, 42, 43). The KD values obtained were 8.5 ± 0.5 × 10−9 m (KD1) and 450.0 ± 65.0 × 10−9 m (KD2). This is in accordance with values obtained by others with immobilized murine receptor (19).
FIGURE 4.
SPR analyses of the smFcRnWT interaction with hIgG1 and mIgG1. Representative sensorgrams of serial dilutions of mIgG1 over immobilized smFcRnWT at pH 6.0 (A) and 7.4 (B), serial dilutions of hIgG1 over immobilized smFcRnWT at pH 6.0 (C) and 7.4 (D). In all experiments smFcRnWT was immobilized by amine coupling to ∼100–200 RU. Dilutions of mIgG1 and hIgG1 were injected over an immobilized smFcRnWT at 25 °C. The flow rate was 50 μl/min.
Cross-species binding to hIgG1 generated responses clearly stronger than those recorded for mIgG1 (Fig. 4C) and derived kinetics gave values of 0.1 ± 0.0 × 10−9 m (KD1) and 63.2 ± 4.8 × 10−9 m (KD2) at pH 6.0. Thus, a >85-fold decreased KD1 was found compared with that of the smFcRn-mIgG1 interaction. At pH 7.4, significant concentration-dependent and reversible binding responses were obtained (Fig. 4D), and the affinity could be calculated with a KD1 of ∼10−6 m. These data are summarized in Table 1. The kinetics of cross-species IgG binding have previously been recorded by SPR with IgG immobilized on the chip (44–46). This receptor:ligand orientation estimates a lower affinity for the interaction than that recorded here, where the receptor is immobilized. The trends were obtained, however.
TABLE 1.
Kinetics of the IgG interactions with smFcRnWT
| Analytea | pHb | ka1 | kd1 | ka2 | kd2 | KD1 | f1c | KD2 | f2c |
|---|---|---|---|---|---|---|---|---|---|
| 104/ms | 10−4/s | 104/ms | 10−3/s | nm | % | nm | % | ||
| mIgG1d | 6.0 | 16.1 ± 0.6 | 13.7 ± 0.4 | 3.8 ± 0.4 | 17.1 ± 0.5 | 8.5 ± 0.5 | 78.9 | 450.0 ± 65.0 | 21.1 |
| mIgG1 | 7.4 | NDe | ND | ND | ND | ND | ND | ND | ND |
| hIgG1d | 6.0 | 41.3 ± 0.6 | 0.5 ± 0.0 | 7.4 ± 0.2 | 4.6 ± 0.4 | 0.1 ± 0.0 | 82.4 | 63.2 ± 4.8 | 17.6 |
| hIgG1d | 7.4 | 4.1 ± 0.3 | 478.5 ± 33.2 | 1.6 ± 0.1 | 2.7 ± 0.3 | 1169.0 ± 0.7 | 84.4 | 168.7 ± 30.0 | 15.6 |
a Dilutions of mIgG1 and hIgG1 were injected over immobilized smFcRnWT as shown in Fig. 4.
b The binding measurements were performed at pH 6.0 or 7.4.
c Fractional occupancies, f1 and f2, of the two independent, parallel interactions.
d The kinetic rate constants were obtained using the heterogeneous ligand binding model, which gave the best global fit using the BIAevaluation 4.1 software. The model assumes two independent, parallel reactions with immobilized smFcRn-GST. The kinetic values represent the average of triplicates.
e ND, not determined due to no or very low binding responses.
To evaluate binding of MSA to smFcRnWT, dilutions of monomeric MSA were injected over the immobilized receptor at pH 6.0, and the representative sensorgram demonstrates reversible binding (Fig. 5A). The data fitted well to a simple first-order bimolecular interaction model applied with the BIAevaluation software and gave a KD of 9.3 ± 0.4 × 10−6 m (Table 2). No binding of MSA to smFcRnWT was obtained at pH 7.4 (Fig. 5B).
FIGURE 5.
SPR analyses of the shFcRnWT and smFcRnWT interaction with HSA and MSA. Representative sensorgrams of serial dilutions of MSA injected over immobilized smFcRnWT at pH 6.0 (A) and 7.4 (B), serial dilutions of HSA injected over immobilized smFcRnWT at pH 6.0 (C), and serial dilutions of MSA injected to immobilized shFcRnWT at pH 6.0 (D). In all experiments shFcRnWT and smFcRnWT were immobilized by amine coupling to ∼500–800 RU. Dilutions of MSA and HSA were injected over an immobilized receptor at 25 °C. The flow rate was 50 μl/min.
TABLE 2.
Kinetics of the albumin interactions with FcRn variants
| Albumin speciesa | FcRn species | FcRn variant | ka | kd | KD | KD Reqb |
|---|---|---|---|---|---|---|
| 103/Ms | 10−3/s | μm | μm | |||
| MSAc | Mouse | WT | 4.2 ± 0.5 | 39.4 ± 3.1 | 9.3 ± 0.4 | NDd |
| MSAc | Human | WT | 3.8 ± 0.0 | 3.1 ± 0.1 | 0.8 ± 0.2 | ND |
| HSAb | Mouse | WT | NAe | NA | NA | 86.2 ± 4.1 |
| HSAf | Human | WT | 2.7 ± 1.3 | 12.2 ± 5.9 | 4.5 ± 0.1 | 4.6 ± 0.5 |
| HSAb | Mouse | L166R/G167E | NA | NA | NA | 26.8 ± 0.1 |
| MSAc | Human | R164L/E165G | 0.7 ± 0.1 | 3.4 ± 0.1 | 4.8 ± 0.1 | ND |
| MSAc | Mouse | L166R/G167E | 2.7 ± 0.2 | 18.5 ± 0.5 | 6.8 ± 1.8 | ND |
| HSAc | Human | R164L/E165G | 3.2 ± 0.1 | 26.3 ± 0.2 | 8.2 ± 0.1 | ND |
b The steady-state affinity constant was obtained using an equilibrium (Req) binding model supplied by the BIAevaluation 4.1 software. The kinetic values represent the average of triplicates.
c The kinetic rate constants were obtained using a simple first-order (1:1) bimolecular interaction model.
d ND, not determined.
e NA, not acquired because of fast kinetics.
f The kinetic values have been published in Ref. 10.
Monomeric HSA bound smFcRnWT, but very weakly and with fast kinetics at pH 6.0 (Fig. 5C), while no binding at pH 7.4 was observed (supplemental Fig. 3A). Injection of higher concentrations of HSA increased binding responses, but aggregation of HSA obscured the results. However, these data could be fitted to a steady-state binding model and gave rise to an estimated KD of 86.2 ± 4.1 × 10−6 m. The results are not affected by the chimeric composition of smFcRn, because the same weak binding responses were obtained using a fully murine form of FcRn (supplemental Fig. 2).
shFcRnWT interacted with MSA at pH 6.0 (Fig. 5D) and not at pH 7.4 (supplemental Fig. 3B). The estimated KD at pH 6.0 was 0.8 ± 0.2 × 10−6 m. The kinetic measurements are summarized in Table 2. When comparing kinetics, the dissociation rates were found to differ dramatically and increased in the following order: smFcRn:MSA > shFcRn:HSA > shFcRn:MSA. No data could be obtained for the smFcRn:HSA pair, due to fast kinetics. Taken together, shFcRnWT bound more strongly than smFcRnWT to both albumin species, and MSA bound more strongly than HSA to both receptor variants. Thus, an affinity hierarchy appears as follows; shFcRn:MSA > shFcRn:HSA > smFcRn:MSA > smFcRn:HSA.
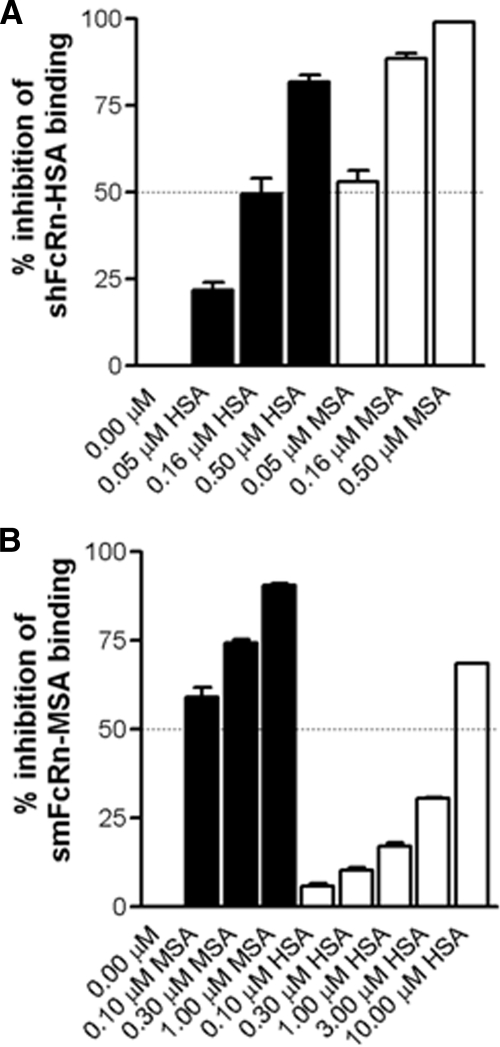
Cross-species Competitive Binding
To investigate the functional impact of cross-species binding, a constant amount of each receptor was preincubated with titrated amounts of MSA or HSA and injected over immobilized HSA or MSA. The percent inhibition of FcRn binding was calculated in each case. MSA preincubated with shFcRnWT inhibited receptor binding to immobilized HSA more efficiently than HSA, because 3-fold more HSA than MSA was required to reach 50% inhibition (0.16 versus 0.05 μm) (Fig. 6A). Furthermore, HSA was shown to inhibit smFcRnWT binding to MSA rather poorly, as in this case ∼10-fold more HSA than MSA was required to reach 50% inhibition (Fig. 6B).
FIGURE 6.
Competitive FcRn-albumin binding across species. A, serial dilutions of HSA (0.5–0.05 μm) and MSA (0.5–0.05 μm) were preincubated with shFcRnWT (0.05 μm) and injected over immobilized HSA (∼2600 RU). B, serial dilutions of HSA (10.0–0.1 μm) and MSA (1.0–0.1 μm) were preincubated with smFcRnWT (0.10 μm) and injected over immobilized MSA (∼2000 RU). The representative binding data are presented as percent inhibition of the FcRn binding to immobilized albumin. Injections were performed at 25 °C, and the flow rate was 50 μl/min.
Mapping the Differences in Albumin Binding Properties
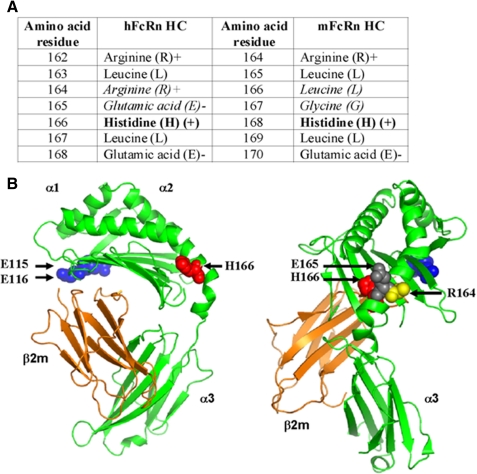
Previously, we reported that the conserved His-166, located to the α2-domain of human heavy chain, is crucial for binding to HSA, because mutation of this residue to alanine completely eliminate binding to HSA at acidic pH, whereas binding to hIgG is retained (10). This residue is conserved in all known FcRn sequences, including the mouse and rat HCs (Fig. 7A) (10). Mutation to alanine of the corresponding residue in the mouse counterpart (His-168) completely eliminated binding to HSA (100 μm), and only weak binding was detected for MSA (20 μm) when injected over a high density surface with immobilized mutant at acidic pH (supplemental Fig. 4, A and B).
FIGURE 7.
The crystal structure of shFcRn. A, amino acids flanking His-166 (hFcRn) and His-168 (mFcRn) located within the heavy chain α2-domain are shown. His-166 and His-168 are shown in bold. The non-conserved Arg-164 and Glu-165 of hFcRn, and Leu-167 and Gly-168 of mFcRn are shown in italic. B, the crystal structure of shFcRn shown in two orientations. The localization of amino acids essential for IgG (Glu-115 and Glu-116) and albumin (His-166) binding are highlighted as blue and red spherical balls. The non-conserved Arg-164 and Glu-165 (human) are highlighted with yellow and gray spherical balls, respectively. The FcRn heavy chains are shown in green and the β2m in orange. The figures were designed using PyMOL (DeLano Scientific) with the crystallographic data of shFcRn (37).
We speculated whether species differences in binding kinetics may be caused by non-conserved amino acids found in proximity to His-166 in the folded molecule. Interestingly, inspection of the flanking amino acids revealed major non-conserved variations, because the neighboring exposed basic Arg-164 and acidic Glu-165 in humans are replaced by the hydrophobic leucine and glycine residues in rodents, respectively (Fig. 7, A and B). His-166 and the non-conserved amino acid residues (Arg-164 and Glu-165) are highlighted in the human crystal structure shown in Fig. 7B. To explore the putative role of these residues in ligand binding, Arg-164 and Glu-165 were mutated to leucine and glycine in shFcRn, whereas Leu-166 and Gly-167 were mutated to arginine and glutamic acid in smFcRn.
Titrated amounts of monomeric HSA and MSA were again injected over immobilized receptor variants at pH 6.0. Representative sensorgrams demonstrate reversible binding responses at acidic pH, and the calculated binding kinetic values differ from that of the wild types (Fig. 8, A–D, and Table 2). The humanized smFcRnL166R/G167E variant bound HSA ∼3-fold more strongly than the wild-type mouse form, and the binding affinity for MSA was also slightly increased. Furthermore, rodentized shFcRnR164L/E165G bound HSA with an affinity of ∼2-fold weaker and MSA with a ∼6-fold weaker affinity than shFcRnWT. Thus, exchange of human-mouse amino acids in the vicinity of the key histidine residue decreased the differences in albumin-binding properties at acidic pH. However, the affinity of rodentized shFcRnR164L/E165G was not completely reduced to that of smFcRnWT, and the affinity of humanized smFcRnL166R/G167E did not totally reach the binding affinity of shFcRnWT.
FIGURE 8.
SPR analyses of albumin binding to rodentized and humanized FcRn variants. Representative sensorgrams of serial dilutions of HSA (A) and MSA (B) injected over immobilized rodentized shFcRnR164L/E165G at pH 6.0. Serial dilutions of MSA (C) and HSA (D) injected over smFcRnL166R/G167E. In all experiments the receptor variants were immobilized by amine coupling to ∼1000–2000 RU. Dilutions of MSA and HSA were injected over immobilized receptors at 25 °C. The flow rate was 50 μl/min. E, binding of shFcRnWT, shFcRnH166A, shFcRnR164L/E165G, and shFcRnE115A/E116A to hIgG1 at pH 6.0 in ELISA. F, binding of smFcRnWT, smFcRnH166A, and smFcRnL166R/G167E to hIgG1 at pH 6.0 in ELISA. The numbers given represent the mean of triplicates.
Also, the impact of the mutations on hIgG1 binding was investigated by ELISA. No differences in binding were detected for shFcRnH166A and shFcRnR164L/E165G compared with shFcRnWT (Fig. 8E). Mutation of two conserved glutamic acids, Glu-115 and Glu-116 (highlighted in Fig. 7B), to alanines (shFcRnE115A/E116A), completely eliminated binding to hIgG1 (Fig. 8E). This result supports a key role for these negatively charged residues in IgG binding, as previously shown by others (30, 47). Both smFcRnH168A and smFcRnL166R/G167E bound hIgG1 like the wild-type receptor (Fig. 8F). Thus, mutation of amino acids close to the conserved histidine did not influence binding to hIgG1.
Bifunctional FcRn Ligand Binding
shFcRn has been shown to bind both hIgG and HSA simultaneously in a pH-dependent manner (11). Cross-species binding of both ligands to FcRn may reveal how they are transported and protected from degradation in WT and transgenic mouse strains. We investigated the effect of each ligand on the binding of the other by injecting IgG and albumin, from both species, separately or together as a preincubated sample, over surfaces immobilized with smFcRn or shFcRn at acidic pH. Fig. 9(A and B) shows the resulting responses for binding to shFcRn and smFcRn, respectively. Both receptors bound their native ligands in an independent and additive manner. shFcRn ignored binding to mIgG, and smFcRn bound very weakly to HSA. In all cases, however, for both receptors, neither ligand (IgG or albumin from both species) interfered with binding of the other.
FIGURE 9.
SPR analyses of the interaction of shFcRnWT and smFcRnWT with human and mouse ligands. A, HSA, hIgG1, MSA, and mIgG1 injected over shFcRnWT at pH 6.0. B, MSA, mIgG1, HSA, and hIgG1 injected over shFcRnWT at pH 6.0. HSA was injected at 10 μm, MSA at 5 μm, and both IgG variants at 100 nm. In all experiments shFcRnWT and smFcRnWT were immobilized by amine coupling to ∼600 RU. Injections were performed at 25 °C and the flow rate was 50 μl/min.
DISCUSSION
Proper folding and cellular transport of the FcRn HC is absolutely dependent on association with β2m in the endoplasmic reticulum (48). Thus, both polypeptides need to be present for generation and secretion of cell bound as well as truncated forms of heterodimeric FcRn. Expression of functional chimeric FcRn has earlier been demonstrated in vivo in mice transgenic for the hFcRn HC (4) or the bovine FcRn HC (49). In both cases, the HC associates with mouse β2m into a functional transmembrane-anchored chimeric FcRn. However, direct interaction studies of soluble forms of chimeric FcRn molecules with ligands have not been reported.
In this study, we show that a truncated mFcRn HC assembles with hβ2m, and that the heterodimer is secreted from HEK 293E cells. The functional integrity of the chimeric smFcRn was extensively investigated by ELISA analyses and revealed binding to mIgG1, mIgG2b, hIgG1 and no detectable binding to a hIgG1H435A mutant. The chimeric receptor performed as a completely murine receptor and was then utilized in a series of ELISA and SPR experiments to obtain new information about cross-species ligand binding to FcRn.
The amino acid sequences of bovine, rat, and mouse β2m showed 73%, 68, and 66% homology with the human counterpart, and the corresponding values for the FcRn HC were 76%, 64 and 66%, respectively (supplemental Table 2 and 3). Thus, FcRn HC from other species may well be co-expressed with hβ2m in HEK 293E cells to produce a variety of chimeric FcRn variants.
To perform SPR, the receptors were immobilized on the chip, and the ligands were injected. Others have immobilized IgG and injected the receptor (45, 46, 50). In this situation, the actual affinities that were calculated were lower than those obtained here using the heterogeneous ligand binding model.
However, the binding hierarchies were the same, with shFcRnWT ignoring mIgG and smFcRnWT binding better to hIgG than to mIgG. The high affinity is in agreement with binding studies of FcRn expressed on cells (51).
Importantly, with IgG immobilized, the interaction between smFcRnWT and hIgG at physiological pH was barely detectable. By immobilizing the receptor, we were able to obtain kinetic data that suggest a difference in KD1 of four logs for the interactions at acidic and physiological pH.
The higher affinity of hIgG1 for smFcRnWT compared with the smFcRn-mIgG interaction at pH 6.0 may indicate that half-life in WT mice could be overestimated. However, the fact that hIgG1 also binds with reasonable affinity at physiological pH could counteract the effect, because it has been shown that such interaction lowers the half-life (52). In any case, half-life estimations of mIgG and hIgG in WT mice show approximately the same values (17).
SPR analyses showed that smFcRnWT interacted pH dependently with MSA with an estimated KD of 9.3 ± 0.4 × 10−6 m at acid pH. This is the first report on in vitro kinetics of the smFcRn-MSA interaction, a finding that supports the role of FcRn in albumin half-life regulation in mice (3, 53).
The remarkably long half-life of albumin was well recognized before its relationship with FcRn was discovered and utilized to enhance the in vivo effect of short-lived therapeutic substances. For instance, HSA-fused interferon α2b is now undergoing Phase 3 trials (54), and other HSA fusions are under study. Importantly, such constructs require animal models for preclinical evaluation.
Recent reports have addressed the in vivo half-life of HSA fused or targeted molecules in mice (25, 28) and argued that the increase in half-life observed is a consequence of FcRn-mediated rescue. Improved tumor imaging in rodents has been obtained using antitumor antigen antibody fragments genetically fused to HSA- or HSA-binding proteins (26, 29, 55). However, no complementary and comparative studies of such mFcRn cross-species binding to HSA have been reported. Here, we demonstrate a large difference in the kinetics of albumin binding to the mouse and human forms of FcRn. smFcRnWT binds MSA with a KD of ∼10 μm. The affinity for the endogenous ligand is 10-fold higher than that for HSA, a fact that nicely correlates with the inhibition data where smFcRnWT was shown to prefer MSA over HSA. This must necessarily affect the in vivo half-life of both HSA and HSA fused molecules in mice in the presence of high amounts of circulating endogenous albumin. When HSA-fused molecules show a moderate increase in half-life in rodents, and not an extended half-life similar to that of endogenous albumin, it may simply be an effect of the increase in molecular weight above the threshold for kidney clearance.
Support for this view is given by studies of HSA and single-chain variable fragment genetically fused to HSA in rats. Neither molecule shows more than half the serum persistence of endogenous rat albumin (56). Notably, mouse and rat FcRn HCs showed high homology (89%) (supplemental Table 3), as did rat and mouse albumin sequences (90%). Thus, the rat FcRn-HSA interaction is likely as weak as the smFcRn-HSA interaction. Albumin-targeted molecules have been described that achieve the same half-life as endogenous albumin. This is the case with human domain antibodies selected to bind albumin (57). Two anti-rat albumin domain antibodies with low (1 μm) and high (13 nm) affinity showed half-lives in rats of 43 and 53 h, respectively. Rat albumin has a half-life of 53 h, similar to the high affinity domain antibody.
Although shFcRnWT ignores mIgG, it interacts strongly with MSA. The affinity for MSA was 100-fold stronger than that of the murine receptor. Based on this, one would predict that the mouse strain transgenic for the hFcRn HC would bind strongly to endogenous MSA and protect it from degradation. This was indeed the case, and a 46% increase of the MSA levels in such mice has been observed (3).
The presence of MSA bound to the human receptor, and also high serum concentrations of MSA, will surely affect rescue of HSA-associated molecules that compete for the same binding site on the receptor. Notably, the off rate of MSA is 10 times lower than that of HSA. Transgenic mice, fortified with serum hIgG, are useful when evaluating serum persistence of engineered hIgGs, but the half-life of HSA variants and conjugates may well be underestimated. The latter is supported by the competitive data presented here where MSA efficiently inhibited shFcRnWT binding to HSA.
The binding sites for hIgG and HSA are distally localized in the α2-domain of the hFcRn HC (10). In line with this, we show that all combinations of ligands bound additively to both receptor forms, and that shFcRnWT ignores mIgG while binding strongly to MSA. A relevant question is whether the absence of one of the ligand affects FcRn trafficking. Notably, the fact that hFcRn HC transgenic mice have a 46% increase in MSA levels predicts that the transgene-encoded hFcRn recycles MSA while ignoring mIgG in acidified endosomal compartments. Furthermore, analbuminemic rats that lack endogenous albumin have almost normal levels of serum proteins with a slightly increased amount of IgGs. This fact supports that FcRn recycles IgG in the absence of albumin (58).
Swapping of two non-conserved amino acids near the conserved histidine residue (shFcRn His-166/smFcRn His-168) influenced the binding kinetics at acidic pH. The humanized smFcRnL166R/G167E gained affinity for HSA and MSA while the rodentized shFcRnR164L/E165G lost affinity for HSA and MSA. Thus, the mouse residues transplanted on shFcRnWT gave the human receptor mouse-like binding properties. However, other residues than the two focused are involved, because the binding kinetics did not fully reach that recorded for the wild-type counterparts.
Supplementary Material
Acknowledgments
We thank John E. Thommesen for introducing the H435A mutation in the pLNOH2/Cγ1 vector, and we are grateful to Kristine Ustgård for help with PCR reactions.
This work was supported by grants from the Steering Board for Research in Molecular Biology, Biotechnology and Bioinformatics at the University of Oslo (to J. T. A.), The Norwegian Research Council (Grant 179573), and the Norwegian Cancer Society (Grant B95078).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Tables 1–3.
- FcRn
- neonatal Fc receptor
- HC
- heavy chain
- HEK
- human embryonic kidney
- hFcRn
- human FcRn
- hIgG
- human IgG
- HSA
- human serum albumin
- hβ2m
- human β2-microglobulin
- mIgG
- mouse IgG
- MSA
- mouse serum albumin
- oriP
- origin of replication
- RU
- resonance unit
- shFcRn
- soluble hFcRn
- smFcRn
- soluble mouse FcRn
- SPR
- surface plasmon resonance
- WT
- wild type
- NIP
- 3-iodo-4-hydroxy-5-nitrophenacetyl
- ELISA
- enzyme-linked immunosorbent assay
- GST
- glutathione S-transferase.
REFERENCES
- 1.Roopenian D. C., Akilesh S. (2007) Nat. Rev. Immunol. 7, 715–725 [DOI] [PubMed] [Google Scholar]
- 2.Qiao S. W., Kobayashi K., Johansen F. E., Sollid L. M., Andersen J. T., Milford E., Roopenian D. C., Lencer W. I., Blumberg R. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9337–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhury C., Mehnaz S., Robinson J. M., Hayton W. L., Pearl D. K., Roopenian D. C., Anderson C. L. (2003) J. Exp. Med. 197, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roopenian D. C., Christianson G. J., Sproule T. J., Brown A. C., Akilesh S., Jung N., Petkova S., Avanessian L., Choi E. Y., Shaffer D. J., Eden P. A., Anderson C. L. (2003) J. Immunol. 170, 3528–3533 [DOI] [PubMed] [Google Scholar]
- 5.Waldmann T. A., Terry W. D. (1990) J. Clin. Invest. 86, 2093–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wani M. A., Haynes L. D., Kim J., Bronson C. L., Chaudhury C., Mohanty S., Waldmann T. A., Robinson J. M., Anderson C. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5084–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burmeister W. P., Huber A. H., Bjorkman P. J. (1994) Nature 372, 379–383 [DOI] [PubMed] [Google Scholar]
- 8.Kim J. K., Firan M., Radu C. G., Kim C. H., Ghetie V., Ward E. S. (1999) Eur. J. Immunol. 29, 2819–2825 [DOI] [PubMed] [Google Scholar]
- 9.Medesan C., Matesoi D., Radu C., Ghetie V., Ward E. S. (1997) J. Immunol. 158, 2211–2217 [PubMed] [Google Scholar]
- 10.Andersen J. T., Dee Qian J., Sandlie I. (2006) Eur. J. Immunol. 36, 3044–3051 [DOI] [PubMed] [Google Scholar]
- 11.Chaudhury C., Brooks C. L., Carter D. C., Robinson J. M., Anderson C. L. (2006) Biochemistry 45, 4983–4990 [DOI] [PubMed] [Google Scholar]
- 12.Bitonti A. J., Dumont J. A., Low S. C., Peters R. T., Kropp K. E., Palombella V. J., Stattel J. M., Lu Y., Tan C. A., Song J. J., Garcia A. M., Simister N. E., Spiekermann G. M., Lencer W. I., Blumberg R. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9763–9768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jazayeri J. A., Carroll G. J. (2008) BioDrugs 22, 11–26 [DOI] [PubMed] [Google Scholar]
- 14.Hinton P. R., Johlfs M. G., Xiong J. M., Hanestad K., Ong K. C., Bullock C., Keller S., Tang M. T., Tso J. Y., Vásquez M., Tsurushita N. (2004) J. Biol. Chem. 279, 6213–6216 [DOI] [PubMed] [Google Scholar]
- 15.Hinton P. R., Xiong J. M., Johlfs M. G., Tang M. T., Keller S., Tsurushita N. (2006) J. Immunol. 176, 346–356 [DOI] [PubMed] [Google Scholar]
- 16.Kenanova V., Olafsen T., Crow D. M., Sundaresan G., Subbarayan M., Carter N. H., Ikle D. N., Yazaki P. J., Chatziioannou A. F., Gambhir S. S., Williams L. E., Shively J. E., Colcher D., Raubitschek A. A., Wu A. M. (2005) Cancer Res. 65, 622–631 [PMC free article] [PubMed] [Google Scholar]
- 17.Petkova S. B., Akilesh S., Sproule T. J., Christianson G. J., Al Khabbaz H., Brown A. C., Presta L. G., Meng Y. G., Roopenian D. C. (2006) Int. Immunol. 18, 1759–1769 [DOI] [PubMed] [Google Scholar]
- 18.Vaccaro C., Zhou J., Ober R. J., Ward E. S. (2005) Nat. Biotechnol. 23, 1283–1288 [DOI] [PubMed] [Google Scholar]
- 19.Ghetie V., Popov S., Borvak J., Radu C., Matesoi D., Medesan C., Ober R. J., Ward E. S. (1997) Nat. Biotechnol. 15, 637–640 [DOI] [PubMed] [Google Scholar]
- 20.Bain V. G., Kaita K. D., Yoshida E. M., Swain M. G., Heathcote E. J., Neumann A. U., Fiscella M., Yu R., Osborn B. L., Cronin P. W., Freimuth W. W., McHutchison J. G., Subramanian G. M. (2006) J. Hepatol. 44, 671–678 [DOI] [PubMed] [Google Scholar]
- 21.Balan V., Nelson D. R., Sulkowski M. S., Everson G. T., Lambiase L. R., Wiesner R. H., Dickson R. C., Post A. B., Redfield R. R., Davis G. L., Neumann A. U., Osborn B. L., Freimuth W. W., Subramanian G. M. (2006) Antivir. Ther. 11, 35–45 [PubMed] [Google Scholar]
- 22.Cox G. N., Smith D. J., Carlson S. J., Bendele A. M., Chlipala E. A., Doherty D. H. (2004) Exp. Hematol. 32, 441–449 [DOI] [PubMed] [Google Scholar]
- 23.Halpern W., Riccobene T. A., Agostini H., Baker K., Stolow D., Gu M. L., Hirsch J., Mahoney A., Carrell J., Boyd E., Grzegorzewski K. J. (2002) Pharm. Res. 19, 1720–1729 [DOI] [PubMed] [Google Scholar]
- 24.Wunder A., Müller-Ladner U., Stelzer E. H., Funk J., Neumann E., Stehle G., Pap T., Sinn H., Gay S., Fiehn C. (2003) J. Immunol. 170, 4793–4801 [DOI] [PubMed] [Google Scholar]
- 25.Müller D., Karle A., Meissburger B., Höfig I., Stork R., Kontermann R. E. (2007) J. Biol. Chem. 282, 12650–12660 [DOI] [PubMed] [Google Scholar]
- 26.Dennis M. S., Jin H., Dugger D., Yang R., McFarland L., Ogasawara A., Williams S., Cole M. J., Ross S., Schwall R. (2007) Cancer Res. 67, 254–261 [DOI] [PubMed] [Google Scholar]
- 27.Dennis M. S., Zhang M., Meng Y. G., Kadkhodayan M., Kirchhofer D., Combs D., Damico L. A. (2002) J. Biol. Chem. 277, 35035–35043 [DOI] [PubMed] [Google Scholar]
- 28.Stork R., Müller D., Kontermann R. E. (2007) Protein Eng. Des. Sel. 20, 569–576 [DOI] [PubMed] [Google Scholar]
- 29.Tolmachev V., Orlova A., Pehrson R., Galli J., Baastrup B., Andersson K., Sandström M., Rosik D., Carlsson J., Lundqvist H., Wennborg A., Nilsson F. Y. (2007) Cancer Res. 67, 2773–2782 [DOI] [PubMed] [Google Scholar]
- 30.Ober R. J., Radu C. G., Ghetie V., Ward E. S. (2001) Int. Immunol. 13, 1551–1559 [DOI] [PubMed] [Google Scholar]
- 31.Wu A. M., Senter P. D. (2005) Nat. Biotechnol. 23, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 32.Berntzen G., Lunde E., Flobakk M., Andersen J. T., Lauvrak V., Sandlie I. (2005) J. Immunol. Methods 298, 93–104 [DOI] [PubMed] [Google Scholar]
- 33.Brüggemann M., Williams G. T., Bindon C. I., Clark M. R., Walker M. R., Jefferis R., Waldmann H., Neuberger M. S. (1987) J. Exp. Med. 166, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norderhaug L., Olafsen T., Michaelsen T. E., Sandlie I. (1997) J. Immunol. Methods 204, 77–87 [DOI] [PubMed] [Google Scholar]
- 35.Michaelsen T. E., Garred P., Aase A. (1991) Eur. J. Immunol. 21, 11–16 [DOI] [PubMed] [Google Scholar]
- 36.Simister N. E., Mostov K. E. (1989) Nature 337, 184–187 [DOI] [PubMed] [Google Scholar]
- 37.West A. P., Jr., Bjorkman P. J. (2000) Biochemistry 39, 9698–9708 [DOI] [PubMed] [Google Scholar]
- 38.Kaplan W., Hüsler P., Klump H., Erhardt J., Sluis-Cremer N., Dirr H. (1997) Protein Sci. 6, 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tudyka T., Skerra A. (1997) Protein Sci. 6, 2180–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurbaxani B., Dela Cruz L. L., Chintalacharuvu K., Morrison S. L. (2006) Mol. Immunol. 43, 1462–1473 [DOI] [PubMed] [Google Scholar]
- 41.Datta-Mannan A., Witcher D. R., Tang Y., Watkins J., Jiang W., Wroblewski V. J. (2007) Drug Metab. Dispos. 35, 86–94 [DOI] [PubMed] [Google Scholar]
- 42.Martin W. L., Bjorkman P. J. (1999) Biochemistry 38, 12639–12647 [DOI] [PubMed] [Google Scholar]
- 43.Vaughn D. E., Bjorkman P. J. (1997) Biochemistry 36, 9374–9380 [DOI] [PubMed] [Google Scholar]
- 44.Vaccaro C., Bawdon R., Wanjie S., Ober R. J., Ward E. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18709–18714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J., Mateos F., Ober R. J., Ward E. S. (2005) J. Mol. Biol. 345, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 46.Zhou J., Johnson J. E., Ghetie V., Ober R. J., Ward E. S. (2003) J. Mol. Biol. 332, 901–913 [DOI] [PubMed] [Google Scholar]
- 47.Martin W. L., West A. P., Jr., Gan L., Bjorkman P. J. (2001) Mol. Cell 7, 867–877 [DOI] [PubMed] [Google Scholar]
- 48.Zhu X., Peng J., Raychowdhury R., Nakajima A., Lencer W. I., Blumberg R. S. (2002) Biochem. J. 367, 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu W., Zhao Z., Zhao Y., Yu S., Zhao Y., Fan B., Kacskovics I., Hammarström L., Li N. (2007) Immunology 122, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Firan M., Bawdon R., Radu C., Ober R. J., Eaken D., Antohe F., Ghetie V., Ward E. S. (2001) Int. Immunol. 13, 993–1002 [DOI] [PubMed] [Google Scholar]
- 51.Mackenzie N. (1984) Immunol. Today 5, 364–366 [DOI] [PubMed] [Google Scholar]
- 52.Dall'Acqua W. F., Woods R. M., Ward E. S., Palaszynski S. R., Patel N. K., Brewah Y. A., Wu H., Kiener P. A., Langermann S. (2002) J. Immunol. 169, 5171–5180 [DOI] [PubMed] [Google Scholar]
- 53.Anderson C. L., Chaudhury C., Kim J., Bronson C. L., Wani M. A., Mohanty S. (2006) Trends Immunol. 27, 343–348 [DOI] [PubMed] [Google Scholar]
- 54.Subramanian G. M., Fiscella M., Lamousé-Smith A., Zeuzem S., McHutchison J. G. (2007) Nat. Biotechnol. 25, 1411–1419 [DOI] [PubMed] [Google Scholar]
- 55.Yazaki P. J., Kassa T., Cheung C. W., Crow D. M., Sherman M. A., Bading J. R., Anderson A. L., Colcher D., Raubitschek A. (2008) Nucl. Med. Biol. 35, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith B. J., Popplewell A., Athwal D., Chapman A. P., Heywood S., West S. M., Carrington B., Nesbitt A., Lawson A. D., Antoniw P., Eddelston A., Suitters A. (2001) Bioconjug. Chem. 12, 750–756 [DOI] [PubMed] [Google Scholar]
- 57.Holt L. J., Basran A., Jones K., Chorlton J., Jespers L. S., Brewis N. D., Tomlinson I. M. (2008) Protein Eng. Des. Sel. 21, 283–288 [DOI] [PubMed] [Google Scholar]
- 58.Esumi H., Sato S., Okui M., Sugimura T., Nagase S. (1979) Biochem. Biophys. Res. Commun. 87, 1191–1199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.