Abstract
c-Myc is an important transcription factor that regulates cellular proliferation, cell growth, and differentiation. A number of transcriptional co-factors for c-Myc have been described that have binding sites within highly conserved regions of the c-Myc transactivational domain (TAD). Given the importance of the c-Myc TAD, we set out to identify new proteins that interact with this region using a yeast two-hybrid assay. HBP1 was identified in our screen as a protein that interacts with full-length c-Myc but not a c-Myc mutant lacking the TAD. HBP1 is a transcriptional repressor and has been shown to negatively regulate the cell cycle. A correlation between HBP1 under-expression and breast cancer relapse has been described, suggesting that HBP1 may be an important tumor suppressor protein. We have found that HBP1 binds c-Myc in cells, and expression of HBP1 inhibits c-Myc transactivational activity at least partly by preventing c-Myc binding to target gene promoters. c-Myc binds to the C terminus of HBP1, a region lost in some breast tumors, and some HBP1 mutants found in breast cancer weakly interact with and/or no longer negatively regulate c-Myc. This work adds to our understanding of c-Myc regulation and mechanisms of tumor suppression by HBP1.
Keywords: Cancer/Breast, DNA/Protein Interaction, Oncogene/Myc, Protein/Protein-Protein Interactions, Transcription/Myc, Tumor/Suppressor
Introduction
c-Myc is a transcription factor whose wide range of functions include promoting cellular proliferation and cell growth, inhibiting differentiation, and inducing apoptosis under growth-restrictive conditions. c-Myc activity is essential for cell cycle progression as cells deleted for c-Myc cease to proliferate and exit the cell cycle (1). Additionally, c-Myc function is required for normal mammalian development as mice homozygously deleted for c-myc die at embryonic day 10.5 (2). Given its extensive role in cell proliferation and cell growth, it is not surprising that c-myc is a potent oncogene and deregulated c-Myc expression is observed in roughly 70% of all human tumors (3).
The c-Myc protein is a basic helix-loop-helix transcription factor that heterodimerizes with its partner protein, Max, to bind E-box sequences in target gene promoters. Together, c-Myc and Max regulate transcription of a number of significant genes including those that encode for cell cycle regulators, such as Cdc25 and E2F2, cell growth regulators, such as eIF4e and Nucleolin, and apoptotic proteins, such as Bax (4–8). c-Myc/Max activity is antagonized by Max binding to the Mad family of proteins which inhibit gene transcription at E-boxes (9). Although the transactivational activity of c-Myc is relatively weak, initially being reported to increase transcription by ∼3-fold (10), recent genomic studies suggest that c-Myc binds up to 15% of the genome, highlighting the importance of its gene regulatory activity (11). In addition to its activation capacity, c-Myc has also been shown to repress gene transcription. For example, c-Myc and Max have been shown to bind the transcription factor Miz1 and prevent transcription from INR elements present in the promoters of the cyclin-dependent kinase inhibitors p15 and p21 (12, 13).
The transactivational domain (TAD)2 of c-Myc is critical for both its activating and repressing activities. The TAD contains two highly conversed domains known as Myc Box I (MBI) and Myc box II (MBII). MBI harbors two phosphorylation sites, threonine 58 and serine 62, which have been shown to regulate c-Myc protein stability (14–16). MBII is important for recruiting a number of co-regulators to c-Myc target genes. For example, c-Myc recruits histone acetyltransferase activity to target gene promoters by MBII-mediated binding of the protein TRRAP, a core component of both the GCN5 and TIP 60 histone acetyltransferase complexes (17–19). These complexes catalyze the acetylation of histones, which promotes an open chromatin structure and allows for transcription of c-Myc target genes. Additionally, in a collaborative effort, we have recently shown that the ribosomal protein L11 interacts with the MBII region of c-Myc and inhibits c-Myc transactivational activity (20). Therefore, both positive and negative regulators of c-Myc activity bind through MBII.
Given the importance of the c-Myc TAD, we used a yeast two-hybrid assay to identify new proteins that interact with the TAD. The HMG-box protein, HBP1, was identified in this screen as a novel c-Myc interacting protein. HBP1 was first identified in a screen for mammalian proteins that rescued a potassium channel defect in yeast (21). Since then it has been described as a binding partner of pRB, and in most cases, HBP1 acts as a transcriptional repressor (22–25). HBP1 has been shown to repress gene expression both by preventing transcriptional activators from binding their target genes as well as by its direct, sequence-specific DNA binding activity. HBP1 direct target genes include the P47PHOX and n-MYC genes (24, 25). In contrast, HBP1 has been shown to negatively regulate Wnt signaling in the absence of DNA binding (23). Specifically, HBP1 binds the transcription factor TCF4 and prevents it from binding to its target genes, including c-MYC and CYCLIN D. Given its suppression of important cell cycle regulators, it is not surprising that overexpression of HBP1 has been shown to induce cell cycle arrest in a number of different cells types (24, 26, 27).
Recent evidence suggests that HBP1 is a tumor suppressor protein. HBP1 maps to chromosome 7q31.1, a region that has been reported to be frequently deleted in numerous cancer types (28–33). Additionally, HBP1 is an important effector in oncogene-induced premature senescence, a tumor suppressing mechanism (34). Finally, a number of natural HBP1 mutants occur in human breast cancer, and under-expression of HBP1 is correlated with poor prognosis in this tumor type (35).
Here we show that HBP1 interacts with both the transactivation domain as well as the C terminus of c-Myc. This binding prevents c-Myc-mediated transcription. Inhibition of c-Myc transactivation involves HBP1-mediated reduction in the binding of c-Myc to its target promoters, resulting in decreased expression of these genes. HBP1 also inhibits c-MYC expression through inhibition of β-catenin/TCF4-mediated c-MYC gene activation, providing a secondary mechanism for HBP1-mediated down-regulation of c-Myc activity. Importantly, some HBP1 mutants identified in breast cancer no longer strongly interact with and/or negatively regulate c-Myc transactivation activity.
EXPERIMENTAL PROCEDURES
Plasmids and shRNA
Generation of CMV-β-gal, pDEST40- c-MycWT, pDEST40-c-MycΔTAD, pDEST40-c-MycΔMB1, and shRNA-scramble as well as the reporter construct E2F2-Luc have been previously described (5, 16, 36). 4xE-box-Luc and pGL2 were kindly provided by Dr. Peter Hurlin (Oregon Health and Sciences University, Portland, OR). pDBLeu and pEXP-AD502 were purchased from Invitrogen. pDEST40-c-MycΔC was generated by PCR amplification (forward, 5′-CACCATGCCCCTCAACGTG-3′; reverse, 5′-GGATCTGGTCACGCAGGG-3′) and Gateway cloning per the manufacturer's instructions. pDEST40-c-MycΔMB1I was generated in the same way except using pcDNA-V5-MycΔMB1I as a template (generous gift from Dr. Mushui Dai, Oregon Health and Sciences University). pEF-BOS-HA-HBP1WT was kindly provided to us by Dr. Amy Yee (Tuffs University School of Medicine, Boston, MA). pEF-BOS-HBP1ΔC and pEF-BOS-HBP1Δ263 were generated using a forward primer to the HA tag (5′-GAGGAATTCTCTAGAATGTACCC-3′) and reverse primers that would result in the deletion of the C-terminal 84 amino acids (5′-CGTCTAGAGCTTAAGTGGCACTCACAG-3′) or the C-terminal 251 amino acids (5′-GCTCTAGAGCTTACTTTAGACCATC-3′). All primers contained XbaI sites (underlined) to allow for cloning back into pEF-BOS. pEF-BOS-HBP1ΔREP was generated as follows; the N-terminal fragment of HBP1ΔREP was amplified using the forward primer to the HA tag and an internal reverse primer containing a BamHI digestion site (5′-CGGGATCCGAAAATGCCAGATTC-3′). The C-terminal fragment was generated by digesting pEF-BOS-HA-HBP1 with XbaI and BglII. The two fragments were ligated together and then ligated in pEF-BOS using T4 DNA ligase (Roche Applied Science). pEF-BOS-HBP1p32-4 was generated as follows: the N-terminal fragment was generated by PCR amplification using the forward primer to the HA tag and an internal primer containing a BamHI site (5′-CGGGATCCGCTTTTAAATGTATC-3′). The C-terminal fragment was amplified using a forward primer containing a BglII site (5′-GAAGATCTAACAGAGCCATAAG-3′) and a HBP1 reverse primer containing a XbaI site (5′-CGTCTAGAGAATTGAGGACAAATGG-3′). The fragments were digested and then ligated together. The resulting construct was digested with XbaI and then ligated into pEF-BOS. pDBLeu-c-Myc/GAL4DB was constructed as follows: The GAL4DB was cloned out of the pDBLeu vector (Invitrogen) by PCR amplification using a forward primer with an BstYI site (5′-CGCAGATCCATGAAGCAAGCCTCCTGAAAG-3′) and a reverse primer with a XbaI site (5′-GCTCTAGACCTCGACGATACAGTCAAC-3′). The PCR product was digested with these restriction enzymes and then ligated into the XbaI and BamH1 sites in pBluescript (pBS), as digesting by BamHI and BstYI results in compatible cohesive ends. The resulting plasmid was designated pBS/GAL4DB. Mouse c-Myc was then cloned into pBS/GAL4DB. Specifically, mouse c-Myc was cloned out of CMV-Myc (5) by first digesting the plasmid with BstYI, which cuts at base pair 1138 of mouse c-Myc1. The fragment was then filled in using the Klenow enzyme (New England Biolabs, Ipswich, MA). The digested and filled product was then further digested with HindIII, which digests the plasmid upstream of the Myc1 start site in CMV-Myc. After purification of the c-Myc fragment, c-Myc was cloned into pBS/GAL4DB into the HindIII and SmaI sites. The c-Myc-Gal4DB fragment was removed from pBS by digestion with NotI and HindIII and cloned into the pDBLeu plasmid. pDBLeu-c-MycΔTAD/GAL4DB was created by digesting pBS/c-Myc/GAL4DB with PstI and religating the linear fragment with T4 ligase. This results in the deletion of amino acids 40–179 of c-Myc. c-MycΔTAD/GAL4DB was cut from pBS by digesting with NotI and HindIII and then ligated into pDBLeu.
The shRNA expression vector to HBP1 (HBP1 shRNA-1) was generated using the targeting sequence previously described (35). Oligos encoding the sense and antisense sequences were annealed and ligated into the pENTR-H1/T0 (Invitrogen) expression vector as described by the manufacturer. HBP1 shRNA-2 was purchased from Sigma.
Yeast Two-hybrid Assay
Yeast two-hybrid yeast strains were from the Proquest Two-Hybrid System with Gateway Technology kit purchased from Invitrogen. Yeast strains were transformed with the human liver cDNA library (Promega, Madison, WI) and/or the c-Myc bait constructs by lithium acetate-mediated transformation. The yeast two-hybrid assay was performed following the Proquest protocol. To rescreen positive interactors, a mating strategy was used. Yeast were first cured of the bait plasmid by plating on cycloheximide plates and then mated to a MaV103 strain previously transformed with pDBLeu-c-Myc/GAL4DB or pDBLeu-c-MycΔTAD/GAL4DB.
Cell Lines and Transfections
HEK-293 cells were maintained in DMEM supplemented with 10% characterized fetal bovine serum, 2 mm l-glutamine, 1× penicillin/streptomycin, nonessential amino acids, and sodium pyruvate at 37 °C and 5% CO2. Cells were plated to achieve 70–80% confluency 24 h post-split for transfection. Transfections were performed using Metafectene (Biontex, Germany), HEK-Fectin (Bio-Rad), or Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. REF52 cells were maintained in DMEM supplemented with 5% defined fetal bovine serum, 5% bovine calf serum. Cells were plated to achieve 50% confluency 24 h post-split. REF52 cells were transfected by the calcium phosphate method as previously described (5). Total transfected DNA was held constant by the addition of empty control plasmids. All transfections included 50–200 ng of CMV-β-gal to determine transfection efficiency. MCF10A and SKBR3 cell lines were purchased from ATCC (American Type Culture Collection). MCF10A cells were grown in 45% DMEM, 45% F-12 hams, 5% horse serum, 2.5 mm l-glutamine, 20 ng/ml epidermal growth factor, 10 mg/ml insulin, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, and 1× penicillin/streptomycin. SKBR3 cells were grown in DMEM with 10% fetal bovine serum and 1× penicillin/streptomycin.
Antibodies
The monoclonal V5 antibody used to detect tagged c-Myc protein was from Invitrogen. The c-Myc antibodies N262 and C-33 as well as the HBP1 antibodies (H-300 and C-20), the Sp1 (PEP-2) antibody, and the Cdk2 (M2) antibody were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The c-Myc antibody Y69 was purchased from Abcam (Cambridge, MA). The monoclonal HA.11 antibody used for Western blotting was from Covance (Berkeley, CA), and the monoclonal HA (G036) antibody used for immunoprecipitations was purchased from Applied Biological Materials, Inc. (Richmond, BC). The β-actin antibody was purchased from Sigma. Normal rabbit IgG used for the control chromatin immunoprecipitation (ChIP) experiments was purchased from Santa Cruz Biotechnology.
Western Blotting and Quantitation
Cell lysates and immunoprecipitations were run on SDS-PAGE gels and transferred to Immobilon-FL membrane (Millipore, Billerica, MA). Membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE). Primary antibodies were diluted in 1:1 Odyssey blocking buffer:phosphate-buffered saline with 0.05% Tween 20. Primary antibodies were detected with secondary antibodies labeled with the near-infrared fluorescent dyes IRDye800 (Rockland, Philadelphia, PA) and Alexa Fluor 680 (Molecular Probes, Eugene, OR) to allow two-color imaging and band overlay. Secondary antibodies were diluted 1:10,000 in 1:1 Odyssey blocking buffer:phosphate-buffered saline. Blots were scanned with a LI-COR Odyssey Infrared Imager to visualize proteins. c-Myc, HBP1, β-actin, and Cdk2 protein levels were quantitated using LI-COR Odyssey Infrared Imager software Version 1.2.
Co-immunoprecipitations
Cells were resuspended in 10× cell pellet volumes of co-IP buffer (20 mm Tris, pH 7.5, 12.5% glycerol, 0.5% Nonidet P-40, 150 mm NaCl, 2 mm EDTA, 2 mm EGTA and 1 mm dithiothreitol) plus protease and phosphatase inhibitors. Cellular lysates were sonicated for 10 pulses (output = 1, 10–15% duty), incubated on ice for 20 min, and cleared by centrifugation at 14,000 rpm for 10 min at 4 °C. Cleared lysates were adjusted for transfection efficiency as measured by β-galactosidase activity and incubated with either 1:1000 dilution of conjugated anti-C33, 1:1000 conjugated anti-SP1, 1:750 conjugated V5, 1:1000 anti-V5, or 1:500 anti-HA (Applied Biological Materials) antibodies. Antibodies were conjugated to either Protein G- or Protein A-Sepharose (GE Healthcare) depending on isotype. Immunoprecipitates were washed 3 times with a 10× volume of co-IP buffer.
Luciferase Assay
Cell pellets were resuspended in 10× volumes of 1.5× reporter lysis buffer (Promega) with protease and phosphatase inhibitors. Cellular lysates were sonicated for 10 pulses at output = 1 and 10% duty and incubated on ice for 20 min. Lysates were cleared by centrifugation at 14,000 rpm for 10 min at 4 °C, and β-galactosidase and luciferase activity were analyzed. Luciferase activity was determined using the Promega Luciferase assay kit and Berthold luminometer (Bundoora, Australia). Luciferase activity was adjusted for β-galactosidase activity. Three or more separate experiments were performed for each luciferase assay. Fold change in luciferase activities were measured relative to empty vector or control transfections, and average fold changes and S.E. were graphed using GraphPad Prism.
Statistics
p values were calculated using a standard Student's t test analysis (two-tailed distribution and two-sample unequal variance) using GraphPad Prism to determine statistical differences as indicated on the graphs.
ChIP Assays
Cells were cross-linked with formaldehyde to a final concentration of 1% in media and incubated at room temperature for 10 min. Glycine was added to a final concentration of 0.125 m, and cells were incubated at room temperature for 5 min. Cells were collected in 1× phosphate-buffered saline, 1 mm EDTA and pelleted by gentle centrifugation. Cells were resuspended in 10× cell pellet volumes of radioimmune precipitation assay buffer (50 mm Tris, pH 8.0, 1% Nonidet P-40, 150 mm NaCl, 0.5% deoxycholate, 5 mm EGTA, and 0.1% SDS) plus protease and phosphatase inhibitors. Cell lysates were sonicated 5× (output = 3.5, 30% duty, 10 pulses) and then cleared by centrifugation at 14,000 rpm for 10 min at 4 °C. Cell lysates was precleared with 25 μl of 50% slurry of protein A beads and 25 μl of sheared salmon sperm DNA for 30 min rotation at 4 °C. Lysates were again cleared by centrifugation at 14,000 rpm for 5 min at 4 °C. Endogenous c-Myc was immunoprecipitated from lysates using 2 μg of Y69 antibody (Abcam) overnight at 4 °C. 2 μg of rabbit IgG (Santa Cruz Biotechnology) was used as a negative control. Immunoprecipitates were washed 2 times with radioimmune precipitation assay buffer, 4 times with IP wash buffer (100 mm Tris, pH 8.5, 500 mm LiCl, 1% Nonidet P-40, and 1% deoxycholate), and an additional 2 times in radioimmune precipitation assay buffer. Samples were rotated 5 min at room temperature in buffer in between each wash. Immunoprecipitates were eluted from beads with elution buffer (50 mm NaHCO3 and 1% SDS) by rotating samples in buffer for 15 min at room temperature. Elution products were transferred to new tubes, and 5 m NaCl was added to a final concentration of 0.2 m. Additionally, 2 μl of 5 mg/ml RNase A was added, and samples were incubated at 65 °C overnight. DNA was precipitated overnight by adding 1 μl of yeast tRNA and 650 μl of 100% ethanol. DNA was isolated by centrifugation at 14,000 rpm for 20 min at 4 °C, air-dried, and resuspended in 100 μl of Tris-EDTA. DNA was purified with the QIAquick PCR purification kit (Qiagen) and used for semiquantitative and quantitative PCR analysis with primers previously described (20). To verify similar input levels, semiquantitative PCR was used to amplify GAPDH, E2F2, and nucleolin in ChIP input samples. For quantitative ChIP experiments, primers to the E2F2 and NUCLEOLIN promoter regions as well as internal GAPDH primers were used to amplify DNA. The internal GAPDH primers were used to normalize each immunoprecipitation for nonspecific binding to the beads. DNA immunoprecipitated by the Y69 antibody relative to the control IP was then calculated.
RNA Isolation and Quantitative Real-time-PCR
RNA was isolated from transfected 293 cells using TRIzol reagent (Invitrogen) according to manufacturer's protocol. cDNA was generated using the QuantiTect reverse transcription kit (Qiagen). Quantitative real-time-PCR analysis was performed using primers previously described (20) and SYBR Green reagent (Invitrogen) on a Step-One Real-Time PCR machine (Applied Biosystems) according to manufacturer's quantitative real-time-PCR cycle conditions.
RESULTS
Identification of HBP1 as a Novel c-Myc Interacting Protein
To identify new proteins that interact with c-Myc and that may help regulate c-Myc activity, we used a yeast two-hybrid assay. Because c-Myc is a transcription factor with an established transactivational domain, we wanted to diminish intrinsic c-Myc activity to only identify true protein-protein interactions in the yeast two-hybrid assay. It was previously shown that a C-terminal-truncated c-Myc protein fused to a GAL4 DNA binding domain (GAL4DB) had greatly diminished transactivation activity in a chloramphenicol acetyl transferase reporter assay (37). Therefore, we truncated the C terminus of c-Myc at amino acid 382 and fused the protein to a GAL4DB (Fig. 1A). This c-Myc fusion construct was used as a bait plasmid in yeast transformed with a human liver cDNA library fused to the GAL4 activation domain (GAL4AD). Approximately 5.4 × 106 transformants were screened for expression of the GAL4-driven HIS3 reporter gene. Colonies were isolated and further screened for expression of two other GAL4-responsive reporter genes.
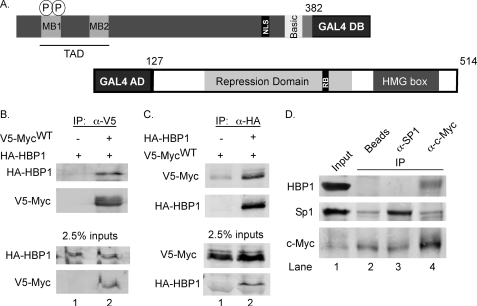
FIGURE 1.
HBP1 is a c-Myc interacting protein. A, shown is a schematic of “bait” and “prey” constructs utilized and isolated in the yeast two-hybrid assay. Amino acids 1–382 of c-Myc were fused to a GAL4 DNA binding domain (GAL4DB) and used to screen for novel interacting proteins. A cDNA encoding amino acids 127–514 of HBP1 was identified in the screen as a positive interactor. Important features and domains of each protein are indicated. B, full-length HBP1 co-immunoprecipitates with full-length c-Myc in mammalian cells. 293 cells were transiently transfected with EF-1α-driven HA-HBP1 and CMV-driven V5-c-Myc or empty vector as indicated. CMV-β-gal was co-transfected to control for transfection efficiency. Lysates were collected, and immunoprecipitation volumes were adjusted based on β-galactosidase activity. Lysates were subjected to immunoprecipitation with anti-V5. Inputs and immunoprecipitates were analyzed by Western blotting with the indicated antibodies. C, c-Myc co-immunoprecipitates with HBP1. 293 cells were transiently transfected with V5-Myc and HA-HBP1 or empty vector along with CMV-β-gal. Lysates were collected, and immunoprecipitation volumes were adjusted based on β-galactosidase activity. Lysates were subjected to immunoprecipitation with anti-HA. Inputs and immunoprecipitates were analyzed by Western blotting with the indicated antibodies. D, endogenous HBP1 interacts with endogenous c-Myc but not the transcription factor Sp1. 293 cleared lysates were immunoprecipitated with Sepharose-conjugated C-33 anti-Myc antibody for c-Myc pulldown or Sepharose-conjugated anti-SP1 for SP1 pulldown. Control immunoprecipitation was done with protein G-Sepharose. Input and immunoprecipitation were analyzed by Western blotting with the indicated antibodies.
We were particularly interested in identifying those proteins that bind within the c-Myc TAD. Both conserved domains Myc box I and Myc box II reside within this region, and these domains have been shown to be important for both c-Myc stability and c-Myc function. To narrow down the clones to only those that express GAL4AD fusion proteins that interact within the TAD of c-Myc, we used a mating strategy. Briefly, the c-Myc bait plasmid was cured from positive yeast clones. These clones were then mated to yeast of the opposite mating type containing the original c-Myc bait plasmid or a c-Myc bait plasmid where the TAD was deleted. The resulting clones were screened again for expression of the HIS3 reporter gene. Only those that interacted with the original bait plasmid but not the ΔTAD plasmid were further considered. Through this process we isolated a cDNA encoding amino acids 127–514 of the HMG box protein 1 (HBP1) as a putative c-Myc interacting protein (Fig. 1A). Re-expression of this HBP1 cDNA with the c-Myc bait plasmid resulted in expression of the HIS3 reporter gene and growth on yeast plates lacking histidine. Conversely, yeast expressing both the HBP1 cDNA and the c-MycΔTAD bait gene did not grow on this plate, confirming that HBP1 cannot interact with c-Myc lacking the TAD in yeast (supplemental Fig. 1).
HBP1 Interacts with c-Myc in Mammalian Cells
HBP1 has been reported to function as a transcriptional repressor both by binding specific DNA sequences and by its direct interaction with other transcription factors (23–25). Therefore, we asked whether HBP1 interacts with c-Myc in mammalian cells by performing co-immunoprecipitation analyses using full-length V5-tagged c-Myc and full-length-HA-tagged HBP1. Specifically, HEK293 cells were transiently transfected with expression constructs for HA-HBP1 and either V5-c-Myc or empty control vector. Cell lysates were subjected to immunoprecipitation using an antibody to the V5 tag, and HBP1 and c-Myc were detected by Western blot analysis. HBP1 co-immunoprecipitated with V5-c-Myc but was not pulled down in the absence of transfected c-Myc, demonstrating specific Myc-dependent pulldown (Fig. 1B, lanes 2 versus 1). To confirm this interaction we preformed the reverse co-immunoprecipitation and found that HBP1 immunoprecipitation specifically co-precipitated c-Myc (Fig. 1C, lane 2 versus 1). Finally, we examined whether the endogenous proteins interact in cells. 293 lysates were incubated with antibodies to c-Myc, the transcription factor Sp1, or Protein G beads alone. Immunoprecipitation of endogenous c-Myc resulted in pulldown of endogenous HBP1 (Fig. 1D, lane 4). No HBP1 pulldown was observed in the beads alone or Sp1 control immunoprecipitations, where only a small amount of nonspecific c-Myc binding was detected (lanes 2 and 3). Together these data demonstrate that HBP1 interacts with c-Myc in mammalian cells.
HBP1 Inhibits c-Myc-induced Expression of Reporter Genes
Given that HBP1 has a described positive role in cell cycle arrest and differentiation, we asked whether HBP1 regulates c-Myc activity (23–27, 38, 39). We first tested this by examining whether HBP1 affected the ability of c-Myc to induce transcription of a reporter gene. A reporter plasmid containing four E-boxes proximal to a minimal SV40 promoter driving luciferase or the minimal promoter-luciferase vector control was transiently transfected into 293 cells with empty vector control or expression vectors for c-Myc, HBP1, or both c-Myc and HBP1. Luciferase activity is shown relative to empty vector control transfection (Fig. 2A). c-Myc induced expression of E-box-driven luciferase ∼4-fold over background levels. This activation is consistent with previous reports (40). Although HBP1 had no significant effect on E-box-driven luciferase expression on its own, it substantially decreased luciferase activity in the presence of c-Myc, and neither c-Myc nor HBP1 significantly affected the minimal promoter without the E-box binding sites. This indicates that overexpression of HBP1 can repress c-Myc-induced transcription from E-box binding sites. We also examined the ability of HBP1 to inhibit c-Myc-mediated activation of luciferase under control of the natural, more complex E2F2 promoter, which harbors three E-box Myc binding sites (5) (Fig. 2B). Similar to the synthetic promoter, overexpression of c-Myc induced E2F2 promoter-driven luciferase activity. HBP1 had no significant effect on the E2F2 promoter alone; however, in the presence of c-Myc, HBP1 reduced expression of luciferase to background levels, demonstrating that HBP1 can also inhibit c-Myc transactivation of a natural promoter.
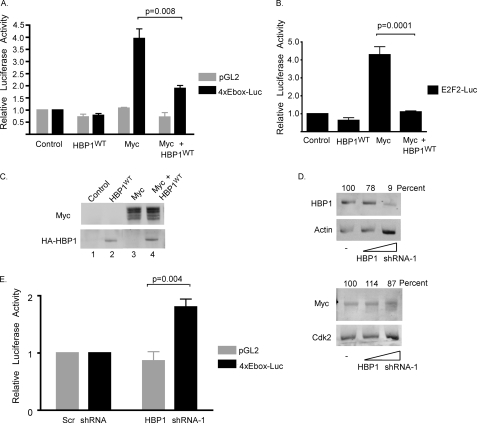
FIGURE 2.
HBP1 inhibits c-Myc-induced transcription. A, HBP1 inhibits c-Myc-induced transcription from a synthetic E-box promoter. 293 cells were transiently transfected with either control pGL2 or 4xE-box-luciferase and CMV-β-galactosidase together with V5-c-Myc and/or HA-HBP1 expression vectors as indicated. 18–20 h post-transfection cells were serum-starved in DMEM supplemented with 0.2% serum for 48 h. Cleared lysates were analyzed for β-galactosidase and luciferase activity. Luciferase activity was normalized to transfection efficiency, as measured by β-galactosidase activity, and is shown relative to empty vector control-transfected cells. The averages of three independent experiments ± S.E. were graphed. Statistical significance is indicated by the p value between c-Myc and c-Myc + HBP1WT. Error bars and p values were calculated using GraphPad Prism. B, HBP1 inhibits c-Myc-induced transcription from the E2F2 promoter. REF52 cells were transiently transfected with CMV-β-gal, E2F2-luciferase, and V5-c-Myc and/or HA- HBP1 expression vectors. 18–20 h post-transfection, cells were serum-starved in DMEM supplemented with 0.1% serum for 48 h. Cleared lysates were assayed for β-galactosidase activity and luciferase activity. Luciferase activity was normalized to β-galactosidase activity and is shown relative to empty vector control-transfected cells. The average of four independent experiments ± S.E. is shown. Statistical significance is indicated by the p value between c-Myc and c- Myc + HBP1WT. C, expression of HBP1 does not affect expression of ectopic c-Myc. Whole cell lysates from the experiment in A were run on a SDS-PAGE gel and visualized by Western blotting using the indicated antibodies. D, knockdown of HBP1 does not significantly change endogenous c-Myc protein levels. 293 cells were transiently transfected with either a scramble control or 2 or 10 μg of shRNA-1 targeting HBP1. Whole cell lysates were run on an SDS-PAGE gel, and protein was analyzed by Western blotting with the indicated antibodies. Blots were scanned with a LI-COR Odyssey Infrared Imager to visualize proteins. Protein levels were quantified using LI-COR Odyssey Infrared Imager software Version 1.2. c-Myc and HBP1 protein levels were normalized to their corresponding loading controls, and values shown were calculated as a percentage of levels with transfected scramble shRNA. E, knockdown of HBP1 results in increased expression of a synthetic reporter gene driven by E-boxes. 293 cells were transiently transfected with CMV-β-gal, either pGL2 or 4xE-box-luc, and either a scramble shRNA control or HBP1 shRNA-1 as indicated. 48 h post-transfection cells were collected, and lysates were assayed for β-galactosidase and luciferase activity. Luciferase activity was normalized to β-galactosidase activity and is shown relative to levels from scramble shRNA transfections. The average of four independent experiments ± S.E. is shown. Statistical significance of the effect of HBP1 shRNA-1 on E-box-mediated transcription is indicated by the p value between the pGL2 control and 4xE-box-Luc.
In the previously described assays, c-MYC expression is controlled by a CMV promoter. Because HBP1 has been reported to inhibit expression from the CMV promoter, we also examined c-Myc protein levels in these assays (24). 293 whole cell lysates were Western-blotted for both c-Myc and HBP1. We consistently saw no appreciable change in ectopic c-Myc protein levels when HBP1 was co-expressed (Fig. 2C, lanes 3–4), indicating that the decreased luciferase expression we observed with HBP1 and c-Myc co-expression is not the result of changes in c-Myc protein levels.
Knockdown of HBP1 Protein Increases c-Myc Transactivational Activity
We next asked whether knockdown of HBP1 affected c-Myc transactivational activity. We first examined the effect of HBP1 knockdown on endogenous c-Myc protein levels in our cells, as HBP1 overexpression has been previously reported to decrease c-MYC expression in some cell types (23, 41). Cells were transiently transfected with shRNA to HBP1 or a scramble control. Both endogenous HBP1 and c-Myc protein levels were quantified and normalized to the indicated loading controls. Percent normalized protein relative to the scramble control is shown for both HBP1 and c-Myc (Fig. 2D). We observed no appreciable effect on c-Myc protein levels in response to a significant knockdown of HBP1 (to 9% of control levels). Therefore, we proceeded to examine the effect of HBP1 knockdown on c-Myc-mediated transcription. 293 cells were transiently transfected with the 4xE-box-luc or the minimal promoter-luciferase vector and either shRNA to HBP1 or a scramble control. As shown in Fig. 2E, we did not observe a significant effect on expression of the minimal promoter-driven luciferase in the presence of shRNA to HBP1. However, we did observe an approximate 1.7-fold increase in the level of E-box-driven luciferase activity when HBP1 protein was reduced. Because this was not a result of increased c-Myc protein expression in our system, our results indicate that endogenous HBP1 has a role in inhibiting endogenous c-Myc activity.
HBP1 Can Prevent c-Myc from Binding Its Target Gene Promoters, Resulting in Decreased Gene Expression
It has previously been demonstrated that HBP1 inhibits TCF4-induced gene expression by preventing binding of TCF4-β-catenin complexes to DNA (23). To investigate whether HBP1 inhibits c-Myc-induced transcription by a similar mechanism, we used ChIP assays to examine c-Myc protein binding to promoters of its target genes in the presence of ectopic HBP1 and with knockdown of endogenous HBP1 protein. For these assays we used a second shRNA to HBP1 (HBP1 shRNA-2) that also significantly knocked down endogenous HBP1 protein (Fig. 3A). Cells were transiently transfected with either an expression vector for HBP1, a control empty expression vector, shRNA to HBP1, or a scramble control. Similar to our previous results, neither overexpression of ectopic HBP1 nor knockdown of endogenous HBP1 appeared to alter endogenous c-Myc protein levels in these experiments (Fig. 3B). After cross-linking, cells were lysed and sonicated, and endogenous c-Myc protein was immunoprecipitated. Co-immunoprecipitated DNA fragments were examined by quantitative PCR using primers to the E2F2 and NUCLEOLIN promoters, both described target genes of c-Myc (5, 42). Additionally, primers within the GAPDH gene were used as a negative control. In the presence of ectopic HBP1, we observed an approximate 40% decrease in binding of c-Myc to the E2F2 promoter and an approximate 65% decrease in binding to the NUCLEOLIN promoter compared with the vector alone transfection when normalized to GAPDH and graphed relative to immunoprecipitation with an IgG control (Fig. 3C, graph). In contrast, when we knocked down HBP1 protein levels, we observed a >3-fold increase in c-Myc binding to the E2F2 promoter and a >2-fold increase in binding to the NUCLEOLIN promoter compared with the scrambled control shRNA. Together these data indicate that HBP1 inhibits c-Myc binding to these promoters.
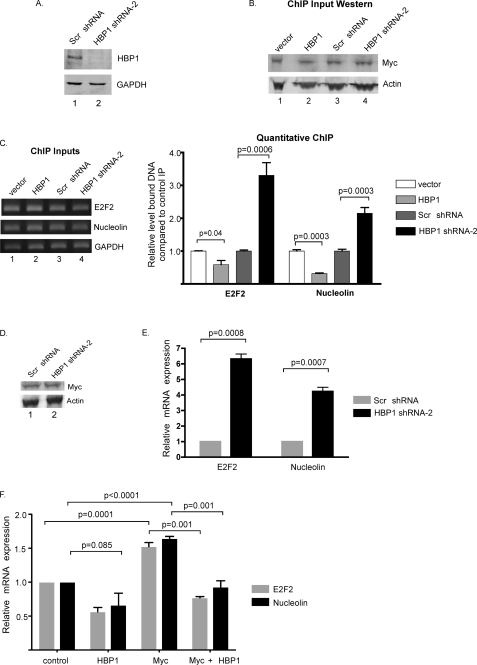
FIGURE 3.
HBP1 inhibits binding of c-Myc to its target gene promoters and decreases expression of c-Myc target genes. A, shown is validation of HBP1 shRNA-2. 293 cells were transiently transfected with HA-HBP1 and either a scrambled shRNA or HBP1-specific shRNA-2. 72 h post-transfection, lysates were harvested and analyzed by Western blotting with the indicated antibodies. B, ectopic overexpression of HBP1 or knockdown of endogenous HBP1 does not affect endogenous c-Myc protein levels. 293 cells were transiently transfected with the indicated plasmids. 72 h post-transfection, cells were cross-linked in formaldehyde, collected, and sonicated to shear DNA. 5% of the pre-cleared lysate was boiled for 30 min in SDS sample buffer and analyzed by Western blot for total c-Myc and β-actin expression. C, overexpression or knockdown of HBP1 alters binding of endogenous c-Myc to the NUCLEOLIN and E2F2 promoters is shown. The remaining cleared lysates from B were immunoprecipitated with an anti-Y69 antibody to immunoprecipitate c-Myc or a normal rabbit IgG as a control immunoprecipitation. DNA-protein complexes were uncross-linked, and DNA was purified. Primers to the E2F2 and NUCLEOLIN promoter regions as well as internal GAPDH primers were used to amplify DNA. ChIP inputs (5%) are shown for each transfection. The graph represents quantitative-PCR quantification of the amount of E2F2 or NUCLEOLIN DNA normalized to GAPDH DNA immunoprecipitated by the Y69 antibody relative to the control IP. Results represent the average of three independent experiments ± S.E. Error bars and p values were calculated using GraphPad Prism. D, knockdown of HBP1 does not alter endogenous c-Myc protein levels. 293 cells were transiently transfected with either a scrambled (scr) shRNA or HBP1 shRNA-2. 72 h post-transfection, lysates were harvested and subjected to Western blot analysis as indicated. E, expression of the c-Myc target genes E2F2 and NUCLEOLIN is increased after HBP1 knockdown. RNA was isolated from duplicate transfection plates from D, cDNA was generated, and the mRNA levels of E2F2, NUCLEOLIN, and ACTIN were measured by real-time quantitative-PCR. The graph shows quantitative-PCR results for relative expression level of E2F2 and NUCLEOLIN, normalized to ACTIN. Results represent the average of three independent experiments ± S.E. Error bars and p values were calculated using GraphPad Prism. F, HBP1 decreases c-Myc-induced expression of the endogenous E2F2 and NUCLEOLIN genes. 293 cells were transiently transfected with c-Myc and/or HBP1 expression vectors as indicated. Transfected cells were collected 48 h post-transfection, and RNA was isolated. cDNA was generated, and message levels of E2F2, NUCLEOLIN, and GAPDH were analyzed by quantitative reverse transcription -PCR. E2F2 and NUCLEOLIN message levels were normalized to GAPDH expression and are shown relative to empty vector control transfection. The averages of three independent experiments ± S.E., as calculated in GraphPad Prism, are shown. p values are indicated.
We next examined whether the effects of HBP1 on c-Myc promoter binding influenced the expression of these c-Myc target genes. To examine this we first looked at the effect of knocking down HBP1 protein on endogenous E2F2 and NUCLEOLIN expression. Cells were transiently transfected with shRNA to HBP1 (HBP1 shRNA-2) or a scramble shRNA control. Analysis of transfected cells from this experiment showed no appreciable change in endogenous c-Myc protein levels (Fig. 3D). Total RNA was isolated and analyzed by quantitative reverse transcription-PCR. Upon knockdown of HBP1 protein, we observed a 6-fold increase in E2F2 mRNA and a 4-fold increase in NUCLEOLIN mRNA (Fig. 3E). This increase in gene expression is consistent with the increased binding of c-Myc to the E2F2 and NUCLEOLIN promoters with knockdown of HBP1. We next examined whether overexpression of HBP1 with or without overexpressed c-Myc decreased expression of E2F2 and NUCLEOLIN. Cells were transfected with empty vector controls, expression vectors for HBP1 alone, c-Myc alone, or c-Myc plus HBP1. As shown in Fig. 3F, overexpression of HBP1 reduced expression of both the E2F2 and NUCLEOLIN genes, consistent with the decrease in endogenous c-Myc binding observed in Fig. 3C. Overexpression of c-Myc resulted in a typical modest induction of these genes, and this induction was eliminated by co-overexpression of HBP1 (Myc + HBP1). This was not due to an effect on 293 cell proliferation in these transient assays with HBP1 or c-Myc expression (data not shown). Taken together, our data indicate that HBP1 inhibits c-Myc from binding the promoters of its target genes, resulting in decreased expression of these genes. Again, this result is similar to HBP1-mediated inhibition of TCF4-β-catenin activity.
Multiple Domains of c-Myc Are Required for Binding to HBP1
To further understand the mechanism behind HBP1-induced repression of c-Myc, we examined the regions of interaction between the two proteins. We first examined the regions of c-Myc responsible for its interaction with HBP1. As we believe HBP1 interacts with the TAD of c-Myc, based on our yeast two-hybrid assay (supplemental Fig. 1), we used a c-Myc deletion mutant lacking the TAD as well as c-Myc mutants where MBI or MBII have been individually deleted (Fig. 4A). Immunoprecipitation of either the TAD or MBII c-Myc deletion mutants resulted in decreased HBP1 binding to c-Myc when compared with its interaction with full-length c-Myc (Fig. 4B, lanes 5 and 6 versus 2). Conversely, deletion of MBI did not appear to alter binding of HBP1 to c-Myc (Fig. 4B, lane 4). These data suggest that HBP1 binds within MBII of c-Myc, a common binding site for c-Myc interacting proteins (18, 20, 43, 44). Interestingly, we still observed some binding of HBP1 to the ΔTAD and ΔMBII c-Myc deletion mutants; therefore, we asked whether the C-terminal region absent in the yeast two-hybrid construct could also be important for HBP1 binding. We created a c-Myc deletion mutant that lacked the C-terminal 57 amino acids, which harbor the leucine zipper and most of the helix-loop-helix region (Fig. 4A). These regions are critical for Max dimerization and DNA binding (45). We found that deletion of this C-terminal region of c-Myc also consistently decreased binding of HBP1 to c-Myc protein even with an intact N-terminal domain (Fig. 4B, lane 3). Therefore, it appears that the C terminus of c-Myc is also important for its ability to interact with HBP1 in mammalian cells. Interestingly, HBP1 was found to bind to two regions of TCF4, an undescribed N-terminal region as well as the HMG box of TCF (24). Moreover, a number of other c-Myc co-factors interact with both the MBII domain and the C terminus of c-Myc (43, 44, 46).
FIGURE 4.
Multiple c-Myc domains are required for binding to HBP1. A, a schematic of full-length c-Myc and c-Myc deletion mutants is shown. c-Myc functional domains are indicated. B, HBP1 interacts with MBII and the C terminus of c-Myc. 293 cells were transiently transfected with CMV-β-galactosidase and HA-HBP1 together with empty vector or one of the indicated c-Myc expression vectors. Cleared lysates were normalized to β-galactosidase and subjected to immunoprecipitation with anti-V5. Inputs and immunoprecipitated complexes were analyzed by Western blotting with the indicated antibodies. NLS, nuclear localization signal; HLH-LZ, helix-loop-helix region leucine zipper.
c-Myc Binds to the C Terminus of HBP1, and Some HBP1 Mutants Identified in Breast Cancer Have Reduced Interaction with c-Myc
We next examined which regions of HBP1 were required for c-Myc binding. To do this we used both a C-terminal HBP1 deletion mutant, which lacks the HMG box (HBP1ΔC), and a deletion mutant lacking the repression domain of HBP1 (HBP1ΔREP) (Fig. 5A). These mutants were chosen because we had previously determined that the N-terminal 127 amino acids of HBP1 were not required for interaction with c-Myc based on the yeast two-hybrid assay (see Fig. 1A). The HMG box is the DNA binding domain of HBP1, and it has previously been shown to be required for sequence-specific repression and activation of HBP1 target genes (24, 25, 39). However, mutation of the HMG box did not prevent HBP1 from inhibiting Wnt signaling (23). Additionally, it was previously shown that TCF4 binds within the HBP1 repression domain, and given the apparently similar mode of HBP1-mediated repression of TCF4 and c-Myc, we were interested in determining if c-Myc binds HBP1 in the same region. However, immunoprecipitation of full-length HBP1 or the HBP1ΔREP mutant resulted in robust pulldown of c-Myc (Fig. 5B, lanes 2 and 4). In contrast, HBP1ΔC only weakly bound to c-Myc, suggesting that this is the main region of interaction (Fig. 5B, lane 3). These data demonstrate that the regions of interaction between c-Myc and HBP1 are different from those for TCF4 and HBP1.
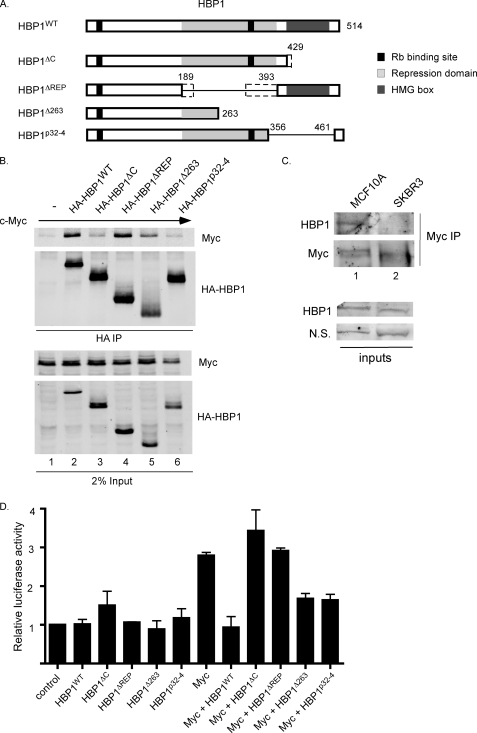
FIGURE 5.
c-Myc shows reduced binding to and/or altered activity in the presence of some HBP1 mutants. A, shown are schematics of HBP1 and HBP1 deletion mutants used in co-immunoprecipitations. HBP1Δ263 and HBP1p32-4 have been previously described as being isolated from human breast tumors (35). Other mutants isolated in breast cancer that are similar to deletion mutants generated for this study are indicated by dotted lines; specifically, a mutant C-terminal truncated at amino acid 431 and a mutant deleted for amino acids 218–314. B, c-Myc binds weakly to the HBP1 C-terminal deletion mutant and has reduced binding to some HBP1 breast cancer-derived mutants. 293 cells were transiently transfected with CMV-β-gal, V5-c-Myc, and either empty vector or one of the indicated HA-HBP1 expression vectors. Cleared lysates were normalized to β-galactosidase and subjected to immunoprecipitation with anti-HA. Inputs and immunoprecipitated complexes were analyzed by Western blotting with the indicated antibodies. C, the interaction between HBP1 and c-Myc is not detected in SKBR3 breast cancer cells. The indicated cells were lysed in co-IP buffer, and c-Myc was immunoprecipitated with C33-conjugated protein A-Sepharose. Immunoprecipitation was visualized by Western blotting with c-Myc (N262) and HBP1 antibodies. Inputs are shown to demonstrate similar HBP1 expression levels in MCF10A and SKBR3 cells. D, some HBP1 mutants fail to repress c-Myc-induced transcription from a synthetic E-box promoter. 293 cells were transiently transfected with either control pGL2 or 4xEbox-luciferase and CMV-β-gal together with V5-c-Myc and/or various HA-HBP1 expression vectors as indicated. 18–20 h post-transfection, cells were serum-starved in DMEM supplemented with 0.2% serum for 48 h. Cleared lysates were analyzed for β-galactosidase and luciferase activity. Luciferase activity was normalized to β-galactosidase activity and is shown relative to control pGL2 activity. The averages of three independent experiments ± S.E., as calculated in GraphPad Prism, are shown.
Recently, a number of naturally occurring HBP1 mutants were found in human breast tumors (35). Many of these mutants lack the C-terminal region of HBP1, suggesting that they would have reduced ability to interact with c-Myc. Indeed, one of these mutants (Δ431) described in Paulson et al. (35) is virtually identical to the HBP1ΔC mutant (Δ429), which shows poor interaction with c-Myc (Fig. 5, A, see the dotted extension, and B, lane 3). To further test the effects of HBP1 breast cancer mutants on c-Myc, we cloned two additional deletion mutants of HBP1 that are found in breast cancer; that is, a mutant deleted for the C-terminal 251 amino acids of HBP1 (Δ263) and a deletion mutant lacking amino acids 356–461 (p32-4). These mutants were then co-expressed with c-Myc. Immunoprecipitation of HBP1Δ263 and HBP1p32-4 both consistently resulted in decreased pulldown of c-Myc when compared with immunoprecipitation with wild type HBP1 (Fig. 5B, lanes 5 and 6). We also examined the association of endogenous c-Myc and HBP1 in the non-transformed MCF10A mammary epithelial cell line and in the SKBR3 breast cancer cell line. We found that, where as HBP1 is able to co-immunoprecipitate with c-Myc in MCF10A cells, in SKBR3 breast cancer cells their interaction was not detected (Fig. 5C). From preliminary non-isotopic RNase cleavage assay mutational analysis, it appears that SKBR3 cells have normal HBP1 mRNA, and further analysis will be required to determine the reason for reduced c-Myc-HBP1 interaction in these breast cancer cells.3 Taken together, these data suggest that reduced or lost interaction between c-Myc and HBP1 may be important in breast cancer cells.
We next examined the effects of the HBP1 deletion mutants shown in Fig. 5A on c-Myc E-box-driven transcription of the luciferase reporter. Neither wild type HBP1 nor any of the deletion mutants had a significant effect on luciferase activity in the absence of overexpressed c-Myc (Fig. 5D). However, consistent with our previous results, wild type HBP1 significantly reduced c-Myc transactivation of the luciferase reporter construct. In contrast, the HBP1ΔC (Δ429) mutant, which is similar to the breast cancer Δ431 mutant and only weakly bound to c-Myc, was unable to inhibit c-Myc activity (Fig. 5D, Myc+HBP1ΔC). Interestingly, the HBP1ΔREP mutant, which was able to interact with c-Myc, was unable to inhibit c-Myc activity, suggesting an important function for this region of HBP1 in c-Myc repression. A deletion mutant that lacks part of the repression domain was also described in breast cancer (Fig. 5A, ΔREP, see the dotted extensions) (35), and because our ΔREP mutant can bind c-Myc but cannot repress its activity, it is possible that this breast cancer mutant would also be ineffective in inhibiting c-Myc activity. In contrast to HBP1ΔC, both HBP1Δ263 and HBP1p32-4, which also had reduced binding to c-Myc, partially retained their ability to repress c-Myc activity (∼50% repression activity relative to HBP1WT). Taken together, our results suggest that some mutations of the tumor suppressor HBP1 in breast cancer may have reduced or lost the ability to repress c-Myc activity and that this correlates with a loss of the C terminus and/or loss of part or all of the repression domain of HBP1. This inability to inhibit c-Myc transactivation could contribute to the cancer phenotype.
DISCUSSION
c-Myc activity drives cellular proliferation and cell growth, inhibits differentiation, and can induce apoptosis in the absence of growth factors. Unchecked c-Myc activity results in tumorigenesis in a number of different cell types; therefore, proper regulation of c-Myc levels and activity is critical for maintaining cells in a differentiated state. To better understand the regulation of c-Myc, we set out to identify new c-Myc interacting proteins by using a yeast two-hybrid assay. In this screen we identified the HMG box transcription factor HBP1 as a novel c-Myc interacting protein. HBP1 is regarded as a tumor suppressor protein, and its expression has been shown to inhibit cell cycle progression in multiple cell types (24, 26, 27). Additionally, HBP1 appears to be an important mediator of oncogene-induced senescence as well as cellular differentiation (26, 27, 34, 38). It was shown that expression of HBP1 results in the induction of differentiation in leukemic cells and in pre-muscle cells HBP1 initiates a necessary cell cycle arrest before differentiation (27, 38). Given that the activities of HBP1 oppose those of c-Myc, a potent oncogene and regulator of cell cycle progression, this is likely to be a significant and important interaction. Indeed, we show here that HBP1 inhibits c-Myc-mediated transactivation of target genes, and some mutant forms of HBP1 similar to those found in human breast cancer no longer interact with and/or negatively regulate c-Myc activity.
Mechanisms of HBP1-mediated Inhibition of c-Myc Transactivation Activity
Our work shows that HBP1 inhibits c-Myc-induced transcription at least in part by preventing binding of c-Myc to its target gene promoters. We mapped the binding of HBP1 to both the MBII region of the c-Myc TAD and to the C terminus of c-Myc, involved in Myc DNA binding. Thus, a straightforward explanation for HBP1 inhibition of c-Myc promoter binding is that HBP1 association with the C terminus of c-Myc inhibits Myc/Max heterodimers from binding DNA. We do not know whether this effect would be to the on- or off-rate of DNA binding by Myc/Max. Although it is possible that HBP1 exerts its effects by interfering with Myc-Max dimerization, we do not believe that this is the case as the Myc-Max interaction is very robust, and other co-factors of c-Myc that bind to the C terminus do not disrupt Max binding (12, 43, 44).
It is also possible that HBP1 could indirectly affect c-Myc binding to target gene promoters via other HBP1 binding partners. HBP1 is a known binding partner of both pRB and the co-repressor SIN3 (22, 24, 51). pRB is recruited to DNA by the E2F transcription factor family, and many c-Myc target gene promoters contain both E-boxes and adjacent E2F binding sites (5, 52, 53). Additionally, SIN3 can be recruited to c-Myc target genes directly through its interaction with Mad/Max heterodimers at E-boxes (54). Although it is not clear whether recruitment of HBP1 to c-Myc target genes occurs through these other partners, it certainly could bring HBP1 in proximity to where c-Myc needs to access its binding sites.
In addition to the effects of HBP1 on c-Myc promoter binding, it is possible that the association of HBP1 with MBII inhibits c-Myc transactivation activity by affecting the interaction of other c-Myc co-regulators with MBII, such as TRRAP. TRRAP is a core subunit of histone acetyltransferase complexes, and recruitment to c-Myc is important for the activation of c-Myc target genes (47–49) by recruiting Gcn4 or Tip60 histone acetyltransferase complexes (17, 50). It has been previously shown that the ribosomal protein L11 binds within MBII of c-Myc and prevents TRRAP binding (20). Therefore, this could be a possible additional mechanism of HBP1 inhibition of c-Myc that will be investigated in future studies.
HBP1 Regulates c-Myc at Multiple Levels
HBP1-mediated repression of c-Myc transactivation activity shares some similarities to HBP1-mediated inhibition of Wnt signaling. Signaling by Wnt proteins regulate a number of important processes including cell proliferation and cell fate decisions, and mutants within the Wnt signaling pathway have been implicated in numerous tumor types (55). Sampson et al. (23) demonstrated that HBP1 could inhibit transcription downstream of activation of the Wnt pathway, and this inhibition was dependent on HBP1 binding to the TCF4 transcription factor. Specifically, binding of HBP1 to TCF4 results in the inhibition of DNA binding by TCF4-β-catenin complexes. HBP1 DNA binding activity was dispensable for this function, as a triple point mutant unable to bind DNA was still able to repress Wnt signaling. Taken together with our data demonstrating that HBP1 inhibits c-Myc binding to its target gene promoters, these results indicate that, in addition to its sequence-specific mediated repression, interference with transcription factor DNA binding is another important mechanism of HBP1-mediated gene repression.
One important aspect of HBP1-mediated inhibition of Wnt signaling is that it was shown to inhibit c-MYC gene expression in multiple cell types (23, 27, 41). This observation coupled with our data suggest that HBP1 may work in multiple ways to inhibit c-Myc activity 1) by directly inhibiting c-Myc activity as a transcription factor as shown here and 2) by inhibiting c-MYC expression via inhibition of TCF4-β-catenin complexes (see Fig. 6). This further solidifies HBP1 as a cell cycle inhibitor and a regulator of differentiation as c-Myc expression is required for cell cycle progression, and prolonged c-Myc expression results in the inhibition of differentiation.
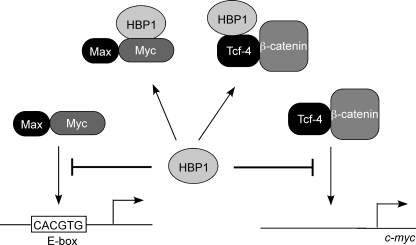
FIGURE 6.
HBP1 inhibits c-Myc activity by multiple mechanisms. HBP1 binds c-Myc and prevents its interaction with DNA thereby preventing activation of c-Myc target genes. Additionally, HBP1 can bind TCF4 and prevent its binding to promoters of target genes including c-MYC (23). Therefore, HBP1 can inhibit both the activity and expression of c-Myc.
Inhibition of c-Myc Activity as a Mechanism of Tumor Suppression by HBP1
As previously described, HBP1 is an emerging tumor suppressor protein. Paulson et al. (35) found that HBP1 expression is reduced in a subset of invasive human breast tumors, and this correlated with a poor prognosis. In addition, the authors found a number of naturally occurring HBP1 mutants in human breast tumors. A majority of these mutants lacked the C-terminal region of HBP1, suggesting that they would be unable to bind c-Myc and, therefore, would be unable to inhibit c-Myc activity. Here we show that a number of these mutants have reduced binding to c-Myc (Fig. 5B). Interestingly, some of the mutants, although unable to bind c-Myc well, could still partially suppress c-Myc-induced expression from a reporter plasmid (Fig. 5D), indicating that interaction alone is not sufficient for inhibition of c-Myc activity. A partially intact HBP1 repression domain appears to be required, as the HBP1ΔREP mutant that does bind c-Myc is defective in its ability to repress c-Myc-induced gene expression. This suggests that some mutants found in breast cancer with intact C termini may also be unable to inhibit c-Myc activity. All of these mutants found in breast cancer were defective in their ability to inhibit Wnt signaling, suggesting that loss of wild-type HBP1 may affect both c-Myc levels and activity (35). It was also demonstrated that these mutants are defective in their ability to repress colony formation by MDA-MB-231 cells, unlike wild-type HBP1, which significantly decreased colony formation when overexpressed. MDA-MB-231 cells have high c-Myc protein expression, and it was previously shown that knockdown of c-Myc in these cells resulted in decreased cell proliferation and cell migration (56). Therefore, it is possible that some of the HBP1 anti-tumorigenic activity, when overexpressed in these cells, is a result of its ability to inhibit c-Myc activity. Together with the ability or HBP1 to repress Wnt signaling, inhibition of c-Myc is likely to contribute substantially to HBP1 tumor suppressor activity.
Supplementary Material
Acknowledgments
We thank Amy Yee for the pEF-BOS-HBP1 plasmid, Peter Hurlin for the 4x-Ebox-Luc plasmid, and William Dewitt, Moon Yoon, and Kristi Piehl for excellent technical support with the yeast two-hybrid screen.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA100855 (to R. S.), R01 CA100855-05S1 (a minority supplement to J. R. E.-P.), and T32-CA106195 (to A. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
A. Yee, personal communication.
- TAD
- transactivational domain
- MBI and MBII
- Myc Box I and Box II, respectively
- ChIP
- chromatin immunoprecipitation
- HBP1
- HMG box protein 1
- HMG
- high mobility group
- shRNA
- short hairpin RNA
- CMV
- cytomegalovirus
- HA
- hemagglutinin
- DMEM
- Dulbecco's modified Eagle's medium
- IP
- immunoprecipitation
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- WT
- wild type
- pBS
- pBluescript.
REFERENCES
- 1.Mateyak M. K., Obaya A. J., Adachi S., Sedivy J. M. (1997) Cell Growth Differ. 8, 1039–1048 [PubMed] [Google Scholar]
- 2.Davis A. C., Wims M., Spotts G. D., Hann S. R., Bradley A. (1993) Genes Dev. 7, 671–682 [DOI] [PubMed] [Google Scholar]
- 3.Nesbit C. E., Tersak J. M., Prochownik E. V. (1999) Oncogene 18, 3004–3016 [DOI] [PubMed] [Google Scholar]
- 4.Galaktionov K., Chen X., Beach D. (1996) Nature 382, 511–517 [DOI] [PubMed] [Google Scholar]
- 5.Sears R., Ohtani K., Nevins J. R. (1997) Mol. Cell. Biol. 17, 5227–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones R. M., Branda J., Johnston K. A., Polymenis M., Gadd M., Rustgi A., Callanan L., Schmidt E. V. (1996) Mol. Cell. Biol. 16, 4754–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell K. O., Ricci M. S., Miyashita T., Dicker D. T., Jin Z., Reed J. C., El-Deiry W. S. (2000) Cancer Res. 60, 6318–6325 [PubMed] [Google Scholar]
- 8.Dang C. V., O'Donnell K. A., Zeller K. I., Nguyen T., Osthus R. C., Li F. (2006) Semin. Cancer Biol. 16, 253–264 [DOI] [PubMed] [Google Scholar]
- 9.Ayer D. E., Kretzner L., Eisenman R. N. (1993) Cell 72, 211–222 [DOI] [PubMed] [Google Scholar]
- 10.Kretzner L., Blackwood E. M., Eisenman R. N. (1992) Nature 359, 426–429 [DOI] [PubMed] [Google Scholar]
- 11.Patel J. H., Loboda A. P., Showe M. K., Showe L. C., McMahon S. B. (2004) Nat. Rev. Cancer 4, 562–568 [DOI] [PubMed] [Google Scholar]
- 12.Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massagué J., Hänel F., Eilers M. (2001) Nat. Cell Biol. 3, 392–399 [DOI] [PubMed] [Google Scholar]
- 13.Wu S., Cetinkaya C., Munoz-Alonso M. J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L. G. (2003) Oncogene 22, 351–360 [DOI] [PubMed] [Google Scholar]
- 14.Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J. R. (2000) Genes Dev. 14, 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutterbach B., Hann S. R. (1994) Mol. Cell. Biol. 14, 5510–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W. C., Stukenberg P. T., Shenolikar S., Uchida T., Counter C. M., Nevins J. R., Means A. R., Sears R. (2004) Nat. Cell Biol. 6, 308–318 [DOI] [PubMed] [Google Scholar]
- 17.McMahon S. B., Wood M. A., Cole M. D. (2000) Mol. Cell. Biol. 20, 556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon S. B., Van Buskirk H. A., Dugan K. A., Copeland T. D., Cole M. D. (1998) Cell 94, 363–374 [DOI] [PubMed] [Google Scholar]
- 19.Park J., Kunjibettu S., McMahon S. B., Cole M. D. (2001) Genes Dev. 15, 1619–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007) EMBO J. 26, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesage F., Hugnot J. P., Amri E. Z., Grimaldi P., Barhanin J., Lazdunski M. (1994) Nucleic Acids Res. 22, 3685–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavender P., Vandel L., Bannister A. J., Kouzarides T. (1997) Oncogene 14, 2721–2728 [DOI] [PubMed] [Google Scholar]
- 23.Sampson E. M., Haque Z. K., Ku M. C., Tevosian S. G., Albanese C., Pestell R. G., Paulson K. E., Yee A. S. (2001) EMBO J. 20, 4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tevosian S. G., Shih H. H., Mendelson K. G., Sheppard K. A., Paulson K. E., Yee A. S. (1997) Genes Dev. 11, 383–396 [DOI] [PubMed] [Google Scholar]
- 25.Berasi S. P., Xiu M., Yee A. S., Paulson K. E. (2004) Mol. Cell. Biol. 24, 3011–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih H. H., Xiu M., Berasi S. P., Sampson E. M., Leiter A., Paulson K. E., Yee A. S. (2001) Mol. Cell. Biol. 21, 5723–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao C. J., Works K., Romagnoli P. A., Austin G. E. (2005) Leukemia 19, 1958–1968 [DOI] [PubMed] [Google Scholar]
- 28.Driouch K., Briffod M., Bièche I., Champème M. H., Lidereau R. (1998) Cancer Res. 58, 2081–2086 [PubMed] [Google Scholar]
- 29.Koike M., Tasaka T., Spira S., Tsuruoka N., Koeffler H. P. (1999) Leuk. Res. 23, 307–310 [DOI] [PubMed] [Google Scholar]
- 30.Liang H., Fairman J., Claxton D. F., Nowell P. C., Green E. D., Nagarajan L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3781–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zenklusen J. C., Thompson J. C., Klein-Szanto A. J., Conti C. J. (1995) Cancer Res. 55, 1347–1350 [PubMed] [Google Scholar]
- 32.Zenklusen J. C., Thompson J. C., Troncoso P., Kagan J., Conti C. J. (1994) Cancer Res. 54, 6370–6373 [PubMed] [Google Scholar]
- 33.Zenklusen J. C., Weitzel J. N., Ball H. G., Conti C. J. (1995) Oncogene 11, 359–363 [PubMed] [Google Scholar]
- 34.Zhang X., Kim J., Ruthazer R., McDevitt M. A., Wazer D. E., Paulson K. E., Yee A. S. (2006) Mol. Cell. Biol. 26, 8252–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulson K. E., Rieger-Christ K., McDevitt M. A., Kuperwasser C., Kim J., Unanue V. E., Zhang X., Hu M., Ruthazer R., Berasi S. P., Huang C. Y., Giri D., Kaufman S., Dugan J. M., Blum J., Netto G., Wazer D. E., Summerhayes I. C., Yee A. S. (2007) Cancer Res. 67, 6136–6145 [DOI] [PubMed] [Google Scholar]
- 36.Arnold H. K., Sears R. C. (2006) Mol. Cell. Biol. 26, 2832–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. (1990) Mol. Cell. Biol. 10, 5914–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shih H. H., Tevosian S. G., Yee A. S. (1998) Mol. Cell. Biol. 18, 4732–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemercier C., Duncliffe K., Boibessot I., Zhang H., Verdel A., Angelov D., Khochbin S. (2000) Mol. Cell. Biol. 20, 6627–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurlin P. J., Quéva C., Eisenman R. N. (1997) Genes Dev. 11, 44–58 [DOI] [PubMed] [Google Scholar]
- 41.Kim J., Zhang X., Rieger-Christ K. M., Summerhayes I. C., Wazer D. E., Paulson K. E., Yee A. S. (2006) J. Biol. Chem. 281, 10865–10875 [DOI] [PubMed] [Google Scholar]
- 42.Greasley P. J., Bonnard C., Amati B. (2000) Nucleic Acids Res. 28, 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K. I., Söderberg O., Kerppola T. K., Larsson L. G. (2003) Mol. Cell 11, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 44.Kim S. Y., Herbst A., Tworkowski K. A., Salghetti S. E., Tansey W. P. (2003) Mol. Cell 11, 1177–1188 [DOI] [PubMed] [Google Scholar]
- 45.Blackwood E. M., Eisenman R. N. (1991) Science 251, 1211–1217 [DOI] [PubMed] [Google Scholar]
- 46.Qi Y., Gregory M. A., Li Z., Brousal J. P., West K., Hann S. R. (2004) Nature 431, 712–717 [DOI] [PubMed] [Google Scholar]
- 47.Nikiforov M. A., Chandriani S., Park J., Kotenko I., Matheos D., Johnsson A., McMahon S. B., Cole M. D. (2002) Mol. Cell. Biol. 22, 5054–5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank S. R., Schroeder M., Fernandez P., Taubert S., Amati B. (2001) Genes Dev. 15, 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouchard C., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M., Lüscher B. (2001) Genes Dev. 15, 2042–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank S. R., Parisi T., Taubert S., Fernandez P., Fuchs M., Chan H. M., Livingston D. M., Amati B. (2003) EMBO Rep 4, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson K. A., Knoepfler P. S., Huang K., Kang R. S., Cowley S. M., Laherty C. D., Eisenman R. N., Radhakrishnan I. (2004) Nat. Struct. Mol. Biol. 11, 738–746 [DOI] [PubMed] [Google Scholar]
- 52.Bolognese F., Forni C., Caretti G., Frontini M., Minuzzo M., Mantovani R. (2006) Gene 366, 109–116 [DOI] [PubMed] [Google Scholar]
- 53.Suzuki S., Adachi A., Hiraiwa A., Ohashi M., Ishibashi M., Kiyono T. (1998) Gene 216, 85–91 [DOI] [PubMed] [Google Scholar]
- 54.Ayer D. E., Lawrence Q. A., Eisenman R. N. (1995) Cell 80, 767–776 [DOI] [PubMed] [Google Scholar]
- 55.Polakis P. (2007) Curr. Opin. Genet. Dev. 17, 45–51 [DOI] [PubMed] [Google Scholar]
- 56.Cappellen D., Schlange T., Bauer M., Maurer F., Hynes N. E. (2007) EMBO Rep. 8, 70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.