Abstract
ADAMTS (A disintegrin and metalloproteinase with thrombospondin motifs)-like (ADAMTSL) proteins, a subgroup of the ADAMTS superfamily, share several domains with ADAMTS proteinases, including thrombospondin type I repeats, a cysteine-rich domain, and an ADAMTS spacer, but lack a catalytic domain. We identified two new members of ADAMTSL proteins, ADAMTSL-6α and -6β, that differ in their N-terminal amino acid sequences but have common C-terminal regions. When transfected into MG63 osteosarcoma cells, both isoforms were secreted and deposited into pericellular matrices, although ADAMTSL-6α, in contrast to -6β, was barely detectable in the conditioned medium. Immunolabeling at the light and electron microscopic levels showed their close association with fibrillin-1-rich microfibrils in elastic connective tissues. Surface plasmon resonance analyses demonstrated that ADAMTSL-6β binds to the N-terminal half of fibrillin-1 with a dissociation constant of ∼80 nm. When MG63 cells were transfected or exogenously supplemented with ADAMTSL-6, fibrillin-1 matrix assembly was promoted in the early but not the late stage of the assembly process. Furthermore, ADAMTSL-6 transgenic mice exhibited excessive fibrillin-1 fibril formation in tissues where ADAMTSL-6 was overexpressed. All together, these results indicated that ADAMTSL-6 is a novel microfibril-associated protein that binds directly to fibrillin-1 and promotes fibrillin-1 matrix assembly.
Keywords: Extracellular Matrix, Extracellular Matrix/Connective Tissue, Extracellular Matrix/Elastin, Extracellular Matrix/Fibrillin, Protease/ADAM/ADAMTS, Protein/Connective Tissues, Tissue/Organ Systems/Connective Tissue
Introduction
Microfibrils are multifunctional fibrillar extracellular matrices (ECMs)2 that contribute to the elastic properties of connective tissues and also control the bioavailabilities of transforming growth factor-β (TGF-β) superfamily molecules (1, 2). The major component of microfibrils is fibrillin-1, a 350-kDa cysteine-rich glycoprotein, which has a modular structure composed of arrays of calcium-binding epidermal growth factor-like domains separated by 8-cysteine domains (also designated TB domain) and hybrid domains (3, 4). This modular domain structure gives rise to an extended 150-nm rod with flexible regions (5). Linear and lateral self-association of fibrillin-1 monomers creates the backbone of the microfibril, onto which various combinations of microfibril-associated proteins assemble, followed by binding and cross-linking of elastin in elastic tissues (6). Electron microscopic observations revealed that tissue microfibrils display periodic beaded string structures with a mean diameter of 10–12-nm and 50–56-nm intervals (7, 8). Several models have been proposed for fibrillin-1 microfibril assembly, including parallel unstaggered and staggered models (8–10), but the mechanism by which fibrillin-1 monomers assemble into microfibrils remains controversial.
Accumulated evidence indicates that fibrillin-1 fibrils elongate in a parallel, head-to-tail manner. Epitope mapping of monoclonal antibodies directed to the N- and C-terminal regions of fibrillin-1 supports an N-to-C linear alignment in tissue microfibrils (7, 9). Consistent with this alignment, N-terminal recombinant fragments exhibit high affinity binding to C-terminal fragments, typically containing the last three calcium-binding epidermal growth factor-like domains (11, 12). Not only N-to-C interactions but also N-to-N and C-to-C interactions have been suggested to occur in microfibril assembly (11–15). Linear and lateral associations of fibrillin-1 monomers do not seem to proceed independently, as lateral association into disulfide-bonded multimers strongly potentiates the binding affinity of the C-terminal half of fibrillin-1 to the N-terminal half adsorbed onto a solid surface (12). Furthermore, multimers, but not monomers, of the C-terminal fragment are capable of inhibiting fibrillin-1 fibril formation by cultured dermal fibroblasts (12), corroborating the importance of lateral association of the C-terminal region in fibrillin-1 fibril formation. N-terminal fragments may also undergo disulfide-bonded dimer formation as a very early event in fibrillin-1 assembly (13, 15).
Fibrillin-1 binds to a variety of ECM molecules, and some of these may play a role in microfibril assembly. Fibrillin-1 and -2 have been shown to bind to fibronectin at their C-terminal regions (16). Interaction of fibrillin-1 with cell surface fibronectin deposits is critical for fibrillin-1 microfibril assembly, as inhibition of fibronectin matrix assembly results in diminished fibrillin-1 microfibril assembly (16, 17). Fibrillin-1 also binds to heparin and heparan sulfate chains at multiple sites, of which the N-terminal region encoded by exons 1–11 exhibits the highest affinity for heparin (18, 19). Interactions of fibrillin-1 with cell surface heparan sulfate proteoglycans may also be involved in microfibril assembly, because exogenously added heparin strongly inhibits fibrillin-1 microfibril assembly, as is also the case when heparan sulfate attachment to core proteins or sulfations of glycosaminoglycan chains are inhibited (20, 21). Fibrillin-1 also binds to other microfibril-associated proteins, including latent TGF-β-binding proteins (22, 23), microfibril-associated glycoproteins (24, 25), and fibulins (23, 26), thereby contributing to tissue-specific heterogeneity of microfibril composition (22, 27). However, none of these associated proteins have been demonstrated to regulate fibrillin-1 matrix assembly so far.
Mutations in the fibrillin-1 gene (FBN1) cause fibrillinopathies, heritable disorders of connective tissues that include Marfan syndrome and related diseases (1). The Marfan syndrome demonstrates pleiotropic features in multiple organ systems. Major features include cardiovascular, ocular, and skeletal anomalies (OMIM 154700). Fibroblasts from various types of Marfan patients show reduced and/or aberrant depositions of fibrillin-1 (28, 29). Most missense mutations in Marfan syndrome affect calcium-binding epidermal growth factor-like domains, frequently disrupting one of the six conserved cysteine residues or the residues involved in calcium binding, and hence leading to aberrant conformation of fibrillin-1 monomers and enhanced proteolytic degradation of microfibrils (4, 30, 31). FBN1 is the gene responsible not only for Marfan syndrome but also for autosomal dominant Weill-Marchesani syndrome (OMIM 608328), a disorder characterized by short stature, acromelic dysplasia, brachydactyly, joint stiffness, and lens abnormalities (32, 33).
ADAMTSL proteins are a subgroup of ADAMTS superfamily proteins, composed of an N-terminal thrombospondin type I repeat (TSR), a cysteine-rich (Cys-rich) repeat, an ADAMTS spacer, and C-terminal tandem TSRs, occasionally with a PLAC domain at their C-terminal ends, but lacking the catalytic domain that is shared by ADAMTS and ADAM proteinases (34). Currently, four ADAMTSL proteins are known (see Fig. 1A), i.e. ADAMTSL-1/punctin (35), ADAMTSL-2 (36), ADAMTSL-3/punctin-2 (37), and ADAMTSL-4/TSRC1 (38). A putative protein containing a netrin-like domain instead of C-terminal TSRs has been registered in GenBank as ADAMTSL-5 (accession number NP001107020). Various ADAMTSL and ADAMTS proteins have been shown to associate with ECM molecules through their TSR domains after secretion (35, 37, 39–44). ADAMTS-1 and -4 are capable of binding to sulfated glycosaminoglycan chains via the first TSR (39, 44), and ADAMTSL-2 was recently shown to bind to LTBP-1 (42). Although the physiological functions of ADAMTSL proteins remain only poorly defined, mutations in ADAMTSL2 have been found in patients of geleophysic dysplasia, an autosomal recessive disorder characterized by short stature, brachydactyly, thick skin, and cardiac valvular anomalies (42). Mutations in ADAMTSL4 have been associated with ectopia lentis, a rare autosomal-recessive disorder characterized by subluxation of the lens resulting from the disruption of the zonular fibers (45). Hence, genetic evidence indicates tissue-specific roles for ADAMTSL proteins in organs in which fibrillin-1 also performs regulatory or mechanical functions.
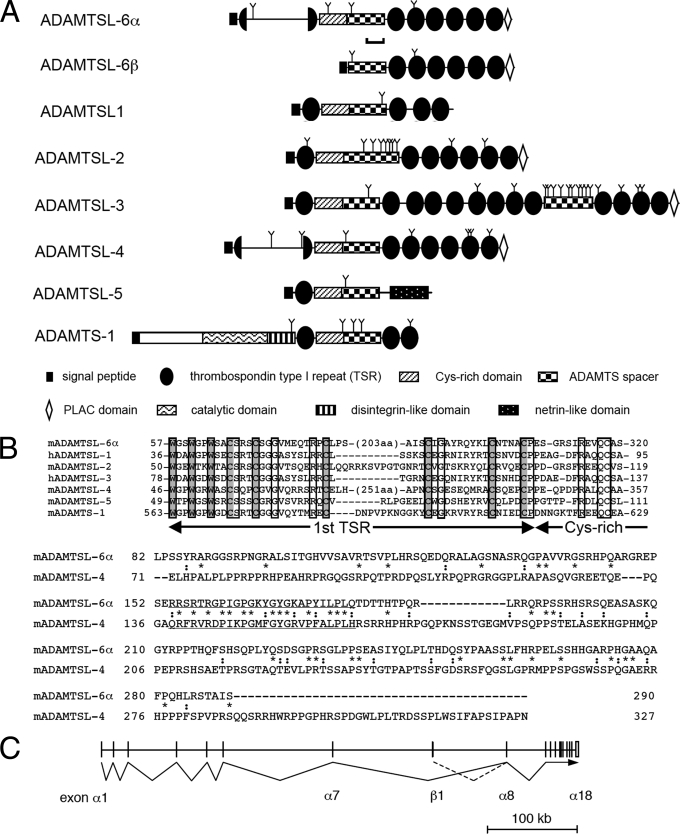
FIGURE 1.
Structures of ADAMTSL-6 proteins. A, schematic representations of the domain structures of ADAMTSL-6α and -6β and other ADAMTS-like proteins. The domain structure of ADAMTS-1 proteinase is included as a reference. Note that the first TSR domains of ADAMTSL-6α and ADAMTSL-4 are split by ∼200-amino acid insertions (detailed in B). The region used as an immunogen for antibody production is indicated by a bracket. The putative N-linked glycosylation sites are marked with Y. B, alignment of amino acid sequences of the first TSR domain and a part of the following Cys-rich domain of ADAMTSL-6α and other ADAMTS-like proteins (upper panel). Species origins are indicated by prefixes m (mouse) and h (human). The conserved residues are boxed. Three tryptophans and six cysteines, which have been shown to be critical in the layered fold of TSR domains (59), are boxed with gray background. Residue numbers are shown on the left and right ends. The insertion sequences that split the first TSR domain of ADAMTSL-6α and ADAMTSL-4 into N- and C-terminal halves are aligned using the ClustalW algorithm. Conserved and semiconserved residues are indicated by “*” and “:”, respectively. A stretch of 26 amino acid residues that are highly homologous between ADAMTSL-6α and ADAMTSL-4 are underlined. C, schematic diagram of exon-intron structure of Adamtsl6 gene on mouse chromosome 9E. Exon numbers encoding ADAMTSL-6α are shown below the diagram. The first exon encoding the ADAMTSL-6β-specific sequence, including the signal peptide, is located between exon 7 and exon 8 of ADAMTSL-6α.
Recently, we developed a methodology for comprehensive identification of unknown ECM proteins by combining in silico screening of secreted proteins with in vitro functional screening and immunohistochemical analysis (46). We identified 16 previously unknown ECM proteins, among which we found one with a domain structure homologous to ADAMTSL proteins. Here, we show that this protein, now designated ADAMTSL-6, is associated with microfibrils through direct interaction with fibrillin-1 and promotes fibrillin-1 matrix assembly in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Computational Screening for Novel ECM Proteins
ADAMTSL-6 cDNAs were isolated by our systematic computational screening for ECM proteins from a RIKEN mouse full-length cDNA collection (46). cDNAs encoding putative secreted proteins were selected based on the presence of an N-terminal signal sequence and the absence of any transmembrane domains using PSORT II (47) and SOSUI (48), respectively. A homology search using FASTY (49) was performed to eliminate any functionally known proteins. Two ADAMTSL-6 cDNAs were identified as those encoding putative ECM proteins with an N-terminal signal sequence and C-terminal tandem TSR repeats.
Cell Culture
MG63 cells were obtained from Health Science Research Resources Bank (Tokyo, Japan) and maintained in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal bovine serum supplemented with penicillin and streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. FreeStyleTM 293F cells (Invitrogen) were maintained in FreeStyleTM 293 Expression Medium (Invitrogen) according to the manufacturer's instructions.
Animals
ICR and C57BL/6 mice were purchased from Japan SLC Inc. (Hamamatsu, Japan). Adamtsl6 transgenic mice were generated as described below. The animals and procedures used in this study were in accordance with the guidelines of Aichi Medical University and Osaka University Animal Care and Use Committees.
RT-PCR
Mouse tissue total RNA was purchased from Clontech. Total RNA of transfected MG63 cells was isolated by using RNeasy kit (Qiagen, Valencia, CA). cDNAs were reverse-transcribed from total RNA and amplified by PCR with target-specific primers using RedTaq polymerase (Sigma). Transcripts encoding β-actin, fibrillin-1, and transfected ADAMTSL-6α and -6β were amplified using the following pairs of primers: 5′-GCGGTTCCGATGCCCTGAGGC-3′ and 5′-CGCCTAGAAGCACTTGCGGTGC-3′ (for β-actin); 5′-GGAGAAGCACAAACGAAACT-3′ and 5′-CCCCAATGGAAATACACGTC-3′ (for fibrillin-1); 5′-CTGGACTGCCACCTTCTGAGGC-3′ and 5′-CCTGGCAGCCCGTGTTATCACC-3′ (for ADAMTSL-6α); and 5′-GCACTTGGCAGACGGAGTCTCC-3′ and 5′-CCTGGCAGCCCGTGTTATCACC-3′ (for ADAMTSL-6β).
Antibodies
An affinity-purified polyclonal antibody against ADAMTSL-6 was prepared as follows. A cDNA fragment encoding the C-terminal half of the ADAM spacer domain (amino acid residues 534–618 of ADAMTSL-6α), which exhibits the least homology to any known ADAMTS or ADAMTSL proteins, was amplified by PCR with the following primers: 5′-CCGGAATTCAGCCCACAGGTGCCACCACAC-3′ and 5′-AACTCGAGTCAGTCCCGGCTTCGCCTTGGG-3′. The PCR product was cleaved with EcoRI and XhoI and cloned into pGEX4T1 (GE Healthcare), followed by expression as a glutathione S-transferase fusion protein in Escherichia coli BL21. The resulting fusion protein was purified using glutathione-Sepharose (GE Healthcare) according to the manufacturer's instructions and used as an immunogen. The antibody against ADAMTSL-6 was raised in rabbits and affinity-purified on an immunogen-immobilized column, followed by depletion of antibodies against glutathione S-transferase on a glutathione S-transferase-immobilized affinity column, as described previously (50). The specific immunoreactivity of the resulting antibody was verified by enzyme-linked immunosorbent assay and Western blotting using the immunogen and recombinant mouse and human full-length ADAMTSL-6 expressed in mammalian cells. Monoclonal antibodies against FLAG tag (M2) and fibrillin-1 (clone 69) were purchased from Sigma and Chemicon (Temecula, CA), respectively. Rabbit polyclonal anti-fibrillin-1 (pAb9543) antibody was described previously (51). Alexa Fluor 488- or 546-conjugated secondary antibodies were purchased from Molecular Probes (Eugene, OR).
Expression and Purification of Recombinant ADAMTSL-6
A cDNA fragment encoding the entire coding region of ADAMTSL-6β was amplified using KOD-Plus DNA polymerase (TOYOBO, Osaka, Japan) with a primer pair of 5′-GCTAGCGAATTCGCCACCATGTTTGTCAGCTACC-3′ and 5′-GCGGCCGCAGGTCGACCTCGGCTCCCC-3′ so that an artificial Kozak sequence was included before the first codon. The amplified fragment was subcloned into the NheI/SalI sites of a eukaryotic expression vector pSecTag2A (Invitrogen). The vector contains Myc and His6 epitopes in tandem downstream of the SalI site, and therefore, the resulting construct expresses full-length ADAMTSL-6β fused with a Myc-His6 tag at its C terminus. For expression of ADAMTSL-6α, an ADAMTSL-6α-specific sequence was amplified by PCR with a primer pair of 5′-GCTTCGAATTCGCCACCATGGTTTCCTATCTCACGAGC-3′ and 5′-GCGAGAACAGGCAGACCAGGGG-3′ and replaced into the EcoRI/PvuI sites of the ADAMTSL-6β expression construct. The nucleotide sequences of the amplified cDNAs were verified by sequencing from both directions. For purification of ADAMTSL-6β, 293F cells were transfected with the cDNA encoding ADAMTSL-6β and cultured for 3 days. Conditioned media were clarified by centrifugation and then applied to a nickel-agarose column (Qiagen). After washing with 50 mm sodium phosphate buffer, pH 8.0, containing 0.3 m NaCl and 10 mm imidazole, bound proteins were eluted with 50 mm sodium phosphate buffer, pH 8.0, containing 0.3 m NaCl and 250 mm imidazole. The eluates were dialyzed against PBS and stored at −80 °C until use.
Generation of Adamtsl6 Transgenic Mice
A cDNA fragment encoding the multicloning site and the franking FLAG tag sequence of pFLAG-CMV-5a (Sigma) was amplified by PCR using the following pair of primers: 5′-GAACTCGAGGCGGCCGCGAATTCAAGCTTGCGG-3′ and 5′-GAAGAGCTCGCGGCCGCTACTTGTCATCATCG-3′, which contained XhoI-NotI and SacI-NotI sites, respectively, and subcloned into the XhoI/SacI sites of pBlueScriptII KS+ (Stratagene, La Jolla, CA). A cDNA fragment encoding an entire open reading frame of ADAMTSL-6β was amplified using a primer pair of 5′- CCGGAATTCGCCGCCATGTTTGTCAGCTACCTGTTATTAAC-3′ and 5′-TTTTCCTTTTGCGGCCGCCTCGGCTCCCCAGAAAGCCCAC-3′, cleaved with EcoRI and NotI, and then subcloned into EcoRI/PspOMI sites of pEGFPN3 vector (Clontech). The resulting plasmid was cleaved with EcoRI and BamHI, and the fragment containing ADAMTSL-6β cDNA was subcloned into a pBlueScriptII KS+ plasmid containing multicloning sites of pFLAG-CMV-5a described above. The NotI fragment containing an entire coding region of ADAMTSL-6β was inserted into the NotI site of pKN185 (kindly provided by Dr. Noriyuki Tsumaki, Osaka University, Japan) containing the mouse Col2a1 promoter/enhancer sequence (52). A HindIII-NdeI fragment of the resulting construct, which contained the Col2a1 promoter/enhancer followed by ADAMTSL-6β cDNA fused with a FLAG tag, was microinjected into fertilized C57BL/6 oocytes to generate transgenic mice. Genotyping by PCR identified a total of 24 lines of founder mice, of which two lines were arbitrarily chosen and used for immunohistochemical analyses.
Southern Blot Hybridization
Genomic DNA was isolated from adult mouse liver using the DNeasy tissue kit (Qiagen). Serially diluted genomic DNAs digested with EcoRI and BamHI were hybridized with a digoxigenin-labeled probe amplified with a primer pair of 5′-AGCATTGGCTGTGACGACTTCC-3′ and 5′-CTAAGTAGTTGTTGCTCTTGTGC-3′ using PCR digoxigenin probe synthesis kit (Roche Applied Science). Hybridized probe was detected using digoxigenin High Prime DNA labeling and detection starter kit II (Roche Applied Science). Intensities of signals from endogenous Adamtsl6 and the transgene were quantified using ImageJ software.
Immunoblotting
MG63 cells seeded on 35-mm dishes were transfected with pSecTag-based expression plasmids using Lipofectamine 2000 (Invitrogen) and incubated for 2 h, and then the media were replaced with Opti-MEM (Invitrogen). In some experiments, tunicamycin (Calbiochem) was added to the medium to inhibit N-glycosylation. After cultivation for the indicated periods of time, conditioned medium was pooled, and cells adhering to culture dishes were harvested with PBS containing 10 mm EDTA for 30 min at 37 °C. A complete detachment of cells was visually confirmed under a microscope. The ECM remaining on the dishes was extracted with 2× SDS-PAGE sample treatment buffer. Tissue samples were obtained from E15.5 littermates after crossing wild-type female and transgenic male mice. Tissues were homogenized in 10 volumes of 1× SDS-PAGE sample treatment buffer. Aliquots of cell lysates, ECM fractions, conditioned media, and tissue extracts were boiled in the sample treatment buffer with or without 100 mm dithiothreitol and subjected to SDS-PAGE using 5–20% gradient gels. Separated proteins were electrotransferred to HybondECL membranes (GE Healthcare) and probed with anti-ADAMTSL-6 antibody (0.2–0.5 μg/ml). Bound antibodies were detected using horseradish peroxidase-conjugated secondary antibodies and an ECL Plus detection system (GE Healthcare).
Immunohistochemical Analysis
Whole mouse embryos at gestational day 16.5 (E16.5) were embedded in OCT compound (Sakura, Japan) and frozen in 2-methylbutane chilled by liquid nitrogen. For immunolocalization of ADAMTSL-6, 10-μm-thick cryosections were prepared, fixed in 4% formaldehyde in PBS, and blocked with 1.5% goat serum (DAKO) in PBS, followed by incubation with anti-ADAMTSL-6 antibody (0.2 μg/ml) for 1 h. After washing three times with PBS containing 0.05% Tween 20, the specimens were treated with EnVision+ system horseradish peroxidase (DAKO), followed by color development using 3,3′-diaminobenzidine. After counterstaining with hematoxylin, the sections were examined under a Nikon Eclipse E800 microscope. The specificities of the antibody against ADAMTSL-6 were verified by absorption of the immunoreactivities with recombinant full-length ADAMTSL- 6β protein. For double-immunofluorescence staining of ADAMTSL-6 and fibrillin-1, sections were fixed in cold acetone and incubated with anti-fibrillin-1 antibody (pAb9543, 1:200 dilution) for 1 h. After washing, the specimens were treated with Alexa Fluor 546-conjugated goat anti-rabbit IgG (1:500 dilution) for 1 h, washed with PBS, and fixed with 4% formaldehyde in PBS. The specimens were then incubated with anti-ADAMTSL-6 antibody labeled by a Zenon Alexa Fluor 488 rabbit IgG labeling kit (Molecular Probes). Nuclei were visualized with Hoechst 33342 (Molecular Probes). All staining procedures were performed at room temperature. The stained images were recorded using a Zeiss LSM5 confocal microscope.
Immunocytochemistry
MG63 cells were seeded at a density of 2.5 × 104 cells per well on 8-well Lab-Tek chamber slides (Nunc, Roskilde, Denmark) and cultured overnight at 37 °C. The cells were transfected with ADAMTSL-6α or -6β and incubated for 2 h, and then the media were replaced with Opti-MEM with or without soluble heparin. Alternatively, MG63 cells were cultured in Opti-MEM supplemented with the indicated concentrations of purified ADAMTSL-6β. After incubation for the indicated periods of time, cells were fixed with 4% formaldehyde in PBS and then incubated with primary antibodies, followed by incubation with Alexa Fluor 488-conjugated anti-rabbit IgG and Alexa Fluor 546-conjugated anti-mouse IgG antibodies. Nuclei were visualized with Hoechst 33342. Fluorescent images were recorded using a Zeiss LSM 5 PASCAL confocal microscope.
Electron Microscopy
Immunoelectron microscopy was performed as described previously (53). In brief, fresh tissue blocks from neonatal mice were incubated with dilutions of anti-ADAMTSL-6 antibody, followed by a gold-conjugated second antibody, and then embedded and prepared for electron microscopy.
Surface Plasmon Resonance Binding Assay
Recombinant fibrillin-1 fragments rF6 and rF90 (rF90 is identical to rF11, but it has a C-terminal His6 tag) were produced and purified as described previously (7). Binding analysis was performed using BIAcore X (BIAcore AB, Uppsala, Sweden). These fragments were covalently coupled to CM5 sensor chips (research grade; BIAcore AB) using the amine coupling kit following the manufacturer's instructions. Background binding to plain control flow cells was subtracted from total responses to yield binding responses due to analyte interaction with the surface-coupled ligand. Binding assays were performed at 25 °C in 10 mm Hepes buffer, pH 7.4, containing 0.15 m NaCl, 3 mm EDTA, and 0.005% (v/v) Surfactant P20 (HBS-EP buffer; BIAcore AB). ADAMTSL-6β or monoclonal antibody mAb69 was diluted in HBS-EP buffer and then injected at several concentrations and different flow rates over immobilized fibrillin-1 fragments. The surface was regenerated with a pulse of 10 mm glycine, pH 1.7. Kinetic constants were calculated by nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves according to the manufacturer's instructions (BIAevaluation 3.0). Apparent equilibrium dissociation constants were calculated as the ratio of Kd/Ka.
RESULTS
Identification of a New Member of the ADAMTSL Family of Proteins
We performed in silico screening to identify novel ECM proteins from over 60,000 varieties of mouse full-length cDNAs (54). Multiple criteria were applied to screen for secreted proteins, i.e. size >300 amino acid residues, the presence of characteristic domain(s) frequently found in ECM proteins, and the absence of domains exclusively found in non-ECM proteins. Among the clones that were selected, two cDNAs (AK138976 and AK045414), encoding 1018 and 658 amino acids, shared almost the identical sequence over 630 residues in their C-terminal regions, including an ADAMTS spacer domain, six TSRs, and a PLAC domain in tandem. Because this domain structure is similar to that of the C-terminal region of ADAMTSL proteins, we designated them ADAMTSL-6α and -6β (Fig. 1A). The N-terminal structures of ADAMTSL-6α and -6β are distinct from each other and also from those of other ADAMTSL proteins. ADAMTSL-6α contains a signal sequence and a Cys-rich domain but does not contain a typical first TSR domain that is conserved in all ADAMTSL and ADAMTS proteins, with the exception of ADAMTSL-4/TSRC1 (Fig. 1A). Nevertheless, sequence alignment of ADAMTSL-6α with other ADAMTS/ADAMTSL proteins revealed that a consensus tryptophan motif (WXXWXXW) found in the first TSR domain of ADAMTS/ADAMTSL proteins is present in the N-terminal region of ADAMTSL-6α along with the conserved three cysteine residues that follow the tryptophan motif (Fig. 1B). Furthermore, the three cysteine residues preceding the Cys-rich domain are also present in ADAMTSL-6α, indicating that ADAMTSL-6α contains a TSR-like domain, in which the six cysteine residues conserved in the first TSR domain of ADAMTS/ADAMTSL proteins were split into the N- and C-terminal halves with an insertion of ∼200 amino acid residues. A similar TSR-like domain with a long insertion has also been found in ADAMTSL-4/TSRC1. Although the overall sequence homology between ADAMTSL-4 and -6α at the inserted sequences is rather weak, a stretch of 26 amino acid residues that are enriched in arginines, prolines, and glycines is highly conserved between these two ADAMTSL proteins (Fig. 1B).
Unlike ADAMTSL-6α, the N-terminal region of ADAMTSL- 6β contains neither the first TSR/TSR-like domain nor the Cys-rich domain, and hence, the signal sequence is directly followed by an ADAMTS spacer domain (Fig. 1A). Because all ADAMTS/ADAMTSL proteins known to date contain both TSR/TSR-like and Cys-rich domains in their N-terminal region, ADAMTSL-6β is the first example of an ADAMTS/ADAMTSL protein that lacks both of these conserved domains.
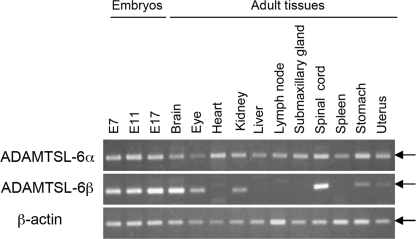
Mapping of ADAMTSL-6α and -6β cDNAs to mouse genomic DNA sequences revealed that both proteins are encoded by an identical gene located on chromosome 9. The mouse Adamtsl6 gene spans more than 500-kb pairs of DNA with the exon/intron structure shown in Fig. 1C. ADAMTSL-6α is encoded by 18 exons, of which the 11 3′ exons are shared with ADAMTSL-6β and the seven 5′ exons are unique to the α form. The ADAMTSL-6β-specific region is encoded by a single exon mapped between the 7th and 8th exons of ADAMTSL-6α, suggesting that transcription of ADAMTSL-6β is driven by a promoter distinct from that for ADAMTSL-6α. To explore the expression patterns of the two variant forms of ADAMTSL-6 in fetal and adult tissues, RT-PCR products of ADAMTSL-6α and -6β transcripts were analyzed. Although both transcripts were detected in mouse embryos from E7 through E17, they differed significantly in their expression levels in different adult tissues (Fig. 2). ADAMTSL-6α transcripts were ubiquitously expressed in all tissues examined, whereas ADAMTSL-6β transcripts were detected in brain, spinal cord, eye, kidney, stomach, and uterus but not in other tissues. Distinct tissue-specific expression levels for these ADAMTSL-6 variants further support the possibility that transcription of these variants is driven by distinct promoters.
FIGURE 2.
Expression levels of ADAMTSL-6α and -6β transcripts in mouse embryos and adult tissues. Transcripts for ADAMTSL-6α and -6β were reverse-transcribed from total RNA extracted from mouse embryos at different gestation periods and from adult tissues and then subjected to PCR amplification (33 cycles) using specific primer sets described under “Experimental Procedures.” Transcripts for β-actin were also amplified by RT-PCR (25 cycles) as a control. Arrows indicate the predicted positions of the PCR products. An apparent discrepancy in the size of PCR products for ADAMTSL-6β was due to distortion of the gel.
ADAMTSL-6 Is an ECM-associated Glycoprotein
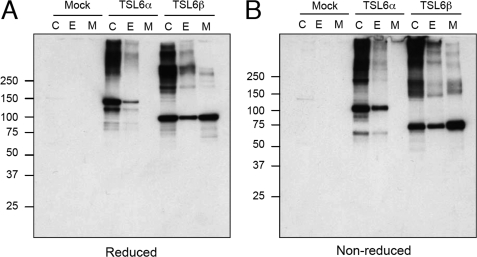
Because both ADAMTSL-6 proteins were predicted to possess typical signal sequences at their N termini, we examined whether ADAMTSL-6 proteins were secreted and deposited in the ECM by transfecting MG63 human osteosarcoma cells with individual cDNAs. Transfected cells were detached with 10 mm EDTA, and the ECMs remaining on the dishes were dissolved in SDS-PAGE sample treatment buffer. The resulting ECM extracts were subjected to Western blotting analyses along with cell lysates and conditioned medium. Under reducing conditions, ADAMTSL-6α and -6β gave major bands of 145 and 95 kDa, respectively (Fig. 3A). Under nonreducing conditions, these major bands were detected at positions slightly lower than those obtained under reducing conditions (Fig. 3B), reflecting the presence of multiple domains with intramolecular disulfide bonds. Multiple higher molecular weight bands were also detected for both variant forms under reducing and nonreducing conditions, suggesting that both proteins tend to form multimers. Under nonreducing conditions, significant amounts of both proteins stayed at the top of gels, and some of these materials moved farther into the gels after reduction, indicating that a part of the multimers are cross-linked by intermolecular disulfide bonds. Nevertheless, some of the high molecular weight materials remained in the reduced gels, suggesting that some multimers are either partially resistant to dissociation under the reducing conditions employed or are covalently cross-linked via non-disulfide bond(s). It should be noted that a fraction of ADAMTSL-6α was recovered in the ECM extracts, but no ADAMTSL-6α was apparent in the conditioned medium, in striking contrast to ADAMTSL-6β, a major fraction of which was recovered in the conditioned medium. The secreted ADAMTSL-6β gave predominantly the 95-kDa monomer band under reducing conditions with smaller amounts of high molecular weight multimers. These results indicate that, although both ADAMTSL-6 proteins tend to form high molecular multimers, ADAMTSL-6β is less prone to do so than ADAMTSL-6α and stays soluble primarily as monomers in the conditioned medium. Secretion of ADAMTSL-6 proteins was dependent on N-glycosylation, because secretion into the conditioned medium was completely blocked when transfected cells were grown in the presence of tunicamycin, an inhibitor of N-glycosylation (supplemental Fig. S1).
FIGURE 3.
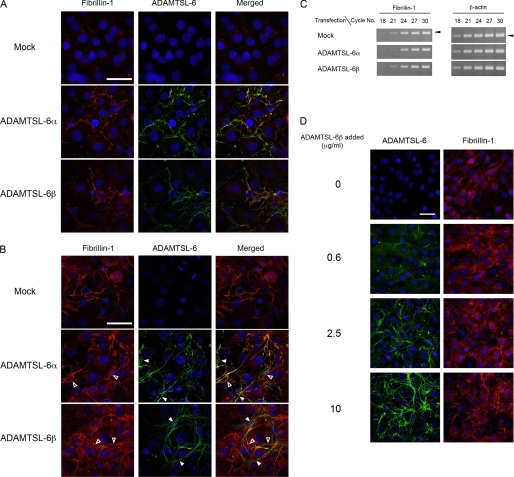
Expression and secretion of recombinant ADAMTSL-6 proteins. ADAMTSL-6α (TSL6α) and -6β (TSL6β) proteins were transiently expressed in MG63 cells for 3 days and then detected by immunoblotting with anti-ADAMTSL-6 antibody under reducing (A) and nonreducing (B) conditions after fractionation into cell lysates (C), extracellular deposits (E), and spent medium (M). Cells transfected with empty vector (Mock) were also analyzed as controls.
ADAMTSL-6 Proteins Are Extracellular Matrix Proteins Associated with Microfibrils
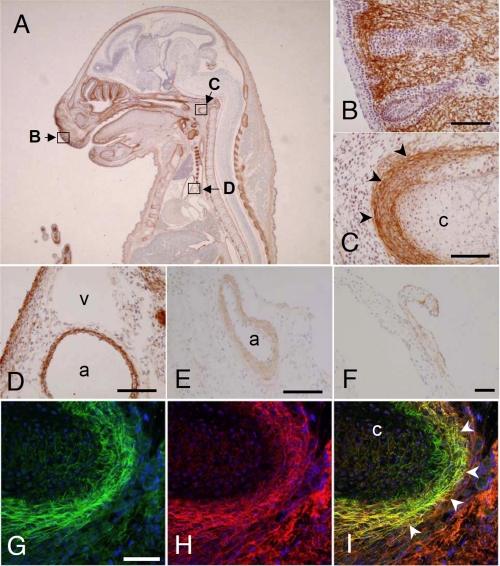
To confirm the secretion and extracellular deposition of ADAMTSL-6 proteins in vivo, we examined their tissue distributions by immunohistochemistry using cryosections of mouse whole embryos and adult tissues. In E16.5 mouse embryos, ADAMTSL-6 proteins were detected in fibrillar structures in various elastic tissues, including developing dermis, perichondria surrounding cartilages, and the vessel walls of aortae (Fig. 4, A–D). ADAMTSL-6 proteins were also localized, although to a lesser extent, in the vessel walls of arteries in adult kidney and the basement membrane zones of mitral valve in adult heart (Fig. 4, E and F, respectively). Because these immunohistochemical data indicate that ADAMTSL-6 proteins are mainly localized in fibrillar ECMs in elastic tissues, we next examined whether the ADAMTSL-6-positive fibrils contained fibrillin-1, a major component of microfibrils and elastic fibers. Double immunofluorescence staining of E16.5 mouse embryos showed that ADAMTSL-6 stained a subset of tissues containing fibrillin-1. In the fibrillar matrices of the perichondrium, ADAMTSL-6 was colocalized with fibrillin-1 (Fig. 4, G–I), as well as in other tissues where ADAMTSL-6 was expressed (data not shown).
FIGURE 4.
Immunolocalization of ADAMTSL-6 in embryos and adult tissues. Cryosections of E16.5 mouse embryos (A–D) and those of kidney and heart from 10-week-old female mice (E and F) were incubated with anti-ADAMTSL-6 antibody, followed by color development as described under “Experimental Procedures.” The following areas in A were magnified in B–D: skin with vibrissa follicles (B); vertebral primordium (c, cartilage; arrowheads, perichondrium) in C; aorta (a) and vein (v) in D). E and F, arterial wall (a) in the kidney and the basement membrane zones of mitral valve in the heart were positively stained, respectively. G–I, double immunofluorescence staining of ADAMTSL-6 and fibrillin-1 in the fetal cartilage. Cryosections of E16.5 mouse embryos were double immunostained with anti-ADAMTSL-6 (G; green) and anti-fibrillin-1 (H; red) antibodies. Merged image is shown in I. The staining patterns of ADAMTSL-6 overlapped with that of fibrillin-1 in the perichondrium (arrowheads). Other tissues (e.g. skeletal muscle shown in H and I) demonstrated staining for fibrillin-1, without staining for ADAMTSL-6. Bars represent 100 μm.
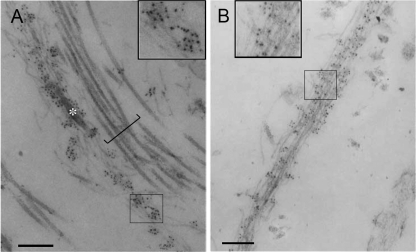
To further analyze the localization of ADAMTSL-6, we employed immunoelectron microscopy. Neonatal elastic cartilage and perichondrium were labeled with anti-ADAMTSL-6 antibody, followed by incubation with gold-conjugated secondary antibody (Fig. 5). Gold labeling was detected along microfibrils in both tissues irrespective of the presence or absence of amorphous elastin. No labeling was detected on collagen fibers or amorphous elastin. Similar labeling of microfibrils was observed in the skin and the tendon of newborn mice (data not shown). These results provide further evidence that the ADAMTSL-6 proteins are microfibril-associated proteins.
FIGURE 5.
Electron microscopic immunolocalization of ADAMTSL-6 in neonatal mouse cartilage and perichondrium. Immunogold particles were localized on microfibrils associated with amorphous elastin cores (asterisk) in the ear cartilage (A) and on microfibril bundles in perichondrium (B). There was no immunogold labeling of collagen fibers (bracket). Insets in A and B are magnified views of boxed areas. Bars represent 500 nm.
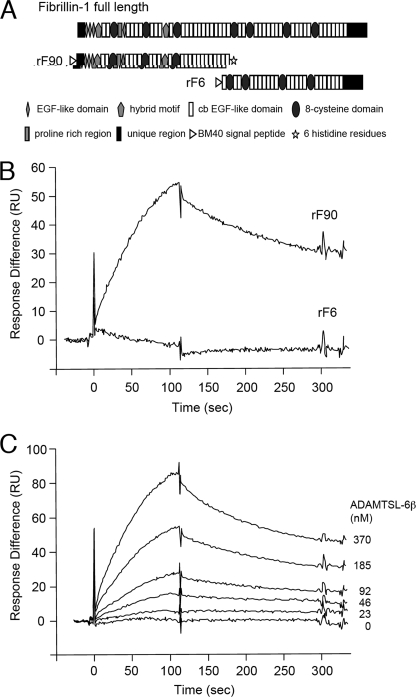
ADAMTSL-6β Binds to the N-terminal Half of Fibrillin-1
Because immunolocalization analyses indicated that ADAMTSL-6 proteins are associated with fibrillin-1-containing microfibrils, we examined whether ADAMTSL-6 could directly bind to fibrillin-1. Recombinant ADAMTSL-6β was expressed with a His6 tag in 293F cells and purified from the conditioned medium using nickel-agarose columns. Binding of the recombinant ADAMTSL-6 to fibrillin-1 was assessed by surface plasmon resonance analyses using fibrillin-1 fragments, rF90 and rF6, which represent the N- and C-terminal halves of fibrillin-1, respectively (Fig. 6A). Recombinant ADAMTSL-6β was found to bind to rF90, but not to rF6, demonstrating that ADAMTSL-6β is capable of binding to the N-terminal half of fibrillin-1 (Fig. 6B). Proper immobilization of rF6 on sensor chips was verified by the significant binding of a monoclonal antibody against the C-terminal region of fibrillin-1. These data indicate the inability of ADAMTSL-6β to bind to the C-terminal half of fibrillin-1. The binding of ADAMTSL-6β to the N-terminal half of fibrillin-1 was dose-dependent with a dissociation constant of 78.8 nm (Fig. 6C). The direct binding of ADAMTSL-6β to the N-terminal half of fibrillin-1 was also confirmed in solid phase enzyme-linked immunosorbent assay binding assays (data not shown).
FIGURE 6.
Binding of ADAMTSL-6β to the N-terminal fragment of fibrillin-1. A, schematic domain structures of fibrillin-1 and its recombinant halves. B, sensorgrams of binding of ADAMTSL-6β to fibrillin-1 fragments. Purified ADAMTSL-6β (200 nm) was infused over sensor chips on which recombinant fibrillin-1 fragments representing either the N-terminal half (rF90) or C-terminal half (rF6) were immobilized. Binding was recorded as resonance units (RU). C, increasing concentrations of purified ADAMTSL-6β were infused over a sensor chip on which rF90 was immobilized. The dissociation constant was calculated to be 78.8 × 10−9 m.
ADAMTSL-6 Promotes Fibrillin-1 Matrix Assembly in Vitro
Direct binding of ADAMTSL-6β to fibrillin-1, together with colocalization of ADAMTSL-6 proteins with fibrillin-1 in vivo, raised the possibility that ADAMTSL-6 proteins are involved in the assembly of fibrillin-1 into microfibrils. To address this possibility, we examined the effects of exogenous expression of ADAMTSL-6 proteins on fibrillin-1 matrix assembly in MG63 cells, which have been shown to produce fibrillin-1-containing fibrillar matrices (55). When we transfected MG63 cells with expression vectors encoding ADAMTSL-6 proteins, both ADAMTSL-6α and -6β proteins were detected in fibrillar matrices where fibrillin-1 fibrils were colocalized (Fig. 7A). No ADAMTSL-6 fibrils were detectable in mock-transfected cells, indicating that the endogenous expression of ADAMTSL-6 proteins was negligible in MG63 cells. It was noted that fibrillin-1 fibrils were barely detectable in mock-transfected cells but clearly detected in ADAMTSL-6-transfected cells 3 days after transfection, suggesting that the forced expression of ADAMTSL-6 proteins promotes fibrillin-1 fibril formation. Such effects of ADAMTSL-6 proteins to promote fibrillin-1 fibril formation were not clearly discernible 5 days after transfection, when endogenous fibrillin-1 fibril formation became evident in mock-transfected cells (Fig. 7B). No differences were detected in the expression levels of fibrillin-1 transcripts between mock-transfected and ADAMTSL-6-transfected cells, irrespective of variant type (Fig. 7C), indicating that the promotion of fibrillin-1 fibril formation by forced expression of ADAMTSL-6 proteins was not due to the up-regulation of fibrillin-1 expression, but likely due to the promotion of fibrillin-1 fibrillogenesis per se.
FIGURE 7.
Promotion of fibrillin-1 microfibril assembly by ADAMTSL-6. MG63 cells were transfected with cDNAs encoding ADAMTSL-6α or -6β or with empty vector (Mock) and then labeled with antibodies against fibrillin-1 and ADAMTSL-6 3 (A) or 5 (B) days after transfection. Merged images are shown in the right column. Nuclei were visualized with Hoechst 33342. Closed and open arrowheads in B point to the fibrils in which ADAMTSL-6 and fibrillin-1 were predominantly stained, respectively. Note that clear fibrillin-1 depositions, which were not detected until day 5 in mock-transfected cells, were detected on day 3 after transfection in ADAMTSL-6-transfected cells. C, expression levels of fibrillin-1 transcripts in cells transfected with cDNAs encoding ADAMTSL-6α or -6β or with empty vector (Mock) were determined by RT-PCR. Total RNA was collected from cells on day 3 after transfection and subjected to RT-PCR amplification of the fibrillin-1 transcripts for indicated reaction cycles. Transcripts for β-actin were also subjected to RT-PCR amplification as controls. D, MG63 cells were incubated with increasing concentrations of ADAMTSL-6β for 3 days to examine the effects of exogenously added ADAMTSL-6β on fibrillin-1 matrix assembly. Cells were immunolabeled with antibodies against ADAMTSL-6 (left, green) and fibrillin-1 (right, red). Scale bar, 50 μm.
To further address the role of ADAMTSL-6 proteins in fibrillin-1 fibril formation, MG63 cells were incubated with increasing concentrations of purified ADAMTSL-6β for 3 days. The exogenously added ADAMTSL-6β was found incorporated into fibrillar matrices in a dose-dependent manner (Fig. 7D). Concomitant with the assembly of ADAMTSL-6β into fibrils, fibrillin-1 fibril formation became more evident as more exogenous ADAMTSL-6β was added to the culture medium. These results support the possibility that ADAMTSL-6 proteins promote fibrillin-1 fibril formation.
ADAMTSL-6 Promotes Fibrillin-1 Matrix Assembly in Vivo
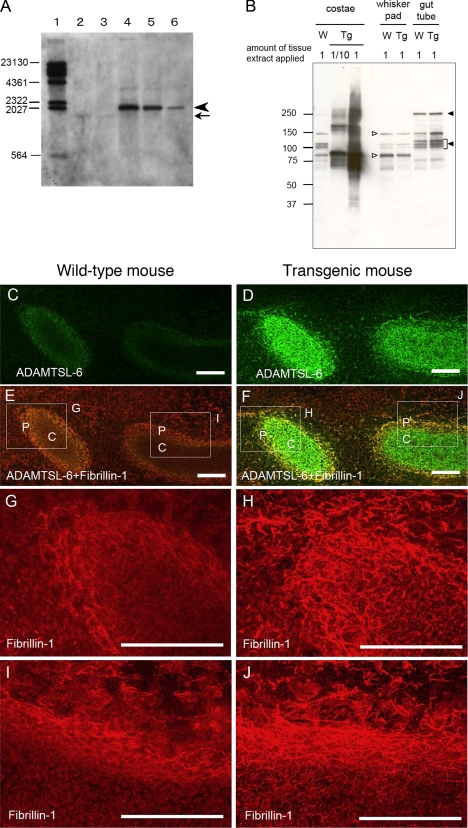
To further explore the role of ADAMTSL-6 in fibrillin-1 microfibril assembly in vivo, we generated transgenic mice overexpressing FLAG-tagged ADAMTSL-6β under the control of the type II collagen promoter/enhancer, which restricts the transgene expression in cartilaginous tissues. Southern blot analyses demonstrated that more than 20 copies of the transgene were integrated into the genome of the mice (Fig. 8A). The type II collagen promoter/enhancer-driven overexpression of the recombinant ADAMTSL-6β was confirmed by Western blotting of tissue extracts prepared from cartilaginous and noncartilaginous tissues of control and transgenic mice. A significant amount of the transgene product was detected in the costae from transgenic mice, when compared with ADAMTSL-6 proteins endogenously expressed in the costae from wild-type mice and the whisker pad and the gut tube from transgenic mice (Fig. 8B). The recombinant ADAMTSL-6β expressed in the costae of transgenic mice gave multiple bands migrating at the positions of monomers, dimers, trimers, and their processed forms under reducing conditions, consistent with the results obtained with recombinant ADAMTSL-6β proteins expressed in MG63 cells (see Fig. 3). No clear difference was detected in the expression level of ADAMTSL-6 proteins between control and transgenic mice in noncartilaginous tissues (i.e. the whisker pad and the gut tube), confirming the specificity of the type II collagen promoter/enhancer. Interestingly, immunoblot band patterns significantly differed between the whisker pad and the gut tube, the former characterized by ∼85- and ∼150-kDa bands and the latter characterized with triplet bands at the 100–120-kDa region and an ∼240-kDa band. Given that the former banding pattern is reminiscent to that of the recombinant ADAMTSL-6β overexpressed in the costae of transgenic mice, it seems likely that the whisker pads express mainly ADAMTSL-6β, whereas ADAMTSL-6α is predominantly expressed in the gut tube.
FIGURE 8.
Promotion of fibrillin-1 fibril formation by ADAMTSL-6 in vivo. A, Southern hybridization of DNA from ADAMTSL-6β transgenic mice. Digestion of genomic DNA with EcoRI and BamHI produced 1.6-kb fragments containing the Adamtsl6 exon 8 from chromosome 9 (indicated with an arrow) and 2.3-kb fragments containing full-length ADAMTSL-6β transgenes (indicated with an arrowhead). Lane 1, digoxigenin-labeled molecular weight markers; lane 2, DNA from wild-type mouse (12 μg); lane 3, DNA from wild-type mouse (3 μg); lane 4, DNA from transgenic mouse (12 μg); lane 5, DNA from transgenic mouse (3 μg); lane 6, DNA from transgenic mouse (0.75 μg). The 2.3-kb band was detected only with the DNA from transgenic mice, whereas the 1.6-kb band, although very faint, was detected with the DNA from wild-type and transgenic mice. Quantification of band intensities using ImageJ software indicated that more than 20 copies of the transgene were integrated into the genomic DNA of transgenic mice. The sizes of DNA molecular weight markers are shown in the left margin. B, tissue extracts from E15.5 wild-type (W) and ADAMTSL-6β transgenic mice (Tg) were subjected to SDS-PAGE under reducing conditions, followed by blotting with anti-ADAMTSL-6 antibody. 10-Fold diluted extracts were also subjected to Western blotting for the costae from transgenic mice, where the transgene was highly expressed. Positions of molecular mass markers (kDa) are shown in the left margin. Open and closed triangles point to the bands characteristic of the whisker pads and the gut tube, respectively. C–J, immunofluorescence detection of ADAMTSL-6 proteins and fibrillin-1. Sagittal cryosections were prepared from E12.5 wild-type (C, E, G, and I) and transgenic littermates (D, F, H, and J) and subjected to double immunostaining with antibodies against ADAMTSL-6 (green) and fibrillin-1 (red). The regions of rib cartilage (C) and surrounding perichondrium (P) are labeled in E and F. G and I, magnified views in the boxed areas in E. H and J, magnified views of the boxed areas in F. The ADAMTSL-6β transgene was highly expressed in the rib cartilage and surrounding perichondrium of transgenic embryos (D and F), where fibrillin-1 depositions were also enhanced (H and J). Scale bar, 100 μm.
Overexpression of the transgene was further corroborated by immunohistochemistry using anti- ADAMTSL-6 antibody. ADAMTSL-6 proteins were highly expressed in the cartilage and the surrounding perichondrium of the transgenic embryos, where the type II collagen promoter/enhancer was expected to be highly active, although endogenous ADAMTSL-6 was only faintly detected in these cartilaginous tissues of wild-type embryos at this stage of development (E12.5) (Fig. 8, C and D). In line with the overexpression of ADAMTSL-6β in these cartilaginous tissues, fibrillin-1 deposition in the ECM was significantly enhanced in both the cartilage and perichondrium of transgenic embryos (Fig. 8, E–J). Fibrillin-1 was barely detected in the cartilage matrix of wild-type embryos but was clearly detectable as pericellular deposits in the cartilage of transgenic embryos, overlapping with the ADAMTSL-6 deposits. Similarly, fibrillin-1 was detected as short fibrillar deposits in the perichondrium of wild-type embryos, but it had coalesced into thick fibrils in the transgenic embryos. No clear difference in the staining intensity of fibrillin-1 was observed between wild-type and transgenic embryos in tissues where transgene expression was not induced. These results further support a role for ADAMTSL-6 in promoting fibrillin-1 matrix assembly in vivo.
DISCUSSION
Our data show that ADAMTSL-6 is a new member of the ADAMTSL protein family, which is secreted and deposited on fibrillin-1-containing microfibrils. There are two forms of Adamtsl6 gene products, ADAMTSL-6α and -6β; these isoforms share the same domain structure comprising an ADAMTS spacer domain, six TSR repeats, and the C-terminal PLAC domain but differ in their N-terminal regions where ADAMTSL-6α, but not ADAMTSL-6β, contains a TSR repeat split by an insertion of ∼200 amino acid residues and the following Cys-rich domain. Immunohistochemistry demonstrated that ADAMTSL-6 proteins are localized to fibrillar structures in various elastic tissues, where they codistribute with fibrillin-1. Association of ADAMTSL-6 proteins with microfibrils was further confirmed by immunoelectron microscopy. When expressed in MG63 cells, ADAMTSL-6 proteins assemble into fibrillar matrices where they colocalize with fibrillin-1. We also provide evidence that ADAMTSL-6 proteins bind directly to the N-terminal half of fibrillin-1 with moderate affinity (a dissociation constant of ∼80 nm). These results point to the conclusion that ADAMTSL-6 is an ECM protein that associates with microfibrils through direct binding to fibrillin-1.
ADAMTSL-6 Promotes Fibrillin-1 Matrix Assembly
Several lines of evidence indicate that ADAMTSL-6 proteins promote the assembly of fibrillin-1 microfibrils. Transfection of MG63 cells with cDNAs encoding ADAMTSL-6 proteins resulted in an accelerated assembly of fibrillin-1 into fibrillar matrices, to which the expressed ADAMTSL-6 proteins were also aligned. The expression levels of fibrillin-1 transcripts remained unaffected in the transfected cells, indicating that the accelerated matrix assembly of fibrillin-1 was not due to the up-regulation of fibrillin-1 gene expression but to the enhancement of the fibrillin-1 matrix assembly per se. In support of this conclusion, matrix assembly of fibrillin-1 was promoted by the exogenous addition of ADAMTSL-6β in a dose-dependent manner. Further support for the role of ADAMTSL-6 in fibrillin-1 matrix assembly was obtained from transgenic mice in which recombinant ADAMTSL-6β was overexpressed under the control of the type II collagen promoter/enhancer. Fibrillin-1 deposition was significantly enhanced in the cartilage and perichondrium of the ADAMTSL-6β transgenic mice, where the expression of the ADAMTSL-6β transgene was selectively induced.
Although we provide evidence for the role of ADAMTSL-6 in fibrillin-1 matrix assembly, the mechanisms by which ADAMTSL-6 accelerates the assembly process remain to be explored. Our data show that the promotion of fibrillin-1 matrix assembly by forced expression of ADAMTSL-6 proteins was evident in cells 3 days after cDNA transfection but became barely discernible after longer cultivation, e.g. in cells 5 days after transfection. These results indicate that ADAMTSL-6 proteins exert their effects in the early but not later stage of fibrillin-1 matrix assembly. It should be also noted that, in cells 5 days after transfection, the fibrils positively stained with anti-ADAMTSL-6 antibody were often undetected with anti-fibrillin-1 antibody, suggesting that ADAMTSL-6 is capable of assembling into fibrillar matrices without accompanying fibrillin-1 microfibril assembly. The possible association of ADAMTSL-6 with fibrillin-2 is currently under investigation.
Although it remains poorly understood how fibrillin-1 monomers begin to assemble into microfibrils, the interactions of fibrillin-1 with other ECM or cell surface components, e.g. fibronectin (16, 17), have been implicated in the initial stage of fibrillin-1 microfibril assembly. Thus, depletion of fibronectin fibrils, by either small interfering RNA knockdown or the peptide that blocks fibronectin matrix assembly, inhibited fibrillin-1 microfibril formation, although exogenously added fibronectin restored fibrillin-1 fibrils to the cells in which endogenous expression of fibronectin was knocked down (16, 17). Fibrillin-1 is capable of binding to fibronectin through its C-terminal region, where it forms multimers (12) and thereby potentiates its fibronectin binding activity (16). Given that self-interaction of fibrillin-1 in a head-to-tail fashion is the initial step in fibrillin-1 microfibril assembly, it is conceivable that local accumulation of multimerized fibrillin-1 on fibronectin fibrils facilitates the subsequent head-to-tail linear association of fibrillin-1. Because ADAMTSL-6 binds to the N-terminal region of fibrillin-1 and it forms dimers and possibly oligomers (see Figs. 3 and 8), ADAMTSL-6 may promote the head-to-tail association of fibrillin-1 by rendering the N-terminal region of fibrillin-1 competent for this initial step in microfibril formation either by conferring multivalency or by conformational activation.
Not only N-to-C interactions but also N-to-N association has been suggested to occur in fibrillin-1 microfibril assembly (11, 13, 15). Pulse-chase analyses demonstrated that recombinant fragments containing the N-terminal proline-rich region are capable of forming disulfide-bonded dimers shortly after de novo synthesis (13, 15). An unpaired cysteine residue in the first hybrid motif, Cys-204, which is not included in the aforementioned fragments, has also been implicated in intermolecular disulfide bond formation of fibrillin-1 (56). Consistent with potential dimer formation, recombinant N-terminal fragments bind to themselves with affinities in the same order of magnitude as those for the binding to C-terminal fragments (11). Given that ADAMTSL-6 proteins form dimers and possibly oligomers, binding of ADAMTSL-6 to the N-terminal region of fibrillin-1 may facilitate the coalescence of fibrillin-1 dimers into multimers at their N-terminal region, thereby promoting the subsequent sequential N-to-C interactions.
Distinctive Function of ADAMTSL-6α-specific Region
We identified two variants of ADAMTSL-6 transcripts, ADAMTSL-6α and -6β. Although both forms are capable of assembling into fibrillar structures and promoting fibrillin-1 microfibril assembly upon transfection to MG63 cells, only ADAMTSL-6β, but not -6α, was detected in the conditioned medium. Because both variants were almost equally deposited into the pericellular fibrillar ECM upon transfection (see Fig. 7, A and B), the failure to detect ADAMTSL-6α in the conditioned medium was not due to the inability of ADAMTSL-6α to be secreted, but rather to its reduced solubility and/or enhanced matrix assembling activity, although the expression and/or secretion of ADAMTSL-6α could be less efficient than those of ADAMTSL-6β. These differences between ADAMTSL-6α and -6β are likely due to their distinctive N-terminal regions. It should be noted that the insertion in the first TSR-like domain in ADAMTSL-6α is enriched in arginine residues (∼13% of a total of 209 residues) and contains two RRXR sequences (see Fig. 1B), which are putative heparin-binding motifs (57). Thus, the insertion in the first TSR-like domain may bind to heparan sulfate and other glycosaminoglycan chains and may thereby contribute to the removal of ADAMTSL-6α from the conditioned medium. The Cys-rich domain may also be involved in the removal of ADAMTSL-6α, as recombinant fragments of ADAMTS-1 and ADAMTS-6 that harbor only the Cys-rich and ADAMTS spacer domains were recovered only in the ECM deposits but not in the conditioned medium (39, 58).
Although the precise structure of the first TSR of ADAMTSL-6α with its insertion remains to be defined, it is interesting to note that the inserted sequence is mapped between strands B and C, which in TSR domains of thrombospondin-1 are connected by a relatively long loop that is secured by a conserved disulfide bond (59). Given that the tryptophan motif (WXXWXXW) and six cysteine residues, including those that secure the loop between strands B and C, which together contribute to the core structure of TSRs (59), are conserved in the first TSR of ADAMTSL-6α, it seems likely that the inserted sequence loops out of the core structure and folds separately. A similar long insertion with >200 amino acid residues has been found in the first TSR of ADAMTSL-4/TSRC1 (38). Although overall homology in the inserted sequence between ADAMTSL-4 and -6 is only moderate, a stretch of 26 amino acid residues, which are enriched in basic residues, prolines, and glycines, are highly conserved between two ADAMTSL proteins, indicative of their roles in biological functions of these proteins, e.g. binding to glycosaminoglycan chains.
ADAMTSL6 as a Potential Candidate Gene Associated with Fibrillinopathies
Mutations in the fibrillin-1 gene cause fibrillinopathies, inherited disorders of connective tissues typically represented by the Marfan syndrome. Because ADAMTSL-6 proteins associate with fibrillin-1 microfibrils and promote microfibril assembly in vitro and in vivo, it is tempting to speculate that mutations in the ADAMTSL-6 gene could lead to aberrant microfibril assembly in elastic and nonelastic tissues and thereby cause conditions similar to fibrillinopathies. Consistent with this possibility, mutations in the ADAMTSL2 gene have been associated with an acromelic dysplasia called geleophysic dysplasia, a rare autosomal recessive disorder characterized by short stature, brachydactyly, thick skin, and cardiac valvular anomalies (42). These clinical features are similar to those of Weill-Marchesani syndrome, which is also an acromeric dysplasia associated with mutations in fibrillin-1. Recently, mutations in the ADAMTSL4 gene were found in patients with ectopia lentis (45), displacement of the lens due to abnormalities in the ciliary zonule, a cardinal feature of the Marfan syndrome. In addition, familial forms of isolated ectopia lentis are also caused by mutations in fibrillin-1, further corroborating the potential involvement of ADAMTSL proteins in fibrillinopathies.
Although it remains to be defined how mutations in ADAMTSL2 and ADAMTSL4 genes lead to the symptoms associated with fibrillinopathies, aberrant microfibril assembly and the resulting effects on TGF-β bioavailability may be an integral part of the pathogenetic processes. Le Goff et al. (42) observed a significant increase in active TGF-β in the culture medium of fibroblasts derived from individuals with geleophysic dysplasia, along with an increased phosphorylation of Smad2 and its nuclear localization, indicative of increased TGF-β signaling events. They also showed that ADAMTSL-2 associates with LTBP-1, which performs a major role in the storage of latent TGF-β in the ECM and is similar to fibrillins in its domain structure (60). Although it is unknown whether ADAMTSL-2 directly binds to fibrillin-1, our results that ADAMTSL-6 binds to fibrillin-1 and promotes its assembly into microfibrils raise the possibility that ADAMTSL-6 is also involved in the ECM-based network regulating TGF-β bioavailability. It remains an open question whether any defects in the ADAMTSL-6 gene dysregulate the bioavailability of TGF-β and hence lead to conditions associated with fibrillinopathies. Mouse models in which Adamtsl6 is mutated should provide important insight into the physiological functions of ADAMTSL-6 and other ADAMTSL family proteins and their relevance to pathogenesis of fibrillinopathies.
Supplementary Material
Acknowledgments
We are grateful to Dr. Yoshihide Hayashizaki for mouse ADAMTSL-6 cDNAs and Dr. Noriyuki Tsumaki for the Col2a1 promoter/enhancer DNA.
This work was supported, in whole or in part, by National Institutes of Health Grant P01AR049698 (to L. Y. S.). This work was also supported by Grants-in-aid for Scientific Research 17082005 and 20370046 (to K. S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and from the Shriners Hospitals for Children (to D. R. K. and to L. Y. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ECM
- extracellular matrix
- ADAMTSL
- ADAMTS-like
- TGF
- transforming growth factor
- TSR
- thrombospondin type I repeat
- RT
- reverse transcription
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Charbonneau N. L., Ono R. N., Corson G. M., Keene D. R., Sakai L. Y. (2004) Birth Defects Res. C Embryo Today 72, 37–50 [DOI] [PubMed] [Google Scholar]
- 2.Chaudhry S. S., Cain S. A., Morgan A., Dallas S. L., Shuttleworth C. A., Kielty C. M. (2007) J. Cell Biol. 176, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai L. Y., Keene D. R., Engvall E. (1986) J. Cell Biol. 103, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handford P. A., Downing A. K., Reinhardt D. P., Sakai L. Y. (2000) Matrix Biol. 19, 457–470 [DOI] [PubMed] [Google Scholar]
- 5.Sakai L. Y., Keene D. R., Glanville R. W., Bächinger H. P. (1991) J. Biol. Chem. 266, 14763–14770 [PubMed] [Google Scholar]
- 6.Kielty C. M., Sherratt M. J., Shuttleworth C. A. (2002) J. Cell Sci. 115, 2817–2828 [DOI] [PubMed] [Google Scholar]
- 7.Reinhardt D. P., Keene D. R., Corson G. M., Pöschl E., Bächinger H. P., Gambee J. E., Sakai L. Y. (1996) J. Mol. Biol. 258, 104–116 [DOI] [PubMed] [Google Scholar]
- 8.Kuo C. L., Isogai Z., Keene D. R., Hazeki N., Ono R. N., Sengle G., Bächinger H. P., Sakai L. Y. (2007) J. Biol. Chem. 282, 4007–4020 [DOI] [PubMed] [Google Scholar]
- 9.Baldock C., Koster A. J., Ziese U., Rock M. J., Sherratt M. J., Kadler K. E., Shuttleworth C. A., Kielty C. M. (2001) J. Cell Biol. 152, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S. S., Knott V., Jovanoviæ J., Harlos K., Grimes J. M., Choulier L., Mardon H. J., Stuart D. I., Handford P. A. (2004) Structure 12, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marson A., Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., Shuttleworth C. A., Baldock C., Kielty C. M. (2005) J. Biol. Chem. 280, 5013–5021 [DOI] [PubMed] [Google Scholar]
- 12.Hubmacher D., El-Hallous E. I., Nelea V., Kaartinen M. T., Lee E. R., Reinhardt D. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6548–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trask T. M., Ritty T. M., Broekelmann T., Tisdale C., Mecham R. P. (1999) Biochem. J. 340, 693–701 [PMC free article] [PubMed] [Google Scholar]
- 14.Lin G., Tiedemann K., Vollbrandt T., Peters H., Batge B., Brinckmann J., Reinhardt D. P. (2002) J. Biol. Chem. 277, 50795–50804 [DOI] [PubMed] [Google Scholar]
- 15.Ashworth J. L., Kelly V., Wilson R., Shuttleworth C. A., Kielty C. M. (1999) J. Cell Sci. 112, 3549–3558 [DOI] [PubMed] [Google Scholar]
- 16.Sabatier L., Chen D., Fagotto-Kaufmann C., Hubmacher D., McKee M. D., Annis D. S., Mosher D. F., Reinhardt D. P. (2009) Mol. Biol. Cell 20, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinsey R., Williamson M. R., Chaudhry S., Mellody K. T., McGovern A., Takahashi S., Shuttleworth C. A., Kielty C. M. (2008) J. Cell Sci. 121, 2696–2704 [DOI] [PubMed] [Google Scholar]
- 18.Cain S. A., Baldock C., Gallagher J., Morgan A., Bax D. V., Weiss A. S., Shuttleworth C. A., Kielty C. M. (2005) J. Biol. Chem. 280, 30526–30537 [DOI] [PubMed] [Google Scholar]
- 19.Cain S. A., Baldwin A. K., Mahalingam Y., Raynal B., Jowitt T. A., Shuttleworth C. A., Couchman J. R., Kielty C. M. (2008) J. Biol. Chem. 283, 27017–27027 [DOI] [PubMed] [Google Scholar]
- 20.Tiedemann K., Bätge B., Müller P. K., Reinhardt D. P. (2001) J. Biol. Chem. 276, 36035–36042 [DOI] [PubMed] [Google Scholar]
- 21.Ritty T. M., Broekelmann T. J., Werneck C. C., Mecham R. P. (2003) Biochem. J. 375, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isogai Z., Ono R. N., Ushiro S., Keene D. R., Chen Y., Mazzieri R., Charbonneau N. L., Reinhardt D. P., Rifkin D. B., Sakai L. Y. (2003) J. Biol. Chem. 278, 2750–2757 [DOI] [PubMed] [Google Scholar]
- 23.Ono R. N., Sengle G., Charbonneau N. L., Carlberg V., Bächinger H. P., Sasaki T., Lee-Arteaga S., Zilberberg L., Rifkin D. B., Ramirez F., Chu M. L., Sakai L. Y. (2009) J. Biol. Chem. 284, 16872–16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen S. A., Reinhardt D. P., Gibson M. A., Weiss A. S. (2001) J. Biol. Chem. 276, 39661–39666 [DOI] [PubMed] [Google Scholar]
- 25.Penner A. S., Rock M. J., Kielty C. M., Shipley J. M. (2002) J. Biol. Chem. 277, 35044–35049 [DOI] [PubMed] [Google Scholar]
- 26.El-Hallous E., Sasaki T., Hubmacher D., Getie M., Tiedemann K., Brinckmann J., Bätge B., Davis E. C., Reinhardt D. P. (2007) J. Biol. Chem. 282, 8935–8946 [DOI] [PubMed] [Google Scholar]
- 27.Charbonneau N. L., Dzamba B. J., Ono R. N., Keene D. R., Corson G. M., Reinhardt D. P., Sakai L. Y. (2003) J. Biol. Chem. 278, 2740–2749 [DOI] [PubMed] [Google Scholar]
- 28.Aoyama T., Francke U., Dietz H. C., Furthmayr H. (1994) J. Clin. Invest. 94, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godfrey M., Raghunath M., Cisler J., Bevins C. L., DePaepe A., Di Rocco M., Gregoritch J., Imaizumi K., Kaplan P., Kuroki Y., Silberbach M., Superti-Furga A., Van Thienen M., Vetter U., Steinmann B. (1995) Am. J. Pathol. 146, 1414–1421 [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson P. N., Arteaga-Solis E., Baldock C., Collod-Béroud G., Booms P., De Paepe A., Dietz H. C., Guo G., Handford P. A., Judge D. P., Kielty C. M., Loeys B., Milewicz D. M., Ney A., Ramirez F., Reinhardt D. P., Tiedemann K., Whiteman P., Godfrey M. (2006) J. Med. Genet. 43, 769–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Handford P. A. (2000) Biochim. Biophys. Acta 1498, 84–90 [DOI] [PubMed] [Google Scholar]
- 32.Faivre L., Mégarbané A., Alswaid A., Zylberberg L., Aldohayan N., Campos-Xavier B., Bacq D., Legeai-Mallet L., Bonaventure J., Munnich A., Cormier-Daire V. (2002) Hum. Genet. 110, 366–370 [DOI] [PubMed] [Google Scholar]
- 33.Faivre L., Gorlin R. J., Wirtz M. K., Godfrey M., Dagoneau N., Samples J. R., Le Merrer M., Collod-Beroud G., Boileau C., Munnich A., Cormier-Daire V. (2003) J. Med. Genet. 40, 34–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porter S., Clark I. M., Kevorkian L., Edwards D. R. (2005) Biochem. J. 386, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirohata S., Wang L. W., Miyagi M., Yan L., Seldin M. F., Keene D. R., Crabb J. W., Apte S. S. (2002) J. Biol. Chem. 277, 12182–12189 [DOI] [PubMed] [Google Scholar]
- 36.Koo B. H., Le Goff C., Jungers K. A., Vasanji A., O'Flaherty J., Weyman C. M., Apte S. S. (2007) Matrix Biol. 26, 431–441 [DOI] [PubMed] [Google Scholar]
- 37.Hall N. G., Klenotic P., Anand-Apte B., Apte S. S. (2003) Matrix Biol. 22, 501–510 [DOI] [PubMed] [Google Scholar]
- 38.Buchner D. A., Meisler M. H. (2003) Gene 307, 23–30 [DOI] [PubMed] [Google Scholar]
- 39.Kuno K., Matsushima K. (1998) J. Biol. Chem. 273, 13912–13917 [DOI] [PubMed] [Google Scholar]
- 40.Kashiwagi M., Enghild J. J., Gendron C., Hughes C., Caterson B., Itoh Y., Nagase H. (2004) J. Biol. Chem. 279, 10109–10119 [DOI] [PubMed] [Google Scholar]
- 41.Somerville R. P., Longpre J. M., Jungers K. A., Engle J. M., Ross M., Evanko S., Wight T. N., Leduc R., Apte S. S. (2003) J. Biol. Chem. 278, 9503–9513 [DOI] [PubMed] [Google Scholar]
- 42.Le Goff C., Morice-Picard F., Dagoneau N., Wang L. W., Perrot C., Crow Y. J., Bauer F., Flori E., Prost-Squarcioni C., Krakow D., Ge G., Greenspan D. S., Bonnet D., Le Merrer M., Munnich A., Apte S. S., Cormier-Daire V. (2008) Nat. Genet. 40, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashimoto G., Shimoda M., Okada Y. (2004) J. Biol. Chem. 279, 32483–32491 [DOI] [PubMed] [Google Scholar]
- 44.Tortorella M., Pratta M., Liu R. Q., Abbaszade I., Ross H., Burn T., Arner E. (2000) J. Biol. Chem. 275, 25791–25797 [DOI] [PubMed] [Google Scholar]
- 45.Ahram D., Sato T. S., Kohilan A., Tayeh M., Chen S., Leal S., Al-Salem M., El-Shanti H. (2009) Am. J. Hum. Genet. 84, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manabe R., Tsutsui K., Yamada T., Kimura M., Nakano I., Shimono C., Sanzen N., Furutani Y., Fukuda T., Oguri Y., Shimamoto K., Kiyozumi D., Sato Y., Sado Y., Senoo H., Yamashina S., Fukuda S., Kawai J., Sugiura N., Kimata K., Hayashizaki Y., Sekiguchi K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12849–12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakai K., Horton P. (1999) Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 48.Hirokawa T., Boon-Chieng S., Mitaku S. (1998) Bioinformatics 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 49.Pearson W. R., Wood T., Zhang Z., Miller W. (1997) Genomics 46, 24–36 [DOI] [PubMed] [Google Scholar]
- 50.Koff A., Giordano A., Desai D., Yamashita K., Harper J. W., Elledge S., Nishimoto T., Morgan D. O., Franza B. R., Roberts J. M. (1992) Science 257, 1689–1694 [DOI] [PubMed] [Google Scholar]
- 51.Pereira L., Andrikopoulos K., Tian J., Lee S. Y., Keene D. R., Ono R., Reinhardt D. P., Sakai L. Y., Biery N. J., Bunton T., Dietz H. C., Ramirez F. (1997) Nat. Genet. 17, 218–222 [DOI] [PubMed] [Google Scholar]
- 52.Tsumaki N., Tanaka K., Arikawa-Hirasawa E., Nakase T., Kimura T., Thomas J. T., Ochi T., Luyten F. P., Yamada Y. (1999) J. Cell Biol. 144, 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakai L. Y., Keene D. R. (1994) Methods Enzymol. 245, 29–52 [DOI] [PubMed] [Google Scholar]
- 54.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Yamanaka I., Kiyosawa H., Yagi K., Tomaru Y., Hasegawa Y., Nogami A., Schönbach C., Gojobori T., Baldarelli R., Hill D. P., Bult C., Hume D. A., Quackenbush J., Schriml L. M., Kanapin A., Matsuda H., Batalov S., Beisel K. W., Blake J. A., Bradt D., Brusic V., Chothia C., Corbani L. E., Cousins S., Dalla E., Dragani T. A., Fletcher C. F., Forrest A., Frazer K. S., Gaasterland T., Gariboldi M., Gissi C., Godzik A., Gough J., Grimmond S., Gustincich S., Hirokawa N., Jackson I. J., Jarvis E. D., Kanai A., Kawaji H., Kawasawa Y., Kedzierski R. M., King B. L., Konagaya A., Kurochkin I. V., Lee Y., Lenhard B., Lyons P. A., Maglott D. R., Maltais L., Marchionni L., McKenzie L., Miki H., Nagashima T., Numata K., Okido T., Pavan W. J., Pertea G., Pesole G., Petrovsky N., Pillai R., Pontius J. U., Qi D., Ramachandran S., Ravasi T., Reed J. C., Reed D. J., Reid J., Ring B. Z., Ringwald M., Sandelin A., Schneider C., Semple C. A., Setou M., Shimada K., Sultana R., Takenaka Y., Taylor M. S., Teasdale R. D., Tomita M., Verardo R., Wagner L., Wahlestedt C., Wang Y., Watanabe Y., Wells C., Wilming L. G., Wynshaw-Boris A., Yanagisawa M., Yang I., Yang L., Yuan Z., Zavolan M., Zhu Y., Zimmer A., Carninci P., Hayatsu N., Hirozane-Kishikawa T., Konno H., Nakamura M., Sakazume N., Sato K., Shiraki T., Waki K., Kawai J., Aizawa K., Arakawa T., Fukuda S., Hara A., Hashizume W., Imotani K., Ishii Y., Itoh M., Kagawa I., Miyazaki A., Sakai K., Sasaki D., Shibata K., Shinagawa A., Yasunishi A., Yoshino M., Waterston R., Lander E. S., Rogers J., Birney E., Hayashizaki Y. (2002) Nature 420, 563–57312466851 [Google Scholar]
- 55.Dzamba B. J., Keene D. R., Isogai Z., Charbonneau N. L., Karaman-Jurukovska N., Simon M., Sakai L. Y. (2001) J. Invest. Dermatol. 117, 1612–1620 [DOI] [PubMed] [Google Scholar]
- 56.Reinhardt D. P., Gambee J. E., Ono R. N., Bächinger H. P., Sakai L. Y. (2000) J. Biol. Chem. 275, 2205–2210 [DOI] [PubMed] [Google Scholar]
- 57.Hileman R. E., Fromm J. R., Weiler J. M., Linhardt R. J. (1998) BioEssays 20, 156–167 [DOI] [PubMed] [Google Scholar]
- 58.Gendron C., Kashiwagi M., Lim N. H., Enghild J. J., Thøgersen I. B., Hughes C., Caterson B., Nagase H. (2007) J. Biol. Chem. 282, 18294–18306 [DOI] [PubMed] [Google Scholar]
- 59.Tan K., Duquette M., Liu J. H., Dong Y., Zhang R., Joachimiak A., Lawler J., Wang J. H. (2002) J. Cell Biol. 159, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rifkin D. B. (2005) J. Biol. Chem. 280, 7409–7412 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.