Abstract
MicroRNAs (miRNAs or miRs) are small, non-coding RNAs that modulate mRNA stability and post-transcriptional translation. A growing body of evidence indicates that specific miRNAs can affect the cellular function of cardiomyocytes. In the present study, miRNAs that are highly expressed in the heart were overexpressed in neonatal rat ventricular myocytes, and cellular ATP levels were assessed. As a result, miR-15b, -16, -195, and -424, which have the same seed sequence, the most critical determinant of miRNA targeting, decreased cellular ATP levels. These results suggest that these miRNAs could specifically down-regulate the same target genes and consequently decrease cellular ATP levels. Through a bioinformatics approach, ADP-ribosylation factor-like 2 (Arl2) was identified as a potential target of miR-15b. It has already been shown that Arl2 localizes to adenine nucleotide transporter 1, the exchanger of ADP/ATP in mitochondria. Overexpression of miR-15b, -16, -195, and -424 suppressed the activity of a luciferase reporter construct fused with the 3′-untranslated region of Arl2. In addition, miR-15b overexpression decreased Arl2 mRNA and protein expression levels. The effects of Arl2 siRNA on cellular ATP levels were the same as those of miR-15b, and the expression of Arl2 could restore ATP levels reduced by miR-15b. A loss-of-function study of miR-15b resulted in increased Arl2 protein and cellular ATP levels. Electron microscopic analysis revealed that mitochondria became degenerated in cardiomyocytes that had been transduced with miR-15b and Arl2 siRNA. The present results suggest that miR-15b may decrease mitochondrial integrity by targeting Arl2 in the heart.
Keywords: Diseases/Muscular, Gene, Metabolism/Energy, RNA/MicroRNA, Subcellular Organelles/Mitochondria, Translation
Introduction
MicroRNAs (miRNAs or miRs)2 are small, non-coding RNAs that modulate mRNA stability and post-transcriptional translation. A growing body of evidence indicates that miRNAs are involved in basic cell functions, including early development and oncogenesis. In the heart, microarray analysis has shown that the expression profile of miRNAs is altered in human heart disease and animal models of cardiac hypertrophy or heart failure (1–3). Numerous studies have revealed that various subcellular organelles, such as the extracellular matrix, myofibrils, sarcoplasmic reticulum, nucleus, and mitochondria, undergo various changes in biochemical composition and structure in cardiac diseases (4–8, 10, 11). These findings imply an association between miRNAs and subcellular organelles, and the identification of the miRNAs and their target genes that affect subcellular organelles may lead to new understandings in cardiac pathophysiology.
The present study focused on the relationship in cardiomyocytes between specific miRNAs, cellular ATP levels, and mitochondria, which are highly abundant and constitute ∼40% of the total cardiomyocyte volume in the heart (9) and mostly generate cellular ATP. To identify miRNAs that can affect cellular ATP levels and mitochondria, the cellular ATP levels of cardiac myocytes, in which a specific miRNA was overexpressed, were assessed. A series of highly expressed miRNAs in the heart were individually overexpressed in neonatal rat cardiomyocytes using a lentiviral vector, and miR-15b was found to decrease cellular ATP levels without affecting cell viability. miR-16, -195, and -424, which have the same seed sequence as miR-15b, also decreased cellular ATP levels. Seed sequences are arranged between the second and eighth nucleotides of the sequence of miRNAs and are the most critical determinant of miRNA targeting (12). Therefore, it was hypothesized that these miRNAs could down-regulate the same target genes and consequently decrease cellular ATP levels.
First we screened a combination of bioinformatics tools for miRNA target prediction, including Target ScanTM, Pic TarTM, and MicroCosmTM, and found that ADP-ribosylation factor-like 2 (Arl2) was a potential target of these miRNAs that may be directly associated with cellular ATP and mitochondria. Arl2 is notable for several unusual features within the ADP-ribosylation factor (Arf) family, including the apparent lack of N-terminal myristoylation, rapid and phospholipid-independent binding of GTP, and association with mitochondria (13, 14). Arl2 forms a complex with Binder of Arl2 (BART), an Arl2-specific effector, and localizes with adenine nucleotide transporter 1 (ANT1) (13). ANT exchanges matrix ATP for cytosolic ADP across the inner mitochondrial membrane and plays a central role in oxidative phosphorylation (15).
The present study shows that 1) miR-15b, -16, -195, and -424, which have the same seed sequence, reduced ATP levels in neonatal cardiac myocytes, 2) miR-15b controls ATP levels by targeting Arl2 in cardiac myocytes, and 3) miR-15b overexpression and Arl2 knockdown changed the mitochondrial morphology.
EXPERIMENTAL PROCEDURES
Cell Culture
Neonatal rat ventricular myocytes (NRVMs) were isolated from 1-day-old Sprague-Dawley rats as described previously (16). These cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin and plated in MULTI-WELLTM PRIMARIATM 6-well or 24-well plates (BD Biosciences) or Lab-TekTM chamber slides in 37 °C in a 5% CO2 incubator. DNA transduction was carried out 48 h after the cells were plated.
Plasmids
miRNA-expressing vectors were constructed using a BLOCK-iTTM Pol II miR RNAi Expression Vector kit (Invitrogen) according to the manufacturer's instructions. Let-7d, miR-1, -15b, -16, -21, -22, -23a, -27a, -29a, -125a, -126–3p, -133a, -133b, -143, -144, -146, -149, -155, -181, -195, -199a, -214, and -424 expression plasmids were made. A control miRNA-expressing vector (miR-cont.) was obtained from the kit. For the construction of anti-miR-15b, double-stranded oligonucleotides containing three or six tandem sequences that were completely complementary to miR-15b were inserted into a pMIR-REPORTTM vector (Ambion) at the PmeI site (Luc-15b decoy). EmGFP-miR-15b decoy was constructed by replacing the luciferase gene with an EmGFP gene. siRNA vectors were constructed from pSINsi-mU6 DNATM (Takara Bio). Double-stranded oligonucleotides were inserted into pSINsi-mU6 DNATM at BamHI/ClaI sites. The oligonucleotides that targeted specific genes and scrambled control (scrambled siRNA) were as follows: Arl2 (5′-GGATTCAAGCTGAACATCT-3′ (Arl2 siRNA1) and 5′-CTGACCATTCTGAAGAAGA-3′ (Arl2 siRNA2)); Bcl-2 (5′-GAGATCGTGATGAAGTACA- 3′ (Bcl-2 siRNA1) and 5′-GGATGACTGAGTACCTGAA-3′ (Bcl-2 siRNA2)); BART (5′-CGCTGGCGACATCTTTGAC- 3′ (BART siRNA1) and 5′ -CGCTTCCCAGAACAACCTA-3′ (BART siRNA2)); Scrambled control (5′-AATAATAATGGGGGGATCC-3′. All of these constructs were inserted into a pLenti6/V5-D-TOPOTM vector (Invitrogen). The rat Arl2 gene was amplified and cloned into a pLenti6/V5-D-TOPO vector using the following primers: forward (5′-CACCATGGGGCTTCTGACCATTCT-3′) and reverse (5′-TCAGTCGGCAGTAAAGACAC-3′). The following primers were used to amplify and clone the 3′-UTR of the mouse Arl2 gene into a pMIR-REPORTTM luciferase vector at the SpeI/HindIII sites (Arl2–3′-UTR Luc) according to the manufacturer's instructions: forward (5′-GGACTAGTGCTTCTTCAGTGTCCCCAGG-3′) and reverse (5′-CCCAAGCTTGTGAAGAGTGCTTTATTCT-3′). The forward primer included a SpeI restriction site, and the reverse primer had a HindIII site (underlined).
Lentivirus Production and DNA Transduction
Lentiviral stocks were produced in 293FT cells according to the manufacturer's protocol (Invitrogen). In brief, virus-containing medium was collected 48 h post-transfection and filtered through a 0.45-μm filter. One round of lentiviral infection was performed by replacing the medium with virus-containing medium that contained 8 μg/ml Polybrene followed by centrifugation at 2500 rpm for 30 min at 32 °C.
RNA Extraction, Quantitative Real-time PCR, and Reverse Transcription-PCR
Total RNA was isolated and purified from NRVMs using TRIzolTM reagent (Invitrogen), and cDNA was synthesized from 5 μg of total RNA using SuperScriptIITM reverse transcriptase (Invitrogen) according to the manufacturer's instructions. For quantitative real-time-PCR, specific genes were amplified for 40 cycles using SYBRTM Green PCR Master Mix (Applied Biosystems). Expression was normalized to the housekeeping gene GAPDH. The primers used were as follows: GAPDH, rat forward (5′-TTGCCATCAACGACCCCTTC-3′), human forward (5′-TTGCCATCAATGACCCCTTC-3′), and reverse (5′-TTGTCATGGATGACCTTGGC-3′); Arl2, rat forward (5′-GAAGCAGAAAGAGCGAGA-3′) and reverse (5′- CAGTAAAGACACGACTGGA-3′); Arl2, human forward (5′-GAAGCAGAAAGAGCGGGA-3′) and reverse (5′-CTGTGAAAATGCGGCTGGA-3′); EmGFP, forward (5′-AGCAAAGACCCCAACGAGAA-3′) and reverse (5′-GGCGGCGGTCACGAA-3′). For reverse transcription-PCR, the rat BART gene was amplified for 25 cycles using TaqDNA polymerase (New England Biolabs, Ipswich, MA) according to the manufacturer's instructions with specific primers as follows: BART, forward (5′-ATGGACAAGTACTACCAGGA-3′) and reverse (5′-CTTACACAGAGAAGTCACCA-3′). The 318-bp fragment was electrophoresed on 1.5% agarose gels and stained using SYBRTM Green I (Invitrogen).
Electron Microscopy
NRVMs on 6-well plates were fixed overnight in phosphate-buffered 2.5% glutaraldehyde (pH 7.4) and then post-fixed for 1 h in 1% osmium tetroxide. The samples were then prepared conventionally for transmission electron microscopy (H-800, Hitachi). We performed a morphometric analysis under an electron microscope using the method described previously with some modification (17). In brief, a uniform sampling of 10 electron micrographs was used for the morphometric assay of each group. Five random fields micrographed at 20,000× from each of five blocks were printed at a final magnification of 50,000× and analyzed on composite grids as described previously to calculate the density of mitochondria (number/μm2), the percentage of shrunken or degenerated mitochondria (%), and the size of mitochondria (μm2) within a cardiomyocyte.
Measurement of Cellular ATP
The amount of ATP in the cell lysates was measured using a luciferin-luciferase ATP assay system (Toyo Ink Co.) in accordance with the manufacturer's instructions. Genomic DNA or protein was extracted from the same samples as used for the ATP assay, and the amount of DNA or protein was used to normalize the cell number.
Cell Viability
Total lactate dehydrogenase activity in the culture medium was measured using a lactate dehydrogenase Cytotoxicity Assay KitTM (Cayman Chemical Co.) in accordance with the manufacturer's instructions. The values were standardized to the amount of protein extracted from the same samples. Viable or dead cells were determined by calcein AM or ethidium homodimer (EthD-1) dyes, respectively, using a LIVE/DEADTM Viability/Cytotoxicity kit (Invitrogen). Five random fields, micrographed at 20×, from each well were printed, and the numbers of viable and dead cells were counted and calculated to give the percentages of each cell group.
Luciferase Assay
For luciferase reporter assays, constructs were transiently transfected into 293FT cells using FuGENE 6TM (Roche Applied Science) or into NRVMs using Lipofectamine 2000TM (Invitrogen) at the following concentrations: 0.1 μg of firefly luciferase reporter gene (Arl2–3′-UTR Luc or Luc-miR-15b decoy), 0.01 μg of pRL-TKTM Renilla reniformis luciferase control plasmid (Promega), and 0.1 μg of BLOCK-iT Pol II miR RNAi Expression Vector encoding the appropriate miRNA or the control. At 24 h after transfection, both luciferase activities were measured using a dual luciferase reporter assay system (Toyo Ink Co.). Firefly luciferase activity was normalized for transfection efficiency by measuring Renilla reniformis luciferase control activity according to the manufacturer's instructions.
Western Immunoblotting Analysis
Immunoblotting analysis was performed using standard procedures as described previously (18). Cultured cells were homogenized in lysis buffer consisting of 100 mm Tris-HCl, pH 7.4, 75 mm NaCl, and 1% TritonTM X-100 (Nacalai Tesque). The buffer was supplemented with Complete MiniTM protease inhibitor (Roche Applied Science), 0.5 mm NaF, and 10 μm Na3VO4 just before use. The protein concentration was determined using a BCA protein assay kit (Bio-Rad). A total of 50 μg of protein was fractionated using NUPAGETM 4–12% Bis-Tris (Invitrogen) gels and transferred to a PROTRANTM nitrocellulose transfer membrane (Whatman). The membrane was blocked using 1× PBS containing 5% nonfat milk for 1 h and incubated with the primary antibody overnight at 4 °C. After being washed in 0.05% T-PBS (1×PBS and 0.05% Tween 20), the membrane was incubated with the secondary antibody for 1 h at 4 °C. After the membrane was washed again in 0.05% T-PBS, the immune complexes were detected using ECL-plusTM chemiluminescent detection reagent (Amersham Biosciences). The primary antibodies used were: anti-GAPDH (Cell Signaling Technology), 1:1000; anti-Arl2 (Protein Tech Group, Inc.), 1:500; anti-Bcl-2 (Assay Designs, Ann Arbor, MI), 1:1000. As secondary antibodies, anti-rabbit IgG (GE Healthcare) was used at a dilution of 1:2000. Immunoblots were detected using LAS-1000 (FUJI FILM).
Mitochondria Isolation and ANT Activity
Mitochondria were isolated from NRVMs 72 h after transduction with a lentiviral vector. Isolated mitochondria were loaded with ATP, and ADP/ATP exchange was measured after the addition of external ADP as described previously (19). Carboxy-atractyloside was purchased from Sigma.
Statistics
Data are presented as the means ± S.E. Statistical comparisons were performed using unpaired two-tailed Student's t tests and a one-way analysis of variance with Bonferroni's or Dunnett's post hoc test where appropriate, and a probability value of <0.05 was taken to indicate significance.
RESULTS
Screening of miRNAs That Alter Cellular ATP Levels
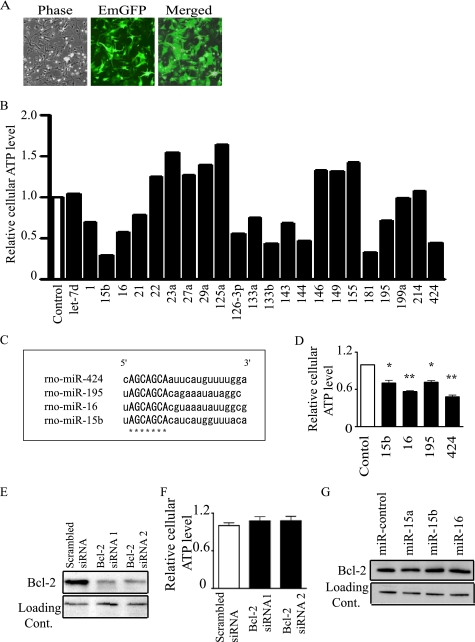
First, lentiviral vectors that expressed let-7d, miR-1, -15b, -16, -21, -22, -23a, -27a, -29a, -125a, -126–3p, -133a, -133b, -143, -144, -146, -149, -155, -181, -195, -199a, -214, and -424 were constructed. Most of these miRNAs have been reported to be highly expressed in the heart (3). To identify miRNAs that can affect cellular ATP levels and mitochondria in the heart, these miRNAs were overexpressed in NRVMs using a lentiviral vector, and the cellular ATP levels were compared. The transduction efficiency was always >90% (Fig. 1A). The lentiviral vectors of miR-15b and -195 could express miR-15b or -195, respectively, in NRVMs at levels that greatly exceeded those of endogenous miRNAs in the control. Moreover, miR-15b and -195 could be overexpressed at the same time by a mixture of multiple miRNA vectors consisting of miR-15b, -16, -195, and -424 (supplemental Fig. S1). After these miRNAs were screened, we found that some miRNAs changed cellular ATP levels (Fig. 1B). Among these, the mechanisms by which miR-15b, -16, -195, and -424 acted on cardiomyocytes were further examined because these miRNAs share a common seed sequence, which is the most critical determinant of miRNA targeting (12). The common seed sequence, which is arranged between the second and eighth nucleotides, is indicated by asterisks in Fig. 1C. We strongly suspected that miR-15b, -16, -195, and -424 could down-regulate the same target genes, which were specifically involved in the regulation of cellular ATP. Moreover, it has been reported that miR-15 and -16 play critical roles in apoptosis of hematopoietic cancer cells (21), and the cardiac-specific overexpression of miR-195 results in cardiac dysfunction in vivo (1). Therefore, we considered whether miR-15b, -16, -195, and -424 might play important roles in the heart. In fact, miR-15b, -16, -195, and -424 were found to decrease cellular ATP levels in NRVMs (Fig. 1D). A previous study indicated that miR-15 and miR-16 down-regulate Bcl-2, an anti-apoptotic protein, post-transcriptionally in hematopoietic cancer cells (21). Therefore, we examined whether the down-regulation of Bcl-2 could affect cellular ATP levels. Knockdown of Bcl-2 by siRNA did not change cellular ATP levels in NRVMs at 72 h after transduction (Fig. 1, E and F). Moreover, the overexpression of miR-15 and -16 did not change Bcl-2 protein expression in NRVMs at 72 h after transduction (Fig. 1G). These results suggest that other target genes are involved in this ATP reduction by miR-15b, -16, -195, and -424 in NRVMs. Absolute amounts of ATP were also measured. As shown in supplemental Fig. 2, the levels were within an anticipated range (22).
FIGURE 1.
miR-15b, -16, -195, and -424 decrease cellular ATP levels. A, shown is expression of the EmGFP gene in NRVMs. B, cellular ATP levels of miR-control or individual miRNAs overexpressing NRVMs 72 h after transduction by a lentiviral vector in serum-containing medium are shown. C, structure of the mature forms of miR-15b, -16, -195, and -424 is shown. The sequence marked by asterisks is the seed sequence. D, cellular ATP levels of miR-control or miR-15b, -16, -195, and -424 overexpressing NRVMs 72 h after transduction by a lentiviral vector in serum-containing medium are shown. E and F, immunoblots of Bcl-2 (E) and cellular ATP levels (F) in NRVMs transduced with Bcl-2 siRNAs are shown. G, immunoblots of Bcl-2 in NRVMs transduced with miR-15 and -16 are shown. The bar graph indicates values expressed as relative ATP levels compared with that of miR-control (D) or scrambled siRNA (F). In D and F, data are presented as the mean ± S.E. of three independent experiments (*, p < 0.05; **, p < 0.01 versus control).
miR-15b Targets Arl2
To identify the targets of miR-15b that modulate cellular ATP levels, we searched for genes that were predicted to be targets of miR-15b by multiple bioinformatics tools for miRNA target prediction, including Target ScanTM, Pic TarTM, and MicroCosmTM. After this combined screening, 28 genes were picked up as candidates (supplemental Fig. S3A). Among these predicted targets of miR-15b, a second search was made for genes for which the function is known to be associated with mitochondria. The six candidates selected were Arl2, glutamate dehydrogenase 1, mitogen-activated protein kinase kinase 1, protein phosphatase 1, regulatory (inhibitor) subunit 11, parathyroid hormone, and TATA box-binding protein. The number of conserved binding sites in their 3′-UTR was then compared. Arl2 had four conserved binding sites, glutamate dehydrogenase 1 and TATA box-binding protein had two sites, and mitogen-activated protein kinase kinase 1, regulatory (inhibitor) subunit 11, and parathyroid hormone had one site according to Target ScanTM (supplemental Fig. S3B). Thus, Arl2 was further evaluated as a potential target of miR-15b. Arl2 is a 21-kDa GTPase that forms a complex with BART, an Arl2-specific effector, and localizes to ANT1 (13). ANT exchanges the matrix ATP for cytosolic ADP across the inner mitochondrial membrane and plays a central role in oxidative phosphorylation (15). Therefore, it was hypothesized that miR-15b, -16, -195, and -424 could down-regulate Arl2, which decreases cellular ATP levels via ANT1. Four evolutionarily conserved binding sites of these miRNAs are indicated in Fig. 2A. To test whether the putative target sequence in the Arl2 3′-UTR could mediate the repression of Arl2 gene expression, the wild-type or mutated full-length 3′-UTR of the Arl2 transcript was inserted into a luciferase reporter construct (wt/mut Arl2–3′-UTR Luc), which was transfected into 293FT cells. In the mutated luciferase reporter constructs, each of the UGCUGCU sequences in the proximal region, which is complementary to the seed sequence of miR-15b, was mutated to UGCUGGG (Fig. 2B). Cytomegalovirus-driven miR-15b, -16, -195, and -424 resulted in a decrease in luciferase activity, whereas similar amounts of miR-214 had no effect (Fig. 2C). Moreover, miR-15b, -16, -195, and -424 did not decrease the luciferase activities of mutated Arl2–3′-UTR constructs (Fig. 2C). The predicted binding site of miR-15b in the distal region of the Arl2 3′-UTR did not mediate gene suppression by miR-15b, -16, -195, and -424 (supplemental Fig. S4, A and B). miR-15b down-regulated both mRNA and protein of Arl2 (Fig. 2, D and E). These results indicated that miR-15b could target Arl2. A mixture of miR-15b, -16, -195, and -424 also decreased Arl2 mRNA and protein levels (Fig. 2, D and E) as well as cellular ATP levels to a similar extent as with miR-15b (Fig. 2F).
FIGURE 2.
miR-15b targets Arl2. A and B, shown is the structure of wild-type- or mutated-Arl2 3′-UTR luciferase construct (wt/mut-Arl2–3′-UTR Luc). The mouse Arl2 3′-UTR was predicted to contain four binding sites of miR-15b, -16, -195, and -424 by TargetScanTM (A). These binding sites are highly conserved among species (B). The gray background indicates the same sequence between human, rat, and mouse. C, 293FT cells were transfected with a wt/mut-Arl2–3′-UTR Luc and expression plasmid for individual miRNAs or miR-control. (***, p < 0.001 versus wt-control; ##, p < 0.01; ###, p < 0.001 versus wt-miR) NRVMs were transduced with miR-15b or miR-control using a lentiviral vector. D and E, Arl2 mRNA and protein expression levels were detected 72 h after transduction of miR-control, -15b, or a mixture of multiple miRNAs such as miR-15b, -16, -195, and -424 (mix) by quantitative real-time-PCR (D) and immunoblotting (E), respectively. F, cellular ATP levels 72 h after transduction of the mixture or miR-control are shown. In D and F data are presented as the mean ± S.E. of three independent experiments (*, p < 0.05; **, p < 0.01 versus control).
miR-15b, -16, -195, and -424 Have Similar Effects in the Recognition and Down-regulation of Arl2 mRNA
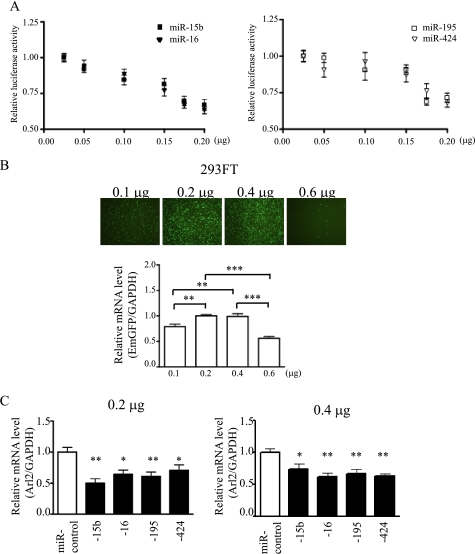
Although the seed sequences of miR-15b, -16, -195, and -424 are identical, the rest of the sequences are quite divergent. To test whether these miRNAs have an identical function in the regulation of Arl2, the dose-response effects of each of these miRNAs were examined. Cytomegalovirus-driven miR-15b, -16, -195, and -424 decreased the luciferase activities of Arl2 3′-UTR Luc constructs in a dose-dependent manner, and the degree of suppression was similar (Fig. 3A). Next, the effects of these miRNAs on Arl2 mRNA down-regulation were evaluated in 293FT cells. The transfection efficacy of miRNA expressing plasmids into 293FT cells was highest at a dose of 0.2 or 0.4 μg, at which Arl2 mRNA levels were maximally decreased to almost similar levels among miR-15b, -16, -195, and -424 (Fig. 3C). These results suggested that miR-15b, -16, -195, and -424 have similar effects in the recognition and down-regulation of Arl2 mRNA.
FIGURE 3.
miR-15b, -16, -195, and -424 have similar effects in the recognition and down-regulation of Arl2 mRNA. A, the luciferase activity is shown of Arl2 3′-UTR Luc construct in response to increasing doses of expression plasmids for individual miRNAs ranging from 0.025 to 0.2 μg. The values are luciferase activities relative to that with a dose of 0.025 μg. B, the transfection efficacy of expression plasmids for miRNAs at a dose of 0.1, 0.2, 0.4, or 0.6 μg was detected by quantitative real-time-PCR for EmGFP transduced into 293FT cells using Fugene6TM. The bar graph indicates values expressed as relative EmGFP levels compared with that with a dose of 0.2 μg (**, p < 0.01; ***, p < 0.001). C, mRNA levels are shown of Arl2 in 293FT cells 24 h after transfection with expression plasmids for individual miRNAs at a dose of 0.2 (left) and 0.4 μg (right). The values are expression levels of Arl2 mRNA relative to that of miR-control. GAPDH was used as an internal control. Data are presented as the mean ± S.E. of three independent experiments (*, p < 0.05; **, p < 0.01 versus control).
Arl2 siRNA Decreases Cellular ATP Levels without Affecting Cell Viability
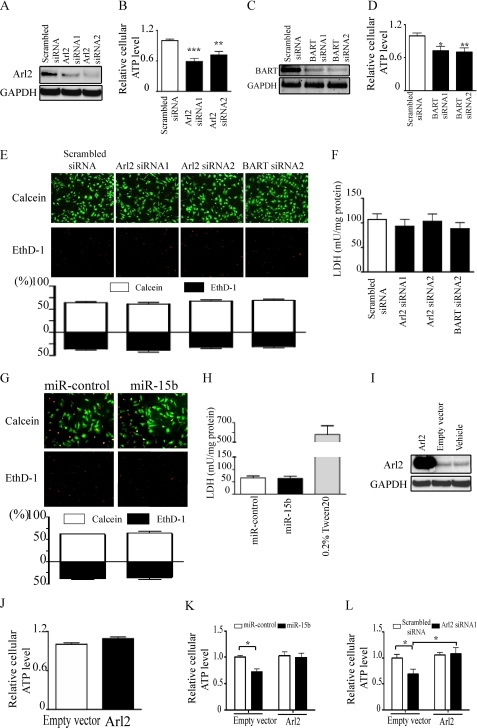
Next, the effect of Arl2 siRNA on cellular ATP levels of cardiac myocytes was examined. Cellular ATP levels were reduced by Arl2 siRNA compared with scrambled siRNA (Fig. 4, A and B). Because it is known that Arl2 forms a complex with BART, an Arl2-specific effector, and localizes with ANT1 (13), BART was suppressed by siRNA in NRVMs, and the cellular ATP levels were measured. siRNAs against BART clearly suppressed cellular ATP levels (Fig. 4, C and D). To test whether Arl2 siRNA or BART siRNA affected cell viability, the proportions of viable and dead cells were evaluated using calcein AM and ethidium homodimer dyes, respectively, and lactate dehydrogenase levels in the culture medium were measured. When Arl2 siRNA, BART siRNA, or scrambled siRNA was transduced into NRVMs, there was no difference in the proportions of viable and dead cells or lactate dehydrogenase levels in the medium of Arl2 siRNA, BART siRNA, or scrambled siRNA-transduced cardiomyocytes (Fig. 4, E and F). These results indicated that Arl2 siRNA and BART siRNA decreased cellular ATP levels without affecting cell viability. Transduction of miR-15b also did not influence cell survival (Fig. 4, G and H).
FIGURE 4.
Down-regulation of Arl2 decreases cellular ATP levels. Assays were performed 72 h after transduction using a lentiviral vector in serum-containing medium. A and B, immunoblots of Arl2 (A) and cellular ATP levels (B) of NRVMs transduced with Arl2 siRNAs are shown. C and D, reverse transcription-PCR of BART (C) and cellular ATP levels (D) of NRVMs transduced with BART siRNAs are shown. In B and D data are presented as the mean ± S.E. of three independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001 versus scrambled siRNA). E–H, NRVMs stained by calcein AM (green) or ethidium homodimer (red) dyes (E and G) are shown. The bar graph indicates the proportions of live or dead cells. Lactate dehydrogenase (LDH) activity in the cultured medium was released from NRVMs (F and H). The values were standardized to the amounts of protein of the same samples. I and J show immunoblots of Arl2 (I) and cellular ATP levels (J) of NRVMs transduced with the Arl2 gene. K and L, cellular ATP levels were measured. NRVMs were co-transduced with either Arl2 gene, an empty vector, or vehicle along with miR-15b or miR-control (K) or along with either Arl2 siRNA1 or scrambled siRNA (L). The bar graph indicates values expressed as relative ATP levels compared with that with transduction with miR-control (K) or scrambled siRNA (L) along with an empty vector. Data are presented as the mean ± S.E. of four independent experiments (*, p < 0.05 versus control).
To further confirm the specific effect of miR-15b on Arl2, cellular ATP levels were analyzed in cardiomyocytes transduced with miR-15b with or without Arl2. Transduction of Arl2 alone did not change cellular ATP levels (Fig. 4, I and J). Cellular ATP levels in miR-15b-transduced cardiomyocytes recovered to the same level as in control cardiomyocytes after the transduction of Arl2 (Fig. 4K). The same results were also seen for the transduction of Arl2 siRNA with or without Arl2 (Fig. 4L). Because Arl2 alone did not increase cellular ATP levels (Fig. 4J), the present data indicate that the down-regulation of Arl2 protein is the main upstream mechanism that explains how miR-15b reduces cellular ATP levels without affecting cell viability in NRVMs.
Endogenous miR-15b Modulates Cellular ATP Levels
To assess the functional consequences of silencing endogenous miR-15b and miRNAs with the same seed sequence in vitro, cardiac myocytes infected with a lentiviral vector were used in which a 3′-UTR with three or six tandem sequences complementary to miR-15b was linked to the luciferase reporter gene or EmGFP (Luc/EmGFP-15b decoy*3 or -*6, Fig. 5A). The complementary sequences acted as a decoy and sequestered endogenous miR-15b and miRNAs that had the same seed sequence. When miR-15b, -16, -195, and -424 were transfected into 293FT cells along with Luc-15b decoy*3 or *6, luciferase activity was significantly reduced (Fig. 5B). On the other hand, miR-1, -133a, -133b, and -21, which have different seed sequences, did not affect the luciferase activity induced by Luc-15b decoy*3 or *6 (Fig. 5B). Next, Luc-15b decoy*3 or *6 was transfected into primary cardiomyocytes and 293FT cells. The reduction of luciferase activity indicated that cardiomyocytes expressed miR-15b or a related miRNA(s), and 15b decoys can react with endogenous miRNAs (Fig. 5C). Next, we confirmed that an increased concentration of GFP-15b decoy*6 reduced the effect of miR-15b on the luciferase activity of Arl2–3′-UTR Luc (Fig. 5D). The cellular Arl2 protein and cellular ATP levels also increased after the transduction of 15b decoy*6 (Fig. 5, E and F). The present data indicate that miR-15b, -16, -195, and -424 can modulate cellular ATP levels in the heart by targeting Arl2.
FIGURE 5.
Loss of miR-15b, -16, -195, and -424 function increases both Arl2 gene expression and cellular ATP levels. A, the structure of “15b decoy” is shown. CMV, cytomegalovirus. B, 293FT cells were transfected with a luciferase decoy construct (Luc-15b decoys or Luc-control decoy) along with an expression plasmid for individual miRNAs or miR-control (control). C, Luc-15b decoys and Luc-control decoy were transfected into 293FT cells or NRVMs. Data are presented as the mean ± S.E. of three independent experiments (***, p < 0.001 versus Luc-control decoy). D, 293FT cells were co-transfected with Arl2–3′-UTR Luc (0.1 μg) and either miR-15b or miR-control (0.1 μg) along with increasing doses of EmGFP-15b decoy*6 (from 0.05 to 0.1 μg) using FuGENE 6TM. The bar graph indicates values expressed as relative luciferase activity compared with that with miR-control. Data are presented as the mean ± S.E. of three independent experiments (*, p < 0.05; **, p < 0.01). NRVMs were transduced with 15b decoys by a lentiviral vector in serum-containing medium. Assays were performed 24 h after transduction. Shown are immunoblots of Arl2 (E) and cellular ATP levels of NRVMs (F). In F, data are presented as the mean ± S.E. of four independent experiments (*, p < 0.05).
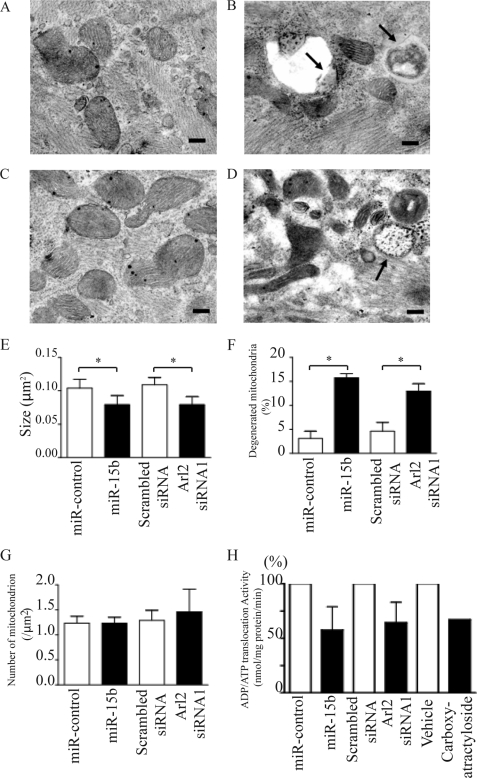
miR-15b and Knockdown of Arl2 Changes Mitochondrial Morphology
Changes in the morphology of mitochondria after transduction of miR-15b and Arl2 siRNA were examined further. When compared with cardiomyocytes that had been transduced with miR-control (Fig. 6A) or scrambled siRNA (Fig. 6C), cells that expressed miR-15b (Fig. 6B) and Arl2 siRNA (Fig. 6D) had small mitochondria (Fig. 6E) and much more degenerated mitochondria (Fig. 6F), whereas the number of mitochondria was not significantly different (Fig. 6G). These findings suggest that miR-15b is involved in the maintenance of mitochondrial morphology via Arl2. Moreover, ANT activities, and particularly the exchange of ADP/ATP, were suppressed in mitochondria that had been isolated from NRVMs transduced with miR-15b and Arl2 siRNA (Fig. 6H). miR-15b and Arl2 siRNA did not change the expression of ANT1 at mRNA or protein levels (data not shown).
FIGURE 6.
Mitochondria were degenerated by miR-15b or Arl2 siRNA in NRVMs. Shown are electron microscopy images of NRVMs 72 h after transduction with miR-control (A) or miR-15b (B) and with scrambled siRNA (C) or Arl2 siRNA1 (D) using a lentiviral vector. E–G, the size (E), degenerated proportions (F), and number/μm2 (G) of mitochondria in NRVMs are shown. Mitochondria are small, electron-dense, and deformed, and some are completely collapsed or degenerated (arrows) in the experimental groups. Bars = 200 nm. *, p < 0.05. H, ANT activities in mitochondria isolated from NRVMs transduced with miR-15b or miR-control and with Arl2 siRNA1 or scrambled siRNA are shown. The values are ADP/ATP exchange activities relative to that of miR-control or scrambled siRNA. Carboxy-atractyloside (0.5 μm) was added 1 min before the addition of external ADP.
DISCUSSION
A growing body of evidence indicates that metabolic and cellular signals influence mitochondrial dynamics, and this could lead to a new understanding of how changes in mitochondria can have a profound impact on their functional output (23). Previously, reduced size, increased number, and structural alterations of cardiac mitochondria have been described in the failing heart (24). It has also been shown that mitochondrial injury is correlated with indices of ventricular dysfunction, such as ejection fraction and end-diastolic pressure, and with the degree of excess orthosympathetic stimulation in congestive heart failure (25). Moreover, reduced expression levels of mitochondrial proteins implicated in ADP rephosphorylation due to reduced gene transcription have been detected during the transition from left ventricular hypertrophy to failure (26). The mechanisms that regulate mitochondrial morphology, size, and number in heart failure are not completely understood, but evidence to date suggests that the morphology of the machinery is modulated through the use of post-translational modifications (27, 28). Therefore, it was hypothesized that miRNAs may influence mitochondrial morphology or function, especially in failing hearts. The present study focused on the relationship between miRNAs and mitochondria.
In the present study miRNAs were transduced into NRVMs by a lentiviral vector. The transduction efficiency was always >90%, and the lentiviral vectors could express miRNAs at abundant levels that exceeded those of endogenous miRNAs (supplemental Fig. S1). This result implied that these lentiviral vectors could express each miRNA beyond the maximal levels that suppressed target gene expression to minimal levels. Therefore, the expression of each target gene could be suppressed equivalently even if the absolute expression levels of overexpressed miRNAs were different. Several groups have reported that in mice treated with transverse aortic coarctation, an after-loaded cardiac hypertrophy model, miR-15a, -15b, -195, and -424 were up-regulated (1–3). miR-16 is relatively highly expressed in NRVMs (29). Recently, it was reported that miR-195 transgenic mice showed cardiac dysfunction, and the severity depended on the miR-195 expression level (1). Seed sequences of miRNAs, arranged between the second and eighth nucleotides, are the most critical determinant of miRNA targeting as well as evolutionary conservation (12). miR-15a, -15b, -16, -195, -424, and -497 possess the same seed sequence, which implies that these miRNAs have the same target genes, and they are expected to strongly influence their target genes if these miRNAs are regulated in the same manner. These findings suggest that miR-15b, -16, 195, and -424 play important roles in maintaining cardiac integrity. The present results showed that miRNAs, such as miR-15b, -16, -195, and -424, could decrease cellular ATP levels in NRVMs. Although a decoy construct acts by binding and sequestering a specific miRNA, which is fully complementary to the sequence in the decoy, miR-16, -195, and -424, which have the same seed sequence as miR-15b, could bind 15b decoys. This result was the same as that in a previous report (30), which suggests that the seed sequence plays a critical role in recognition of the target sequence. Therefore, these miRNAs could be blocked by 15b decoys, which could result in an increase in cellular ATP levels. These results suggest that endogenous miR-15b, -16, -195, and -424 modulate cellular ATP levels in NRVMs. In human hematopoietic cancer cells, it has been reported that miR-15 and miR-16 down-regulate Bcl-2 expression post-transcriptionally (21). However, in the present study, miR-15 and miR-16 did not down-regulate Bcl-2 in NRVMs. This discrepancy may be due to the difference between species, tissues, or assay time points. Furthermore, Bcl-2 siRNA did not decrease cellular ATP levels in the present study.
In the present study, we searched for potential target genes by using a combination of three different bioinformatics tools: Target ScanTM, PicTarTM, and MicroCosmTM. miRNAs are fundamentally preserved beyond species, and if the binding site of a miRNA is conserved, the miRNA may regulate target genes in other species. However, miRNA targeting is not based only on conservation of the binding sequence, and the predicted genes are not always the actual targets. Therefore, experimental validations are always required. In this study we found that miR-15b can target Arl2, which is ubiquitously expressed in rodent and human tissues and shows 95% amino acid sequence identity between human and rat (13). Arl2 is a 21-kDa GTPase that associates with a β-tubulin-specific chaperone protein known as cofactor D (31). Moreover, Arl2 forms a complex with BART, an Arl2-specific effector, and localizes with ANT1 (13). Two different isoforms of ANT (ANT1 and ANT2) have been identified in rodents, whereas four isoforms (ANT1–4) are known in humans (32–34). In rodents, ANT1 is the major isoform present in skeletal and cardiac muscle and is also abundant in brain (13, 15, 34, 35). In contrast, murine ANT2 is more widespread than ANT1 but is less prevalent in the heart and skeletal muscle (13, 15, 34, 35). The predominant binding partner for the BART·Arl2 complex is ANT1, whereas the structurally homologous ANT2 protein does not bind to this complex (13).
We found that miR-15b, -16, -195, and -424 have an identical function in the regulation of Arl2. Moreover, a mixture of these miRNAs could decrease mRNA and protein levels of Arl2 and cellular ATP levels in NRVMs as equally as miR-15b, which suggests that these miRNAs did not interfere with each other in Arl2 gene suppression or in the reduction of cellular ATP. The present results also showed that down-regulation of Arl2 or BART leads to cellular ATP reduction in NRVMs. ATP is generated mostly by oxidative phosphorylation via the mitochondrial respiratory chain. ANT exchanges matrix ATP for cytosolic ADP across the inner mitochondrial membrane and plays a central role in oxidative phosphorylation (15). The inhibition of ADP/ATP exchange should deprive ATP synthase of substrate, resulting in a depletion of cellular ATP (36, 37). ANT1 is the predominant mitochondrial inner-membrane protein that is capable of ADP/ATP exchange (15). Cardiac and skeletal muscle mitochondria from ANT1-deficient mice have been shown to have increased levels of Arl2 relative to that seen in mitochondria from wild-type animals (13). The amount of Arl2 in mitochondria is subject to regulation via an ANT1-sensitive pathway in muscle tissues (13). These findings suggest that the interaction between Arl2 and ANT1 could regulate cellular ATP levels.
ANT1-deficient mice and human diseases such as chronic progressive external ophthalmoplegia and Senger syndrome, which can impair the function and expression of ANT1, respectively, are accompanied by hypertrophic cardiomyopathy and various mitochondrial abnormalities in function or morphology (15, 20, 36, 38). This suggests that the dysregulation of ANT1 could cause cardiac hypertrophy associated with impaired mitochondrial integrity. In the present study, miR-15b and Arl2 siRNA caused mitochondrial degeneration and a decrease in ANT activity in NRVMs without affecting ANT1 expression. These findings imply that miR-15b and Arl2 siRNA could decrease cellular ATP levels via ANT1. However, it is still unknown whether the function of Arl2 in ATP production is only due to ANT activity by direct binding because NRVMs transduced with miR-15b and Arl2 siRNA did not enhance the rate of glucose uptake, which would be expected to be accelerated if ATP production is compromised at the level of adenine nucleotide exchange across the mitochondrial membrane (data not shown).
In conclusion, miR-15b, -16, -195, and -424 modulate cellular ATP levels in neonatal rat cardiac myocytes by targeting Arl2. Up-regulation of these miRNAs affects mitochondrial integrity, which may contribute to cardiac dysfunction. A further examination of Arl2 and ANT1 function may lead to a new understanding and therapeutic options for heart diseases.
Supplementary Material
Acknowledgments
We thank Naoya Sowa and Akemi Fukumoto for excellent technical assistance and Akiko Tsujimoto at Gifu University for the electron microscopy analysis.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K. O., K. H., T. K., and T. K.) and by Grant R31-10069 (WCU Program) from the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1–S4.
- miRNA or miR
- microRNA
- ANT1
- adenine nucleotide transporter 1
- Arl2
- ADP ribosylation factor-like 2
- BART
- Binder of Arl2
- Bcl-2
- B-cell CLL/lymphoma 2
- EmGFP
- emerald green fluorescent protein
- siRNA
- small interfering RNA
- Luc
- luciferase
- NRVM
- neonatal ventricular myocyte
- UTR
- untranslated region
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2- (hydroxymethyl)propane-1,3-diol
- PBS
- phosphate-buffered saline
- wt
- wild type
- mut
- mutant.
REFERENCES
- 1.van Rooij E., Sutherland L. B., Liu N., Williams A. H., McAnally J., Gerard R. D., Richardson J. A., Olson E. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sayed D., Hong C., Chen I. Y., Lypowy J., Abdellatif M. (2007) Circ. Res. 100, 416–424 [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y., Ji R., Yue J., Yang J., Liu X., Chen H., Dean D. B., Zhang C. (2007) Am. J. Pathol. 170, 1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sordahl L. A., McCollum W. B., Wood W. G., Schwartz A. (1973) Am. J. Physiol. 224, 497–502 [DOI] [PubMed] [Google Scholar]
- 5.Heyliger C. E., Ganguly P. K., Dhalla N. S. (1985) Can. J. Cardiol. 1, 401–408 [PubMed] [Google Scholar]
- 6.Mercadier J. J., Lompré A. M., Wisnewsky C., Samuel J. L., Bercovici J., Swynghedauw B., Schwartz K. (1981) Circ. Res. 49, 525–532 [DOI] [PubMed] [Google Scholar]
- 7.Rupp H., Elimban V., Dhalla N. S. (1992) FASEB J. 6, 2349–2353 [DOI] [PubMed] [Google Scholar]
- 8.Scheuer J., Malhotra A., Hirsch C., Capasso J., Schaible T. F. (1982) J. Clin. Invest. 70, 1300–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barth E., Stämmler G., Speiser B., Schaper J. (1992) J. Mol. Cell. Cardiol. 24, 669–681 [DOI] [PubMed] [Google Scholar]
- 10.Johnson D. T., Harris R. A., French S., Blair P. V., You J., Bemis K. G., Wang M., Balaban R. S. (2007) Am. J. Physiol. Cell Physiol. 292, C689–C697 [DOI] [PubMed] [Google Scholar]
- 11.Schulz T. J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. (2007) Cell Metab. 6, 280–293 [DOI] [PubMed] [Google Scholar]
- 12.van Rooij E., Sutherland L. B., Qi X., Richardson J. A., Hill J., Olson E. N. (2007) Science 316, 575–579 [DOI] [PubMed] [Google Scholar]
- 13.Sharer J. D., Shern J. F., Van Valkenburgh H., Wallace D. C., Kahn R. A. (2002) Mol. Biol. Cell 13, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Van Valkenburgh C., Fang H., Green N. (1999) J. Biol. Chem. 274, 37750–37754 [DOI] [PubMed] [Google Scholar]
- 15.Graham B. H., Waymire K. G., Cottrell B., Trounce I. A., MacGregor G. R., Wallace D. C. (1997) Nat. Genet. 16, 226–234 [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa K., Meyers M. B., Kitsis R. N. (1997) J. Biol. Chem. 272, 20049–20054 [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa M., Takemura G., Kanamori H., Goto K., Maruyama R., Tsujimoto A., Ohno T., Okada H., Ogino A., Esaki M., Miyata S., Li L., Ushikoshi H., Aoyama T., Kawasaki M., Nagashima K., Fujiwara T., Minatoguchi S., Fujiwara H. (2008) Circ. Res. 103, 98–106 [DOI] [PubMed] [Google Scholar]
- 18.Yanazume T., Hasegawa K., Morimoto T., Kawamura T., Wada H., Matsumori A., Kawase Y., Hirai M., Kita T. (2003) Mol. Cell. Biol. 23, 3593–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passarella S., Ostuni A., Atlante A., Quagliariello E. (1988) Biochem. Biophys. Res. Commun. 156, 978–986 [DOI] [PubMed] [Google Scholar]
- 20.Dörner A., Schultheiss H. P. (2007) Trends Cardiovasc. Med. 17, 284–290 [DOI] [PubMed] [Google Scholar]
- 21.Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., Rassenti L., Alder H., Volinia S., Liu C. G., Kipps T. J., Negrini M., Croce C. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das A. M., Harris D. A. (1990) Biochem. J. 266, 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soubannier V., McBride H. M. (2009) Biochim. Biophys. Acta 1793, 154–170 [DOI] [PubMed] [Google Scholar]
- 24.Schaper J., Froede R., Hein S., Buck A., Hashizume H., Speiser B., Friedl A., Bleese N. (1991) Circulation 83, 504–514 [DOI] [PubMed] [Google Scholar]
- 25.Sabbah H. N., Sharov V., Riddle J. M., Kono T., Lesch M., Goldstein S. (1992) J. Mol. Cell. Cardiol. 24, 1333–1347 [DOI] [PubMed] [Google Scholar]
- 26.Ning X. H., Zhang J., Liu J., Ye Y., Chen S. H., From A. H., Bache R. J., Portman M. A. (2000) J. Am. Coll. Cardiol. 36, 282–287 [DOI] [PubMed] [Google Scholar]
- 27.Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. (2007) J. Biol. Chem. 282, 11521–11529 [DOI] [PubMed] [Google Scholar]
- 28.Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H. M. (2007) J. Cell Sci. 120, 1178–1188 [DOI] [PubMed] [Google Scholar]
- 29.Duisters R. F., Tijsen A. J., Schroen B., Leenders J. J., Lentink V., van der Made I., Herias V., van Leeuwen R. E., Schellings M. W., Barenbrug P., Maessen J. G., Heymans S., Pinto Y. M., Creemers E. E. (2009) Circ. Res. 104, 170–178, 6p following 178 [DOI] [PubMed] [Google Scholar]
- 30.Ebert M. S., Neilson J. R., Sharp P. A. (2007) Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhamidipati A., Lewis S. A., Cowan N. J. (2000) J. Cell Biol. 149, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dörner A., Olesch M., Giessen S., Pauschinger M., Schultheiss H. P. (1999) Biochim. Biophys. Acta 1417, 16–24 [DOI] [PubMed] [Google Scholar]
- 33.Dolce V., Scarcia P., Iacopetta D., Palmieri F. (2005) FEBS Lett. 579, 633–637 [DOI] [PubMed] [Google Scholar]
- 34.Levy S. E., Chen Y. S., Graham B. H., Wallace D. C. (2000) Gene 254, 57–66 [DOI] [PubMed] [Google Scholar]
- 35.Stepien G., Torroni A., Chung A. B., Hodge J. A., Wallace D. C. (1992) J. Biol. Chem. 267, 14592–14597 [PubMed] [Google Scholar]
- 36.Wallace D. C. (2001) Am. J. Med. Genet. 106, 71–93 [DOI] [PubMed] [Google Scholar]
- 37.Murdock D. G., Boone B. E., Esposito L. A., Wallace D. C. (1999) J. Biol. Chem. 274, 14429–14433 [DOI] [PubMed] [Google Scholar]
- 38.Jordens E. Z., Palmieri L., Huizing M., van den Heuvel L. P., Sengers R. C., Dörner A., Ruitenbeek W., Trijbels F. J., Valsson J., Sigfusson G., Palmieri F., Smeitink J. A. (2002) Ann. Neurol. 52, 95–99 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.