Abstract
The rare neurodegenerative disease Niemann-Pick Type C (NPC) results from mutations in either NPC1 or NPC2, which are membrane-bound and soluble lysosomal proteins, respectively. Previous studies have shown that mutations in either protein result in biochemically indistinguishable phenotypes, most notably the hyper-accumulation of cholesterol and other cargo in lysosomes. We comparatively evaluated the kinetics of [3H]dextran release from lysosomes of wild type, NPC1, NPC2, and NPC1/NPC2 pseudo-double mutant cells and found significant differences between all cell types examined. Specifically, NPC1 or NPC2 mutant fibroblasts treated with NPC1 or NPC2 siRNA (to create NPC1/NPC2 pseudo-double mutants) secreted dextran less efficiently than did either NPC1 or NPC2 single mutant cell lines, suggesting that the two proteins may work independently of one another in the egress of membrane-impermeable lysosomal cargo. To investigate the basis for these differences, we examined the role of NPC1 and NPC2 in the retrograde fusion of lysosomes with late endosomes to create so-called hybrid organelles, which is believed to be the initial step in the egress of cargo from lysosomes. We show here that cells with mutated NPC1 have significantly reduced rates of late endosome/lysosome fusion relative to wild type cells, whereas cells with mutations in NPC2 have rates that are similar to those observed in wild type cells. Instead of being involved in hybrid organelle formation, we show that NPC2 is required for efficient membrane fission events from nascent hybrid organelles, which is thought to be required for the reformation of lysosomes and the release of lysosomal cargo-containing membrane vesicles. Collectively, these results suggest that NPC1 and NPC2 can function independently of one another in the egress of certain membrane-impermeable lysosomal cargo.
Keywords: Cell/Trafficking, Diseases/Neurodegeneration, Lipid/Cholesterol, Membrane/Fusion, Amine Transport, NPC1, NPC2, Retrograde Transport
Introduction
Niemann-Pick type C (NPC)2 is a rare neurodegenerative disorder that has been traced to mutations in one of two genes (1). The first gene, NPC1, encodes a late endosomal/lysosomal membrane protein of 1278 amino acids that has 13 transmembrane domains (2). The second gene, NPC2, encodes a glycosylated, soluble 151-amino acid lysosomal protein (3).
Loss of function mutations in either NPC1 or NPC2 has been shown to cause the hyper-accumulation of several molecules including, but not limited to, cholesterol, glycosphingolipids, sphingosine, and sphingomyelin (4–7). Clinically indistinguishable diseases have been documented regardless of which of the aforementioned NPC proteins is mutated (8–9). Based on these observations, it has been proposed that NPC1 and NPC2 function in concert in a common pathway to facilitate the trafficking of cholesterol and other cargo from lysosomes (10).
It is a fact that cholesterol homeostasis is significantly disturbed in patients with NPC disease; however, we reason that any molecule that accumulates in lysosomes that cannot readily permeate out by passive or active transport mechanisms will experience excessive accumulation and potentially altered cellular homeostasis in patients with NPC disease. An example of such cargo includes lysosomotropic endogenous amines that accumulate into lysosomes according to an ion-trapping mechanism (11). We have recently reported that NPC1 plays an important functional role in regulating amine accumulation in lysosomes (12). Specifically, amine-induced vacuolization of lysosomes was shown to require functional NPC1 and involve retrograde fusion of lysosomes with late endosomes to create hybrid organelles. Certain lysosomotropic amines were shown to significantly stimulate late endosome/lysosome fusion events as well as the overall rate of cell secretion of lysosomal cargo. Importantly, these events were only observed in cells with functional NPC1.
Considering that mutations in NPC1 and NPC2 have been shown to be phenotypically indistinguishable in nearly all assays examined, we sought to determine here if NPC2 functions like NPC1 in the secretion of membrane-impermeable lysosomal cargo and in the regulation of lysosomal amine content. We comparatively examined wild type and NPC1 and NPC2 mutant fibroblasts as well as cells mimicking a NPC1/NPC2 double mutant, which were created by treating NPC1 or NPC2 mutant fibroblasts with NPC1 or NPC2 siRNA. Surprisingly, we found significant differences between all cell types examined.
Our results suggest that NPC1 functions independently of NPC2 in late endosome/lysosome retrograde fusion that leads to the creation of hybrid organelles. We also provide evidence to support the notion that NPC2 is required for fission of membrane vesicles from the hybrid organelles, which most likely leads to the reformation of lysosomes as well as the subsequent trafficking of lysosomal cargo to other cellular destinations. The relationship between these findings and amine trafficking and regulation in late endocytic compartments is addressed.
EXPERIMENTAL PROCEDURES
Reagents
AlexaFluor (AF) 647 succinimidyl ester, AF555 succinimidyl ester, and biotinylated dextran amine (Mr 10,000) were obtained from Invitrogen. [3H]dextran (Mr 70,000) was obtained from American Radiolabeled Chemicals (St. Louis, MO). Carboxylate-modified streptavidin-labeled latex beads (∼800 nm), dextran (70,000 Mr), neutral red (NR), nocodazole, chloroquine (CQ), and TLC plates were all purchased from Sigma.
Cell Culture and Conditions
Wild type human foreskin fibroblasts (CRL-2076, Coriell Cell Repository, Camden, NJ) and human foreskin fibroblasts with a mutant form of NPC1, denoted NPC1−/− (CCR, catalogue ID GM03123), were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Two human fibroblast lines with mutant forms of the NPC2 protein, denoted NPC2−/− GM17910 (CCR, catalogue ID GM17910) and NPC2−/− GM18455 (Coriell, catalogue ID GM18455), were grown in Earl's modified Eagle's medium supplemented with 15% FBS. Chinese hamster ovary cells containing a deletion of the NPC1 locus, denoted NPC1-null (M12 cells) (13), were a kind gift from Dr. D. Ory (Washington University, St. Louis, MO) and were grown in DMEM/F-12 with 5% FBS. Mouse embryonic fibroblasts (MEFs, wild type, NPC1+/−/NPC2+/−, NPC1+/−/NPC2+/+, NPC1−/−/NPC2+/+, and NPC1−/−/NPC2−/−) were a kind gift from Dr. P. Lobel (Rutgers University, Piscataway, NJ) and were grown in DMEM + 10% FBS. Fibroblasts with Sandhoff disease (Coriell, catalogue ID GM11707) and with mucolipidosis type IV (Coriell, catalogue ID GM02408) were grown in Earl's modified Eagle's medium supplemented with 15% FBS. All cells and experiments were maintained at 37 °C in a humidified 5% CO2 atmosphere unless otherwise stated. All fibroblasts were grown to 80–90% confluency before experiments were performed. To deplete wild type fibroblasts of LDL-derived cholesterol, cells were incubated in serum-free DMEM for 72 h before use.
Dextran Release Experiments
The indicated fibroblasts were plated at 50,000 cells/well at 37 °C in 6-well plates 24 h before release experiments. Cells were incubated with a total of 1 mg/ml 70,000 Mr dextran including 0.1 mg/ml [3H]dextran (25 μCi/ml) for a total of 6 h at 37 °C. Monolayers were then washed 4 times with ice-cold Dulbecco's phosphate-buffered saline (D-PBS) and replaced with dextran-free DMEM at 37 °C. The supernatant was subsequently removed and replaced with DMEM at the indicated time points. The supernatant was assayed for radioactivity by mixing with 5 ml of ScintiVerse BD Mixture (Fisher) using a liquid scintillation counter (Beckman; LS 60001C). To account for nonspecific [3H]dextran-associated release, the experiment was repeated at 4 °C, and these values were subtracted from the total amount released at 37 °C for all time points. For drug-treated fibroblasts, U18666A (10 μm) and progesterone (10 μg/ml) were incubated with cells for 6 h before experimentation and for the duration of the experiment.
Lysosome/Late Endosome Retrograde Fusion Assay
A fluorescence resonance energy transfer (FRET)-based assay was employed to evaluate rates of late endosome/lysosome retrograde fusion as previously described (12, 14). Briefly, fibroblasts were plated on 4-well cell culture slides (1.7 cm2) at 50,000 cells per well at 37 °C 1 day before testing. Before experimentation, biotinylated dextran amine was conjugated to AF647, and the carboxylate-modified streptavidin-labeled latex beads were conjugated to AF555 using previously reported protocols (14). Cell monolayers were incubated with 2.5 mg/ml biotinylated dextran amine-AF647 for 2 h and chased in DMEM for 6 h. Monolayers were then washed once with D-PBS, pulsed with AF555-latex beads for 20 min, and washed 4 times with DMEM before obtaining FRET measurements. This point was defined as the initial FRET ratio for all FRET assays and is reported as such in subsequent figures. FRET measurements were made using a Photon Technologies International Ratiomaster microscope-mounted spectrofluorometer with photomultiplier tube detection (Birmingham, NJ). Multiple measurements were made: a control for bead loading, (donor excitation/emission, AF555, excitation 550 nm; emission 610/40 nm), the FRET signal (donor excitation/acceptor emission, FRET, excitation 550 nm; emission 710/60 nm), and a control for dextran loading (acceptor excitation/emission, AF647; excitation 650 nm; emission 670/40 nm). All dichroic and emission filters used were from Chroma (Rockingham, VT).
siRNA Knockdown Experiments
NPC1 or NPC2 protein expression was silenced using previously described siRNA constructs (15). Briefly, NPC1−/− (GM03123) or NPC2−/− (GM17910) fibroblasts were plated at 50,000 cells per well at 37 °C on a 4-well cell culture slide 24 h before transfections. siRNA constructs as well as scrambled controls (Ambion, Austin, TX) were transfected into cells using serum-free DMEM and siPORT-Amine (Ambion) per the manufacturer's instructions. Cells were used 72 h post-transfection to allow for sufficient NPC1 or NPC2 knockdown. Knockdown was confirmed by Western blot analysis using a polyclonal rabbit NPC1 antibody (Novus Biologicals, Littleton, CO), polyclonal goat NPC2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), donkey anti-goat IgG-horseradish peroxidase (HRP), and goat anti-rabbit IgG-HRP antibody (Santa Cruz).
DNA Transfections
Indicated fibroblasts were grown to ∼80% confluency in 4-well cell culture slides and transfected with the fluorescent fusion proteins NPC1-GFP (provided by Dr. M. Scott, Stanford University, Stanford, CA) and Rab9-YFP (provided by Dr. S. Pfeffer, Stanford University, Stanford, CA) using the transfection reagent FuGENE 6 per the manufacturer's protocol (Roche Applied Science). Cells were transfected for 48 h to achieve sufficiently high protein expression. Treated cells were incubated with 100 μm CQ for 3 h.
Specificity of siRNA-mediated NPC1 Knockdown
To confirm the specificity of the NPC1 siRNA for the NPC1 gene, we utilized an orthologous form of NPC1, namely NPC1-GFP. The NPC1 siRNA used previously targeted human-specific mRNA to achieve knockdown. NPC1-GFP is derived from a mouse origin and contains six different nucleotides in the siRNA binding region (as compared with human NPC1) and, therefore, is not effectively suppressed by the NPC1 siRNA. Wild type fibroblasts were transfected with NPC1-GFP 48 h before treatment with NPC1 siRNA. [3H]Dextran secretion was then measured 72 h post-transfection with siRNA.
Amine-induced Vacuole Sizing
Wild type, NPC1−/−, (GM03123), NPC2−/− (GM17910), and NPC2−/− (GM18455) fibroblasts were plated on 4-well cell culture slides (BD Biosciences) at 50,000 cells/well at 37 °C 24 h before use. On the day of experimentation, cells were incubated with the lysosomotropic agents NR and CQ (70 and 100 μm, respectively) in DMEM for 6 h at 37 °C. The indicated cells were treated with 1 μm nocodazole for 1 h before and concurrent to NR treatment. Cells were then washed 3 times at 37 °C with D-PBS and subsequently prepared for microscopy. Images were captured on a Zeiss Meta 510 laser scanning confocal microscope (Thornwood, NY) with an argon laser at 458 nm. All size and area measurements were done using ImageJ software (available from the NIH online at rsbweb.nih.gov) using a background subtraction followed by the Threshold algorithm to define vacuole dimensions and the Analyze algorithm to size all identified vacuoles. The average number of sized vacuoles per cell ranged from 106 to 1744 with an average of 463. A minimum of five cells was evaluated for each treatment.
Immunofluorescence
Fibroblasts were plated on 8-well cell culture slides at 25,000 cells per well at 37 °C 24 h before microscopy. Fibroblasts were washed once with D-PBS and fixed with a 4% paraformaldehyde solution in D-PBS for 7 min at room temperature. Fibroblasts were then permeabilized with 0.1% saponin in a 10% FBS in D-PBS solution for 30 min. Primary antibodies for lysosomal-associated membrane protein 1 (LAMP1, rabbit, monoclonal, Developmental Studies Hybridoma, Iowa City, IA) and the cation-independent mannose 6-phosphate receptor (MPR, mouse, monoclonal; Developmental Studies Hybridoma; Iowa City, IA) diluted 1:50 in 0.05% saponin in a 10% FBS solution of D-PBS were applied to the cells at room temperature for 2 h. Cells were then washed twice with 0.1% saponin in a 10% FBS in D-PBS solution. Next, fibroblasts were incubated for 2 h at room temperature with secondary antibodies (goat anti-rabbit AF488 and goat anti-mouse AF647; Invitrogen) diluted 1:500 in a 0.05% saponin in 10% FBS solution. Cells were then washed twice with D-PBS and prepared for imaging. CQ-treated cells were incubated with 100 μm for 3 h before immunofluorescence analysis. Images were acquired with an Olympus/3I Spinning Disk Confocal/TIRF inverted microscope with laser lines (Coherent Inc., Santa Clara, CA) at 488 and 635 nm. Images were acquired using a Hamamatsu ImagEM CCD Camera. Co-localization was quantified in ImageJ using the Colocalization plug-in.
Cholesterol Efflux
Cholesterol efflux was measured similarly to a previously published protocol (16). Briefly, fibroblasts were plated on 6-well plates at 50,000 cells per well and allowed to adhere overnight. The next day media were replaced with DMEM containing lipoprotein-free serum for 48 h. Cells were then washed once with lipoprotein-free DMEM and refrigerated at 4 °C for 30 min. Cells were incubated with 20 μg/ml [14C]cholesteryl oleate-labeled LDL (LDL was labeled with [14C]cholesteryl oleate according to previous protocols (17)) in lipoprotein-free DMEM and incubated for 4 h at 18 °C. Cells were washed with PBS three times and then incubated for 2 additional hours in lipoprotein-free DMEM. 4% methyl-β-cyclodextrin in lipoprotein-free DMEM was then applied for 10 min to extract membrane cholesterol, and media fractions were pooled. Cells were lysed with 0.1 n NaOH. Cholesterol was extracted using a 2:1 mixture of chloroform/methanol after the addition of 50 μg of cholesterol, 50 μg of cholesteryl oleate, and 0.001 μCi of [3H]cholesteryl oleate. Samples were then dried under nitrogen, and TLC was run. TLC running buffer was a 90:30:1 mixture of heptane/ethyl ether/acetic acid, and visualization was done using iodine. Lipids were quantified using liquid scintillation counting after scraping of the appropriate spots off the TLC plates. Efflux was expressed as a fraction of radioactivity in media divided by total radioactivity in media and cells.
Cholesterol Esterification
Esterification assays were based on a previously published report (16). Cells were plated on 6-well plates at 100,000 cells per well and allowed to adhere overnight. The next day cells were fed lipoprotein-free DMEM for 24 h. 50 μg/ml LDL was then added and incubated for 16 h. Afterward, ∼1 μCi of [3H]oleate was pulsed for 2 h, cells were then washed 3 times with PBS, and lipids were extracted using a 3:2 mixture of hexane and isopropyl alcohol. After the addition of 50 μg of cholesteryl oleate and 0.001 μCi of [14C]cholesteryl oleate, samples were dried under nitrogen, and TLC was run. TLC running conditions were identical to cholesterol efflux TLC conditions. Lipids were quantified using liquid scintillation counting, and data were normalized to protein content.
RESULTS
Roles of NPC1 and NPC2 in the Cellular Efflux of Membrane-impermeable Lysosomal Cargo
We and others have shown that mutations in NPC1 impair vesicle-mediated egress of a variety of membrane-impermeable cargo from lysosomes, including dextran, sucrose, and small molecular weight amines (12, 18–19). More recently, Tang et al. (20) have shown that NPC1 is required for the efficient trafficking of HIV-1 viral proteins from late endosomes/lysosomes after infection. To our knowledge, no prior investigations have evaluated how mutations in NPC2 influence the release of such membrane-impermeable cargo from cells.
We were interested in determining if NPC2-deficient cells had impaired lysosomal release kinetics similar to what had previously been observed in cells with mutations in NPC1 (12). More importantly, we questioned whether or not cells with deficiencies in both proteins had release profiles that were similar to those from cells with single protein mutations.
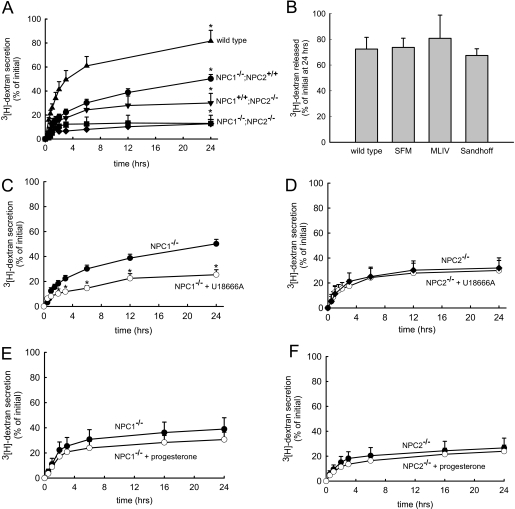
To examine this we evaluated the release of [3H]dextran (70,000 Mr) from human skin fibroblasts that were appropriately pulsed-chased to ensure specific localization within lysosomes (data not shown). We comparatively analyzed wild type and NPC1 and NPC2 mutant fibroblasts. The cumulative percent of dextran released into the culture medium is plotted as a function of time for each of the cell lines. As previously reported (12), wild type fibroblasts release ∼80% of the total dextran in the cells at 24 h post-dextran incubation (Fig. 1A). NPC1 mutant fibroblasts had significantly reduced dextran secretion relative to wild type fibroblasts (∼45% at 24 h, Fig. 1A). Interestingly, we found that NPC2 mutant cells exocytosed even less dextran than did NPC1 mutant cells at the 24-h time point (∼25%, Fig. 1A).
FIGURE 1.
Release of lysosomal [3H]dextran polymers from cells with or without functional mutations in NPC1 and NPC2. A, wild type cells (▴) released about 80% of their initial dextran over a 24-h time period. Cells with mutations in NPC1 (NPC1−/−/NPC2+/+, GM03123) (●) had significantly compromised dextran release relative to wild type cells. Cells with functional mutations in NPC2 (NPC1+/+/NPC2−/−, GM18455) (▾) secreted dextran less efficiently than cells with mutation in NPC1. Pseudo-double mutants (designated NPC1−/−/NPC2−/−) were created from NPC2−/− (GM18455) cells that had been transfected with NPC1 siRNA (■) or NPC1−/− fibroblasts (GM03123) that had been transfected with NPC2 siRNA (♦). Both pseudo-double mutant fibroblasts exhibited less efficient dextran secretion than either NPC1 or NPC2 single mutant cell lines. B, wild type cells released about 80% of their total dextran at 24 h. Cells incubated in serum-free media (SFM) for 72 h exhibited similar cumulative dextran secretion at the 24-h time point. Similarly, mucolipidosis type IV (MLIV) and Sandhoff-diseased cells exhibited no changes in dextran secretion at 24 h. C and D, dextran secretion profiles observed for NPC1−/− or NPC2−/− fibroblasts with (○) or without (●) 10 μm U18666A in the culture medium. The addition of U18666A significantly decreased dextran secretion in cells with functional mutations in NPC1 but not in fibroblasts with mutations in NPC2. E and F, dextran secretion profiles were observed with fibroblasts harboring mutations in NPC1 or NPC2 that were (○) or were not (●) treated with 10 μg/ml progesterone in the cell culture medium. Progesterone treatment did not significantly influence dextran secretion in either cell line. Data shown are the means ± S.E. from experiments run in triplicate (*, p < 0.05 by unpaired t test).
We subsequently sought to determine whether deficiencies in NPC1 and NPC2 in the same cell resulted in dextran secretion phenotypes that were similar to single mutants or if the defects in secretion were additive. If the dextran secretion profiles were similar to that of either single mutant profile, it would support the notion that the two proteins work together at the same step in retrograde trafficking. Alternatively, if the secretion was significantly less than either single mutant, this would suggest that both NPC1 and NPC2 function in the egress of lysosomal dextran but possibly in different steps. Because we could not readily acquire fibroblasts with loss of function mutations in both NPC1 and NPC2, we used siRNA to silence the expression of NPC1 in NPC2 mutant fibroblasts and to silence the expression of NPC2 in NPC1 mutant fibroblasts. These mutant cells treated with siRNA are referred to as NPC1−/−/NPC2−/− pseudo-double mutants. According to densitometric analysis of Western blots, we were able to suppress NPC1 and NPC2 expression by ∼95% (data not shown). Unexpectedly, both pseudo-double mutants released about half the dextran of non-transfected NPC2 mutant cells (∼15% at 24 h, Fig. 1A). Because it is possible that the transfection procedure itself could have negatively impacted dextran release, we also examined mutant cells that were transfected with scrambled siRNA and found that the dextran release was not significantly different from non-transfected mutant cells (supplemental Fig. S1A). Additionally we tested the specificity of the siRNA to NPC1 on cells expressing an orthologous NPC1 (mouse NPC1-GFP), which lacks the human siRNA target sequence. As shown in supplemental Fig. 1B, cells with functional NPC1-GFP treated with NPC1 siRNA showed significantly greater dextran release at the 24-h time point as compared with cells treated with NPC1 siRNA alone.
It is possible that the aforementioned alterations in dextran secretion profiles associated with the cells harboring mutations in NPC proteins could be due to alterations in lipid homeostasis and not directly due to mutations in the NPC proteins themselves. Indeed, it has been shown that the ability of late endosomes and lysosomes to fuse can depend on lipid accumulation in these compartments (21). To examine this, we evaluated dextran release in cells that had functional NPC proteins but had lysosomal lipid storage disorders. Specifically, we tested fibroblasts from patients with mucolipidosis type IV and from patients with Sandhoff disease. Mucolipidosis type IV cells harbor mutations in mucolipin, a protein responsible for subcellular lipid transport (22). These cells accumulate sphingolipids, phospholipids, and acid mucopolysaccharides. Sandhoff disease is characterized by GM2 ganglioside hyper-accumulation (23–24). As shown in Fig. 1B, there was no statistically significant change in dextran secretion at 24 h for both lipid storage disorders as compared with wild type fibroblasts. Additionally, we incubated wild type fibroblasts in serum-free media for 72 h (conditions which deplete LDL-derived cholesterol from late endosomes and lysosomes) and then tested their dextran release. Again, no difference in dextran secretion was seen at 24 h. Collectively, these data suggest that significant alterations in late endosomal/lysosomal lipid homeostasis cannot alone account for the alterations in dextran secretion observed with our studies.
Additionally, we evaluated the effects of a well known inducer of the NPC disease phenotype, U18666A, on dextran secretion in wild type cells or those with functional mutations in either NPC1 or NPC2. In wild type cells, treatment with 10 μm U18666A significantly inhibited dextran secretion (supplemental Fig. S2A), consistent with previously published results from this laboratory (12). Interestingly, cells with single mutations in NPC2 that were treated with U18666A had dextran secretion profiles that were indistinguishable from untreated NPC2 mutant fibroblasts (Fig. 1D). Conversely, cells with single mutations in NPC1 experienced a significantly reduced dextran secretion when treated with U18666A compared with untreated NPC1 mutant fibroblasts (Fig. 1C).
The finding that cells with mutations in NPC1 or NPC2 respond differently to U18666A treatment is interesting and suggests that U18666A may preferentially inhibit NPC2-mediated events involved in the retroendocytic release of the dextran molecules. To explore this possibility further, we evaluated the effects of a second NPC mimetic, progesterone, on dextran release. Similar to the effects of U18666A in normal cells, treatment with 10 μg/ml progesterone impaired dextran release significantly (supplemental Fig. S2B). Cells with mutations in NPC1 treated with progesterone exhibited slightly decreased dextran release (Fig. 1E), although this decrease was not statistically significant. Cells with mutations in NPC2 exhibited insensitivity toward progesterone treatment (Fig. 1F). Although progesterone might exert its effects on cells via a different mechanism than U18666A, the seemingly insensitive response of NPC2 mutant cells against NPC phenotype inducers is consistent.
These results suggest that NPC mimetics may preferentially target the NPC2 pathway. This result appears to contradict the work of Ko et al. (25), who have shown that the overexpression of NPC1 could rescue U18666A-induced hyper-accumulation of cholesterol in late endocytic compartments, as evidenced by filipin staining. In this work the authors demonstrated that the rescue effect was dependent on the concentration of U18666A used (i.e. at higher concentrations of U18666A the rescue was not evident). Based on our work it appears that the NPC2 pathway may be more sensitive than the NPC1 pathway to the effects of U18666A at the concentrations utilized. It is possible that NPC1 and NPC2 have distinctly different dose-response relationships to NPC mimetics, with the NPC2 pathway being more sensitive at lower U18666A concentrations. It is probable that at higher concentrations, both cell types would exhibit significantly impaired dextran release profiles. Unfortunately, we were not able to incubate cells with such high concentrations as the cells succumb to the toxic effects of the compounds. It is likely that the concentrations of U18666A or progesterone required to interfere with cholesterol trafficking from late endocytic compartments is much less than the concentrations required to interfere with dextran release. Based on this reasoning, we feel that it may be premature to definitively argue that NPC mimetics have specific effects on NPC2-mediated events but not on those specifically mediated by NPC1.
Roles of NPC1 and NPC2 in Late Endosome/Lysosome Fusion
The vesicle-mediated release of lysosomal cargo from cells can in theory occur by two separate pathways. First, lysosomes could directly fuse with the plasma membrane to release their cargo. When damage occurs to the plasma membrane, cytosolic calcium levels rise, and lysosomes have been shown to fuse with the plasma membrane to reseal the injury (26, 27). However, under normal circumstances (i.e. without injury) this pathway has been shown to contribute minimally to the secretion of lysosomal contents (28). A second pathway involves a multistep retroendocytic pathway. Despite the fact that much is unknown regarding the molecular details of this transport pathway, it has been shown that lysosomes fuse with late endosomes in a retrograde fashion to create hybrid organelles, and this is likely the first step in retrograde transport of lysosomal cargo (29–30).
We have previously developed a quantitative fluorescence resonance energy transfer (FRET)-based approach to evaluate the kinetics of this fusion event in live cells (14). Briefly, cells are pulse-chased with biotinylated dextran containing a FRET pair acceptor (AlexaFluor 647) to localize them specifically within lysosomes. The dextran polymers were confirmed to be in lysosomes using immunofluorescence co-localization with the lysosomal protein, LAMP1 (data not shown). Subsequently, cells are incubated with latex beads (∼800 nm) conjugated to streptavidin and a FRET pair donor (AlexaFluor 555). Because of their size, these beads are endocytosed but do not progress beyond late endosomes to lysosomes (14, 31, 32). Accordingly, only retrograde fusion of lysosomes with late endosomes will allow the FRET signal to be observed, which is facilitated by streptavidin-biotin binding bringing the two fluorophores in close proximity. It is important to note that this assay represents a cumulative record of hybrid organelle fusion events over the indicated period of time.
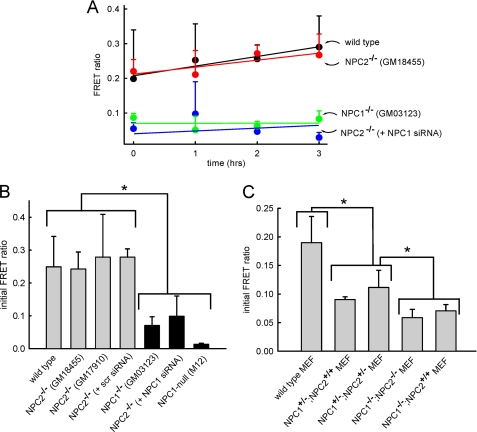
Using this assay we have previously shown and have repeated here that fibroblasts with mutated NPC1 have hybrid organelle formation rates that are much slower than the rates observed with wild type cells (12). Contrary to expectations, fibroblasts with mutant NPC2 had rates of hybrid organelle formation that were indistinguishable from wild type fibroblasts (Fig. 2, A and B). We considered the possibility that the NPC2 mutant cell line initially evaluated may not be representative of other fibroblasts with loss of function mutations in the NPC2 protein. We, therefore, evaluated the hybrid organelle formation rate in a different NPC2 mutant cell line (GM17910) and found that this cell line also had virtually identical hybrid organelle formation rates compared with wild type cells (Fig. 2B). The NPC2 mutant fibroblasts transfected with NPC1 siRNA had hybrid organelle fusion rates that were much slower than wild type cells and nearly identical to NPC1 mutant cells (Fig. 2, A and B). The transfection protocol did not appear to have any significant influence on hybrid organelle formation rates as NPC2 mutant cells transfected with scrambled siRNA had rates that were not statistically different from those observed with wild type cells (Fig. 2B).
FIGURE 2.
NPC1, but not NPC2, is required for efficient retrograde fusion of lysosomes with late endosomes. A, the appearance of a FRET signal indicates retrograde fusion of lysosomes with late endosomes to create hybrid organelles. The appearance of FRET signal as a function of time is shown for wild type fibroblasts (black circles), NPC2−/− fibroblasts (red circles), NPC1−/− fibroblasts (green circles), and NPC2−/− fibroblasts treated with siRNA against NPC1 (blue circles). B, measured FRET in all cell types at the zero time point (see “Experimental Procedures”) is shown. The FRET signal observed with wild type cells is not significantly different from that of cells with mutations in NPC2 (both GM17910 and GM18455 cell lines). Alternatively, cells with mutations in NPC1 have significantly reduced hybrid organelle formation. The efficiency of retrograde fusion of cells with mutations in NPC2 can be decreased to levels comparable with NPC1 mutant cells by treating them with NPC1 siRNA. As a control, a scrambled version of the NPC1 siRNA has no significant impact on the rate of hybrid organelle formation in cells with mutations in NPC2. Additionally, NPC1-null Chinese hamster ovary cells (M12) exhibit similarly low fusion profiles. C, wild type MEFs exhibited normal rates of fusion, whereas MEFs heterozygous in NPC1 (NPC1+/− /NPC2+/+ and NPC1+/− /NPC2+/−) showed intermediary fusion kinetics. NPC1−/−/NPC2−/− MEFs showed fusion kinetics similar to NPC1−/−/NPC2+/+ cells. Data points and bars represent the average ± S.E. from three independent experiments (*, p < 0.05 by unpaired t test).
These results would suggest that there are phenotypic differences in cells with mutations in NPC1 or NPC2. According to the work of Sleat et al. (8), mice with mutations in NPC1, NPC2, or both showed virtually identical lipid accumulation phenotypes and NPC disease progression. To address this apparent discrepancy, we obtained MEFs from the mice used in these earlier studies to determine whether we could observe the same phenotypic differences as were previously observed for fibroblasts examined in Figs. 2, A and B. The MEFs for these evaluations were obtained from the laboratory of Peter Lobel (Dept. of Pharmacology, Robert Wood Johnson Medical School, Rutgers University) and were characterized as having homozygous or heterozygous mutations in either NPC1 or NPC2. Wild type MEFs had shown hybrid organelle formation kinetics similar to wild type fibroblasts (Fig. 2C). Unexpectedly, we found that MEFs with heterozygous mutations in NPC1 had significantly reduced hybrid organelle formation rates relative to wild type MEFs (Fig. 2C). This was contrary to expectations as previous studies have shown that cells with heterozygous mutations in NPC1 behave similarly to wild type cells with respect to disease progression. Nevertheless, we examined if the functional status of NPC2 had any impact on hybrid organelle formation rates in the MEFs that were heterozygous in NPC1. Consistent with our previous observations (Fig. 2A), there was no significant difference in hybrid organelle formation rates between MEFs regardless of the presence or absence of functional NPC2 (Fig. 2C). Moreover, MEFs with homozygous mutations in NPC1 had significantly compromised hybrid organelle formation rates, again independent of NPC2 functional status (Fig. 2C). We additionally obtained Chinese hamster ovary cells containing a deletion in the NPC1 locus but with normal NPC2, denoted as M12 cells, that were originally described by Millard et al. (13). As expected, M12 cells also exhibited decreased hybrid organelle formation similar to other cells with functional mutations in NPC1 (Fig. 2B).
NPC2 and Lysosomal Amine Regulation
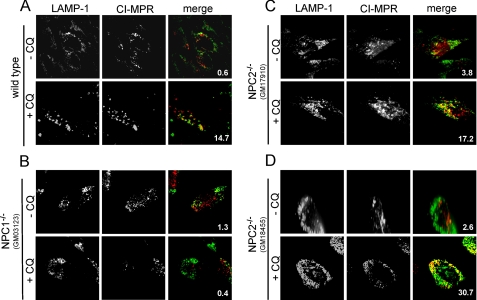
When fibroblasts are grown under typical culture conditions, we observe very little co-localization of lysosomes and late endosomes, as observed through immunofluorescence analysis of the lysosome- and late endosome-specific proteins, LAMP1 and MPR, respectively. It is important to point out that this does not imply that lysosomes are not continuously fusing with late endosomes; rather, this lack of co-localization suggests that the steady-state population of hybrid organelles is low. We have previously shown that exposing cells to lysosomotropic amines facilitates the co-localization of lysosomes with late endosomes in an NPC1-dependent fashion (12). In this report we examined the role of NPC2 in this amine-induced late endosome/lysosome fusion. Consistent with our previously published results, wild type cells have limited late endosome/lysosome co-localization without amines in the culture medium (Fig. 3A). However, when the lysosomotropic amine CQ was added at high concentrations (100 μm), the LAMP1/MPR co-localization was significantly enhanced (Fig. 3A). The amount of co-localization was quantified in ImageJ and is indicated in each merged image. With cells lacking functional NPC1, we did not observe significant co-localization between late endosomes and lysosomes regardless of CQ treatment (Fig. 3B). These results are consistent with results obtained in Fig. 2 that show that cells with mutations in NPC1 have inefficient lysosome/late endosome fusion using the FRET assay. We subsequently examined how fibroblasts with mutations in NPC2 (both GM17910 and GM18455) responded to high concentrations of CQ. Interestingly, we observed significant amine-induced late endosome/lysosome co-localization in cells lacking functional NPC2, similar to the observations using wild type cells (Fig. 3, C and D).
FIGURE 3.
NPC1, but not NPC2, is required for amine (CQ)-induced co-localization of lysosomes with late endosomes. A, under normal cell culture conditions very few lysosomes co-localize with late endosomes, as was observed from immunofluorescence analysis of cells probed for the cation-independent (CI)-MPR and LAMP1 (late endosome- and lysosome-specific proteins, respectively). When wild type cells are exposed to high concentrations of the lysosomotropic amine (100 μm CQ for 3 h), the degree of lysosome co-localization with late endosomes was significantly enhanced. B, cells with mutations in NPC1 did not show enhanced late endosome/lysosome co-localization when exposed to CQ. C and D, cells with mutations in NPC2 (both GM17910 and GM18455 cell lines) behaved similarly to wild type cells in their response to CQ treatment. Images are representative of cells observed from at least three independent experiments. The percent co-localization is shown in each merge panel.
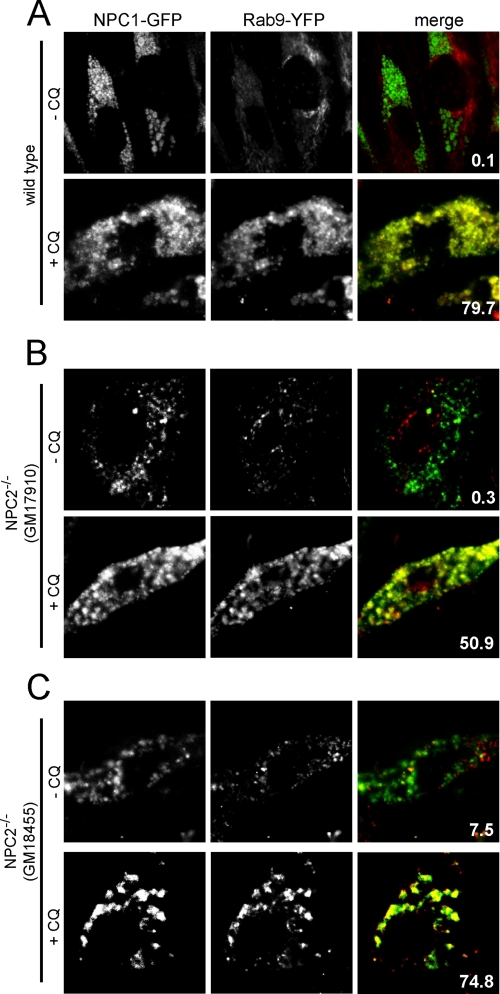
In an effort to support our results presented in Fig. 3, we used a different experimental approach to evaluate the influence of amine treatment on late endosome/lysosome co-localization. To accomplish this we transiently transfected fibroblasts with functional NPC1-GFP and with Rab9-YFP and again used the amine CQ. NPC1 has been reported to exist on both lysosomes and late endosomes (25), whereas Rab9 has been shown to be specific to late endosomes (33). Despite the fact that NPC1 has been reported to reside on both late endosomes and lysosomes, we observed little co-localization with Rab9-YFP in wild type cells with no CQ added to the culture medium (Fig. 4A). When the CQ was added to wild type fibroblasts, we observed significant co-localization of the two fluorescent fusion proteins (Fig. 4A). As with previous results, amines induced significant co-localization of late endosomes with lysosomes in cells with dysfunctional NPC2 regardless of the location of the mutation (Fig. 4, B and C). Collectively, these analyses are consistent with our quantitative FRET-based late endosome/lysosome fusion assay results (Fig. 2) that support the notion that NPC1 is required for amine-induced hybrid organelle formation, which is not the case for NPC2.
FIGURE 4.
Amine-induced co-localization of NPC1 and Rab9 fluorescent fusion proteins. Fibroblasts were transfected with plasmids to express NPC1-GFP (a late endosome/lysosomal protein) and Rab9-YFP (a late endosomal protein) and, where indicated, incubated with 100 μm CQ for 3 h to observe the influence of the amine on the degree of co-localization of these two fluorescent fusion proteins in living cells. A, Rab9-YFP did not significantly co-localize with NPC1-GFP in wild type cells without amine treatment. When wild type cells were exposed to CQ, the fluorescent fusion proteins became significantly co-localized. B and C, the amine CQ enhanced Rab9-YFP/NPC1-GFP co-localization in cells with mutations in NPC2 (both GM17910 and GM18455 cell lines), as was the case with wild type cells. Images are representative of cells observed from at least three independent experiments. The percent co-localization is shown in each merge panel.
NPC2 and Amine-induced Vacuolization of Lysosomes
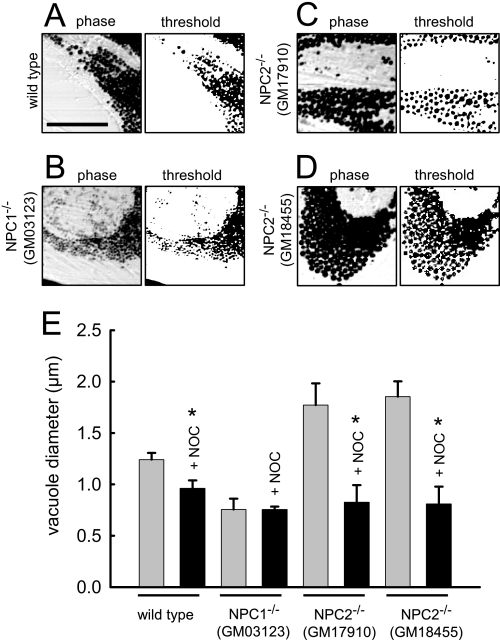
It has been well established that certain lysosomotropic amines induce the formation of large vacuoles when added to culture medium in high concentrations (34–36). We have previously shown that the amine-induced lysosome vacuolization involves fusion of lysosomes with late endosomes and requires functional NPC1 (12). Here we investigated if NPC2, like NPC1, is required for amine-induced vacuolization of lysosomes. To examine this we incubated fibroblasts with 70 μm NR for 6 h and visualized the cells using microscopy. As shown in Fig. 5A, the accumulation of NR in the vacuoles darkens their appearance, which improves our ability to evaluate their size. We utilized ImageJ software to estimate the average diameter of amine-induced vacuoles. The software identifies areas in micrographs with large changes in contrast, which represent NR-containing vacuoles. The computer software-generated threshold images of cells are shown to the right of all representative phase contrast images. We selected parameters in the software program that only identified vacuoles for sizing that had well defined circularity (>90%). This analysis allowed us to quantitatively estimate the average NR-containing vacuole diameter in the cells after amine treatment. Using this approach we estimated that the mean diameter of NR-containing vacuoles was 1.24 ± 0.07 μm in wild type fibroblasts (Fig. 5, A and E). Using a different experimental approach to estimate amine-containing vacuole size (i.e. manual sizing), we had previously reported that the average diameter of vacuoles from fibroblasts with mutated NPC1 were significantly smaller than those observed in wild type fibroblasts (12). Consistent with these results, we show here that cells with the mutant NPC1 form vacuoles that are 0.75 ± 0.11 μm, which is significantly smaller than vacuoles from normal cells (Fig. 5, B and E).
FIGURE 5.
Evaluation of amine-induced vacuole size in fibroblasts with mutations in NPC1 and NPC2. Indicated fibroblasts were incubated with 70 μm NR for 6 h and visualized by phase contrast microscopy. To the right of each phase image is the computer software-generated threshold image of the amine-induced vacuoles that was used to estimate their average diameter. A, wild type cells are shown. B, cells with mutations in NPC1 are shown. C and D, cells with mutations in NPC2 (both GM17910 and GM18455 cell lines) are shown. E, bars represent the average diameter of NR-containing vacuoles in the indicated cell type. Indicated cells (nocodazole (+NOC), black bars) were treated with the microtubule depolymerizing agent nocodazole (1 μm, 1 h before and concurrent with the administration of NR). Bars represent the average ± S.E. of six different cells obtained in three independent experiments (*, p < 0.05 by unpaired t test).
Based on our findings in this report, we anticipated that NPC2 mutant cells would form amine-induced vacuoles that were of similar size to wild type cells when treated with amines. Unexpectedly, both of the NPC2 mutant cell lines (GM18455 and GM17910) formed significantly larger amine-induced vacuoles than were formed in wild type cells (Fig. 5, C and D). A summary of the amine-induced vacuole sizes for each cell line examined is shown in Fig. 5E. To illustrate that the formation of enlarged amine-induced vacuoles is a vesicle-mediated event, we have repeated each of the aforementioned experiments in cells that were preincubated with the microtubule depolymerizing agent nocodazole. When in the presence of nocodazole, the size of amine-induced vacuoles is relatively small and not significantly different from the size of amine-induced vacuoles obtained from cells with mutated NPC1 without nocodazole pretreatment.
The finding that cells with mutations in NPC2 formed amine-induced vacuoles that were significantly larger than those observed in wild type cells was intriguing. After amine incubation, we propose that the observed size of the amine-induced vacuoles is determined by their rate of formation and their rate of depletion. Accordingly, NPC2 mutant cells could have enhanced vacuole formation rates and/or impaired ability to undergo the necessary fission events that would be required for their reduction in size.
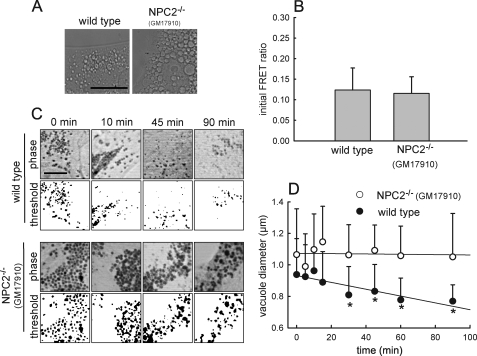
Our data presented in Fig. 2 indicated that hybrid organelle formation rates were nearly identical between NPC2 mutant and wild type fibroblasts; however, these rates were obtained in cells without amine treatment. Accordingly, we examined the possibility that the addition of amines had a different influence on the rates of hybrid organelle formation in wild type versus NPC2 mutant cells. We utilized the amine CQ for these evaluations because NR has fluorescence that interferes with the FRET-based late endosome/lysosome fusion assay. Similar to the results obtained with NR, cells treated with CQ that harbored mutations in NPC2 formed significantly larger amine-induced vacuoles compared with wild type cells (Fig. 6A). We observed no significant differences in hybrid organelle formation rates between wild type and NPC2 mutant cells in the presence of the amine CQ (Fig. 6B).
FIGURE 6.
Kinetics of amine-induced vacuole formation and size reduction. A, phase contrast images are shown of wild type and NPC2 mutant fibroblasts incubated for 3 h with 100 μm CQ to form amine-induced vacuoles. The size of the amine-induced vacuoles is greater in cells with mutations in NPC2 relative to wild type cells (images are representative of three independent experiments; the scale bar is 20 μm). B, kinetics are shown of retrograde fusion of lysosomes with late endosomes using the FRET assay in wild type and NPC2 mutant fibroblasts treated with 100 μm CQ for 30 min before data analysis. Data points represent the average ± S.D. for three independent experiments. C, wild type and NPC2 mutant fibroblasts incubated with 70 μm NR and visualized by phase contrast microscopy are shown. The indicated times represent how long the cells have been in cell culture medium devoid of NR (sink conditions). Below each phase image is the computer software-generated threshold image of the amine-induced vacuoles that were used to estimate their average diameter. Images are representative of at least 10 independent observations. The scale bar represents 10 μm. D, kinetic analysis is shown of the NR-containing vacuole diameter reduction as a function of time in NR-free medium in normal (●) and NPC2 mutant (○) fibroblasts. Data points represent the average ± S.E. for 10 independent experiments (*, p < 0.05 by unpaired t test).
In light of the previous results, we postulated that NPC2 fibroblasts may have impaired hybrid organelle reduction rates relative to rates observed with wild type cells. To evaluate this we investigated the kinetics of amine-induced hybrid organelle diameter reduction as a function of time after the amine was removed from the cell culture medium. At the indicated times after amine depletion, the cells were fixed with formaldehyde and viewed using microscopy. ImageJ software was utilized to quantitatively estimate the vacuole size as a function of time. Representative images of both NPC2 mutant and wild type fibroblasts illustrate qualitatively that NPC2 mutant fibroblasts have vacuoles that retain their large diameter for longer periods of time relative to wild type fibroblasts (Fig. 6C). When the images were quantitatively evaluated with the software, we observed significantly slower rates of vesicle diameter reduction in NPC2 mutant cells relative to wild type cells even though errors were quite large (Fig. 6D, p < 0.05, n = 10). It is important to note that we utilized NR for vacuole size reduction studies because it enabled efficient vacuole identification using the software quantitation approach. We qualitatively observed similar slow vacuole reduction rates with NPC2 mutant cells treated with CQ (data not shown). These observations are consistent with the notion that NPC2 mutant fibroblasts yield excessively large amine-induced vacuoles because they have an impaired ability to release vesicles from the nascent hybrid organelles.
DISCUSSION
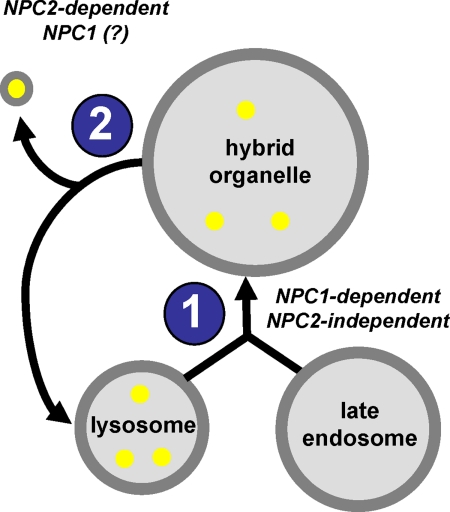
Luzio et al. (29) have previously proposed a model for lysosome dynamics that is relevant to this work. They suggest that lysosomes continuously fuse with late endosomes to form hybrid organelles, which are larger in size compared with the lysosomes. It is also thought that hybrid organelles must continuously reform into lysosomes so as to avoid their consumption in this dynamic process (37–39). Because lysosomes are denser than hybrid organelles, it is reasonable to assume that membrane fission events must take place during the lysosome reformation process. We propose that it is likely that membrane vesicles released from hybrid organelles during these fission events may contain cargo that is to be directed to different cellular locations, including the plasma membrane. A model illustrating these events is shown in Fig. 7. Included in this illustration are the steps in which NPC1 and NPC2 function, consistent with the observations reported in this work.
FIGURE 7.
Proposed model illustrating the initial steps involved in the retrograde transport of membrane-impermeable lysosomal cargo. Lysosomes containing membrane-impermeable cargo (yellow circles) fuse with late endosomes to create hybrid organelles (labeled as step 1). Fission of membrane vesicles from hybrid organelles leads to the reformation of lysosomes and the release of impermeable cargo-containing transport vesicles (labeled as step 2). The suggested role of NPC proteins in each step is indicated.
We have previously shown that certain lysosomotropic amines stimulate late endosome/lysosome fusion and result in the formation of visibly expanded hybrid organelles in normal fibroblasts, a process that was shown to be impaired in cells lacking functional NPC1 (12). In this work we evaluated how cells with mutations in NPC2 responded to the addition of lysosomotropic amines in the culture medium. Interestingly, cells with mutations in NPC2 not only formed hybrid organelles but formed significantly larger ones than those found in wild type cells. We propose that the resultant size of amine-induced hybrid organelles is a function of their rate of formation (step 1, Fig. 7) and the rate at which membrane vesicles are removed from them (step 2, Fig. 7). We found that cells with mutations in NPC2 had rates of hybrid organelle formation that were not statistically different from wild type cells (see Fig. 2). Interestingly, we found that the hybrid organelle size reduction, upon removal of the amines, was impaired in cells with mutations in NPC2 (see Fig. 6). Accordingly, our data are consistent with the notion that cells with mutations in NPC2 form larger hybrid organelles because they are less efficient at membrane fission events associated with hybrid organelle size reduction.
Importantly, these observations suggested to us that NPC1 and NPC2 may function in separate events involved in the retroendocytic transport of membrane-impermeable cargo such as protonated lysosomotropic amines. We and others have shown that NPC1 is indeed involved in the retrograde transport of multiple types of membrane impermeable cargo (12, 18); however, to our knowledge, no prior studies have comparatively evaluated if cells with functional mutations in NPC2 function comparatively to cells harboring functional mutations in NPC1 in the cellular egress of this type of lysosomal cargo. To evaluate this further, we investigated the cell efflux kinetics of membrane-impermeable dextran polymers specifically localized to lysosomes in cells with deficiencies in NPC1, NPC2, or both. As previously shown, cells with mutations in NPC1 had significantly impaired dextran secretion profiles relative to wild type cells (Fig. 1A). Interestingly, cells with functional mutations in NPC2 secreted dextran less efficiently than did cells with mutations in NPC1 (Fig. 1A). Moreover, cells mimicking the NPC1/NPC2 double mutant had shown significantly less dextran secretion compared with either single mutant cell line (Fig. 1A). These data suggest that these proteins both contribute to the vesicle-mediated egress of membrane-impermeable lysosomal cargo but in distinctly separate steps.
This observation may appear to contradict the work of others that have evaluated the roles of NPC1 and NPC2 in cholesterol trafficking from late endocytic compartments. Most noteworthy, Sleat et al. (8) established that NPC1 and NPC2 function concertedly and non-redundantly in cholesterol trafficking in mice. More recently, Infante et al. (40) have provided evidence to support the mechanism by which NPC1 and NPC2 could function in a concerted mechanism to mobilize cholesterol from lysosomes. It is important to state that this would only truly present a contradiction if cholesterol and other lipids follow the exact same intracellular trafficking pathway as do dextran molecules. To address this possibility, we performed both cholesterol esterification and cholesterol efflux assays using the same cells evaluated in this study to see if we could observe phenotypic differences in cells with mutations in either NPC1 or NPC2. Results from these evaluations were virtually identical to previously established trends (16). Specifically, both NPC1- and NPC2-deficient cells have shown marked impairments in both cholesterol esterification and efflux (see supplemental Figs. 3 and 4). Importantly, we observed no differences between cells with mutations in NPC1 compared with those with mutations in NPC2. Comparing these experimental findings with our results presented here strongly suggests that dextran and cholesterol do not appear to precisely follow the same NPC protein-mediated retroendocytic trafficking pathway. Accordingly, we do not believe that the findings reported here should be interpreted as providing evidence that NPC1 and NPC2 do not function cooperatively in the mobilization of cholesterol and possibly other lipids from late endocytic compartments.
It is highly possible that NPC proteins aid in the trafficking of different cargo through different mechanisms. In support of this notion, Tang et al. (20) have recently investigated the involvement of NPC1 in HIV-1 post-entry replication. Importantly, the authors show that NPC1 was essential for proper trafficking of HIV-1 Gag protein from late endocytic compartments. The authors demonstrated that overexpression of Rab9 in NPC1-deficient cells, which has been shown to promote cholesterol and glycosphingolipid clearance from late endocytic compartments (41, 42), failed to rescue Gag hyper-accumulation in these compartments. This result suggests that NPC protein-mediated trafficking of cholesterol may be separate from the trafficking of Gag (a membrane-impermeable cargo) from late endocytic compartments.
It is important to consider how these results may impact what is currently understood about NPC disease etiology and progression. It is our belief that the progressive neurodegeneration associated with NPC disease likely results from disruption of the cellular homeostasis of multiple lysosomal cargos, both lipid in nature and the soluble membrane impermeable-type examined here. We have recently shown that cells with non-functional NPC1 were more susceptible to the toxic effects of an endogenous lysosomal, membrane-impermeant polyamine metabolite (3-aminopropanal), relative to wild type cells. 3-Aminopropanal has been previously shown to be a potent neurotoxin implicated in neurodegenerative disorders (43–46). It is thought to cause its toxic effects through its ability to react with and disrupt lysosomal membranes once sequestered in this compartment. Accordingly, we hypothesized that NPC cells were more sensitive to 3-aminopropanl because these cells were relatively inefficient in clearing the molecule from late endocytic compartments. In the present work, we show that the egress of membrane-impermeant lysosomal cargo is significantly impaired in cells with functional mutations in either NPC1 or NPC2. Consequently, if neurodegeneration associated with NPC disease is due in part to the hyperaccumulation of toxic lysosomotropic metabolites, it is likely that subjects with mutations in either NPC1 or NPC2 may appear clinically indistinguishable, which is consistent with previous observations (8). It is obvious that more work will be required in this area to elucidate the potential contributions of endogenous lysosomotropic metabolites on NPC disease etiology.
Collectively, the work in this report reveals unique phenotypic differences between NPC1 and NPC2 in specific stages of retrograde transport of lysosomal cargo, namely membrane-impermeable cargo and amines. Currently, the precise molecular basis for NPC disease progression is far from being completely understood. Considering this, we believe that it is imperative that we improve our understanding regarding the fundamental roles that NPC proteins play in retroendocytic intracellular trafficking events of all possible types of lysosomal cargo.
Supplementary Material
Acknowledgments
We thank D. Moore and H. Shinogle at the University of Kansas Microscopy and Analytical Imaging facility for help with cell imaging studies. We thank Suzanne Pfeffer and Mathew Scott (Stanford University) for the Rab9-YFP and NPC1-GFP constructs, respectively. We thank Daniel Ory (Washington University) for the M12 cells. We also thank Dr. Peter Lobel and his group for providing the MEFs and for support.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA106655.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- NPC
- Niemann-Pick type C disease
- NPC1
- Niemann-Pick C1 protein
- NPC2
- Niemann-Pick C2 protein
- LAMP1
- lysosome-associated membrane protein-1
- AF
- AlexaFluor
- MPR
- mannose 6-phosphate receptor
- NR
- neutral red
- CQ
- chloroquine
- FRET
- fluorescence resonance energy transfer
- MEF
- mouse embryonic fibroblast
- HIV
- human immunodeficiency virus
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- LDL
- low density lipoprotein
- D-PBS
- Dulbecco's phosphate-buffered saline
- GFP
- green fluorescent protein
- siRNA
- small interfering RNA
- YFP
- yellow fluorescent protein
- GM2
- GaINAcβ4(Neu5Acα3)Galβ4GlcCer.
REFERENCES
- 1.Liscum L., Sturley S. L. (2004) Biochim. Biophys. Acta 1685, 22–27 [DOI] [PubMed] [Google Scholar]
- 2.Carstea E. D., Morris J. A., Coleman K. G., Loftus S. K., Zhang D., Cummings C., Gu J., Rosenfeld M. A., Pavan W. J., Krizman D. B., Nagle J., Polymeropoulos M. H., Sturley S. L., Ioannou Y. A., Higgins M. E., Comly M., Cooney A., Brown A., Kaneski C. R., Blanchette-Mackie E. J., Dwyer N. K., Neufeld E. B., Chang T. Y., Liscum L., Strauss J. F., 3rd, Ohno K., Zeigler M., Carmi R., Sokol J., Markie D., O'Neill R. R., van Diggelen O. P., Elleder M., Patterson M. C., Brady R. O., Vanier M. T., Pentchev P. G., Tagle D. A. (1997) Science 277, 228–2319211849 [Google Scholar]
- 3.Naureckiene S., Sleat D. E., Lackland H., Fensom A., Vanier M. T., Wattiaux R., Jadot M., Lobel P. (2000) Science 290, 2298–2301 [DOI] [PubMed] [Google Scholar]
- 4.Pentchev P. G., Comly M. E., Kruth H. S., Tokoro T., Butler J., Sokol J., Filling-Katz M., Quirk J. M., Marshall D. C., Patel S. (1987) FASEB J. 1, 40–45 [DOI] [PubMed] [Google Scholar]
- 5.Liscum L., Ruggiero R. M., Faust J. R. (1989) J. Cell Biol. 108, 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Evans E., Morgan A. J., He X., Smith D. A., Elliot-Smith E., Sillence D. J., Churchill G. C., Schuchman E. H., Galione A., Platt F. M. (2008) Nat. Med. 14, 1247–1255 [DOI] [PubMed] [Google Scholar]
- 7.Zervas M., Dobrenis K., Walkley S. U. (2001) J. Neuropathol. Exp. Neurol. 60, 49–64 [DOI] [PubMed] [Google Scholar]
- 8.Sleat D. E., Wiseman J. A., El-Banna M., Price S. M., Verot L., Shen M. M., Tint G. S., Vanier M. T., Walkley S. U., Lobel P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5886–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanier M. T., Millat G. (2003) Clin. Genet. 64, 269–281 [DOI] [PubMed] [Google Scholar]
- 10.Subramanian K., Balch W. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15223–15224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Duve C., de Barsy T., Poole B., Trouet A., Tulkens P., Van Hoof F. (1974) Biochem. Pharmacol. 23, 2495–2531 [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann A. M., Krise J. P. (2008) J. Biol. Chem. 283, 24584–24593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millard E. E., Srivastava K., Traub L. M., Schaffer J. E., Ory D. S. (2000) J. Biol. Chem. 275, 38445–38451 [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann A. M., Goldman S. D., Krise J. P. (2009) Anal. Biochem. 386, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganley I. G., Pfeffer S. R. (2006) J. Biol. Chem. 281, 17890–17899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millard E. E., Gale S. E., Dudley N., Zhang J., Schaffer J. E., Ory D. S. (2005) J. Biol. Chem. 280, 28581–28590 [DOI] [PubMed] [Google Scholar]
- 17.Krieger M., Brown M. S., Faust J. R., Goldstein J. L. (1978) J. Biol. Chem. 253, 4093–4101 [PubMed] [Google Scholar]
- 18.Neufeld E. B., Wastney M., Patel S., Suresh S., Cooney A. M., Dwyer N. K., Roff C. F., Ohno K., Morris J. A., Carstea E. D., Incardona J. P., Strauss J. F., 3rd, Vanier M. T., Patterson M. C., Brady R. O., Pentchev P. G., Blanchette-Mackie E. J. (1999) J. Biol. Chem. 274, 9627–9635 [DOI] [PubMed] [Google Scholar]
- 19.Gong Y., Duvvuri M., Duncan M. B., Liu J., Krise J. P. (2006) J. Pharmacol. Exp. Ther. 316, 242–247 [DOI] [PubMed] [Google Scholar]
- 20.Tang Y., Leao I. C., Coleman E. M., Broughton R. S., Hildreth J. E. K. (2009) J. Virol. 83, 7982–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M., Dwyer N. K., Love D. C., Cooney A., Comly M., Neufeld E., Pentchev P. G., Blanchette-Mackie E. J., Hanover J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4466–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C. S., Bach G., Pagano R. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6373–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolhuis P. A., Oonk J. G., Kamp P. E., Ris A. J., Michalski J. C., Overdijk B., Reuser A. J. (1987) Neurology 37, 75–81 [DOI] [PubMed] [Google Scholar]
- 24.Jatzkewitz H., Pilz H., Sandhoff K. (1965) J. Neurochem. 12, 135–144 [DOI] [PubMed] [Google Scholar]
- 25.Ko D. C., Gordon M. D., Jin J. Y., Scott M. P. (2001) Mol. Biol. Cell 12, 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNeil P. L. (2002) J. Cell Sci. 115, 873–879 [DOI] [PubMed] [Google Scholar]
- 27.Idone V., Tam C., Goss J. W., Toomre D., Pypaert M., Andrews N. W. (2008) J. Cell Biol. 180, 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez A., Webster P., Ortego J., Andrews N. W. (1997) J. Cell Biol. 137, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luzio J. P., Rous B. A., Bright N. A., Pryor P. R., Mullock B. M., Piper R. C. (2000) J. Cell Sci. 113, 1515–1524 [DOI] [PubMed] [Google Scholar]
- 30.Luzio J. P., Pryor P. R., Gray S. R., Gratian M. J., Piper R. C., Bright N. A. (2005) Biochem. Soc. Symp. 72, 77–86 [DOI] [PubMed] [Google Scholar]
- 31.Rabinowitz S., Horstmann H., Gordon S., Griffiths G. (1992) J. Cell Biol. 116, 95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jahraus A., Storrie B., Griffiths G., Desjardins M. (1994) J. Cell Sci. 107, 145–157 [DOI] [PubMed] [Google Scholar]
- 33.Barbero P., Bittova L., Pfeffer S. R. (2002) J. Cell Biol. 156, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morissette G., Moreau E., C-Gaudreault R., Marceau F. (2004) J. Pharmacol. Exp. Ther. 310, 395–406 [DOI] [PubMed] [Google Scholar]
- 35.Solheim A. E., Seglen P. O. (1983) Biochim. Biophys. Acta 763, 284–291 [DOI] [PubMed] [Google Scholar]
- 36.Solheim A. E., Seglen P. O. (1983) Biochem. J. 210, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullock B. M., Bright N. A., Fearon C. W., Gray S. R., Luzio J. P. (1998) J. Cell Biol. 140, 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pryor P. R., Mullock B. M., Bright N. A., Gray S. R., Luzio J. P. (2000) J. Cell Biol. 149, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bright N. A., Reaves B. J., Mullock B. M., Luzio J. P. (1997) J. Cell Sci. 110, 2027–2040 [DOI] [PubMed] [Google Scholar]
- 40.Infante R. E., Wang M. L., Radhakrishnan A., Kwon H. J., Brown M. S., Goldstein J. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15287–15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter M., Davies J. P., Ioannou Y. A. (2003) J. Lipid Res. 44, 243–253 [DOI] [PubMed] [Google Scholar]
- 42.Choudhury A., Dominguez M., Puri V., Sharma D. K., Narita K., Wheatley C. L., Marks D. L., Pagano R. E. (2002) J. Clin. Invest. 109, 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood P. L., Khan M. A., Kulow S. R., Mahmood S. A., Moskal J. R. (2006) Brain Res. 1095, 190–199 [DOI] [PubMed] [Google Scholar]
- 44.Wood P. L., Khan M. A., Moskal J. R. (2007) Brain Res. 1145, 150–156 [DOI] [PubMed] [Google Scholar]
- 45.Virgili M., Crochemore C., Peña-Altamira E., Contestabile A. (2006) Neurochem. Int. 48, 201–207 [DOI] [PubMed] [Google Scholar]
- 46.Morrison L. D., Kish S. J. (1995) Neurosci. Lett. 197, 5–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.