Abstract
Toll-like receptor-4 (TLR4) is the receptor for bacterial lipopolysaccharide, yet it may also respond to a variety of endogenous molecules. Necrotizing enterocolitis (NEC) is the leading cause of death from gastrointestinal disease in newborn infants and is characterized by intestinal mucosal destruction and impaired enterocyte migration due to increased TLR4 signaling on enterocytes. The endogenous ligands for TLR4 that lead to impaired enterocyte migration remain unknown. High mobility group box-1 (HMGB1) is a DNA-binding protein that is released from injured cells during inflammation. We thus hypothesize that extracellular HMGB1 inhibits enterocyte migration via activation of TLR4 and sought to define the pathways involved. We now demonstrate that murine and human NEC are associated with increased intestinal HMGB1 expression, that serum HMGB1 is increased in murine NEC, and that HMGB1 inhibits enterocyte migration in vitro and in vivo in a TLR4-dependent manner. This finding was unique to enterocytes as HMGB1 enhanced migration of inflammatory cells in vitro and in vivo. In seeking to understand the mechanisms involved, TLR4-dependent HMGB1 signaling increased RhoA activation in enterocytes, increased phosphorylation of focal adhesion kinase, and increased phosphorylation of cofilin, resulting in increased stress fibers and focal adhesions. Using single cell force traction microscopy, the net effect of HMGB1 signaling was a TLR4-dependent increase in cell force adhesion, accounting for the impaired enterocyte migration. These findings demonstrate a novel pathway by which TLR4 activation by HMGB1 delays mucosal repair and suggest a novel potential therapeutic target in the amelioration of intestinal inflammatory diseases like NEC.
Keywords: Cell/Migration, Immunology/Innate Immunity, Immunology/LPS, Immunology/Toll Receptors, Receptors/Extracellular Matrix, Receptors/Toll-like, Tissue/Organ Systems/Intestine, Tissue/Organ Systems/Epithelium
Introduction
Toll-like receptor 4 (TLR4)3 is the receptor for bacterial lipopolysaccharide (LPS) on inflammatory cells (1), yet it may also respond to a variety of endogenous molecules that are collectively termed “DAMPS,” for damage-associated molecular patterns (2). Necrotizing enterocolitis (NEC) is the leading cause of death from gastrointestinal disease in newborn infants and is characterized by destruction of the intestinal mucosa (3) and increased TLR4 signaling within enterocytes (4, 5). We have shown that increased enterocyte TLR4 signaling leads to reduced enterocyte migration and impaired mucosal healing, leading to the development of NEC (4, 6). High mobility group box-1 (HMGB1) is a DNA-binding protein that is released from injured cells, where it possesses cytokine-like properties (7), and can be secreted by macrophages in culture (8). Although HMGB1 has been shown to activate TLR4 on immune cells (9), a potential link between HMGB1 and TLR4 signaling in non-immune cells such as enterocytes remains largely unexplored. However, our recent demonstration that NEC is characterized by TLR4-dependent impaired enterocyte migration (4) raises the spotlight on the consequences of TLR4 signaling in the intestinal epithelium and pinpoints the search for potential endogenous molecules that may serve as TLR4 ligands. We now hypothesize that extracellular HMGB1 activates TLR4 on enterocytes in vitro and in vivo, leading to reduced enterocyte migration, and sought to define the pathways involved.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
Intestinal epithelial cell (IEC)-6 cells and RAW264.7 macrophages were obtained from ATCC (Manassas, VA) and maintained as described (6). The source of antibodies is provided in the supplemental methods. RhoA activation was measured using the Rhotekin precipitation assay according to the manufacturer's protocol (Cytoskeleton, Denver, CO). The immunoanalysis of cultured enterocytes, mouse, and human intestine was performed as described previously (6) and evaluated using an Olympus Fluoview 1000 confocal microscope under oil-immersion objectives. SDS-PAGE was performed as described previously (6). The images on radiographic film were quantified using a GS700 Bio-Rad densitometer and Quantity One analysis software. Adenoviruses expressing wild-type or dominant negative (P712H) TLR4 with the C-terminal fusion of GFP were constructed using the Adeno-X Expression System 2 (Clontech) according to the manufacturer's protocol, as described in the supplemental methods and in our previous work (10).
Isolation and Purification of Recombinant HMGB1 from Yeast
To avoid the potential for endotoxin contamination and thus inadvertent activation of TLR4 in experiments involving HMGB1, we elected to purify HMGB1 from yeast. To do so, the full-length cDNA for human HMGB1 was subcloned in-frame to the secretion signal of the FLAG expression vector, YEpFLAG (Sigma), modified to eliminate the FLAG cassette. This vector was then transformed into the protease-deficient yeast strain BJ3505. The transformed yeast were grown at 30 °C for 3 days on a rotary shaker at 175 rpm in 500 ml of expression medium (1% glucose, 3% glycerol, 1% yeast extract, 2% peptone, 100 mm potassium phosphate, pH 6.4) as described (11). The culture was then chilled at 4 °C and centrifuged (10 min, 1000 × g), the supernatant was centrifuged (10,000 × g, 15 min), and ammonium sulfate was added to a final concentration of 65%. After 30 min at 4 °C, the mixture was centrifuged for 15 min, 10,000 × g, and ammonium sulfate was added to bring the final concentration to 80%, which was incubated for 30 min at 4 °C. The mixture was then centrifuged 15 min at 10,000 × g, the supernatant was discarded, and the pellet was dissolved in 20 mm TrisCl, 150 mm, KCl, 1 mm dithiothreitol, pH 8.0, and dialyzed versus 2 × 1 liter of same. The sample was then loaded onto an Econo-Pac High Q column (Bio-Rad), washed with 20 mm TrisCl, 220 mm Kcl, 1 mm dithiothreitol, pH 8.0, and then eluted with a gradient of 220 mm KCl to 700 mm KCl in the same TrisCl-dithiothreitol buffer. Aliquots from the elution profile were run on 12% PAGE and analyzed by Western blot and protein staining (GelCode Blue, Pierce). Pure fractions were pooled and then dialyzed versus 25 mm TrisCl, 150 mm KCl, 2 mm dithiothreitol, pH 8.0. Protein concentration was determined by BCA (Pierce). To further guard against inadvertent contamination with LPS in the HMGB1 preparations, polymyxin B (Sigma-Aldrich) was added to HMGB1 (50 units/μg of recombinant protein).
Enterocyte-Macrophage Co-culture System
To assess whether activated or necrotic macrophages could affect the migration of adjacent enterocytes via release of HMGB1, IEC-6 cells were plated on the bottom of a six-well plate, RAW264.7 macrophages were plated on a 0.4-μm Transwell insert (Corning, Corning, NY), and IEC-6 migration was assessed using the wound-scraping assay as we have described (6) in the presence of either necrotic or activated macrophages. Necrotic macrophages were prepared via freezing (−80 °C) and thawing (37 °C) cycles as described. Macrophages were activated by treatment with IFNγ (100 units/ml, Sigma-Aldrich). Polyclonal HMGB1 antibody was added at 10 μg/ml into the bottom chamber.
Cell Traction Force Microscopy
For single cell determination of cell-matrix adhesiveness, cell traction force microscopy was performed. This technique specifically and sensitively measures cell contraction force based on substrate surface deformations caused by cells spreading on a thin elastic surface (12). In brief, polyacrylamide gels embedded with 0.5-μm red fluorescent Microbeads (Molecular Probes, Eugene, OR) were assembled according to the methods of Chen et al. (13). The gels were then covered with a solution of sulfo-SANPAH (Pierce), exposed to UV for 10 min, and incubated with 100 μg/ml type I collagen solution (Angiotech BioMaterials, Palo Alto, CA) overnight at 4 °C. After rinsing with PBS three times, 4,000 cells, some of which were transfected 48 h earlier with wild-type adenoviral TLR4, dominant negative adenoviral TLR4, or adenoviral GFP as a control, were seeded on a polyacrylamide gel dish and cultured at 37 °C in 5% CO2 for 6 h. To perform cell traction force microscopy, a phase contrast image of individual cells and a fluorescence image of the embedded fluorescent beads were taken under the same view. Cells were then removed by trypsinization, and another fluorescence image of the beads in the same view and same z-plane was taken. Based on this set of images, cell traction forces were obtained by computation using the MATLAB program according to published algorithms (14).
In Vitro and in Vivo Determination of Enterocyte Migration, Macrophage Chemotaxis, and the Induction of Necrotizing Enterocolitis
Detailed information regarding these procedures and the relevant statistical analyses has been published from our group elsewhere (4, 10) and can be found in the supplemental methods.
RESULTS
HMGB1 Expression Is Increased in the Intestinal Mucosa in Humans and Mice with Necrotizing Enterocolitis and Inhibits Enterocyte Migration in a TLR4-dependent Manner
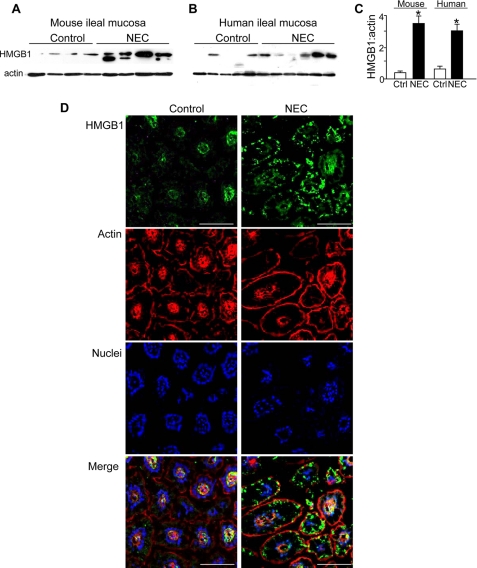
To investigate the effects of HMGB1 on enterocytes, we first examined its expression in the intestinal mucosa in a disease characterized by increased TLR4 signaling and reduced enterocyte migration, namely necrotizing enterocolitis (10). As shown in Fig. 1, HMGB1 expression was significantly increased in the intestinal mucosa of human infants (panels A and C) and mice (panels B and C) with NEC when compared with healthy controls. This finding is in agreement with previous work performed in a rat model of NEC in which HMGB1 was shown to be elevated, yet neither its relevance nor a potential receptor for HMGB1 was explored in that prior work (15). As shown in Fig. 1D, in normal intestinal tissue, HMGB1 was distributed in a pattern that was largely associated with the nuclear compartment. By contrast, in mice with NEC, HMGB1 was found in aggregates scattered at the periphery of the damaged mucosa, consistent with the release of this molecule during intestinal injury. By enzyme-linked immunosorbent assay, the concentration of HMGB1 in the serum of mice with NEC was 15 ± 3 ng/ml when compared with 3 ± 3 ng/ml in breast-fed control mice (n = three separate experiments, p < 0.05).
FIGURE 1.
HMGB1 expression is increased in the ileal mucosa in human and murine necrotizing enterocolitis. A–C, SDS-PAGE showing the expression of HMGB1 and F-actin in terminal ileum from mice (A) and human infants (B) with NEC (NEC) and without NEC (Control). Quantification is shown in C (*, p < 0.05, n = three experiments; error bars indicate S.E.). Ctrl, control. D, confocal microscopic expression of HMGB1 (green), F-actin (phalloidin, red), nuclei (Draq-5, blue), and merged image in terminal ileum of mice with and without NEC, results shown are representative of three separate experiments. Bar = 100 μm.
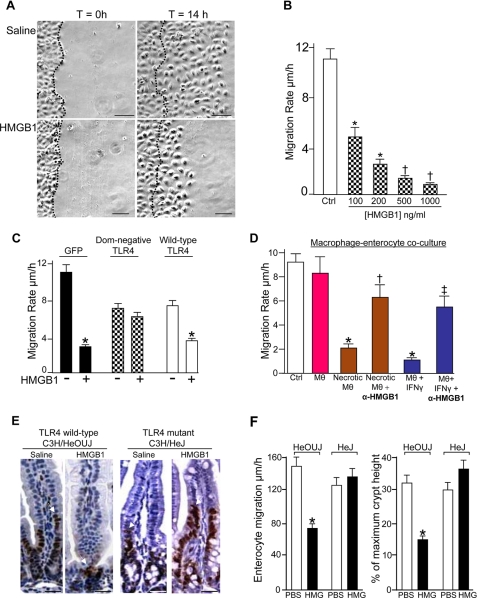
We next sought to determine the effects, if any, of HMGB1 on enterocyte migration and whether TLR4 may be involved. To do so, IEC-6 enterocytes were plated to confluence and allowed to migrate into a scraped wound. As shown in Fig. 2A, control enterocytes healed the wound by ∼14 h, consistent with our previous results (4, 6). Strikingly, HMGB1-treated cells showed a dose-dependent inhibition in the enterocyte migration (Fig. 2B). As a negative control, there was no effect of the addition of CD14 on enterocyte migration, a protein of similar molecular weight to HMGB1 that was synthesized using the same yeast expression system (data not shown).
FIGURE 2.
HMGB1 release inhibits enterocyte migration in vivo and vitro in a TLR4-dependent manner. A, migration of IEC-6 cells in the absence or presence of HMGB1 (100 ng/ml) over 14 h; dashed line indicates leading edge at t = 0, bar = 40 μm. B, effect of HMGB1 on enterocyte migration, n = 5, *, p < 0.05,†, p < 0.005 versus control (Ctrl). C, IEC-6 cell migration after transfection with GFP (black), GFP-dominant negative TLR4 (checkered), or GFP-wild-type TLR4 (white) ± HMGB1 *, p < 0.05 versus paired control. D, IEC-6 migration after co-culture with macrophages (MΘ, pink), being induced to become necrotic (brown), or activation with IFN (blue) ± inhibitory HMGB1 antibodies (α-HMGB1). *, p < 0.05 versus control, †, p < 0.05 versus necrotic macrophages, ‡, p < 0.05 versus IFN-treated macrophages. E, micrographs showing bromodeoxyuridine immunostaining in the terminal ileum of TLR4-wild-type and mutant mice 24 h after injection with saline or HMGB1 (1 μg/kg). Arrows show leading enterocytes. F, the migration rate and percentage of the maximal crypt height achieved by migrating enterocytes as shown; mean ± S.E., 100 cells/experiment, bar = 50 μm.
In consideration of our previous finding that bacterial endotoxin (LPS) inhibits enterocyte migration (6, 16), four separate lines of experimental evidence now exclude the possibility that LPS contamination within the HMGB1 preparation could account for the observation that HMGB1 inhibits enterocyte migration. First, HMGB1 was synthesized in our laboratory from yeast cells, excluding the possibility of bacterial-derived LPS contaminating our preparations. Second, all experiments included the addition of the LPS-neutralizing agent polymyxin B, which would significantly reduce the biological activity of any LPS within any of the buffers. Thirdly, our HMGB1 preparation did not cause either the release of cytokines or the activation of NF-kB in enterocytes, events that do occur in response to even nanomolar concentrations of LPS (not shown). Finally, measurement of our HMGB1 preparation by limulus assay did not demonstrate any measurable endotoxin activity (not shown). Taken together, we are confident that the effects of HMGB1 on enterocyte migration cannot be attributable to LPS contamination, yet rather represent a novel and direct inhibitory effect of this danger molecule on cell movement.
We next sought to define further the initial molecular requirements that mediate the inhibition of enterocyte migration by HMGB1 and focused on the innate immune receptor TLR4, which has been shown to signal in response to HMGB1 in immune cells but not in any other cell type. To do so, we engineered a dominant negative adenovirus with the inhibitory P712H mutation in the cytoplasmic tail of TLR4 based upon the TLR4 mutation in the C3H/HeJ mouse that renders it insensitive to endotoxin (17). As shown in Fig. 2C, treatment of IEC-6 cells with dominant negative TLR4 reversed the inhibitory effects of HMGB1 on enterocyte migration (checkered bars). In two important control experiments, HMGB1 did inhibit enterocyte migration in IEC-6 cells that were transfected with adenoviral-GFP (black bars) and in IEC-6 cells that were transfected with GFP-tagged wild-type TLR4 (white bars), thereby excluding nonspecific effects of either the virus or the infection procedure on enterocyte migration. Taken together, these findings indicate that HMGB1 inhibits enterocyte migration in vitro in a TLR4-dependent manner.
In the next series of studies, we sought to define in greater detail the physiological relevance of the inhibitory effects of HMGB1 on enterocyte migration. HMGB1 was originally shown to be released from necrotic cells or actively secreted by immune cells that had been activated by pro-inflammatory cytokines such as interferon γ (18). To model these parameters in vitro, we utilized a macrophage-enterocyte co-culture system in which macrophages were untreated, induced to become necrotic, or activated to release HMGB1 with the addition of the pro-inflammatory cytokine IFN and then co-incubated with enterocytes in a Transwell apparatus; the enterocytes were then induced to migrate by scraping (see supplemental methods). To define the involvement of extracellular HMGB1 on enterocyte migration, inhibitory antibodies against HMGB1 were added to the upper Transwell. As is shown in Fig. 2D, treatment of enterocytes with necrotic macrophages (brown bars) inhibited enterocyte migration, whereas this effect could be reversed by the addition of inhibitory antibodies to HMGB1. By enzyme-linked immunosorbent assay, necrotic macrophages released >200 ng/ml HMGB1 when compared with live macrophages, which released 2–3 ng/ml. Treatment with IFNγ, which is known to cause the release of HMGB1 from macrophages (18), inhibited enterocyte migration in a manner that could also be reversed by co-treatment with inhibitory antibodies to HMGB1 (blue bars). Taken together, these findings indicate that HMGB1 inhibits enterocyte migration in vitro, either when administered exogenously or when released from adjacent activated macrophages.
To explore the potential in vivo relevance of these findings, we sought to determine whether HMGB1 may affect enterocyte migration in mice, and if so, whether TLR4 signaling may play a role. Enterocyte migration was determined in vivo by assessing the extent of migration of bromodeoxyuridine-labeled enterocytes from the crypts of the terminal ileum along individual villi over time, as we have performed and validated previously (6, 19). As shown in Fig. 2E, treatment of TLR4 wild-type (C3H/HeOUJ) mice with HMGB1 significantly inhibited the rate of enterocyte migration when compared with mice injected with saline alone. A similar inhibition in the rate of migration was observed when half of this concentration was used (not shown). Importantly, injection of TLR4-mutant (i.e. C3H/HeJ) mice with HMGB1 did not affect enterocyte migration (Fig. 2, E and F). These findings indicate that HMGB1 inhibits enterocyte migration in vivo as well as in vitro and suggest a critical role for TLR4 signaling in the process.
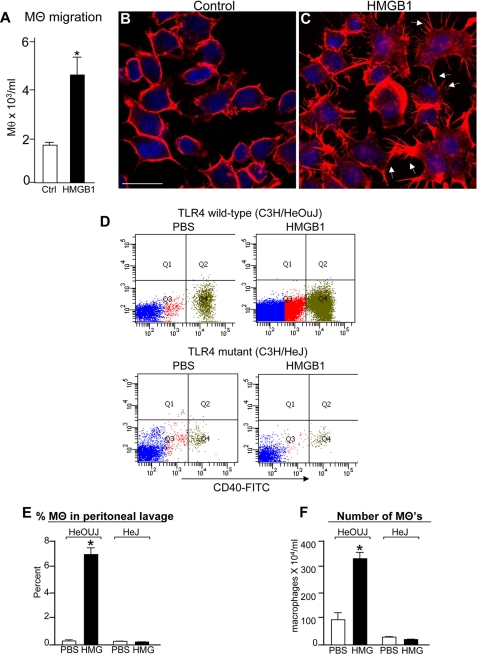
HMGB1 Increases Macrophage Chemotaxis in Vitro and in Vivo
Although we now demonstrate that HMGB1 inhibits enterocyte migration, previous authors have shown that HMGB1 enhances the migration of several cell types at inflammatory sites, especially macrophages (20). Consistent with these prior results, we observed that HMGB1 significantly increased macrophage chemotaxis when compared with PBS (Fig. 3A). Importantly, HMGB1 also caused a dramatic rearrangement in the distribution of F-actin in macrophages, from a uniform appearance within the cytoplasm (Fig. 3B) to the accumulation of actin filopods emanating from the cell periphery, known to be associated with increased macrophage chemotaxis (21) (Fig. 3C, arrows). Furthermore, injection with HMGB1 significantly increased both the proportion (Fig. 3, D and E) and the number (Fig. 3F) of peritoneal macrophages that migrated into the peritoneal cavity of TLR4 wild-type mice (i.e. C3H/HeOUJ) yet had no measurable effect on macrophage migration in TLR4-mutant mice (i.e. C3H/HeJ) (see panel D, quadrant 4 (Q4), and quantification in panel E). These findings indicate that although HMGB1 inhibits enterocyte migration (Fig. 2), HMGB1 serves to enhance the migration of macrophages both in vitro, as well as into the peritoneal cavity, and does so via TLR4. This suggests that the inhibitory effects of HMGB1 on migration may be relatively restricted to enterocytes.
FIGURE 3.
HMGB1 increases macrophage chemotaxis in vitro and in vivo in a TLR4-dependent manner. A, the number of macrophages (MΘ) found to migrate toward formylmethionylleucylphenylalanine by Transwell assay ± HMGB1 (100 ng/ml) *, p < 0.05, three separate experiments. Ctrl, control. B and C, confocal micrographs showing F-actin in RAW264.7 macrophages ± HMGB1 (100 ng/ml). Arrows denote filopod-like extensions, bar = 10 μm, D, flow cytometric analysis of peritoneal lavage effluent obtained from C3H/HeOUJ and C3H/HeJ mice that had been injected with either PBS or HMGB1 3 h earlier. FITC, fluorescein isothiocyanate. E and F, quantification of the percentage (E) and number (F) of macrophages migrating into the peritoneal fluid as indicated, *, p < 0.05 versus by analysis of variance. Error bars indicate S.E.
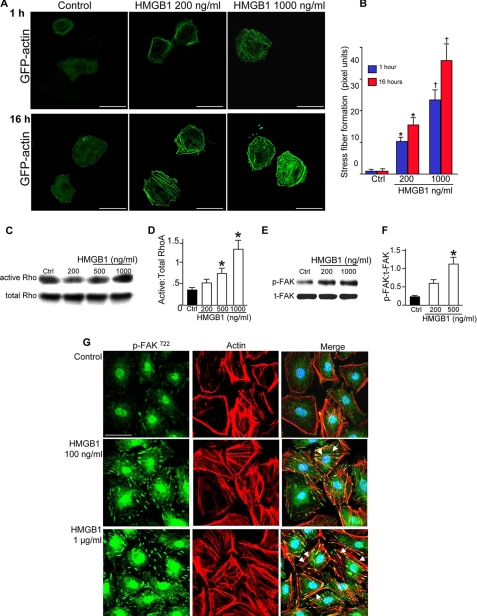
HMGB1 Increases Actin Stress Fiber Formation and Cell-Matrix Adhesion in Enterocytes
We next sought to further explore the mechanisms by which HMGB1 inhibits enterocyte migration and thus investigated whether HMGB1 altered actin polymerization in enterocytes. To measure actin polymerization, IEC-6 cells were transfected with GFP-actin, which becomes incorporated into nascent actin filaments and therefore allows for the assessment of actin polymerization through the detection of the intensity and distribution of the GFP emission. As is shown in Fig. 4A, in untreated cells, GFP-actin appears diffusely throughout the cytoplasm. By contrast, HMGB1 caused a time- and dose-dependent increase in the accumulation of actin stress fibers that traversed throughout the cytoplasm (Fig. 4, panels A and B). This effect on the cytoskeleton led us to assess whether HMGB1 may affect the signaling molecules that are known to be important for stress fiber formation, specifically RhoA, focal adhesion kinase (FAK), and cofilin. As is shown in Fig. 4, C and D, HMGB1 caused a dose-dependent increase in the extent of RhoA activation, as determined using a pull-down assay with the effector molecule Rhotekin. HMGB1 also caused a dose-dependent increase in the phosphorylation of FAK, a kinase that is immediately downstream of RhoA activation and plays a key role in the generation of stable focal adhesions in various cells (Fig. 4, E and F). To measure directly whether HMGB1 affected focal adhesions, we performed confocal microscopy to assess the expression and distribution of phosphorylated FAK (Fig. 4G). Under control settings, p-FAK was detected in a largely cytoplasmic distribution with the occasional sites of colocalization at discreet ends of actin stress fibers (arrows). By contrast, HMGB1 dramatically increased focal adhesion formation with significantly increased colocalization of p-FAK along the lengths of individual actin stress fibers. The increase in focal adhesion formation was dose-dependent as there was a more profound effect on focal adhesion formation at higher concentrations of HMGB1 (Fig. 4G).
FIGURE 4.
HMGB1 leads to rearrangements in the cytoskeleton in enterocytes. A, confocal micrographs showing GFP expression in IEC-6 cells after GFP-actin transfection and treatment with HMGB1 as indicated. Size bar = 10 μm; quantification of stress fiber density by Metamorph in B. *, p < 0.05, †, p < 0.005 versus control (Ctrl) by analysis of variance. C–F, SDS-PAGE showing expression of active and total RhoA (C) or p-FAK and total FAK (t-FAK) (E) in IEC-6 cells lysates treated with HMGB1 as indicated. Quantification is shown in D and F; error bars indicate S.E. G, confocal micrographs showing expression of p-FAK (green) and actin (red), as well as merged images, after treatment with HMGB1 or PBS as indicated. Arrows show focal adhesions, bar = 10 μm.
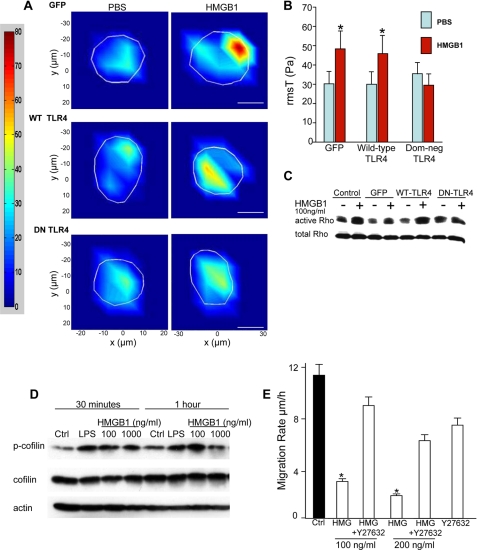
Given that focal adhesions serve to generate increased strength in the interaction of cells with the underlying matrix, we next sought to evaluate whether HMGB1 could affect cell-matrix adhesiveness in enterocytes. Because HMGB1 may act non-uniformly on cells to generate cell-matrix adhesiveness, we utilized a highly robust single cell assay of cell-matrix tension, thereby capturing any potential regional differences in the effects of HMGB1 on cell-matrix binding as described under “Experimental Procedures.” As is shown in Fig. 5, A and B, in IEC-6 cells that were transfected with either GFP-TLR4 or wild-type TLR4, HMGB1 significantly increased cell-matrix adhesiveness, whereas HMGB1 treatment of IEC-6 cells that had been transfected with dominant negative TLR4 cells did not demonstrate an increase in cell-matrix adhesiveness.
FIGURE 5.
HMGB1 increases adhesiveness of enterocytes to the underlying matrix in a TLR4-dependent manner. A, single cell force contraction microscopy of IEC-6 cells after transfection with the indicated construct and treatment with PBS or HMGB1 (100 ng/ml); the region of interest is shown in white. Bar = 5 μm. WT, wild type; DN, dominant negative. Quantification is shown in B. *, p < 0.05 versus PBS paired control. Results shown are representative of three separate experiments. C and D, SDS-PAGE showing expression of active and total RhoA (C) or phosphorylated and total cofilin (D) in IEC-6 cells after transfection with indicated construct ± HMGB1 or LPS (50 μg/ml) as indicated. Results shown are representative of three separate experiments. Ctrl, control. E, IEC-6 migration after treatment with HMGB1 ± Rho kinase inhibitor Y27632. Results shown are representative of three separate experiments. *, p < 0.05 versus control. Error bars indicate S.E.
Given this finding and because RhoA is required for regulating cell-matrix adhesiveness, we next evaluated whether TLR4 signaling was required for the increase in RhoA activation in response to HMGB1. As is shown in Fig. 5C, treatment of either non-transfected or GFP-transfected IEC-6 cells with HMGB1 significantly increased RhoA activation when compared with untreated cells, consistent with Fig. 4C, or when compared with IEC-6 cells that were transfected with wild-type TLR4. However, transfection with dominant negative TLR4 prevented the increase in RhoA activation after exposure to HMGB1, confirming the importance of TLR4 signaling in the molecular events that lead to RhoA activation.
HMGB1 Inhibits Enterocyte Migration through Activation of the RhoA
In the next series of studies, we sought to explore further the mechanisms by which HMGB1 inhibits enterocyte migration. To do so, we first examined whether HMGB1 may alter the phosphorylation of cofilin, a critical actin-binding protein that regulates actin turnover in response to RhoA. The activity of cofilin is critical for cell motility and is inversely regulated by increased phosphorylation such that phosphorylated cofilin completely loses its actin depolymerization activity, leading to increased actin stability (22). As shown in Fig. 5D, HMGB1 time- and dose-dependently increased the extent of phosphorylation of cofilin. This suggested the possibility that RhoA activation may be required for the inhibition in enterocyte migration in response to HMGB1. To determine this experimentally, IEC-6 cells were treated with HMGB1 in the presence or absence of Y27632, a specific Rho kinase inhibitor. As shown in Fig. 5E, HMGB1 dose-dependently inhibited the rate of enterocyte migration, yet this was reversed upon pretreatment with Y27632. Taken in aggregate, these findings indicate that HMGB1-induced activation of TLR4 leads to RhoA signaling, increased focal adhesion formation, and an inhibition in enterocyte migration.
DISCUSSION
We now demonstrate that extracellular HMGB1 can activate TLR4 in enterocytes, leading to increased cell-matrix adhesiveness and impaired migration. This effect was found to occur both in vitro and in vivo and not to occur in macrophages, whose migrations were found to be enhanced by HMGB1. These results may have relevance to the pathogenesis of NEC, a disease that we have shown to require TLR4 signaling in the intestinal mucosa (4), yet for which no prior study has demonstrated the role of endogenous ligands for TLR4 in its development. We acknowledge that we have not definitively proven a role for HMGB1 in the development or progression NEC; rather this study was designed to assess the roles, if any, of extracellular HMGB1 on enterocyte migration. Indeed, the lack of readily available inhibitors to HMGB1 and the early mortality of HMGB1-deficient mice (23) make definitive proof of HMGB1 in NEC pathogenesis difficult to determine. However, given the novel finding that HMGB1 inhibits enterocyte migration both in vivo and in vitro, molecules that are designed to modify HMGB1 function may now be evaluated not merely for their role on immune cells but also on the ability of HMGB1 to affect intestinal healing.
A major finding of the current study relates to the fact that the impairment in enterocyte migration in response to HMGB1 occurs in large part through an increase in focal adhesions and a subsequent increase in the strength with which enterocytes attach to the underlying matrix. To accurately determine the effects of HMGB1 on enterocyte-matrix adhesiveness, we utilized a novel single cell assay device in which the extent of matrix adhesion could be calculated using a complex fluorometric, force detection system. This technique served to remedy potential pitfalls associated with standard, global population studies of cell-matrix adhesiveness in which the degree of binding may be variable and less reliable. Given that increased cell-matrix adhesiveness in response to HMGB1 would be expected to reduce the ability of individual enterocytes to detach from the underlying matrix, the current findings present a plausible mechanism to account for the inhibitory effects of HMGB1 on cell movement and are consistent with previous reports showing that HMGB1 can modulate the actin cytoskeleton of a variety of cell types (21).
In summary, we have now identified that human and murine NEC are associated with an increase in the expression of HMGB1 in the intestine and that the release of HMGB1 leads to an inhibition in enterocyte migration in vitro and in vivo via signaling through TLR4. The current studies serve to complement and advance previous work from our laboratory showing that TLR4 activation by its known agonist bacterial LPS leads to reduced enterocyte migration in part via increased focal adhesion formation (6) and expand our understanding of the role of TLR4 in non-hematopoietic cells as a receptor that can contribute to organ injury rather than cytoprotection. The current findings extend these observations by identifying HMGB1 as a potentially important endogenous molecule that may be involved in TLR4 signaling on enterocytes, possibly playing a role in the development of intestinal inflammation and necrotizing enterocolitis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM078238 and RO1-DK083752 (to D. J. H.) and The Hartwell Foundation, Memphis, TN.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods.”
- TLR4
- Toll-like receptor-4
- HMGB1
- high mobility group box-1
- LPS
- lipopolysaccharide
- IEC
- intestinal epithelial cell
- IFN
- interferon
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline
- FAK
- focal adhesion kinase
- p-FAK
- phosphorylated FAK.
REFERENCES
- 1.Takeuchi O., Akira S. (2009) Immunol. Rev. 227, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchi M. E. (2007) J. Leukoc. Biol. 81, 1–5 [DOI] [PubMed] [Google Scholar]
- 3.Anand R. J., Leaphart C. L., Mollen K. P., Hackam D. J. (2007) Shock 27, 124–133 [DOI] [PubMed] [Google Scholar]
- 4.Leaphart C. L., Cavallo J., Gribar S. C., Cetin S., Li J., Branca M. F., Dubowski T. D., Sodhi C. P., Hackam D. J. (2007) J. Immunol. 179, 4808–4820 [DOI] [PubMed] [Google Scholar]
- 5.Jilling T., Simon D., Lu J., Meng F. J., Li D., Schy R., Thomson R. B., Soliman A., Arditi M., Caplan M. S. (2006) J. Immunol. 177, 3273–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cetin S., Ford H. R., Sysko L. R., Agarwal C., Wang J., Neal M. D., Baty C., Apodaca G., Hackam D. J. (2004) J. Biol. Chem. 279, 24592–24600 [DOI] [PubMed] [Google Scholar]
- 7.Wang H., Bloom O., Zhang M., Vishnubhakat J. M., Ombrellino M., Che J., Frazier A., Yang H., Ivanova S., Borovikova L., Manogue K. R., Faist E., Abraham E., Andersson J., Andersson U., Molina P. E., Abumrad N. N., Sama A., Tracey K. J. (1999) Science 285, 248–251 [DOI] [PubMed] [Google Scholar]
- 8.Abraham E., Arcaroli J., Carmody A., Wang H., Tracey K. J. (2000) J. Immunol. 165, 2950–2954 [DOI] [PubMed] [Google Scholar]
- 9.Park J. S., Gamboni-Robertson F., He Q., Svetkauskaite D., Kim J. Y., Strassheim D., Sohn J. W., Yamada S., Maruyama I., Banerjee A., Ishizaka A., Abraham E. (2006) Am. J. Physiol. Cell Physiol. 290, C917–924 [DOI] [PubMed] [Google Scholar]
- 10.Gribar S. C., Sodhi C. P., Richardson W. M., Anand R. J., Gittes G. K., Branca M. F., Jakub A., Shi X. H., Shah S., Ozolek J. A., Hackam D. J. (2009) J. Immunol. 182, 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ngamkitidechakul C., Twining S. S. (2002) BioTechniques 33, 1296–1300 [DOI] [PubMed] [Google Scholar]
- 12.Wang J. H., Lin J. S. (2007) Biomech. Model. Mechanobiol. 6, 361–371 [DOI] [PubMed] [Google Scholar]
- 13.Chen J., Li H., SundarRaj N., Wang J. H. (2007) Cell Motil. Cytoskeleton 64, 248–257 [DOI] [PubMed] [Google Scholar]
- 14.Butler J. P., Toliã-Nçrrelykke I. M., Fabry B., Fredberg J. J. (2002) Am. J. Physiol. Cell Physiol. 282, C595–605 [DOI] [PubMed] [Google Scholar]
- 15.Zamora R., Grishin A., Wong C., Boyle P., Wang J., Hackam D., Upperman J. S., Tracey K. J., Ford H. R. (2005) Am. J. Physiol. Gastrointest Liver Physiol. 289, G643–652 [DOI] [PubMed] [Google Scholar]
- 16.Qureshi F. G., Leaphart C., Cetin S., Li J., Grishin A., Watkins S., Ford H. R., Hackam D. J. (2005) Gastroenterology 128, 1012–1022 [DOI] [PubMed] [Google Scholar]
- 17.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 18.Rendon-Mitchell B., Ochani M., Li J., Han J., Wang H., Yang H., Susarla S., Czura C., Mitchell R. A., Chen G., Sama A. E., Tracey K. J., Wang H. (2003) J. Immunol. 170, 3890–3897 [DOI] [PubMed] [Google Scholar]
- 19.Leaphart C. L., Qureshi F., Cetin S., Li J., Dubowski T., Baty C., Beer-Stolz D., Guo F., Murray S. A., Hackam D. J. (2007) Gastroenterology 132, 2395–2411 [DOI] [PubMed] [Google Scholar]
- 20.Rouhiainen A., Kuja-Panula J., Wilkman E., Pakkanen J., Stenfors J., Tuominen R. K., Lepäntalo M., Carpén O., Parkkinen J., Rauvala H. (2004) Blood 104, 1174–1182 [DOI] [PubMed] [Google Scholar]
- 21.Orlova V. V., Choi E. Y., Xie C., Chavakis E., Bierhaus A., Ihanus E., Ballantyne C. M., Gahmberg C. G., Bianchi M. E., Nawroth P. P., Chavakis T. (2007) EMBO J. 26, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailly M., Jones G. E. (2003) Curr. Biol. 13, R128–130 [DOI] [PubMed] [Google Scholar]
- 23.Calogero S., Grassi F., Aguzzi A., Voigtländer T., Ferrier P., Ferrari S., Bianchi M. E. (1999) Nat. Genet. 22, 276–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.