Abstract
The retinoic acid receptor-related orphan receptors α and γ (RORα (NR1F1) and RORγ (NR1F3)) are orphan nuclear receptors and perform critical roles in regulation of development, metabolism, and immune function. Cholesterol and cholesterol sulfate have been suggested to be RORα ligands, but the physiological significance is unclear. To date, no endogenous RORγ ligands have been described. Here, we demonstrate that 7-oxygenated sterols function as high affinity ligands for both RORα and RORγ by directly binding to their ligand-binding domains (Ki ∼20 nm), modulating coactivator binding, and suppressing the transcriptional activity of the receptors. One of the 7-oxygenated sterols, 7α-hydroxycholesterol (7α-OHC), serves as a key intermediate in bile acid metabolism, and we show that 7α-OHC modulates the expression of ROR target genes, including Glc-6-Pase and phosphoenolpyruvate carboxykinase, in an ROR-dependent manner. Furthermore, glucose output from hepatocytes is suppressed by 7α-OHC functioning as an RORα/γ ligand. Thus, RORα and RORγ are ligand-regulated members of the NR superfamily and may serve as sensors for 7-oxygenated sterols.
Keywords: Hormones/Steroid, Lipid/Bile Acid, Lipid/Cholesterol, Lipid/Sterol, Metabolism/Gluconeogenesis, Metabolism/Glucose, Receptors/Nuclear
Introduction
In the late 1980s, as the canonical domain structure and conserved sequence of members of the nuclear hormone receptor (NHR)3 superfamily became apparent, several laboratories began to isolate additional members of this superfamily that had no identified ligands. Many of these so-called orphan receptors still have no identified ligands. The first member of the ROR subfamily of receptors (RORα) was identified in the early 1990s based on sequence similarities to the retinoic acid receptor and the retinoid X receptor, hence the name “retinoic acid receptor-related orphan receptor” (1, 2). The highly similar receptors, RORβ and RORγ, were identified soon after (3, 4). Like all NHRs, the RORs display the conserved domain structure with a variable amino-terminal A/B domain, a central, highly conserved DNA-binding domain containing two zinc fingers (C domain) and hinge region (domain D), and a carboxyl-terminal ligand-binding domain (LBD; E domain). The three RORs display significant sequence similarities, and each ROR gene generates multiple isoforms based on alternative promoter usage and splicing. The RORs exhibit distinct patterns of expression. RORα is expressed in the liver, skeletal muscle, skin, lungs, adipose tissue, kidney, thymus, and brain (5, 6). RORβ (NR1F2) exhibits the most restricted pattern of expression and is limited to the central nervous system (7, 8). RORγ is most highly expressed in the thymus, but significant expression is also found in the liver, skeletal muscle, adipose tissue, and kidney (9). All RORs recognize and bind to specific sequences of DNA termed ROR-response elements (ROREs) as monomers, and these ROREs typically consist of an AGGTCA “half-site” with a 5′ AT-rich extension (2–4). When bound to their element within the promoter of a target gene, all three RORs constitutively recruit coactivators resulting in constitutive activation of transcription of their target genes.
Another orphan NHR subclass, the REV-ERBs (REV-ERBα (NR1D1) and REV-ERBβ (NR1D2)), also recognize the RORE sites and are often coexpressed with RORs (10). In contrast to the RORs, the REV-ERBs are repressors of transcription. Until recently, the REV-ERBs were thought to be constitutive repressors of transcription; however, our recent work as well as that from the Lazar laboratory demonstrated that the porphyrin heme functions as a ligand for both REV-ERBs and is required for repression of transcription (11, 12). Both REV-ERBs and ROR share a number of target genes indicating that they coordinately regulate physiological processes.
The coordinate regulation of biological processes by the RORs and REV-ERBs can be demonstrated by the manner in which they regulate the mammalian clock. Both RORs and REV-ERBs are major regulators of the cyclic expression of the aryl hydrocarbon receptor nuclear translocator-like 1 (ARNTL also known as BMAL1) (13, 14). The Bmal1 promoter contains two ROREs, and Bmal1 transcription is directly activated by RORs and repressed by REV-ERBs. Consistent with these receptors playing a critical role in regulation of the circadian rhythm, mice deficient in either RORα, RORβ, or REV-ERBα display abnormal circadian behavior patterns (14–16).
RORs have also been shown to play a significant role in the regulation of metabolism, and much has been revealed by studies in the staggerer (RORαsg/sg) mouse. This mutant mouse strain carries an intragenic deletion within the RORα gene that results in a frameshift and premature stop codon rendering RORα inactive (5). The staggerer mouse was initially identified due to cerebellar defects resulting in ataxia, later revealing a critical role for RORα in cerebellar development. Detailed examination of the staggerer mice exposed significant alterations in lipid metabolism evidenced by low levels of total plasma cholesterol, triglycerides, apolipoprotein CIII, high density lipoprotein, and apolipoprotein AI (17). RORα directly regulates the expression of the genes for both apoA1 and apoCIII (18, 19). Reduced expression of ATP-binding cassette transporter A1, a key transporter for reverse cholesterol transport, has been noted in these mice that is consistent with the abnormally low levels of high density lipoprotein, which are essential for reverse cholesterol transport (20). Genes important in the regulation of triglyceride homeostasis are dysregulated in the liver and skeletal muscle of the staggerer mouse such as sterol regulatory element-binding protein-1c and fatty-acid synthase (20). staggerer mice are less susceptible to hepatic steatosis and have a reduced body fat index relative to wild type mice despite higher food consumption. The size of both brown and white adipose cells are smaller in these animals, and hepatic triglyceride content is lower. Consistent with this phenotype, the animals are less susceptible to high fat diet-induced obesity and hepatic steatosis (20).
RORγ null mice exhibit normal levels of plasma cholesterol and triglycerides (21). An interesting metabolic phenotype was revealed when staggerer mice were crossed with RORγ null mice effectively creating an RORα/γ double knock-out. Although neither individual strain showed significant alterations in plasma glucose levels, the double knock-out was hypoglycemic illustrating a role for these receptors in maintaining glucose homeostasis (21). This study also demonstrated that RORα and RORγ display significant redundancy in function, which is consistent with plasma glucose levels remaining unaffected unless both receptors were lost. Even more recently, a role for RORα in regulation of glucose metabolism was characterized when Chopra et al. (22) found that loss of the NHR coactivator SRC-2 in mice led to a phenotype similar to von Gierke disease, which is associated with severe hypoglycemia and abnormal accumulation of glycogen in the liver. Loss of expression of the enzyme glucose-6-phosphatase (Glc-6-Pase) is responsible for 80% of the diagnosed von Gierke disease cases, and it was demonstrated that SRC-2 was required for RORα to regulate this gene in a normal manner (22).
Although several molecules have been suggested to be ligands for one or more of the RORs, identification of a bona fide functional, physiologically relevant ligand has remained elusive. Retinoids such as all-trans retinoic acid have been shown to suppress the activity of RORβ selectively, but the physiological relevance of this activity has not yet been demonstrated (23). Some of the most interesting work on putative ROR ligands was performed by Kallen et al. (24), where x-ray crystallographic studies identified cholesterol in the ligand binding pocket of RORα. A later study by the same group demonstrated that cholesterol sulfate also binds within the pocket (25). Although the possibility of RORα, which plays such a prominent role in lipid metabolism, functioning as a receptor for cholesterol is attractive, it remains unclear if cholesterol or a derivative of this sterol is truly a physiological ligand for RORα. Recently, we demonstrated that synthetic ligands can bind with high affinity and repress the transcriptional output of both RORα and RORγ, thus functioning as inverse agonists (26). This finding suggests that there might also be natural product ligands that also behave as inverse agonists.
EXPERIMENTAL PROCEDURES
Reagents
Oxysterols were obtained from Sigma or Steraloids (Newport, RI). Gal4-RORαLBD, Gal4-RORγLBD, pTrex-RORα, and pTrex-RORγ were gifts from Phenex Pharmaceuticals AG (Ludwigshafen, Germany). pSPORT-SRC2 was provided by Cell-based Screening Core at Scripps (Jupiter, FL). pGL4-G6PC promoter reporter was kindly provided by Bert O'Malley (Baylor College of Medicine). pG5-Luc and pGL4.73 reporters were obtained from Promega (Madison, WI). RORα was tagged with FLAG and subcloned into pAd/CMV/V5-DEST vector through the GatewayTM technique (Invitrogen). The adenovirus with FLAG-RORα was produced according to the manufacturer's instructions. Unless otherwise noted, oxysterols were used at a final concentration of 10 μm.
Cell Culture and Cotransfections
HEK293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. HepG2 cells were maintained and routinely propagated in minimum essential medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. In experiments where lipids and sterols were depleted, cells were maintained on charcoal-treated serum (10% fetal bovine serum) and treated with 7.5 μm lovastatin and 100 μm mevalonic acid. 24 h prior to transfection, HepG2 or HEK293 cells were plated in 96-well plates at a density of 15 × 103 cells/well. Transfections were performed using LipofectamineTM 2000 (Invitrogen). 16 h post-transfection, the cells were treated with vehicle or compound. 24 h post-treatment, the luciferase activity was measured using the Dual-GloTM luciferase assay system (Promega). The values indicated represent the means ± S.E. from four independently transfected wells. The experiments were repeated at least three times. Data were analyzed using GraphPad Prizm software, and IC50 values were determined by nonlinear regression analysis.
cDNA Synthesis and Quantitative PCR
Total RNA extraction and cDNA synthesis were performed as described previously (11).
Chromatin Immunoprecipitation (ChIP) and ReChIP
HepG2 cells were infected with adenovirus for 24 h and then treated with vehicle, 7α-hydroxycholesterol, 7β-hydroxycholesterol, or 7-ketocholesterol (10 μm) for another 24 h. ChIP assays were performed according to the manufacturer's instructions (Millipore, Bedford, MA). Immunoprecipitation with the following antibodies was performed at 4 °C overnight: anti-mouse IgG, anti-acetyl histone H3, anti-SRC2 (Bethyl Laboratories, Montgomery TX). ReChIP assays were performed by using the kit from Active Motif®. Anti-FLAG (Sigma) was used to do the first immunoprecipitation for all the samples. The second immunoprecipitation was performed by using anti-mouse IgG (Millipore), anti-RNA polymerase II (Millipore), or anti-SRC2 (Bethyl Laboratories). The Glc-6-Pase primers used in PCR are CCCTGAACATGTTTGCATCA (forward) and CATTCCTTCCTCCATCCTCA (reverse).
Mouse Primary Hepatocytes, siRNA, and Glucose Assay
Mouse primary hepatocytes were purchased from Celsis (Chicago). The cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. The control siRNA, mouse RORα, and RORγ siRNA (Thermo Scientific, Waltham, MA) were transfected into the cells using LipofectamineTM RNAiMAX (Invitrogen). After 24 h, cell media were switched to glucose-free Dulbecco's modified Eagle's medium with 20 mm sodium lactate, plus vehicle (DMSO), or 10 μm 7α-hydroxycholesterol. Six hours later, the media were collected and measured for glucose content using Amplex® red glucose/glucose oxidase assay kit (Invitrogen).
Radioligand Binding Assay
RORα and RORγ LBDs were PCR-amplified and cloned into a pGEX-2T (GE Healthcare) encoding an amino-terminal glutathione S-transferase tag following the manufacturer's instructions. The protein was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside in BL21 gold (DE3) cells (Invitrogen) and purified by affinity chromatography with Protino GST/4B column (Macherey-Nagel) followed by size exclusion chromatography with HiLoad 26/60 Superdex 200 column (GE Healthcare). The protein was eluted, concentrated, and stored in 20 mm Tris, pH 8.0, 150 mm NaCl, 2 mm dithiothreitol, and 10% glycerol. Forty five or 90 ng of purified GST-RORα or GST-RORγ was used per assay, and the assay was performed in the following buffer: 50 mm HEPES, pH 7.4, 0.01% bovine serum albumin, 150 mm NaCl, and 5 mm MgCl2. Receptors were incubated with various concentrations of 25-[3H]hydroxycholesterol to determine the Kd values (RORα = 3.3 ± 0.89 nm and RORγ = 5.1 ± 0.71 nm). Nonspecific binding was defined in the absence of protein as well as excess of cold 25-hydroxycholesterol and were shown to be identical. The assays were terminated by rapid filtration through pre-soaked Whatman GF/B filters (0.5% polyethyleneimine in phosphate-buffered saline) in Multiscreen plates (Millipore) and were washed three times with ice-cold assay buffer (1 ml). For the competition assay, various concentrations of sterols were incubated with receptor in the presence of 3 nm 25-[3H]hydroxycholesterol. Results were analyzed using GraphPad Prism software, and the Ki was determined using the Cheng-Prusoff equation.
Coactivator Interaction Assay
Mutiplexed coactivator interaction assay (NR box and CoRNR box peptide interaction assay) was performed as described previously (11). The peptide sequences used have also been described previously (11). RORα produced for this assay was produced in Escherichia coli using high pressure refolding as described previously for farnesoid X receptor and LRH-1 (27).
Hydrogen/Deuterium Exchange (HDX) Mass Spectrometry
Solution phase amide HDX experiments were performed with a LEAP technologies (Carroboro, NC) CTC HTS twin PAL autosampler interfaced with an electrospray ionization Orbitrap mass spectrometer (Exactive, Thermo Fisher, San Jose, CA). 4 μl of the 10 μm RORα-LBD stock solution (1× phosphate-buffered saline, pH 7.4) were diluted with 16 μl of an equivalent composition D2O buffer and incubated for 10, 30, 60, 900, or 3600 s to allow hydrogen/deuterium exchange to occur (28). Following the allotted time period, the sample was diluted to 50 μl with cold (1.5 °C) 5 m urea containing 1% trifluoroacetic acid. Protein was then passed through an immobilized pepsin column (1 mm × 2 cm made in-house) held at 1.5 °C, and the resultant peptides were captured on a C8 peptide trap (1 mm inner diameter × 10 mm). Peptides were then eluted across a high pressure liquid chromatography column (C18 50 × 1 mm, 1.9 μm) into the electrospray ionization source with a gradient of 4% CH3CN increasing to 50% CH3CN over 5 min. For the RORα plus ligand data, 10 μm ROR stock solution was incubated with 50 μm ligand prior to mixing with D2O buffer. Data represent the average of four replicates and were processed with in-house developed software (29, 30).
The primers for quantitative PCR are as follows: human RORα, AAACAAGCAGCGGGAGGTGA (forward) and TGGCAAACTCCACCACATAC (reverse); human Glc-6-Pase, TGAGGATGGAGGAAGGAATG (forward) and GGGGAAGAGGACGTAGAAGG (reverse); human phosphoenolpyruvate carboxykinase, GGAATTCTGGGAGAAGGAGG (forward) and TTGCTTCAAGGCAAGGATCT (reverse); mouse RORα, GGGAAGAGCTCCAGCAGATA (forward) and AGCTGCCACATCACCTCTCT (reverse); mouse RORγ, CAGCAGCAAGTGATGGAGAA (forward) and CCTGGATTTATCCCTGCTGA (reverse); mouse Glc-6-Pase, CTGTGTGCTTGCATTCCTGT (forward) and CAAGTGGAGAATTCTGGGGA (reverse); mouse phosphoenolpyruvate carboxykinase, TTTGAGATAGCGGCACAATG (forward) and ATTTGCCCCTAGCCTGTTCT (reverse).
RESULTS
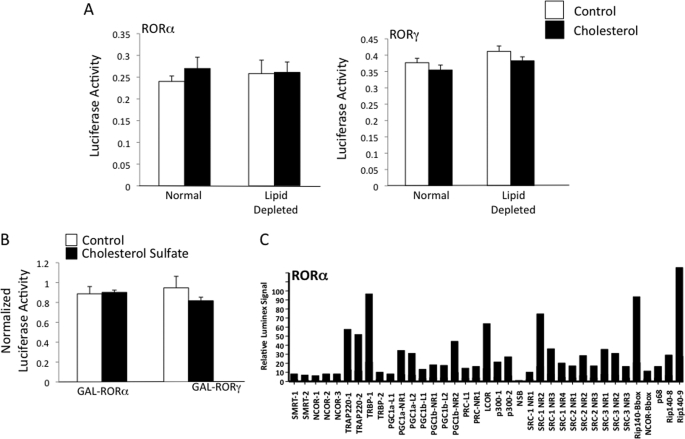
Cholesterol is an essential component of biological membranes and is a precursor for biosynthesis of important lipids and hormones such as bile acids, steroid hormones, and some vitamins. Cholesterol levels are tightly regulated, and the liver plays an essential role in elimination of excess cholesterol via conversion of this sterol to bile acids. X-ray crystallography studies demonstrated that the LBD of RORα is able to bind to cholesterol (24) and cholesterol sulfate (25); however, the physiological relevance of binding of these sterols to the receptor is unclear. It is also unclear whether RORα is active in the absence of any ligand or if cholesterol or another sterol may be constitutively bound conferring this activity. We found that neither depletion of lipid using cyclodextrin-treated serum and inclusion of a statin and mevalonate to the media had an effect on the transcriptional activity of RORα or RORγ in a GAL4-ROR transfection assay nor did addition of cholesterol (500 μm) (Fig. 1A). We also noted no effect of cholesterol sulfate (500 μm) in the same assay (Fig. 1B) suggesting that neither sterols are functional ligands.
FIGURE 1.
Modulation of cholesterol or cholesterol sulfate levels does not alter the transcriptional activity of RORα or RORγ. A, Gal4-ROR LBD cotransfection assay in HEK293 cells. Cholesterol was added at 500 μm. Ethanol was used as the solvent, and an equivalent amount was used in the control. Lipid depletion was performed as described under “Experimental Procedures.” B, Gal4-ROR LBD cotransfection assay in HEK293 cells. Cholesterol sulfate was added at 500 μm. DMSO was used as the solvent, and an equivalent amount was used in the control. C, biochemical assay analyzing of cofactor peptide binding reveals that apo-RORα LBD binds to a large number of coactivator LXXLL peptides. The peptides and methods (Luminex) are described under “Experimental Procedures.”
Previous studies using RORα LBD purified from insect cells found cholesterol, 7-dehydrocholesterol, and an unidentifiable oxysterol bound to receptor (31). These authors presumed that the inability to produce RORα in E. coli was due to the lack of cholesterol in the bacteria (32) and the potential requirement for cholesterol to stabilize the receptor. During one of our first attempts to express the LBD of RORα in E. coli, we also found that the protein was completely insoluble and was contained within inclusion bodies. However, we were able to reconstitute functional RORα LBD using high pressure denaturation-renaturation, a procedure that involves purification of the insoluble protein by ultracentrifugation and extensive washing in a sterol/lipid-free environment (27). Thus, in addition to the fact that E. coli lack sterols, the procedure to reconstitute the LBD would likely eliminate binding of an endogenous ligand. The ability of the refolded LBD to bind to LXXLL peptides derived from a variety of coactivators was tested (11) and was found to be effective in recruiting a number of coactivator NR boxes (Fig. 1C). Based on these results, it is apparent that cholesterol is not required for LBD stability and that the apo-receptor is in an active conformation and displays constitutive activity in the absence of a sterol ligand. As discussed below, we later optimized the expression of RORα and RORγ LBDs in E. coli allowing for production of soluble proteins (for radioligand binding assays), which is also consistent with sterols not being required for solubility of the proteins.
7α-Hydroxycholesterol, a Bile Acid Precursor, Regulates the Activity of RORα
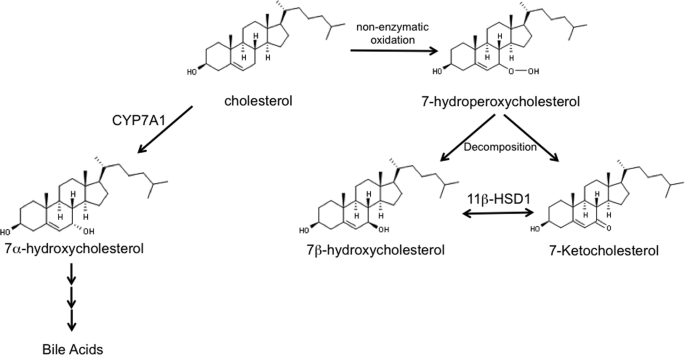
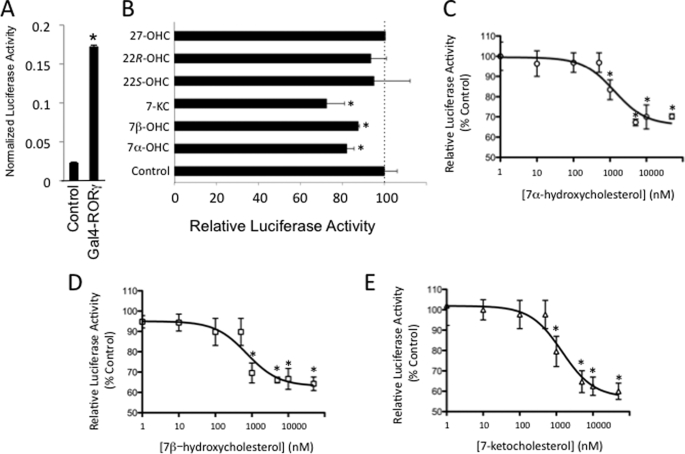
Recent studies demonstrating a role for both RORα and RORγ (21, 33) as well as REV-ERBα (34, 35) in regulation of bile acid metabolism intrigued us. Thus, we began to examine the possibility that a key sterol in the bile acid pathway might function as a physiological ligand for the RORs. We tested the ability of a key sterol intermediate in the bile acid synthetic pathway, 7α-OHC (structure shown in Fig. 2), for its ability to modulate RORα activity in a cotransfection assay. 7α-OHC is produced in the first step of bile acid biosynthesis by the cytochrome P-450 cholesterol 7α-hydroxylase (CYP7A1) using cholesterol as the substrate. This step is the rate-limiting step in bile acid biosynthesis, and the expression of CYP7A1 is highly regulated, thus the levels of 7α-OHC reflect the degree of activation of this pathway (36–39). Based on the importance of this particular oxysterol in bile acid synthesis, we hypothesized that it may modulate the activity of RORα. We used a cotransfection assay to test the activity of this particular sterol where HEK293 cells are cotransfected with a chimeric ROR receptor, where the yeast GAL4-DNA-binding domain is fused to the LBD of RORα, and a reporter responsive to GAL4. As shown in Fig. 3A, addition of GAL4-RORα to the cells results in significant activation of transcription (∼7-fold) consistent with the constitutive activity described for this receptor. When 7α-OHC is included, a dose-dependent inhibition of the transcriptional activity of RORα was observed with the maximal efficacy reaching approximately a 40% reduction in RORα-stimulated transcription (Fig. 3B) with an IC50 of 1.3 μm. This level of reduction of transcriptional activity would be expected to have a significant physiological impact on pathways regulated by an RORα target gene such as Glc-6-Pase. Interestingly, levels of 7α-OHC in human plasma have been measured and range from ∼0.2 to ∼2 μm (40–42).
FIGURE 2.
Schematic illustrating the pathways for production of 7-oxygenated sterols.
FIGURE 3.
7-Oxygenated sterols suppress the transcriptional activity of RORα. A, Gal4-RORα LBD displays constitutive transcriptional activation activity. HEK293 were transfected with vectors directing the expression of either a Gal4-RORα LBD fusion protein (Gal4-RORα) or Gal4 alone (control) along with a luciferase reporter responsive to Gal4 (pG5-luc). Luciferase values were normalized using Renilla luciferase. B, Gal4-RORα LBD transfection assay illustrating dose-dependent repression of the constitutive transactivation activity of RORα by 7α-OHC (IC50 = 1.3 μm). C, Gal4-RORα LBD transfection assay demonstrating that 7-oxygenated sterols repress RORα transactivation activity. Oxysterols were tested at a final concentration of 10 μm. *, p < 0.05. D, Gal4-RORα LBD transfection assay illustrating dose-dependent repression of the constitutive transactivation activity of RORα by 7β-OHC (IC50 = 0.7 μm). E, Gal4-RORα LBD transfection assay illustrating dose-dependent repression of the constitutive transactivation activity of RORα by 7-KC (IC50 = 1.4 μm).
Other 7-Oxygenated Sterols Also Regulate RORα Transcriptional Activity
Several 7-oxygenated sterols are found in biological systems. In addition to 7α-OHC, the diastereomer 7β-OHC (Fig. 2) as well as 7-ketocholesterol (Fig. 2) are produced in significant levels in adipose tissue (43) and liver (44). After 27-OHC, the most enriched oxysterols in atherosclerotic plaques are 7-KC, 7β-OHC, and 7α-OHC, and these oxysterols are predominantly concentrated within foam cells (45). We tested these three 7-oxygenated sterols in the GAL4-RORα cotransfection assay along with 27-OHC, 22(S)-OHC, and 22(R)-OHC. 27-OHC and 22(R)-OHC function as LXRα/β agonists, but the 7-substituted oxysterols are devoid of LXR activity (43, 46, 47) (supplemental Fig. 1). As shown in Fig. 3C, all three of the 7-oxygenated sterols inhibited the transcriptional activity of RORα, whereas 27-, 22(S)-, and 22(R)-OHCs had no effect. Dose-dependent inhibition of RORα transactivation activity was observed for both 7β-OHC and 7-KC with maximal efficacy of ∼40% inhibition and IC50 values in the range of 0.7 to 1.4 μm (Fig. 3, D and E).
7-Oxygenated Sterols Regulate RORγ Transcriptional Activity
RORγ displays an overlapping pattern of expression with RORα, and these two receptors cooperate to regulate diverse physiological functions, including glucose metabolism (21) and Th17 cell development (48). Thus, we reasoned that both of these receptors might respond to similar ligands. We tested this first using the cotransfection assay utilizing GAL4-RORγ LBD cotransfected into HEK293 cells along with a GAL4-responsive promoter. The RORγ LBD displayed constitutive transactivation activity as shown in Fig. 4A. Similar to RORα, the 7-oxygenated sterols suppressed the constitutive activity of GAL4-RORγ LBD, although other oxysterols did not (Fig. 4B). All three 7-oxygenated sterols (7α-OHC, 7β-OHC, and 7-KC) dose-dependently repressed the transactivation activity of GAL4-RORγ with IC50 values in the range of 0.62 to 2.2 μm (Fig. 4, C and D).
FIGURE 4.
7-Oxygenated sterols suppress the transcriptional activity of RORγ. A, Gal4-RORγ LBD displays constitutive transcriptional activation activity. HEK293 were transfected with vectors directing the expression of either a Gal4-RORγ LBD fusion protein (Gal4-RORγ) or Gal4 alone (control) along with a luciferase reporter responsive to Gal4 (pG5-luc). Luciferase values were normalized using Renilla luciferase. B, Gal4-RORγ LBD transfection assay demonstrating that 7-oxygenated sterols repress RORγ transactivation activity. Oxysterols were tested at a final concentration of 10 μm. C, Gal4-RORγ LBD transfection assay illustrating dose-dependent repression of the constitutive transactivation activity of RORγ by 7α-OHC (IC50 = 1.6 μm). D, Gal4-RORγ LBD transfection assay illustrating dose-dependent repression of the constitutive transactivation activity of RORγ by 7β-OHC (IC50 = 2.2 μm). E, Gal4-RORγ LBD transfection assay illustrating dose-dependent repression of the constitutive transactivation activity of RORγ by 7-KC (IC50 = 0.6 μm). *, p < 0.05.
7-Oxygenated Sterols Are High Affinity RORα Ligands
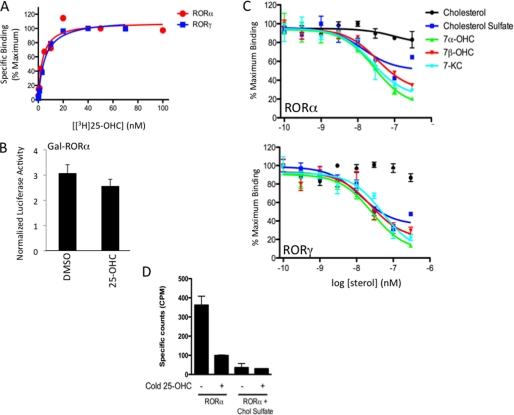
In addition to cholesterol and cholesterol sulfate, 25-OHC has been shown to bind to RORα using mass spectroscopy, but no functional ROR activity has been attributed to this oxysterol (31). Modulators of RORs not only need to bind to the receptor, they must make contacts within the ligand binding pocket that leads to conformational changes on the receptor surface. These conformational changes modulate interaction with coactivators and corepressors. Taking advantage of this finding, we developed a radioligand binding assay for RORα using tritiated 25-OHC and noted high affinity saturable binding of the 25-[3H]OHC after correction for nonspecific binding (Kd = 3.3 ± 0.89 nm) (Fig. 5A). Interestingly, 25-OHC slightly reduced Gal4-RORα activity, but the effect was not statistically significant (Fig. 5B) suggesting that this sterol can bind tightly to the receptor but is inefficient in inducing conformational change to the receptor. To directly compare the relative affinities of the various sterols to RORα, we performed competition radioligand binding assays using 25-[3H]OHC as the radioligand and competing with unlabeled 7α-OHC, 7β-OHC, 7-KC, and cholesterol and cholesterol sulfate. Fig. 5C shows that the 7-substituted oxysterols display similar affinities for RORα (Ki values ranged from 12 to 18 nm), whereas cholesterol exhibited only very limited ability to displace the radiolabeled oxysterol. Cholesterol sulfate bound to RORα, but the degree of displacement of 25-[3H]OHC was less than for the 7-oxygenated sterols (Fig. 5C), and as was the case for 25-OHC, cholesterol sulfate was ineffective in modulating the transcriptional activity of RORα (Fig. 1B).
FIGURE 5.
Radioligand binding assay for RORα and RORγ using tritiated 25-OHC reveals high affinity binding to 7-oxygenated sterols. A illustrates that RORα and RORγ LBDs display saturable specific binding to the labeled oxysterol. B, results from a GAL4-RORα LBD cotransfection assay in HEK293 cells demonstrating that 25-OHC (10 μm) does not significantly affect the activity of the receptor. C, competition radioligand binding assay illustrating the direct binding of the 7-oxygenated sterols to the LBD of RORα (left) and RORγ (right) relative to cholesterol and cholesterol sulfate. Bile acids, chenodeoxycholic acid and cholic acid, did not display binding (data not shown). D, saturation of RORα-LBD with cholesterol sulfate prior to size exclusion chromatography yields protein that does not bind detectable levels of radiolabeled 25-OHC.
Although E. coli does not produce sterols (32), we examined the effect of an exogenously added sterol to the bacterially expressed RORα-LBD to confirm that endogenous sterol was not affecting the activity of the radioligand binding assay. Following affinity purification, the RORα-LBD was treated either with 1 mm cholesterol sulfate or buffer alone overnight. The following day, both samples were further purified, independently, by size exclusion chromatography. Each sample was then used in the radioligand binding assay. As shown in Fig. 5D, the control buffer-treated RORα-LBD displayed specific binding to 25-[3H]OHC, whereas the cholesterol sulfate-saturated RORα-LBD blocked binding of the trace radiolabel to the receptor. These data suggest that if the purified RORα-LBD was indeed saturated with an endogenous ligand such as cholesterol sulfate, the radioligand binding assay would be unlikely to work.
All three 7-oxygenated sterols were able to effectively bind to the RORγ LBD as illustrated in Fig. 5C (Ki values ranged from 17 to 31 nm) with the sterols displaying a similar binding pattern as that seen for RORα. Thus, the 7-oxygenated sterols are high affinity ligands for both RORα and RORγ and act to repress the constitutive transcriptional output of these receptors. The apparent difference between the binding affinity (biochemical assay on purified LBD) and potency in cell-based assays (cells treated with ligand) is not unexpected because it is also seen with the LXR class of oxysterol receptors. For example 24,25-dihydroxycholesterol binds to LXRα with a Ki of 200 nm but displays a 4 μm EC50 in a cotransfection assay (46).
7-Oxygenated Sterols Alter RORα LBD Conformation and Regulate Coactivator Recruitment
The ability of the 7-oxygenated sterols to modulate the activity of full-length RORα and RORγ was examined in a cotransfection assay using a Glc-6-Pase promoter-luciferase reporter construct. The importance of RORα in the regulation of Glc-6-Pase was recently demonstrated by Chopra et al. (22) where both RORα and the coactivator SRC-2 were shown to be critical for normal regulation of this enzyme in mice. Transfection of HEK293 cells with either RORα or RORγ resulted in increased expression of the reporter (Fig. 6A), and coexpression of SRC-2 resulted in a further significant increase in luciferase expression (Fig. 6A). Treatment of the cells with 7-oxygenated sterols resulted in a suppression of the activity of RORα and RORγ (30–40%) consistent with the results from the GAL4-ROR LBD studies (Fig. 6A). In the absence of exogenous SRC-2, the 7-oxygenated sterols were also able to suppress the constitutive activity of RORα (Fig. 6B). The ability of the 7-oxygenated sterols to suppress Glc-6-Pase reporter transcription was ROR-dependent because mutation of the RORE within the Glc-6-Pase promoter resulted in loss of the ability of these oxysterols to suppress transcription (Fig. 6C). 7α-OHC dose-dependently inhibited RORα and RORγ activity on the Glc-6-Pase promoter as shown in Fig. 6, D and E, with IC50 values ranging from 1.3 to 1.7 μm. We next examined the ability of the 7-oxygenated sterols to repress an ROR target gene in HepG2 cells. Glc-6-Pase was selected based on recent studies indicating the importance of ROR on glucose metabolism. Treatment of the cells with 7- oxygenated sterols leads to a significant decrease in expression of Glc-6-Pase (∼34%; Fig. 6F). We examined the ability of the 7-oxygenated sterols to modulate SRC-2 occupancy of the Glc-6-Pase promoter using ChIP. HepG2 cells were treated for 24 h with 10 μm 7α-OHC, 7β-OHC, or 7-KC followed by assessment of SRC-2 occupancy of the Glc-6-Pase promoter. As shown in Fig. 6G, a 40–48% decrease in SRC-2 occupancy was detected upon treatment of cells with the 7-oxygenated sterols, which is consistent with the magnitude of decrease in Glc-6-Pase expression caused by these ligands (Fig. 6G). The RORα dependence of the decrease in SRC-2 recruitment to the Glc-6-Pase promoter was examined in HepG2 cells using a sequential ChIP assay in which we first performed a ChIP assay against RORα followed by a second immunoprecipitation of SRC-2. This allows for detection of changes in the degree of SRC-2 recruitment by RORα at the Glc-6-Pase promoter. Treatment with 7α-OHC did not affect the level of RORα occupancy of the Glc-6-Pase promoter (Fig. 6H, ChIP); however, in the reChIP experiment using the SRC-2 antibody, a 36% decrease in the amount of SRC-2 occupancy was noted in the presence of the ROR ligand demonstrating that 7α-OHC decreased the ability of RORα to recruit this coactivator to the Glc-6-Pase promoter and thus decreasing the expression of the gene. It is interesting to note that 7α-OHC caused a nearly identical decrease in SRC-2 recruitment to RORα at the Glc-6-Pase promoter (36%) compared with the decrease in Glc-6-Pase mRNA expression (33%; Fig. 6E).
FIGURE 6.
7-Oxygenated sterols repress the activity of full-length RORα and RORγ and inhibit SRC-2 recruitment to a target promoter. A, ability of RORα and RORγ to activate transcription of a reporter gene driven by the Glc-6-Pase (G6Pase) promoter is repressed by 7-oxygenated sterols in HEK293 cells. luc, luciferase. B, ability of RORα (no exogenous SRC-2 added) to activate transcription of a reporter gene driven by the Glc-6-Pase promoter is repressed by 7-oxygenated sterols in HEK293 cells. C, ability of the 7-oxygenated sterols to repress transcription is dependent on ROR because mutation of the RORE in the Glc-6-Pase promoter eliminates the activity of the sterols. D, dose-response curve for 7α-OHC in the RORα Glc-6-Pase promoter luciferase cotransfection assay. The IC50 is 1.3 μm. E, dose-response curve for 7α-OHC in the RORγ Glc-6-Pase promoter luciferase cotransfection assay. The IC50 is 1.7 μm. F, treatment of HepG2 cells with 7-oxygenated sterols significantly decreases the expression of Glc-6-Pase. G, 7-oxygenated sterols reduce the level of SRC-2 occupancy of the Glc-6-Pase promoter in HepG2 cells as determined by ChIP. IgG was used as a negative control, and α-acetylated histone H3 was used as a positive control. H, sequential ChIP (ChIP/reChIP) demonstrates that SRC-2 recruitment by RORα at the Glc-6-Pase promoter is reduced by treatment of HepG2 cells with 7α-OHC. IgG was used as a negative control, and α-RNA polymerase II was used as a positive control. Oxysterols were evaluated at a concentration of 10 μm. Asterisk indicates p < 0.05 versus control or as indicated on the figure.
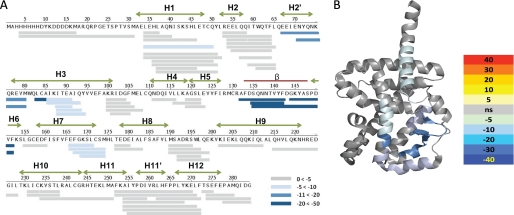
We suspected that the 7-oxygenated sterols were binding to the ROR LBDs and inducing a conformational change altering the ability of the receptors to recruit coactivators. To investigate this, we performed comprehensive differential HDX experiments on the ligand-receptor complex of RORα LBD ± 7α-OHC. As shown in Fig. 7, A and B, receptor-ligand interaction resulted in significant reduction in amide hydrogen exchange kinetics in helices 1, 2′, 3, 6, and 7 and the β-sheet of RORα LBD. The region of the RORα LBD that is protected from amide hydrogen exchange upon binding 7α-OHC is reminiscent of HDX protection on peroxisome proliferator-activated receptor γ LBD upon binding synthetic partial agonists (49) and some endogenous oxidized fatty acids (50). In both of these cases, there is no ligand-induced perturbation in AF2, yet compounds can drive coactivator binding. It is important to note that the HDX profile of the RORα LBD prior to addition of ligand was highly dynamic and very similar to that observed for apo-peroxisome proliferator-activated receptor γ further suggesting that the receptor produced in E. coli is devoid of ligand in its pocket. These data led us to propose a model in which unliganded ROR is in an active conformation constitutively interacting with SRC-2, and binding of an inverse agonist ligand such as 7α-OHC to the LBD leads to a decrease in affinity for the coactivator resulting in decreased transactivation activity and reduced transcriptional output.
FIGURE 7.
7α-OHC induces a conformational change in the RORα LBD. A, differential HDX data for RORα LBD ± 7α-OHC plotted over the linear sequence of the RORα LBD. Ligand binding–induced protection to exchange was observed for residues within H1, H2′, H3, β-sheet, H6, and H7. The magnitude of the reduction in exchange is colored according to the key. B, differential HDX data for RORα LBD ± 7α-OHC are plotted over Protein Data Bank code 1N83. Ligand binding-induced protection to exchange was observed for amino acid residues 273–289, 306–332, 371–381, 383–391, and 405–413. The magnitude of the reduction in exchange is colored according to the key.
7α-Hydroxycholesterol Regulates Hepatocyte Glucose Output Acting as an RORα/γ Inverse Agonist
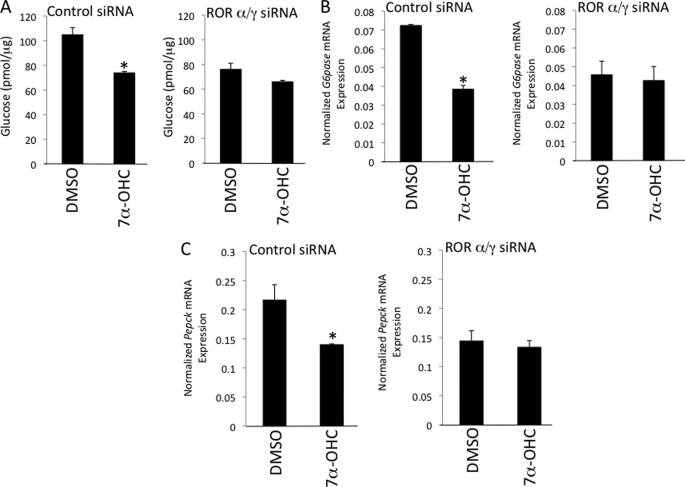
Based on the significant role RORα and RORγ play in regulation of glucose metabolism, as well as our data illustrating 7-oxygenated sterol regulation of Glc-6-Pase expression, we assessed the physiological relevance of 7α-OHC as an RORα/γ ligand in primary mouse hepatocytes. Primary hepatocytes were treated with either control siRNA or a mixture of RORα/γ siRNAs (supplemental Fig. 2) followed by treatment with either DMSO or 7α-OHC (Fig. 8). Glucose output and expression of Glc-6-Pase and phosphoenolpyruvate carboxykinase were measured in these cells 24 h post-treatment. As expected, reduction in the expression of RORα and RORγ in primary hepatocytes resulted in a 24% decrease in glucose output (Fig. 8A), a result that is consistent with data indicating that mice deficient in both RORα and RORγ display hypoglycemia (21). In addition, Glc-6-Pase and phosphoenolpyruvate carboxykinase gene expression were also reduced (Fig. 8, B and C). More importantly, treatment of control siRNA-transfected cells with 7α-OHC reduced glucose output, Glc-6-Pase expression, and phosphoenolpyruvate carboxykinase expression (Fig. 8, A–C), whereas treatment of RORα/γ siRNA-transfected cells displayed a severely blunted response to the oxysterol. These data indicate that 7α-OHC repression of Glc-6-Pase and phosphoenolpyruvate carboxykinase expression and suppression of glucose output is RORα- and RORγ-dependent, which is consistent with this oxysterol functioning as a direct endogenous RORα/γ ligand.
FIGURE 8.
7α-Hydroxycholesterol modulates glucose metabolism in hepatocytes functioning as an ROR ligand. A, treatment of primary mouse hepatocytes with 7α-OHC reduces glucose output in an RORα/γ-dependent manner. B, 7α-OHC reduces the expression of the ROR target gene Glc-6-Pase (G6pase) in primary mouse hepatocytes in an RORα/γ-dependent manner. C, 7α-OHC reduces the expression of the phosphoenolpyruvate carboxykinase (Pepck) in primary mouse hepatocytes in an RORα/γ-dependent manner. 7α-OHC was evaluated at a concentration of 10 μm. Asterisk indicates p < 0.05 versus DMSO.
DISCUSSION
Previous studies examining the crystal structure of RORα identified cholesterol and cholesterol sulfate in the ligand binding pocket (24, 25). Although our radioligand binding assay detected high affinity cholesterol sulfate binding to both RORα and RORγ, there was only minimal displacement of the radioligand by cholesterol. Cholesterol may indeed bind (albeit at a lower affinity), but the radioligand binding assay has limited dynamic range due to the low solubility of the sterols. However, our data are consistent with that of Kallen et al. (25) who showed that cholesterol sulfate had higher affinity for RORα than cholesterol using differential scanning calorimetry. Our binding data suggest that cholesterol sulfate has a binding affinity several orders of magnitude higher than cholesterol. Interestingly, we found that 7-oxygenated sterols (7α-OHC, 7β-OHC, and 7-KC) bound to RORα and RORγ with higher affinity than cholesterol sulfate. In cell-based assays, in contrast to cholesterol and cholesterol sulfate that did not modulate RORα or RORγ activity, the 7-oxygenated sterols acted as inverse agonists decreasing the transcriptional activity of these two nuclear hormone receptors. It has been proposed that cholesterol binds to RORα and acts as an agonist enhancing the transcriptional activity of the receptor (24); thus one might predict that the 7-oxygenated sterols are competing for binding of cholesterol and inhibiting receptor transcriptional activity. Although this might be the case, our data suggest, however, that apo-RORα is constitutively active and retains the ability to interact with a range of coactivator NR box peptides in the absence of binding to any ligand. Given our data indicating that 7α-OHC binds to RORα and induces a conformational change, we propose a model where the receptor binds to coactivator constitutively, and binding of 7α-OHC reduces the level of this constitutive interaction resulting in repression of the transcriptional output of the receptors.
7-Oxygenated sterols play important roles in the organism functioning as intermediates of bile acid metabolism and key lipids in the atherosclerotic process. We demonstrate that 7α-OHC modulates a key physiological RORα-regulated pathway, glucose metabolism, in an RORα/γ-dependent manner. Although the 7-oxygenated sterols only repress ROR transcription by 30–40% in the Gal4 cotransfection assay, this level of efficacy is consistent in all of the assays and, most importantly, underlies the ability of 7α-OHC to suppress hepatocyte glucose output by 30%, a physiologically significant decrease. This limited level of inverse agonist efficacy appears to be intrinsic to the 7-oxygenated sterols, because we recently described a synthetic RORα/γ ligand that has considerably more efficacy (26). An additional question can be raised as to the physiologically relevant level of 7α-OHC in an ROR target tissue such as the liver. End product inhibition is a common mechanism used by enzymes to regulate their activity, and CYP7A1 activity is known to be inhibited by its product 7α-OHC (51, 52). Micromolar levels of 7α-OHC are required to inhibit the activity of this enzyme (35% at 10 μm) (51); thus, if end product inhibition is indeed functional, the levels of 7α-OHC must be in the range required for modulation of RORα/γ activity.
Interestingly, administration of glucose to fasting rats results in a significant increase in the production of 7α-OHC in the liver (53), and it is possible that production of this ROR ligand may play a role in regulation of glucose metabolism in response to feeding. This is supported by data from both animal models and in humans. Our model proposes that when feeding occurs and glucose levels are elevated, an increase in hepatic levels of 7α-OHC results, leading to decreased hepatic glucose output via inhibition of RORα/γ activity. We predict that under conditions where CYP7A1 expression is increased and 7α-OHC levels are elevated, there should be a reduction in plasma glucose levels. Bile acid-binding resins are commonly used to treat hypercholesterolemia by sequestering bile acids in the intestine resulting in their elimination and preventing them from being recirculated to the liver. The net loss in bile acids results in activation of the bile acid biosynthetic pathway via increased levels of CYP7A1. The increased levels of CYP7A1 leads to increased hepatic levels of 7α-OHC. Based on our model, we would predict that bile acid sequestrants would decrease plasma glucose levels, and this is the case both in rodents and in humans. Treatment of diabetic mice with a bile acid sequestrant results in an approximate 4-fold increase in CYP7A1 expression and an ∼50% decrease in phosphoenolpyruvate carboxykinase expression (54). These alterations in gene expression correlate with a significant decrease in both fasting and nonfasting plasma glucose levels (54). Several clinical trials have been performed examining the effects of bile acid sequestrants on glucose levels in type 2 diabetics, and all three demonstrate improvement in plasma glucose levels (decreased plasma glucose levels, postprandial glucose levels, fasting glucose levels, and HbA1c) (55).
The observation that 7-oxygenated sterols function as RORα/γ ligands also has implications for atherosclerosis. staggerer mice develop severe atherosclerosis when placed on a high fat diet (17) suggesting that RORα plays an atheroprotective role. RORα is expressed in human aortic smooth muscle cells and endothelial cells, and there is a significant decrease in expression of this receptor in atherosclerotic arteries (56). The 7-oxygenated sterols are atherogenic and are the most abundant oxysterols in atherosclerotic plaques following 27-OHC (40). 7-KC is the most enriched oxysterol in oxidized low density lipoprotein and in the macrophage foam cell (40, 57) and has been shown to exert atherogenic activity by inhibiting sterol efflux from these cells (58). One mechanism by which the 7-oxygenated sterols may exhibit their atherogenic activity is by decreasing the atheroprotective effects of RORα. The 7-oxygenated sterols may reach low micromolar levels in the plasma of normal individuals and may reach even higher concentrations in foam cells, atherosclerotic plaques, and in the plasma of dyslipidemic patients (40). Plasma levels of both 7β-OHC and 7-KC are elevated as much as 370% in patients with familial combined hyperlipidemia, and treatment with a statin or fibrate significantly reduces these levels in addition to low density lipoprotein, very low density lipoprotein, and triglycerides (59).
RORα and another nuclear receptor, REV-ERBα, coordinately regulate the oscillatory pattern of expression of the key clock gene, BMAL1 (10). Originally, it was believed that neither RORα nor REV-ERBα would be regulated by ligands due to their constitutive transcriptional activator/repressor activity. Recently, our laboratory, as well as others, identified heme as the ligand for REV-ERBα that modulates the ability of this receptor to repress transcription (11, 12). Interestingly, the rate-limiting enzymes for biosynthesis of both heme (60) and 7α-OHC are regulated in a circadian fashion. Aminolevulinic acid synthase 1 has been shown to follow a circadian pattern of expression (60), and intracellular heme levels have also been shown to oscillate (61). CYP7A1 expression is also circadian (34) indicating that its product, 7α-OHC, may also display a circadian pattern of production. Because serum 7α-OHC levels correlate with the level of bile acid synthesis (41), it is possible that this RORα ligand may modulate the mammalian clock in many tissues. These results suggest that there may be an additional degree of complexity added to regulation of the mammalian clock where the activities of both of these receptors (REV-ERBα and RORα) may be modulated by ligands. Thus, we propose that the 7-oxysterols may regulate the transcriptional output of the constitutively active RORs on both hepatic glucose production and circadian rhythms. This allows for regulation of constitutive transcriptional output by transient levels of oxysterols. These observations open opportunities for designing synthetic modulators of the RORs as possible therapeutic agents in metabolic and immune disorders. In this light, we recently identified a synthetic RORα/γ inverse agonist in an NHR specificity screen of known NHR ligands T0901317, a well characterized agonist of the LXRα and LXRβ oxysterol receptors, functions as an inverse agonist of RORα and RORγ displaying the ability to bind with high affinity directly to their LBDs. It is common for NHRs that bind to similar endogenous ligands to also display cross-reactivity of synthetic ligands (e.g. steroid receptors). Thus, it was not unexpected that we found that synthetic ligands for LXR also modulate the activity of the RORs, which we have found also respond to oxysterol ligands.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK080201, NS066417, and NS067589 (to T. P. B.) and GM084041 (to P. R. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- NHR
- nuclear hormone receptor
- ROR
- retinoic acid receptor-related orphan receptor
- LBD
- ligand-binding domain
- ChIP
- chromatin immunoprecipitation
- 7α-OHC
- 7α-hydroxycholesterol
- siRNA
- small interfering RNA
- RORE
- ROR-response element
- HDX
- hydrogen/deuterium exchange
- 7-KC
- 7-ketocholesterol
- LXR
- liver X receptor.
REFERENCES
- 1.Becker-André M., André E., DeLamarter J. F. (1993) Biochem. Biophys. Res. Commun. 194, 1371–1379 [DOI] [PubMed] [Google Scholar]
- 2.Giguère V., Tini M., Flock G., Ong E., Evans R. M., Otulakowski G. (1994) Genes Dev. 8, 538–553 [DOI] [PubMed] [Google Scholar]
- 3.Carlberg C., Hooft van Huijsduijnen R., Staple J. K., DeLamarter J. F., Becker-André M. (1994) Mol. Endocrinol. 8, 757–770 [DOI] [PubMed] [Google Scholar]
- 4.Hirose T., Smith R. J., Jetten A. M. (1994) Biochem. Biophys. Res. Commun. 205, 1976–1983 [DOI] [PubMed] [Google Scholar]
- 5.Hamilton B. A., Frankel W. N., Kerrebrock A. W., Hawkins T. L., FitzHugh W., Kusumi K., Russell L. B., Mueller K. L., van Berkel V., Birren B. W., Kruglyak L., Lander E. S. (1996) Nature 379, 736–739 [DOI] [PubMed] [Google Scholar]
- 6.Steinmayr M., André E., Conquet F., Rondi-Reig L., Delhaye-Bouchaud N., Auclair N., Daniel H., Crépel F., Mariani J., Sotelo C., Becker-André M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3960–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.André E., Conquet F., Steinmayr M., Stratton S. C., Porciatti V., Becker-André M. (1998) EMBO J. 17, 3867–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.André E., Gawlas K., Becker-André M. (1998) Gene 216, 277–283 [DOI] [PubMed] [Google Scholar]
- 9.Medvedev A., Yan Z. H., Hirose T., Giguère V., Jetten A. M. (1996) Gene 181, 199–206 [DOI] [PubMed] [Google Scholar]
- 10.Burris T. P. (2008) Mol. Endocrinol. 22, 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. (2007) Nat. Struct. Mol. Biol. 14, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 13.Guillaumond F., Dardente H., Giguère V., Cermakian N. (2005) J. Biol. Rhythms 20, 391–403 [DOI] [PubMed] [Google Scholar]
- 14.Sato T. K., Panda S., Miraglia L. J., Reyes T. M., Rudic R. D., McNamara P., Naik K. A., FitzGerald G. A., Kay S. A., Hogenesch J. B. (2004) Neuron 43, 527–537 [DOI] [PubMed] [Google Scholar]
- 15.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. (2002) Cell 110, 251–260 [DOI] [PubMed] [Google Scholar]
- 16.Akashi M., Takumi T. (2005) Nat. Struct. Mol. Biol. 12, 441–448 [DOI] [PubMed] [Google Scholar]
- 17.Mamontova A., Séguret-Macé S., Esposito B., Chaniale C., Bouly M., Delhaye-Bouchaud N., Luc G., Staels B., Duverger N., Mariani J., Tedgui A. (1998) Circulation 98, 2738–2743 [DOI] [PubMed] [Google Scholar]
- 18.Raspé E., Duez H., Gervois P., Fiévet C., Fruchart J. C., Besnard S., Mariani J., Tedgui A., Staels B. (2001) J. Biol. Chem. 276, 2865–2871 [DOI] [PubMed] [Google Scholar]
- 19.Vu-Dac N., Gervois P., Grötzinger T., De Vos P., Schoonjans K., Fruchart J. C., Auwerx J., Mariani J., Tedgui A., Staels B. (1997) J. Biol. Chem. 272, 22401–22404 [DOI] [PubMed] [Google Scholar]
- 20.Lau P., Fitzsimmons R. L., Raichur S., Wang S. C., Lechtken A., Muscat G. E. (2008) J. Biol. Chem. 283, 18411–18421 [DOI] [PubMed] [Google Scholar]
- 21.Kang H. S., Angers M., Beak J. Y., Wu X., Gimble J. M., Wada T., Xie W., Collins J. B., Grissom S. F., Jetten A. M. (2007) Physiol. Genomics 31, 281–294 [DOI] [PubMed] [Google Scholar]
- 22.Chopra A. R., Louet J. F., Saha P., An J., Demayo F., Xu J., York B., Karpen S., Finegold M., Moore D., Chan L., Newgard C. B., O'Malley B. W. (2008) Science 322, 1395–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stehlin-Gaon C., Willmann D., Zeyer D., Sanglier S., Van Dorsselaer A., Renaud J. P., Moras D., Schüle R. (2003) Nat. Struct. Biol. 10, 820–825 [DOI] [PubMed] [Google Scholar]
- 24.Kallen J. A., Schlaeppi J. M., Bitsch F., Geisse S., Geiser M., Delhon I., Fournier B. (2002) Structure 10, 1697–1707 [DOI] [PubMed] [Google Scholar]
- 25.Kallen J., Schlaeppi J. M., Bitsch F., Delhon I., Fournier B. (2004) J. Biol. Chem. 279, 14033–14038 [DOI] [PubMed] [Google Scholar]
- 26.Kumar N., Solt L. A., Conkright J. J., Wang Y., Istrate M. A., Busby S. A., Garcia-Ordonez R., Burris T. P., Griffin P. R. (2009) Mol. Pharmacol. 8, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoner B. E., Bramlett K. S., Guo H., Burris T. P. (2005) Mol. Genet. Metab. 85, 318–322 [DOI] [PubMed] [Google Scholar]
- 28.Chalmers M. J., Busby S. A., Pascal B. D., He Y., Hendrickson C. L., Marshall A. G., Griffin P. R. (2006) Anal. Chem. 78, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 29.Pascal B. D., Chalmers M. J., Busby S. A., Griffin P. R. (2009) J. Am. Soc. Mass Spectrom. 20, 601–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pascal B. D., Chalmers M. J., Busby S. A., Mader C. C., Southern M. R., Tsinoremas N. F., Griffin P. R. (2007) BMC Bioinformatics 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bitsch F., Aichholz R., Kallen J., Geisse S., Fournier B., Schlaeppi J. M. (2003) Anal. Biochem. 323, 139–149 [DOI] [PubMed] [Google Scholar]
- 32.Volkman J. K. (2003) Appl. Microbiol. Biotechnol. 60, 495–506 [DOI] [PubMed] [Google Scholar]
- 33.Wada T., Kang H. S., Angers M., Gong H., Bhatia S., Khadem S., Ren S., Ellis E., Strom S. C., Jetten A. M., Xie W. (2008) Mol. Pharmacol. 73, 891–899 [DOI] [PubMed] [Google Scholar]
- 34.Duez H., van der Veen J. N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Baugé E., Havinga R., Bloks V. W., Wolters H., van der Sluijs F. H., Vennström B., Kuipers F., Staels B. (2008) Gastroenterology 135, 689–698 [DOI] [PubMed] [Google Scholar]
- 35.Le Martelot G., Claudel T., Gatfield D., Schaad O., Kornmann B., Sasso G. L., Moschetta A., Schibler U. (2009) PLoS Biol. 7, e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang J. Y. (2004) J. Hepatol. 40, 539–551 [DOI] [PubMed] [Google Scholar]
- 37.Chiang J. Y. (2009) J. Lipid Res. 50, 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell D. W. (2009) J. Lipid Res. 50, S120–S125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell D. W. (2003) Annu. Rev. Biochem. 72, 137–174 [DOI] [PubMed] [Google Scholar]
- 40.Brown A. J., Jessup W. (1999) Atherosclerosis 142, 1–28 [DOI] [PubMed] [Google Scholar]
- 41.Hahn C., Reichel C., von Bergmann K. (1995) J. Lipid Res. 36, 2059–2066 [PubMed] [Google Scholar]
- 42.Teng J. I., McGehee M. F., Smith L. L. (1981) J. Steroid Biochem. Mol. Biol. 14, 569–573 [DOI] [PubMed] [Google Scholar]
- 43.Wamil M., Andrew R., Chapman K. E., Street J., Morton N. M., Seckl J. R. (2008) Endocrinology 149, 5909–5918 [DOI] [PubMed] [Google Scholar]
- 44.Maeda Y., Nagatomo H., Uchiyama F., Nagatomo J., Yamada M., Shiotsuki H., Ohta Y., Sato S., Kai M. H., Kondo K. H., Higashi S., Setoguchi T. (2002) Steroids 67, 703–708 [DOI] [PubMed] [Google Scholar]
- 45.Jessup W., Brown A. J. (2005) Rejuvenation Res. 8, 9–12 [DOI] [PubMed] [Google Scholar]
- 46.Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W., Chen G., Head D. L., Mangelsdorf D. J., Russell D. W. (2007) Cell Metab. 5, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X. O., Pappu B. P., Nurieva R., Akimzhanov A., Kang H. S., Chung Y., Ma L., Shah B., Panopoulos A. D., Schluns K. S., Watowich S. S., Tian Q., Jetten A. M., Dong C. (2008) Immunity 28, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruning J. B., Chalmers M. J., Prasad S., Busby S. A., Kamenecka T. M., He Y., Nettles K. W., Griffin P. R. (2007) Structure 15, 1258–1271 [DOI] [PubMed] [Google Scholar]
- 50.Itoh T., Fairall L., Amin K., Inaba Y., Szanto A., Balint B. L., Nagy L., Yamamoto K., Schwabe J. W. (2008) Nat. Struct. Mol. Biol. 15, 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz M. A., Margolis S. (1983) J. Lipid Res. 24, 28–33 [PubMed] [Google Scholar]
- 52.Bostrom H. (1983) J. Biol. Chem. 258, 15091–15094 [PubMed] [Google Scholar]
- 53.Uchida K., Takeuchi N., Yamamura Y. (1975) Lipids 10, 473–477 [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi M., Ikegami H., Fujisawa T., Nojima K., Kawabata Y., Noso S., Babaya N., Itoi-Babaya M., Yamaji K., Hiromine Y., Shibata M., Ogihara T. (2007) Diabetes 56, 239–247 [DOI] [PubMed] [Google Scholar]
- 55.Staels B., Kuipers F. (2007) Drugs 67, 1383–1392 [DOI] [PubMed] [Google Scholar]
- 56.Besnard S., Heymes C., Merval R., Rodriguez M., Galizzi J. P., Boutin J. A., Mariani J., Tedgui A. (2002) FEBS Lett. 511, 36–40 [DOI] [PubMed] [Google Scholar]
- 57.Brown A. J., Leong S. L., Dean R. T., Jessup W. (1997) J. Lipid Res. 38, 1730–1745 [PubMed] [Google Scholar]
- 58.Gelissen I. C., Brown A. J., Mander E. L., Kritharides L., Dean R. T., Jessup W. (1996) J. Biol. Chem. 271, 17852–17860 [DOI] [PubMed] [Google Scholar]
- 59.Arca M., Natoli S., Micheletta F., Riggi S., Di Angelantonio E., Montali A., Antonini T. M., Antonini R., Diczfalusy U., Iuliano L. (2007) Free Radic. Biol. Med. 42, 698–705 [DOI] [PubMed] [Google Scholar]
- 60.Kaasik K., Lee C. C. (2004) Nature 430, 467–471 [DOI] [PubMed] [Google Scholar]
- 61.Rogers P. M., Ying L., Burris T. P. (2008) Biochem. Biophys. Res. Commun. 368, 955–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.