Abstract
We previously found that a population of colonic stromal cells that constitutively express high levels of prostaglandin-endoperoxide synthase 2 (Ptgs2, also known as Cox-2) altered their location in the lamina propria in response to injury in a Myd88-dependent manner (Brown, S. L., Riehl, T. E., Walker, M. R., Geske, M. J., Doherty, J. M., Stenson, W. F., and Stappenbeck, T. S. (2007) J. Clin. Invest. 117, 258–269). At the time of this study, the identity of these cells and the mechanism by which they expressed high levels of Ptgs2 were unknown. Here we found that these colonic stromal cells were mesenchymal stem cells (MSCs). These colonic MSCs expressed high Ptgs2 levels not through interaction with bacterial products but instead as a consequence of mRNA stabilization downstream of Fgf9 (fibroblast growth factor 9), a growth factor that is constitutively expressed by the intestinal epithelium. This stabilization was mediated partially through a mechanism involving endogenous CUG-binding protein 2 (CUGbp2). These studies suggest that Fgf9 is an important factor in the regulation of Ptgs2 in colonic MSCs and may be a factor involved in its constitutive expression in vivo.
Keywords: Cyclooxygenase (COX) pathway, ERK, Fibroblast, MRNA, Stem cell, CUGbp2, Cox-2, Fgf, PGE2, Mesenchymal stem cell
Introduction
Homeostasis in the mammalian colon relies on proper function of the epithelium and underlying mucosal immune system. Both of these cellular components are challenged by a local environment of a robust and diverse array of microbes that normally reside in the lumen of the colon (1–3). An important feature of the epithelial barrier is that it undergoes constant renewal during the lifetime of the organism. This process can be enhanced in response to either infection or injury (4, 5). Proliferative colonic epithelial progenitors (ColEPs)2 located in the basal one-third of the crypts of Lieberkühn are central players in the maintenance of the barrier epithelium in health and during injury response (6). ColEPs serve as a continuous source of maturing epithelial cells that migrate up crypts and terminally differentiate into one of three lineages: absorptive enterocytes, mucus-secreting goblet cells, and enteroendocrine cells.
The source of the signals responsible for regulating ColEP proliferation and differentiation of their daughter cells both during homeostasis and in response to injury is the local microenvironment surrounding the crypt base termed the progenitor cell niche. This niche is partially composed of the surrounding mesenchyme (reviewed in Ref. 7) that includes myofibroblasts, endothelial cells, neural cell extensions from the autonomic enteric nervous system, and mobile stromal cells capable of trafficking into and within the mesenchyme, such as immune cells (8, 9). Signals released from these many varied cell types interact with each other and the epithelium to regulate ColEPs and their progeny in homeostasis and in response to injury.
One additional cell type that we found appears to be an important component of the ColEP niche is a Ptgs2-expressing stromal cell (10). The key signaling molecule secreted by these cells is one of the downstream synthetic products of Ptgs2: prostaglandin E2 (PGE2) (10, 11). The immune modulatory properties of this and other downstream products of this gene family (Ptgs1 and Ptgs2), have been well established in vitro (12). In the gut, prostaglandins, most notably PGE2, have been shown to improve inflammation, ulceration, and other common measures of disease in model injury systems (9, 13–16).
Our goal in the current studies was to define and identify these Ptgs2-expressing colonic stromal cells and determine the mechanism by which their expression of Ptgs2 is mediated. We identified Ptgs2-expressing stromal cells as consistent with tissue-resident mesenchymal stem cells (MSCs). We isolated colonic MSCs using previously established protocols (17, 18) and found that their properties further supported the hypothesis that the Ptgs2-expressing stromal cells were colonic MSCs. We found that the high level of Ptgs2 expression in the colonic MSCs was not dependent upon exposure to bacterial products but rather upon local growth factors, specifically Fgf9. We found that Fgf9 signaling was sufficient to maintain the high levels of Ptgs2 expression in colonic MSCs (cMSCs) and that it appears to do so partially by ERK-mediated increase in mRNA-binding protein CUGbp2 and subsequent stabilization of Ptgs2 mRNA. These studies suggest further the recognized role of various growth factors in stabilizing Ptgs2 mRNA and suggest part of the mechanism in play in constitutive, as opposed to induced, expression of this important enzyme.
EXPERIMENTAL PROCEDURES
Mice
All animal experiments were performed in accordance with approved protocols from the Washington University School of Medicine Animal Studies Committee. Mice involved in this study were housed in microisolator cages, in a specified pathogen-free barrier facility following a 12-h light cycle and fed a standard irradiated chow diet (PicoLab Rodent Chow 20, Purina Mills) and water ad libitum. Ptgs2−/− mice (19) were generated previously on a C57Bl/6 background. C57Bl/6 mice were obtained from NCI (National Institutes of Health), and C57Bl/6 germ-free mice were obtained from the Digestive Disease Research Core Facility (Washington University).
Isolation of Organ-specific MSCs
Lung, stomach, colon, and bone marrow (flushed from isolated tibias and femurs) were removed from the mouse and were rinsed thoroughly with 1:1 Hanks' balanced salt solution/Dulbecco's modified Eagle's medium (DMEM). Each tissue was separately minced with scissors. The tissue fragments were placed in 20 ml of 10 mm HEPES-DMEM and 5 μg/ml type I collagenase (Invitrogen) and were incubated in an oscillating shaker at 225 rpm at 37 °C for 35 min. Dithiothreitol (Sigma) was then added to a final concentration of 20 mm, and incubation was continued for an additional 20 min. The resulting cell suspension was passed through a 70-μm filter, pelleted at ∼400 × g for 5 min, washed once in complete medium (low glucose DMEM with 10 mm HEPES and 10% fetal bovine serum with penicillin/streptomycin), and plated on standard tissue culture plates (VWR) in complete medium. Cells were cultured in a humidified chamber at 37 °C and 5% CO2. After 1 h, non-adherent cells were removed, and adherent cells were maintained in culture, feeding every 3–4 days in complete medium and were passaged (1:3) when they reached 90–100% confluence.
Isolation of Bone Marrow-derived Macrophages
Bone marrow-derived macrophages were cultured by flushing femurs and tibias of mice and culturing the cell suspension in standard medium supplemented with L-cell supernatant containing Csf-1 (20) for 7 days before replating and experimentation.
Splenocyte Proliferation Assay
This assay was performed by isolation and activation of 5 × 104 splenocytes in a 96-well dish by incubation with anti-CD3ϵ and anti-CD28 antibodies (21) in the presence or absence of cMSCs at a ratio of 5:1 to 50:1 splenocytes/cMSC or in the presence of cMSC culture supernatant. Cells were incubated for 72 h and pulsed with [3H]thymidine. Plates were harvested, and [3H]thymidine was measured. All conditions were performed in triplicate for each experiment.
Manipulation of Cell Lines
To differentiate MSCs into adipocytes, isolated MSCs were treated with 10−8 dexamethasone and 5 μg/ml insulin for 21 days (22). Verification of lipid stores was done by staining with Oil Red O. To differentiate MSCs into osteocytes, MSCs were treated with 10−8 dexamethasone, 5 μg/ml ascorbic acid 2-phosphate, 10 mm β-glycerophosphate for 21 days (22). Verification of Ca2+ deposits was performed by Alizarin Red S staining.
Serum starvation experiments were performed with slight modifications based on specific application. Experiments were carried out on cells plated in 24-well or 6-well dishes using 5 × 104 or 1 × 105 cells/well, respectively. For Fgf9 treatment, cells were starved for 2–3 h in low glucose DMEM with 10 mm HEPES before treatment with Fgf9 (Peprotech) (1–500 ng/ml) for 1 h before cells were lysed for RNA and protein isolation. For transcriptional inhibition coupled with starvation, treatment with actinomycin D (4 μg/ml) (Sigma) was begun simultaneously with or without 250 ng/ml Fgf9, and RNA was analyzed at 3 h. For MEK inhibition, treatments with PD98059 (100–300 μm) (Enzo Life Sciences) was performed in the same method as actinomycin D, and RNA and protein were collected at 3 h. LPS (Sigma) treatment of cells was carried out on cells plated at 105 cells/6-well dish well at a concentration of 1–100 ng/ml.
Prostaglandin E2 Assay
Cells were cultured with or without LPS (1–100 ng/ml) (Sigma) in 6-well plates at a density of 105 cells/well. Following 21–22 h of incubation, supernatants were collected for analysis using a PGE2 enzyme-linked immunosorbent assay kit (Cayman Chemical) according to the manufacturer's protocol. Cells were also counted, and RNA was isolated for quantitative reverse transcription-PCR (qRT-PCR) analysis.
Cell Transfection for shRNA Treatment
HEK293T cells were transfected with Mission shRNA constructs specific for CUGbp2 and non-targeting control (CUGbp2 clones NM_010160.1-131s1c1 and NM_010160.1-305s1c1; Sigma) and lentiviral packaging plasmids using FuGene HD reagent (Roche Applied Science). Following a 1-day transfection, cell medium was refreshed and allowed to accumulate virus for 24 h. Virus-containing supernatant was used undiluted directly on plated cMSCs for 24 h before adding puromycin (Sigma)-containing medium for selection of positively infected cells.
qRT-PCR Analysis
Cells were treated as noted in various experiments before RNA isolation. RNA was isolated and purified using QiaShredders and a Qiagen RNEasy minikit according to the manufacturer's protocol. cDNAs were synthesized using a Superscript III reverse transcriptase (Invitrogen), primed using random primers. qRT-PCRs were performed in triplicate for each sample using SYBR Green master mix (Clontech) and analyzed by either a Stratagene Mx3000P or Eppendorf Realplex Mastercycler. The following primers were used: 18S (AACCCGTTGAACCCCATT, CCATCCAATCGGTAGTAGCG); Ptgs2 (TGCCTGGTCTGATGATGTATG, GCCCTTCACGTTATTGCAGATG); human Ptgs2 (ATATGTTCTCCTGCCTACTGGAA, GCCCTTCACGTTATTGCAGATG); Primer 1 (Fig. 6D; AGCGAGGACCTGGGTTCAC); Primer 2 (Fig. 6D; TGGGTGTGATTTGTTTGGCATGG); Primer 3 (Fig. 6D; GGATACACCTCTCCACCAATGAC); Fgfr1b (GGGAATTAATAGCTCGGATG, CCACAGGTCTGGTGACAGTG); Fgfr1c (CCAGATCCTGAAGACTGCTG, GAGTCCGATAGAGTTACCCG); Fgfr2b (CTCGGGGATAAATAGCTCCA, GGAAGCCGTGATCTCCTTCT); Fgfr2c (GGGAATCGCTAGAGTTGCAG, TGTCGTCCTCATCATCTCCA); Fgfr3b (CAAGTTTGGCAGCATCCGGCAGAC, TCTCAGCCACGCCTATGAAATTGGTG); Fgfr3c (CAAGTTTGGCAGCATCCGGCAGAC, CACCACCAGCCACGCAGAGTGATG); Fgfr4 (GCCCTTCACGTTATTGCAGATG, CCTCTCCAACCCCGTACTC); Ptgs1 (AGTGCGGTCCAACCTTATCC, GCAGAATGCGAGTATAGTAGCTC); CD14 (CTCTGTCCTTAAAGCGGCTTAC, GTTGCGGAGGTTCAAGATGTT); Tlr4 (GCCTTTCAGGGAATTAAGCTCC, AGATCAACCGATGGACGTGTAA); Myd88 (AGGACAAACGCCGGAACTTTT, GCCGATAGTCTGTCTGTTCTAGT); MD-2 (CGCTGCTTTCTCCCATATTG, GTCTTATGCAGGGTTCAGAAC); IRAK-M (CTGGCTGGATGTTCGTCATATT, GGAGAACCTCTAAAAGGTCGC); tumor necrosis factor-α (CCCTCACACTCAGATCATCTTCT, GCTACGACGTGGGCTACAG); I-Ab (CCCTCATCAGGGAGAGGTCTA, CCGATGCCGCTCAACATCT); and CUGbp2 (GCTGCTTCAACCCCCAATTC, CGCCATACCTGCTAGTGCAT).
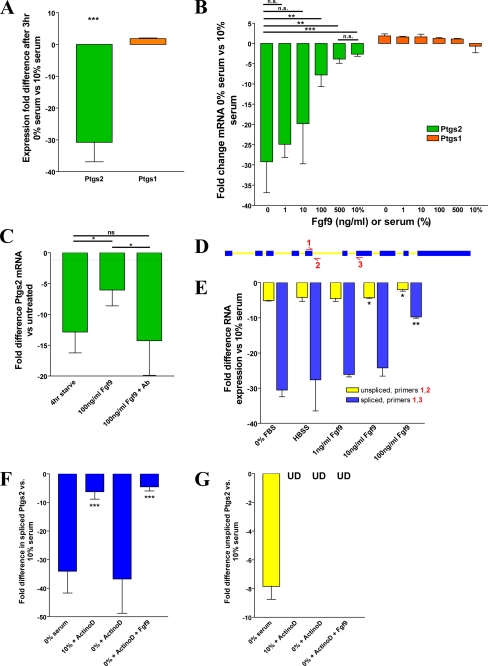
FIGURE 6.
Fgf9 is sufficient to stabilize Ptgs2 mRNA. qRT-PCR analysis (A–C and E–G) of cMSCs is shown. A, graph of the -fold difference for Ptgs2 and Ptgs1 expression of serum-starved cMSCs versus cMSCs grown in 10% serum. B, graph of the -fold difference for Ptgs2 and Ptgs1 expression in cMSCs after 1 h of Fgf9 treatment (range = 1–500 ng/ml) following 3 h of serum starvation. C, graph of the -fold difference for Ptgs2 expression after treatment with Fgf9 or Fgf9 plus anti-Fgf9 neutralizing antibody following 3 h of serum starvation. D, schematic illustrating the location of primers 1, 2, and 3 on the Ptgs2 gene. E, graph of the -fold difference for the spliced and unspliced forms of Ptgs2 isolated from 3-h serum-starved cMSCs additionally treated for 1 h with Fgf9. F, graph showing the -fold difference for Ptgs2 spliced RNA after treatment with or without serum starvation, 4 μg/ml actinomycin D, and 250 ng/ml Fgf9. The asterisks show comparison of data with -fold difference in cells treated with 0% serum and actinomycin D. G, graph showing the -fold difference for Ptgs2 unspliced RNA with or without serum starvation, 4 μg/ml actinomycin D, and 250 ng/ml Fgf9 (UD, undetectable). All data are representative of three independent experiments. Error bars, S.D. Statistical analysis by Student's t test is shown: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
Immunoblotting
Colonic MSCs were treated as noted. Following treatment, cells were lysed in radioimmune precipitation buffer (Sigma) containing protease and phosphatase inhibitor mixtures (Sigma) and frozen at −80 °C. Protein content was reduced by heating to 95 °C for 5 min in a ratio of 1:1 with 2× Laemmli buffer. Protein was loaded onto 10% or 4–12% BisTris gels (Invitrogen) and electrophoresed with MOPS buffer (Invitrogen). Samples were transferred to a 0.45-μm nitrocellulose membrane (BD Pharmingen) and blocked in 5% milk with 0.05% Tween 20, TBS (TBST) overnight at 4 °C. Blots were incubated for 2 h at room temperature in primary antibody, washed three times in TBST, and incubated for 1 h in horseradish peroxidase-conjugated anti-rabbit secondary (1:10,000) (Bio-Rad) before development using the SuperSignal West Dura chemiluminescent kit (Pierce). Primary antibodies were specific for phospho-ERK (diluted 1:500) (Cell Signaling Technologies), CUGbp2, and actin (each diluted 1:500) (Sigma). Quantitation was performed using ImageJ software.
FACS Analysis
For analysis of extracellular molecules, cultured cells were treated with trypsin/EDTA, pelleted, and resuspended in 100 μl of FACS buffer (1% bovine serum albumin in PBS) with the appropriate primary antibody for 1 h, washed three times in FACS buffer, incubated in 100 μl of FACS buffer with the appropriate secondary antibody, washed again, and analyzed on a FACSCalibur flow cytometer. Analysis was performed using FlowJo software. For analysis of intracellular enzymes, cells were fixed after suspension in 50% methanol in PBS for 10 min at −20 °C, followed by 90% methanol overnight at −20 °C. Cells were then stained and analyzed by the same methodology for extracellular molecules. Antibodies employed at a 1:100 dilution were CD3ϵ (BD Pharmingen), CD11b (BD Pharmingen), CD11c (BD Pharmingen), CD29 (BD Pharmingen), CD31 (BD Pharmingen), CD34 (eBioscience), CD44 (BD Pharmingen), CD45 (BD Pharmingen), CD54 (eBioscience), CD90 (Caltag Laboratories), CD105 (eBioscience), CD106 (eBioscience), Sca-1 (BioLegend), B220 (BD Pharmingen), F4/80 (Caltag Laboratories), NK1.1 (BD Pharmingen), Gr-1 (BD Pharmingen), I-Ab (BioLegend), and Ptgs2/Cox-2 (BD Pharmingen).
Global Gene Array Analysis
RNA from two cultured cell lines each of cMSCs and bone marrow MSCs (bmMSCs) was isolated using QiaShredders and the Qiagen RNEasy minikit according to instructions. RNA was measured on a nanodrop machine and amplified in one step using RiboAmp Plus from MDS Analytical Technologies according to the provided protocol. Amplified RNA was labeled with Turbo Labeling Biotin (MDS Analytical Technologies) and fragmented before hybridization onto MOE430A microarray chips from Affymetrix. Analyses of data were performed using dCHIP software (23) and gene ontology comparisons (24). Significance of differential expression was set at 1.3-fold difference with p < 0.05.
Immunohistochemistry
Mice were sacrificed, and colons were removed, flushed with room temperature PBS, opened, and pinned in 2% paraformaldehyde for 30 min. Tissue was then rinsed in 5% sucrose PBS and blocked in optimum cutting temperature compound. 7-μm sections were cut and stained by blocking in 2% dry milk PBS, incubating for 1 h in primary antibody, washing, incubating for 45 min in secondary antibody, staining with Hoechst nuclear dye, and finally coverslipping with a 1:1 glycerol/PBS solution. Sections were viewed with a Zeiss Axiovert 200 microscope with an Axiocam MRM camera. Antibodies used in staining are listed under “FACS Analysis.”
RESULTS
Ptgs2-expressing Stromal Cells Co-localize with Markers of MSCs
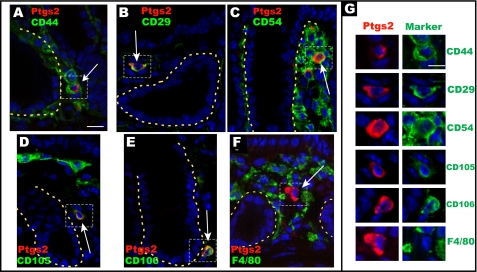
Previously, we found that stromal cells in the colonic mesenchyme expressed high levels of Ptgs2 in stained tissue sections co-labeled with CD44 but not with definitive hematopoietic, myofibroblast, or endothelial cell markers (10). We noted that other investigators found that CD44 was expressed in all MSCs isolated from a wide array of mouse tissues (18). Therefore, we tested the hypothesis that Ptgs2-expressing stromal cells were tissue-resident MSCs by screening a variety of MSC markers. CD29 (integrin β1) and CD44 (hyaluronate-binding protein) were selected as the best current defining markers for MSCs of all tissues (18), whereas CD54 (Icam1), CD105 (endoglin), and CD106 (Vcam1) are commonly described in numerous MSC lines (e.g. see Ref. 12). We confirmed our previous work (10) that demonstrated that Ptgs2-expressing cells co-expressed CD44 (Fig. 1A). Using double label immunofluorescence on colonic sections from WT mice, we also found that Ptgs2-expressing cells co-stained with the remaining MSC markers (Fig. 1, B–E), supporting our hypothesis. We also confirmed our previous analysis indicating that the Ptgs2-expressing stromal cells lacked expression for all hematopoietic markers tested, including F4/80 (macrophages; Fig. 1F), CD68 (macrophages), CD3ϵ (T cells), CD45, B220 (B cells), Gr-1 (granulocytes), and NK1.1 (NK cells) as well as markers of endothelial cells (CD31/Pecam) and prolyl-4-hydroxylase and fibroblast-specific protein (10). Taken together, these data support a hypothesis that the Ptgs2-expressing stromal cells are MSCs.
FIGURE 1.
Ptgs2-expressing stromal cells co-localize with markers of mesenchymal stem cells in vivo. Sections of the rectum from WT C57Bl/6 mice co-stained with Alexa-Fluor 594 Zenon-labeled anti-mouse Ptgs2 IgG1 (red) and various MSC and hematopoietic markers. All MSC markers (A–E) were visualized with an Alexa-Fluor 488-conjugated anti-rat Ig secondary antibody used to detect rat anti-mouse CD44 (A), rat anti-mouse CD29 (B), rat anti-mouse CD54 (C), rat anti-mouse CD105 (D), and rat anti-mouse CD106 (E). The hematopoietic co-staining marker was fluorescein isothiocyanate-conjugated F4/80 (F). The yellow dashed lines denote the basolateral membrane of the epithelium. G, magnified insets of the dashed boxed portions of A–F. Ptgs2 co-labels with all MSC markers but not F4/80. Bars, 20 μm (A–F) and 15 μm (G).
Isolated Colonic Stromal Cells Are Identified as MSCs and Exhibit Marker Expression Similar to Ptgs2-expressing Stromal Cells in Vivo
MSCs are multipotent progenitors for a number of mesenchymal cell types that have been isolated from many mouse and human tissues (18, 22). One of their functions is to mediate tissue repair either through the secretion of various factors or by differentiation into specific cell types (e.g. see Ref. 25). To test our hypothesis that Ptgs2-expressing stromal cells are MSCs, we isolated MSCs from the adult mouse colon by the technique originally developed by Friedenstein et al. (17) and modified for a wide array of mouse organs (18). Briefly, collagenase-dissociated colonic cells were plated for 1 h, and the adherent cells were expanded and analyzed. We have maintained cell lines produced in this manner for at least 40 passages in culture as has been described for MSCs isolated from other organs.
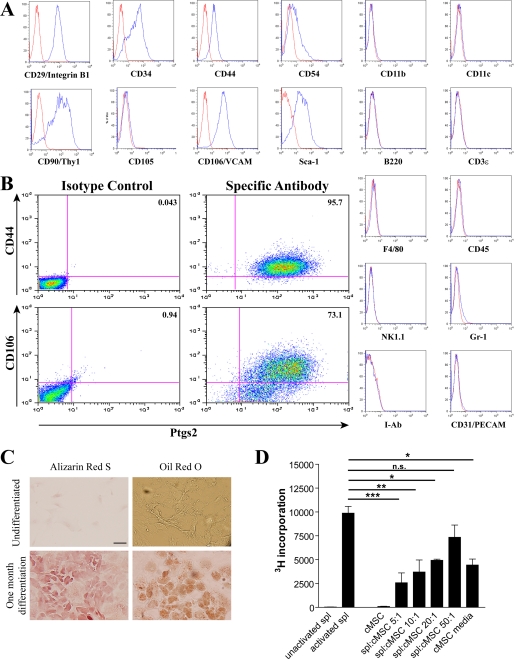
We characterized the putative MSC cell lines isolated from the mouse colon in order to determine whether these cells were representative of the Ptgs2-expressing cells that we identified in vivo. Cells cultured in low glucose DMEM with 10% serum were used to analyze expression of pertinent surface proteins identified in vivo. Single channel flow cytometric analysis of the colonic stromal cells showed expression of CD29, CD44, CD54, and CD106 in all lines isolated from WT colons (n = 5) at early passages (P3–P5; Fig. 2A). Expression of these markers was stably maintained up to P40 (data not shown). Interestingly, we could detect expression of CD105 in cells after initial isolation and expansion, but expression of this marker was undetectable by P3 (Fig. 2A). These putative MSCs also lacked expression of hematopoietic markers (CD11b, CD11c, B220, CD3ϵ, F4/80, CD45, NK1.1, Gr-1, and I-Ab) and endothelial markers (CD31/Pecam). Most significantly, these cells co-expressed Ptgs2 with CD44 and CD106 as demonstrated by dual label flow cytometry (Fig. 2B). Taken together, these data suggested that the isolated and expanded stromal cells were representative of the stromal cells identified by strong Ptgs2 immunofluorescence in vivo (10).
FIGURE 2.
Isolated colonic stromal cells are mesenchymal stem cells and maintain Ptgs2 expression. A, representative histograms of flow cytometric analysis of cultured colonic stromal cells (n = 5 lines) at passages 3–5 stained for the markers shown (blue lines). Antibodies to CD11b, CD11c, CD90, Sca-1, B220, CD3ϵ, F4/80, CD45, NK1.1, Gr-1, and I-Ab were preconjugated fluorescein isothiocyanate- or Alexa-Fluor 488-labeled primary mouse antibodies. Cells labeled with CD29, CD34, CD44, CD54, CD105, CD106, and CD31 were detected with an Alexa-Fluor 488-conjugated anti-rat secondary. Controls were antibodies of the same isotype (red lines). B, representative double label flow cytometric analysis of colonic stromal cells (n = 3 lines) co-stained with either CD44 or CD106 as described above and Ptgs2 detected by Alexa-Fluor 647-conjugated anti-mouse antibody. C, representative histological images of colonic stromal cells (n = 3 lines), which were plated on coverslips and treated for 21 days in appropriate media conditions (see “Experimental Procedures”). Following incubation, cells were fixed and stained for calcium deposition by Alizarin Red S or for lipid deposition by Oil Red O. Undifferentiated cells plated 2 days before staining were likewise visualized. Bar, 50 μm. D, graph of [3H]thymidine incorporation by splenocytes (spl). Splenocytes were plated with or without activation by antibodies to CD3ϵ and CD28 in triplicate in 96-well plates. Colonic stromal cells were similarly plated either alone or at varying ratios with splenocytes. Error bars, S.D. Statistical analysis by analysis of variance and Bonferroni post-test is shown: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
We then performed expression analysis of CD90, Sca-1, and CD34. These proteins are additional markers of various organ-specific MSCs (isolated from spleen, muscle, blood vessels, kidney, lung, liver, brain, and thymus) that are known to show variation in expression (18). The isolated colonic stromal cells expressed all three of these markers (Fig. 2A). This finding recapitulated analysis of marker proteins from bmMSCs with the exception of CD34, which is commonly reported to be absent on bmMSCs (26, 27).
We then determined if the isolated colonic stromal cell lines had properties of bona fide MSCs. Like all other MSCs, the colonic stromal cells were able to differentiate into osteocytes and adipocytes when cultured under appropriate conditions, as demonstrated by staining by Alizarin Red S and Oil Red O, respectively (22, 28) (Fig. 2C). This capacity confirmed the multipotency of these cells. Furthermore, the isolated cells were capable of modulating immune function as has been previously shown for bmMSCs (12, 29). Co-culture of the colonic stromal cells with anti-CD3/CD28-activated splenocytes resulted in significantly decreased splenocyte proliferation (to a ratio of splenocytes/cMSCs of 20:1) in a dose-dependent manner as measured by incorporation of [3H]thymidine (Fig. 2D). As in similar experiments with bmMSCs (12), culture media from the colonic stromal cells also significantly decreased splenocyte proliferation (Fig. 2D). Taken together, these data confirm that our isolated colonic stromal lines have the properties of MSCs; therefore, they were termed cMSCs.
High Constitutive Ptgs2 Expression Is Unique to cMSCs
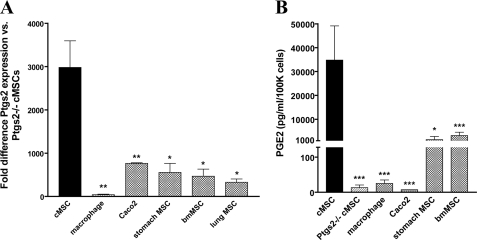
Based on previous experiments in colonic injury repair (9, 13, 14) (reviewed in Ref. 11), our hypothesis was that one key defining characteristic of cMSCs is their expression of high levels of Ptgs2 and consequent production of high levels of PGE2. We first directly compared the expression of Ptgs2 in and secretion of PGE2 from cMSCs isolated from WT versus Ptgs2−/− mice. This analysis showed that Ptgs2 activity accounted for >99.9% of secreted PGE2 (Fig. 3, A and B). We also compared our cMSC lines with other major colonic cell types capable of expressing high Ptgs2 in certain circumstances: macrophages (30) and epithelial tumor cells (31). We found that cMSCs contained 74-fold higher levels of Ptgs2 mRNA when compared with bone marrow-derived macrophages and >1000-fold higher levels of secreted PGE2. In a comparison of gene expression in cMSCs and a model tumor line (Caco-2 cells), we saw that the cMSCs contained 4-fold higher levels of Ptgs2 and produced >4000-fold higher levels of PGE2.
FIGURE 3.
High constitutive Ptgs2 expression is unique to cMSCs. A, graph comparing Ptgs2 expression by qRT-PCR in various cell lines (base line: cMSCs isolated from Ptgs2−/− mice). MSC lines were isolated from the colon (n = 7 lines), stomach (n = 4 lines), bone marrow (n = 4 lines), and lung (n = 2 lines) of WT C57Bl/6 mice and colon of Ptgs2−/− mice (n = 2 lines). Macrophages (n = 2 lines) were derived from bone marrow of C57Bl/6 mice with differentiation in L-cell supernatant. Caco2 cell data represent n = 2 different passages. All cDNAs used in qRT-PCR were synthesized using random hexamer primers. Statistical markings (asterisks) refer to comparison with WT cMSC Ptgs2 expression. B, graph of PGE2 secretion measured by an enzyme-linked immunosorbent assay of various cell lines described above. Statistical markings refer to comparison with WT cMSC PGE2 secretion. Error bars, S.D. Statistical analysis by Student's t test is shown: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
We then assessed whether the high levels of Ptgs2 in cMSCs were a general property of MSCs because bmMSCs have also been reported to produce PGE2 (12). Therefore, we isolated MSCs from a variety of adult mouse organs, including the bone marrow, lung, and stomach, and maintained these MSCs in culture as we did for cMSCs. Interestingly, the cMSCs contained the highest levels of Ptgs2 mRNA (range of 5–10-fold greater message) and produced the highest levels of PGE2 (range of 10–25-fold) in comparison with all of these other MSC lines (Fig. 3, A and B).
High Constitutive Ptgs2 Expression Is Not Due to TLR Stimulation
We considered two possible explanations for the observed high constitutive expression of Ptgs2 in cMSCs: 1) that it is cell-autonomous or 2) that it requires endogenous signal(s) from the local environment of the colon that educate the cells to express abundant Ptgs2. These endogenous signals could be derived from the host and/or from the indigenous microbes.
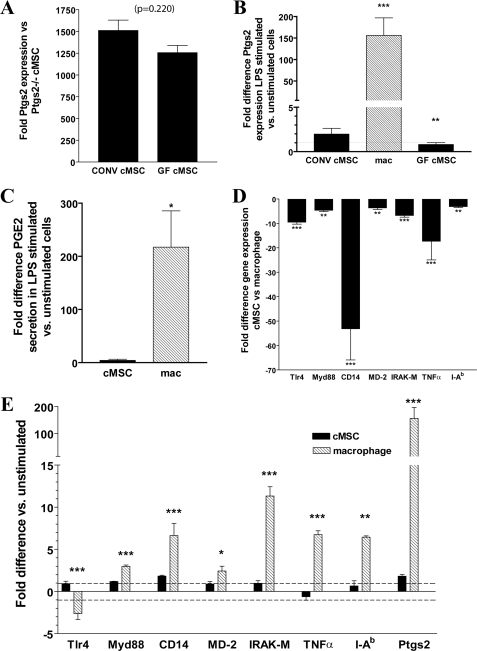
Because cMSCs are located in the colon and would thus potentially be exposed to microbes and/or microbial products, we first tested the hypothesis that the high levels of Ptgs2 expression may be induced/maintained via TLR signaling. To test this idea, we isolated cMSCs from adult C57Bl/6 germ-free (GF) mice that lacked all microbes. Interestingly, GF cMSCs contained similar levels of Ptgs2 expression as conventionally raised (CONV) mice that contained the normal gut microbiota (Fig. 4A), indicating that in vivo TLR stimulation was not necessary during cMSC development to produce high levels of Ptgs2.
FIGURE 4.
High constitutive expression of Ptgs2 is not due to TLR activation. Colonic MSCs from CONV and GF mice and bone marrow-derived macrophages were isolated and analyzed. A, graph of Ptgs2 expression difference in cMSCs isolated from CONV (n = 3 lines) and GF mice (n = 2 lines) measured by qRT-PCR (base line: cMSCs from CONV Ptgs2−/− mice). p = 0.220, Student's t test. B, graph of Ptgs2 expression difference in MSCs and macrophages plated and stimulated by LPS (1–100 ng/ml) for 21 h (base line: unstimulated cells). C, graph of PGE2 secretion -fold difference (base line: secretion from unstimulated cells) from cells stimulated by LPS measured by an enzyme-linked immunosorbent assay of supernatants from cMSC (n = 3 lines) and macrophages (n = 3). D, graph of TLR4 pathway member gene expression -fold difference in cMSCs (n = 3 lines) base-lined to macrophages (n = 3 lines) for each gene measured by qRT-PCR. E, graph of -fold expression difference in members of the TLR4 pathway and target genes measured by qRT-PCR in 21-h LPS-stimulated cells (base line: unstimulated cells). Statistical markings represent significance in the -fold difference of a given gene in cMSCs compared with the -fold difference of that gene in macrophages. Error bars, S.D. Statistical analysis by analysis of variance and Bonferroni post-test (B) or Student's t test (A and E) is shown: *, p < 0.05; **, p < 0.01; ***, p < 0.001. TNFα, tumor necrosis factor-α.
The striking result obtained using GF cMSCs led us to determine if cMSCs could respond to TLR stimulation. In the normal colon, one dominant TLR ligand is lipopolysaccharide (LPS). Therefore, we tested the response of cMSCs isolated from CONV mice to LPS stimulation. We considered the possibility that a lack of response in the CONV mice could be the result of attenuation of TLR signaling due to microbial stimulation of these cells while in the mouse gut (32), so we also evaluated the response of GF cMSCs to account for this possibility. We found that stimulation of either CONV or GF cMSCs with LPS (at concentration ranging from 1 to 100 ng/ml) showed no significant induction of Ptgs2 as compared with untreated CONV and GF cMSCs, respectively. As a positive control, we similarly treated bone marrow-derived macrophages (a cell type known to respond to LPS) isolated from adult C57Bl/6 mice and found that these cells demonstrated >150-fold increase in Ptgs2 expression upon incubation with LPS (Fig. 4B), which corresponded with an increase of >200-fold in PGE2 secretion (Fig. 4C). These results suggested that the high constitutive level of Ptgs2 expression in cMSCs was not the result of an in vivo microbe-dependent maturation.
We next identified a potential mechanism for the lack of cMSC response to LPS by comparing expression of various members of the TLR4 pathway in cMSCs versus bone marrow-derived macrophages. By qRT-PCR analysis, expression levels of many genes important in relaying the LPS signal were statistically significantly lower in untreated cMSCs than untreated macrophages, including Tlr4 (-fold difference = −30) and CD14 (-fold difference = −50; Fig. 4D). The near lack of CD14 expression in cMSCs is probably important because this molecule is a necessary component for recognition of LPS by Tlr4 (for a review, see Ref. 33). Last, macrophages predictably responded to LPS stimulation by alteration of the expression of TLR pathway members, including IRAK-M, major histocompatibility complex class II (I-Ab), and Ptgs2 (Fig. 4E). Analysis of similarly treated cMSCs did not show significant changes in the expression of any of these genes (Fig. 4E). This expression analysis confirms that the high constitutive expression of Ptgs2 in cMSCs was probably not the result of TLR-stimulated maturation.
Comparison of Global Gene Expression between Organ-specific MSCs Reveals Candidate Pathways That Could Enrich Ptgs2 Expression in cMSCs
Because microbially derived signals did not appear to participate in the high endogenous levels of Ptgs2 expression in cMSCs, we evaluated signals from other host cells. A proven method for identifying upstream candidates in a target cell type is global gene expression analysis. To identify cMSC-specific Ptgs2 regulatory pathways, we compared these cells to bmMSCs because 1) they expressed less Ptgs2/PGE2 than cMSCs and 2) they shared important properties with cMSCs (i.e. multipotency and immune modulation).
We probed Affymetrix MOE430A microarrays with cRNA amplified from total RNA procured from two independently isolated lines of cMSCs and bmMSCs. We analyzed the data using dCHIP software (23) and generated a list of 575 probe sets (corresponding to 427 unique genes) that were elevated in the cMSCs as compared with bmMSCs (using a 1.3-fold expression difference cut-off and p < 0.05 (9). Importantly, this list included Ptgs2 (11-fold difference) and CD34 (200-fold difference), which correlated with the observed differences in protein expression between these two cell lines (data not shown).
We then functionally categorized the genes that were enriched in cMSCs and bmMSCs using gene ontology (GO) terms (24). The three most highly represented categories of genes preferentially expressed in cMSCs were “integral to membrane” (fractional representation: 0.284), “protein binding” (fractional representation: 0.275), and “membrane” (fractional representation: 0.275). Conversely, the three most highly represented GO terms in bmMSCs were “protein binding” (fractional representation: 0.354), “membrane” (fractional representation: 0.320), and “nucleus” (fractional representation: 0.296). The most striking feature of this analysis was that the “integral to membrane” GO term was the most highly represented in the cMSC list. This is quite unusual for a stem cell (34), and we therefore focused closely on this subset of genes.
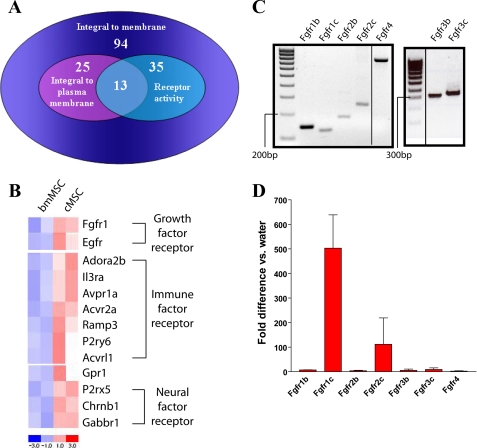
In order to narrow the list of candidates involved in the regulation of high levels of Ptgs2 in cMSCs from the 94 genes in the “integral to membrane” category, we reasoned that genes of interest would also be capable of receiving and transducing an extracellular signal. Therefore, we required that genes of interest also contain the GO terms “integral to plasma membrane” (25 genes) and “receptor activity” (35 genes). The intersection of “integral to membrane” genes that contained these two additional GO terms generated a list of 13 genes (Fig. 5A) that were enriched in cMSCs as compared with bmMSCs (Fig. 5B). Interestingly, these 13 genes could be loosely categorized into the receipt of three general types of signals: growth, immune, and neural (Fig. 5B).
FIGURE 5.
Comparison of cMSCs with bmMSCs reveals possible mechanistic pathways involved in Ptgs2 expression. A, Venn diagram illustrating the intersection of the GO terms “integral to plasma membrane” and “receptor activity” obtained through GO analysis of genes preferentially expressed in cMSCs (compared with bmMSCs). Data for analysis were obtained by isolation of total RNA from cMSCs (n = 2 lines) and bmMSCs (n = 2 lines), amplification and cRNA labeling, hybridization to Affymetrix MOE430A microarrays, and dCHIP analysis of genes preferentially expressed in one cell type or the other at a 1.3-fold level, p < 0.05. B, heat map generated by dCHIP software of genes fitting into the logical intersection of “integral to plasma membrane” and “receptor activity” showing relative expression levels in each cell line. C, electrophoresis gel of PCR amplifying cDNA obtained from whole WT C57Bl/6 mouse intestine for the FGF receptor splice forms: Fgfr1b, Fgfr1c, Fgfr2b, Fgfr2c, and Fgfr4 (annealing temperature = 60 °C) and Fgfr3b and Fgfr3c (annealing temperature = 65 °C). D, graph of qRT-PCR measurements of the various splice forms of the FGF receptors expressed in cMSCs (n = 3 lines) compared with water control. Error bars, S.D.
We further evaluated the possible role of Fgfr1 because previous studies in our laboratory demonstrated that communication between Fgf9 and MSCs in the intestinal tract exists during development (35), thus demonstrating the ability of at least one type of MSC to respond physiologically to FGF signaling. PCR analysis of cDNA isolated from whole WT adult colon showed that all four FGF receptors were expressed, including all “b” and “c” splice forms. The “b” forms are expressed predominantly in epithelial cells, and the “c” forms are expressed predominantly in mesenchymal cells (36) (reviewed in Ref. 37) (Fig. 5C). We then performed qRT-PCR for Fgfr1, -2, -3, and -4 using total RNA isolated from cMSCs and found that cMSCs contained detectable expression of only the “c” form of Fgfr1 and Fgfr2 (Fig. 5D). This expression analysis confirmed that signaling by FGF ligands to cMSCs was possible. We then focused on the role of Fgf9 as it is expressed in the epithelium of the colon (38) and appears to signal to MSCs during development (35).
Fgf9 Is Sufficient to Maintain High Levels of Ptgs2 Expression in cMSCs
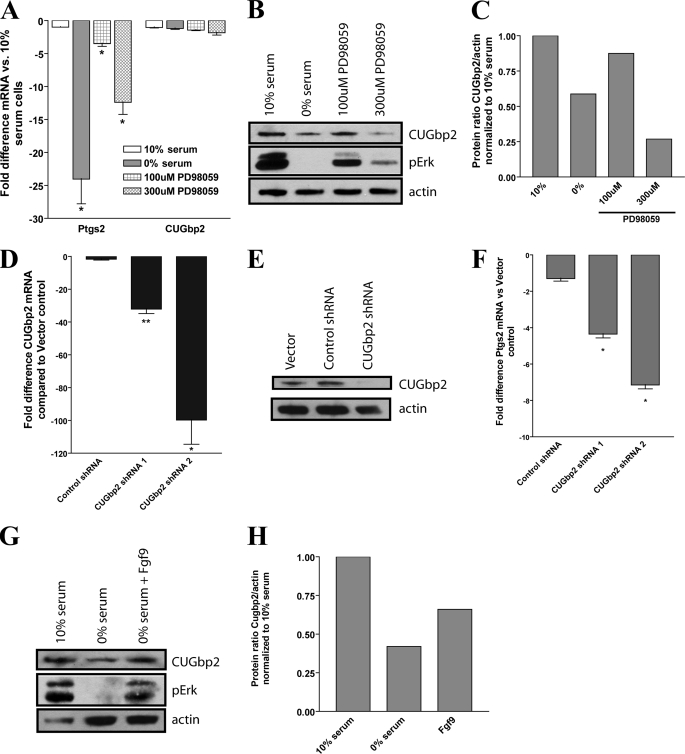
We hypothesized that Fgf9 stimulation may be responsible for induction and/or maintenance of Ptgs2 expression in cMSCs. Therefore, we studied its effects on expression in cMSCs in vitro. Colonic MSCs were serum starved to remove the growth factor signaling that was present during standard tissue culture. As little as 3 h of starvation resulted in an ∼25-fold decrease of Ptgs2 expression, whereas expression of other genes, notably Ptgs1, was unaffected within this time frame (Fig. 6A). This loss of Ptgs2 mRNA was rescued in a dose-dependent manner by the addition of Fgf9 (range of 1–500 ng/ml) to the culture medium for 1 h (Fig. 6B). This treatment had no effect on expression of Ptgs1. The addition of Fgf9 was sufficient to rescue Ptgs2 expression to a similar level as the addition of 10% serum. Furthermore, the effects of exogenous Fgf9 on Ptgs2 mRNA levels could be blocked by the addition of a neutralizing antibody directed against Fgf9 (Fig. 6C).
In order to determine whether Ptgs2 mRNA was increased due to transcriptional activation or post-transcriptional regulation/stabilization, we designed a transcript-specific qRT-PCR assay. Primers were designed to amplify a product specifically from the unspliced (unprocessed) form of Ptgs2 RNA (primers 1 and 2) or to amplify a product specifically from the spliced (mature) form of Ptgs2 RNA (primers 1 and 3; Fig. 6D). Both the unspliced and spliced forms of Ptgs2 RNA expressed by cMSCs showed decreased levels after serum starvation for 3 h, but the effect on spliced mRNA was much more pronounced. Treatment with 100 ng/ml Fgf9 after starvation significantly rescued both the spliced and unspliced forms of Ptgs2 RNA (Fig. 6E). These data suggest that Fgf9 may regulate Ptgs2 RNA expression at either the transcriptional or post-transcriptional level.
In order to better delineate between the two levels of regulation, we devised an experiment to specifically test the effect of Fgf9 on the post-transcriptional regulation of Ptgs2. To do this, we removed the possibility of Fgf9 affecting Ptgs2 expression at the transcriptional level by blocking transcription in cMSCs with actinomycin D treatment (4 μg/ml). Blockade of transcription in cMSCs cultured with 10% serum in the medium had no significant effect on the mature Ptgs2 mRNA levels in 3 h (Fig. 6F). This finding suggested that a growth factor present in the serum was able to maintain the mature form of Ptgs2 mRNA synthesized before transcriptional blockade. As in serum-starved conditions alone, a significant decrease in Ptgs2 mature mRNA was seen when serum starvation was compounded by transcriptional repression by actinomycin D (Fig. 6F). We suspected that Fgf9 would be sufficient to mediate the stabilization of mature Ptgs2 mRNA seen in transcriptional blockade in the presence of 10% serum and tested this hypothesis. We found that serum-starved cMSCs treated with actinomycin D in the presence of Fgf9 (250 ng/ml) showed no loss of mature Ptgs2 mRNA, indicating that Fgf9 is capable of maintaining Ptgs2 expression independent of transcriptional activation (Fig. 6F). Also, because expression of unspliced Ptgs2 should depend completely upon transcription, we showed that Fgf9 was unable to rescue the total loss of unspliced Ptgs2 expression upon blockade of transcription by actinomycin D treatment (Fig. 6G). These series of experiments confirmed that the serum starvation-induced decrease of mature Ptgs2 mRNA expression could be rescued by Fgf9 independent of transcription. Thus, Fgf9 was sufficient to maintain mature Ptgs2 mRNA expression and affected Ptgs2 expression at a post-transcriptional level.
Fgf9 Stabilizes Ptgs2 mRNA in Part through CUGbp2
To investigate the mechanism of Fgf9-mediated Ptgs2 mRNA stabilization, we first tested the role of ERK, a component of the MAPK cascade capable of transducing FGF signaling in many cells (39). ERK activation was inhibited by use of the MEK inhibitor PD98059. We found that an increasing dosage of PD98059 in cMSCs decreased phosphorylation of ERK and that this treatment also corresponded to a decrease in Ptgs2 mRNA expression (Fig. 7, A and B). As expected, ERK phosphorylation is also decreased in the absence of serum. These results support the role for ERK activation in Ptgs2 stabilization.
FIGURE 7.
Fgf9 stabilizes Ptgs2 partially through ERK activation and increased CUGbp2 protein. A, graph of the -fold difference for Ptgs2 and CUGbp2 mRNA following a 3-h treatment of cMSCs with an ERK kinase inhibitor, PD98059 (100 or 300 μm), or 0% serum. B, reducing immunoblot of PD98059-treated cMSCs probed for actin (loading control), phosphorylated ERK, and CUGbp2. C, graph of the quantification of B showing the ratio of CUGbp2/actin protein quantity in cMSCs treated by serum starvation or with PD98059. Base line (1.00) was the ratio of cMSCs grown in 10% serum. D, graph of the -fold difference in CUGbp2 mRNA in cMSCs lentivirally transfected with control shRNA or CUGbp2 shRNAs compared with vector control. E, reducing immunoblot of cMSCs transfected with vector control, control shRNA, or CUGbp2 shRNA showing specific knockdown of CUGbp2 protein. F, graph of the -fold difference for Ptgs2 mRNA in cMSCs transfected with control shRNA and CUGbp2 shRNA versus vector control. Shown is a reducing immunoblot of cMSCs treated with 10% serum, 0% serum, or 0% serum plus Fgf9 (250 ng/ml) (G) and quantification of the ratio of CUGbp2/actin in these cells (H). Base line (1.00) was the ratio of cMSCs grown in 10% serum. Immunoblots and quantifications are representative of three independent experiments. A, D, and F, the asterisks show comparison of data with -fold difference in cells grown in 10% serum. Statistical analysis was performed by Student's t test: *, p < 0.05; **, p < 0.01.
We then undertook a candidate molecule approach to identify possible proteins that could be downstream targets of Fgf9/ERK activation. Our microarray data (Fig. 5) identified multiple mRNA-binding proteins that were preferentially expressed in cMSCs compared with bmMSCs. One of these was CUGbp2, a protein known to stabilize Ptgs2 mRNA (40, 41) and whose activity can be regulated by phosphorylation downstream of growth factors (42). We evaluated this gene at both the mRNA and protein level in the context of ERK inhibition and found that although CUGbp2 mRNA was not significantly altered by either serum starvation or by ERK inhibition (Fig. 7A), the level of endogenous CUGbp2 protein was decreased both in the absence of serum and upon ERK inhibition (Fig. 7, B and C).
This result suggested that CUGbp2 protein was required to stabilize Ptgs2 mRNA in cMSCs. We tested this idea by specifically inhibiting CUGbp2 protein translation through shRNA-mediated knockdown of CUGbp2 expression. Lentiviral transfection of cMSCs with two different CUGbp2-specific shRNAs produced significant knockdown of CUGbp2 mRNA as measured by qRT-PCR (Fig. 7D). Importantly, transfection with control shRNA did not affect CUGbp2 expression. The shRNA-mediated decrease of CUGbp2 mRNA corresponded to a decrease in CUGbp2 protein expression as demonstrated by immunoblot (Fig. 7E). We then assessed the effect of CUGbp2 loss on Ptgs2 expression in cMSCs. We found a significant decrease in Ptgs2 message level in agreement with studies in other cell lines (Fig. 7F) (40, 42).
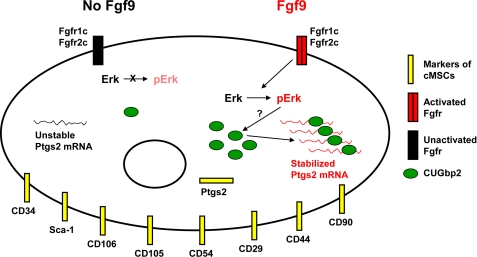
Based on these results, we hypothesized that Fgf9 may act through ERK to increase CUGbp2 protein. To test this hypothesis, we repeated our previous experiments and assessed the phosphorylation of ERK and the quantity of CUGbp2 protein in cells treated in normal serum, in 0% serum, and in 0% serum supplemented only with Fgf9 protein. We observed an increase in both ERK phosphorylation in Fgf9-treated cells as well as increased levels of endogenous CUGbp2 protein (Fig. 7, G and H). Taken together, these observations suggest that Ptgs2 mRNA stabilization by Fgf9 signaling is partially mediated through phosphorylation of ERK, leading to increased CUGbp2 protein, resulting in CUGbp2-mediated stabilization of Ptgs2 mRNA (Fig. 8).
FIGURE 8.
Model of Fgf9 regulation of Ptgs2 expression in cMSCs. Colonic MSCs express CD29, CD34, CD44, CD54, CD90, CD105, CD106, Sca-1, and Ptgs2. When Fgf9 is present, it signals through Fgfr1c and/or Fgfr2c on the cell surface. Transduction of this signal via ERK phosphorylation and activation results in an increase in CUGbp2 protein. This protein in turn binds to Ptgs2 mRNA and stabilizes the message. In the absence of Fgf9, ERK phosphorylation is decreased, resulting in decreased CUGbp2 protein quantity and loss of Ptgs2 mRNA stabilization.
DISCUSSION
Here we define a previously unidentified Ptgs2-expressing cell population within the adult mammalian intestine as mesenchymal stem cells. We demonstrated a novel mechanism by which the observed constitutive expression of Ptgs2 is maintained within these cells downstream of Fgf9, a growth factor expressed in the adult mammalian intestine. Our investigations suggest that the stabilization of Ptgs2 mRNA downstream of Fgf9 is mediated via phosphorylation of the MAPK signaling cascade member ERK, followed by increased abundance of CUGbp2 protein (Fig. 8).
Role of FGFs in Development and Homeostasis
There are at present 22 recognized FGF ligands that can be grouped into seven subfamilies based on phylogenetic analysis. The FGF of interest in these studies, Fgf9, is the namesake for one of these subfamilies that also includes Fgf16 and Fgf20. FGF ligands signal specifically through certain FGF receptors. There are four total receptors with Fgfr1, -2, and -3 expressed as either a “b” or “c” splice form, creating essentially seven different FGF receptors (for more details see Refs. 37 and 43). FGFs are known to play important roles in the morphogenesis and expansion of developing structures, including the lung (44, 45), limb (46), intestine (35, 38, 47), epidermis (48), and nervous system (49).
We have demonstrated an interaction between Fgf9 and Ptgs2 in a mesenchymal cell type unique to the colon, the colonic mesenchymal stem cell. Our findings potentially expand the growing recognition of the role of FGFs in homeostasis and normal physiology within the adult organism. In particular, FGFs have recently been shown to aid regulation of nutrient and mineral homeostasis. For example, Fgf15 can be induced in the small intestinal epithelium and subsequently signal to the liver, where it regulates enzymes of the bile acid synthesis pathway (50). Thus, Fgf15 has an important role in enterohepatic homeostatic regulation of bile acid production. In other studies, Fgf23, produced by the bone, plays a role in regulating calcium and phosphate homeostasis that requires precise control of the kidney, parathyroid, intestine, and bone. Fgf23 appears to have multiple cellular targets in this regulation (51, 52). Also, nutrient availability and composition can impact expression of specific FGFs, including keratinocyte growth factor (also known as Fgf7) (53). Our studies raise the intriguing possibility that another FGF may be acting in a homeostatic role in the adult organism, specifically that Fgf9 may be important in maintaining the constitutive expression of Ptgs2 in this specific subset of mesenchymal cells that have already been implicated in injury response (10).
Growth Factors Affecting Ptgs2 Expression
Our studies have implicated Fgf9 as a factor sufficient to mediate Ptgs2 mRNA stability. However, a variety of other growth factors are also likely to be capable of regulating Ptgs2 expression in this cell type. These factors include FGFs other than Fgf9 (54), epidermal growth factor (55, 56), and insulin-like growth factor (57, 58).
These various growth factors may act via a number of signaling networks that have been implicated in the regulation of Ptgs2, many of which are involved in the induction of gene expression during the inflammatory response rather than in constitutive expression (59). Our in vitro studies strongly indicate that, rather than acting primarily via transcriptional up-regulation, Fgf9 acts at the post-transcriptional level and suggest that the mechanism of this interaction partially involves CUGbp2 protein expression. This protein and others, including HuR and Apobec-1, have been implicated in the regulation of Ptgs2 expression in various tumor cell lines (40, 41) and in an in vivo model of irradiation injury (60). Comparing the ability of Fgf9 to increase CUGbp2 protein with its ability to increase Ptgs2 mRNA stabilization suggests that increased CUGbp2 is only a portion of the overall mechanism downstream of Fgf9 involved in Ptgs2 expression. It is very likely that this growth factor is also signaling through other kinase cascade pathways, including MAPK through c-Jun N-terminal kinase (JNK) and p38, protein kinase C, and phosphatidylinositol 3-kinase (reviewed in Ref. 61). Intriguingly, the Src family of kinases has recently also been implicated in the stabilization of Ptgs2 mRNA downstream of signaling by platelet-derived growth factor (42). There must also be more than one target of kinase activity. There are a variety of further mRNA-binding proteins that are probably involved in this cascade, some well researched, such as HuR (41), and others unknown.
Role of CUGbp2 in Stabilizing Ptgs2 Expression
Previous studies have shown that CUGbp2 is capable of stabilizing Ptgs2 mRNA via a mechanism requiring binding of the protein to the AU-rich region of the 3′-UTR of the Ptgs2 message (40–42). Our findings contribute to this field by recognition of a mechanism of the regulation of Ptgs2 mRNA stability by endogenously expressed CUGbp2. We demonstrated that Ptgs2 stabilization is in part dependent on the quantity of endogenous CUGbp2 protein and appears to be independent of altered transcription of CUGbp2. The question now is the mechanism by which endogenous CUGbp2 protein is increased consequent of ERK activation. The fact that CUGbp2 protein is increased downstream of ERK activation suggests a mechanism involving protein phosphorylation. ERK may directly phosphorylate CUGbp2 itself resulting in stabilization. Alternatively, it may act indirectly through other proteins that then stabilize CUGbp2, disrupt CUGbp2 degradation, or increase CUGbp2 translation from available mRNA. Further studies are needed to understand the regulation of CUGbp2 itself.
Constitutive Expression of Ptgs2
The presence of constitutive Ptgs2 expression in the adult mammal has been well demonstrated (10, 62–64). However, Ptgs2 remains primarily recognized as the inducible cyclooxygenase largely due to its up-regulation in inflammation and in various cancers (reviewed in Ref. 65). However, Ptgs2 knock-out mice, although largely healthy, do exhibit multiple problems that arise from loss of constitutive expression specifically of Ptgs2 that are not found in Ptgs1 knock-out mice. These include defects in kidney development and renal function, in reproduction, and in proper bone maintenance (reviewed in Ref. 66). These data highlight the necessity of understanding the regulation of constitutive Ptgs2 expression in addition to understanding the mechanisms involved in the up-regulation of Ptgs2 in pathological situations. Our colonic MSCs can serve as a prime vessel for investigating the various mechanisms involved in regulating constitutive Ptgs2 expression because they are a primary cell type that naturally maintains extremely high levels of Ptgs2 mRNA. The mechanistic findings can then be evaluated in the whole organism in order to better understand the basic biology underlying Ptgs2 expression.
Colonic MSC, Monitors of the Homeostatic Environment?
Our findings raise the highly intriguing question of the precise role of colon-resident MSCs in homeostasis. We have demonstrated a probable role as an immune modulator through the production of PGE2. Interestingly, cMSCs do not express major histocompatibility complex class II, even after LPS stimulation, indicating that they probably are not acting as non-professional antigen-presenting cells. We followed this line of inquiry from our analysis of receptors whose expression was enriched specifically in cMSCs as compared with bmMSCs. Twelve other receptors were uncovered in this analysis, each of which raises possibilities for future experimental pathways and inquiries into the role of cMSCs.
We propose that cMSCs, due to their location in the complex environment of the mature colonic mesenchyme made up of a great variety of host cell types, are well positioned to act as monitors of the colonic environment. These cells specifically express receptors that are capable of receiving signals from a variety of immune and nervous cells, rendering them ideal for integrating signals indicative of the state of the organism as a whole and of the level of immune activation locally. The cells would thus be poised to combine and interpret neural and immune signals, respond to slight changes in their activity, and mediate responses directly or indirectly to these fluctuations before serious consequences such as full inflammation result.
Receptors on the surface of the cMSC capable of receiving neurally derived signals include a cholinergic receptor (Chrnb1), a γ-aminobutyric acid receptor (Gabbr1), and a purinergic receptor (P2rx5). These are intriguing in colonic immune homeostasis because there is an emerging literature on the cross-talk between the autonomic enteric nervous systems and cellular constituents of the immune system (67, 68). Other receptors on the surface of the cMSC identified by our chip analysis are capable of receiving immune signals either directly derived from immune effector cells or indirectly as a consequence of immune effector cell action. Two of these are activin receptors (Acvrl1 and Acvr2a) that recognize this member of the transforming growth factor-β superfamily, a factor that may be secreted by Th2 cells (69) and by dendritic cells (70). The expressed adenosine receptor, Adora2b, can receive signals important to NK cell activity and regulation (71). A receptor capable of sensing extracellular nucleotides released in inflammation is also present (P2ry6) (72). Other receptors include interleukin-3 receptor (Il3ra), a vasopressin receptor (Avpr1a), and a calcitonin receptor (Ramp3). Indeed, many more receptors capable of sensing other neural, immune, and growth factor signals are also likely to be present; we have only highlighted those that appear to be enriched in cMSCs compared with bmMSCs. The simultaneous expression of these many and varied receptors on cMSCs and the localization of these cells in the complex environment of the colon place them in a prime position to act as monitors of the colonic homeostatic environment. This proposed role suggests a wealth of future investigative pathways that could aid the gastrointestinal field in gaining a greater understanding of the cellular players and mechanisms involved in the complex regulation of colonic homeostasis.
Acknowledgments
We thank Drs. David Ornitz and Jason Mills for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK071619 (to T. S.), DK07161-90251 (to T. S. S.), DK33165 (to W. S.), DK55753 (to W. S.), and P30-DK52574 (to the Washington University Digestive Disease Research Core). This work was also supported by the Crohn's Colitis Foundation of America (to T. S.), the Pew Scholars Foundation (to T. S.), and a Cancer Research Institute Training Grant (Predoctoral Emphasis Pathway in Tumor Immunology) (to M. W.).
- ColEP
- colonic epithelial progenitor
- PGE2
- prostaglandin E2
- MSC
- mesenchymal stem cell
- cMSC
- colonic mesenchymal stem cell
- bmMSC
- bone marrow mesenchymal stem cell
- qRT-PCR
- quantitative reverse transcription-PCR
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MOPS
- 4-morpholinepropanesulfonic acid
- ERK
- extracellular signal-regulated kinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- shRNA
- short hairpin RNA
- FACS
- fluorescence-activated cell sorting
- PBS
- phosphate-buffered saline
- GF
- germ-free
- CONV
- conventional
- LPS
- lipopolysaccharide
- GO
- gene ontology
- MAPK
- mitogen-activated protein kinase
- TLR
- Toll-like receptor
- FGF
- fibroblast growth factor.
REFERENCES
- 1.Savage D. C. (1977) Annu. Rev. Microbiol. 31, 107–133 [DOI] [PubMed] [Google Scholar]
- 2.Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., Relman D. A. (2005) Science 308, 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cliffe L. J., Humphreys N. E., Lane T. E., Potten C. S., Booth C., Grencis R. K. (2005) Science 308, 1463–1465 [DOI] [PubMed] [Google Scholar]
- 5.Seno H., Miyoshi H., Brown S. L., Geske M. J., Colonna M., Stappenbeck T. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang W. W., Leblond C. P. (1971) Am. J. Anat. 131, 73–99 [DOI] [PubMed] [Google Scholar]
- 7.Walker M. R., Stappenbeck T. S. (2008) Curr. Opin. Gastroenterol. 24, 115–120 [DOI] [PubMed] [Google Scholar]
- 8.Kim C. H. (2004) Curr. Drug Targets Immune Endocr. Metabol. Disord. 4, 343–361 [DOI] [PubMed] [Google Scholar]
- 9.Pull S. L., Doherty J. M., Mills J. C., Gordon J. I., Stappenbeck T. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown S. L., Riehl T. E., Walker M. R., Geske M. J., Doherty J. M., Stenson W. F., Stappenbeck T. S. (2007) J. Clin. Invest. 117, 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., Mann J. R., DuBois R. N. (2005) Gastroenterology 128, 1445–1461 [DOI] [PubMed] [Google Scholar]
- 12.Aggarwal S., Pittenger M. F. (2005) Blood 105, 1815–1822 [DOI] [PubMed] [Google Scholar]
- 13.Allgayer H., Deschryver K., Stenson W. F. (1989) Gastroenterology 96, 1290–1300 [DOI] [PubMed] [Google Scholar]
- 14.Morteau O., Morham S. G., Sellon R., Dieleman L. A., Langenbach R., Smithies O., Sartor R. B. (2000) J. Clin. Invest. 105, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tessner T. G., Muhale F., Riehl T. E., Anant S., Stenson W. F. (2004) J. Clin. Invest. 114, 1676–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.North T. E., Goessling W., Walkley C. R., Lengerke C., Kopani K. R., Lord A. M., Weber G. J., Bowman T. V., Jang I. H., Grosser T., Fitzgerald G. A., Daley G. Q., Orkin S. H., Zon L. I. (2007) Nature 447, 1007–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedenstein A. J., Deriglasova U. F., Kulagina N. N., Panasuk A. F., Rudakowa S. F., Luriá E. A., Ruadkow I. A. (1974) Exp. Hematol. 2, 83–92 [PubMed] [Google Scholar]
- 18.da Silva Meirelles L., Chagastelles P. C., Nardi N. B. (2006) J. Cell Sci. 119, 2204–2213 [DOI] [PubMed] [Google Scholar]
- 19.Morham S. G., Langenbach R., Loftin C. D., Tiano H. F., Vouloumanos N., Jennette J. C., Mahler J. F., Kluckman K. D., Ledford A., Lee C. A., Smithies O. (1995) Cell 83, 473–482 [DOI] [PubMed] [Google Scholar]
- 20.Stanley E. R., Cifone M., Heard P. M., Defendi V. (1976) J. Exp. Med. 143, 631–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon B., Yu K. Y., Ni J., Yu G. L., Jang I. K., Kim Y. J., Xing L., Liu D., Wang S. X., Kwon B. S. (1999) J. Biol. Chem. 274, 6056–6061 [DOI] [PubMed] [Google Scholar]
- 22.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 23.Schadt E. E., Li C., Su C., Wong W. H. (2000) J. Cell Biochem. 80, 192–202 [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phinney D. G., Prockop D. J. (2007) Stem Cells 25, 2896–2902 [DOI] [PubMed] [Google Scholar]
- 26.Meirelles Lda S., Nardi N. B. (2003) Br. J. Haematol. 123, 702–711 [DOI] [PubMed] [Google Scholar]
- 27.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. (2006) Cytotherapy 8, 315–317 [DOI] [PubMed] [Google Scholar]
- 28.Pereira R. F., Halford K. W., O'Hara M. D., Leeper D. B., Sokolov B. P., Pollard M. D., Bagasra O., Prockop D. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4857–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krampera M., Glennie S., Dyson J., Scott D., Laylor R., Simpson E., Dazzi F. (2003) Blood 101, 3722–3729 [DOI] [PubMed] [Google Scholar]
- 30.Bonney R. J., Wightman P. D., Davies P., Sadowski S. J., Kuehl F. A., Jr., Humes J. L. (1978) Biochem. J. 176, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuji S., Kawano S., Sawaoka H., Takei Y., Kobayashi I., Nagano K., Fusamoto H., Kamada T. (1996) Prostaglandins Leukot. Essent. Fatty Acids 55, 179–183 [DOI] [PubMed] [Google Scholar]
- 32.Shibolet O., Podolsky D. K. (2007) Am. J. Physiol. Gastrointest Liver Physiol. 292, G1469–G1473 [DOI] [PubMed] [Google Scholar]
- 33.Takeda K., Akira S. (2005) Int. Immunol. 17, 1–14 [DOI] [PubMed] [Google Scholar]
- 34.Doherty J. M., Geske M. J., Stappenbeck T. S., Mills J. C. (2008) Stem Cells 26, 2124–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geske M. J., Zhang X., Patel K. K., Ornitz D. M., Stappenbeck T. S. (2008) Development 135, 2959–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ornitz D. M., Xu J., Colvin J. S., McEwen D. G., MacArthur C. A., Coulier F., Gao G., Goldfarb M. (1996) J. Biol. Chem. 271, 15292–15297 [DOI] [PubMed] [Google Scholar]
- 37.Ornitz D. M., Itoh N. (2001) Genome Biol. 2, REVIEWS3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Stappenbeck T. S., White A. C., Lavine K. J., Gordon J. I., Ornitz D. M. (2006) Development 133, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eswarakumar V. P., Lax I., Schlessinger J. (2005) Cytokine Growth Factor Rev. 16, 139–149 [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay D., Houchen C. W., Kennedy S., Dieckgraefe B. K., Anant S. (2003) Mol. Cell 11, 113–126 [DOI] [PubMed] [Google Scholar]
- 41.Sureban S. M., Murmu N., Rodriguez P., May R., Maheshwari R., Dieckgraefe B. K., Houchen C. W., Anant S. (2007) Gastroenterology 132, 1055–1065 [DOI] [PubMed] [Google Scholar]
- 42.Xu K., Kitchen C. M., Shu H. K., Murphy T. J. (2007) J. Biol. Chem. 282, 32699–32709 [DOI] [PubMed] [Google Scholar]
- 43.Itoh N., Ornitz D. M. (2008) Dev. Dyn. 237, 18–27 [DOI] [PubMed] [Google Scholar]
- 44.Colvin J. S., White A. C., Pratt S. J., Ornitz D. M. (2001) Development 128, 2095–2106 [DOI] [PubMed] [Google Scholar]
- 45.Weaver M., Dunn N. R., Hogan B. L. (2000) Development 127, 2695–2704 [DOI] [PubMed] [Google Scholar]
- 46.ten Berge D., Brugmann S. A., Helms J. A., Nusse R. (2008) Development 135, 3247–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sala F. G., Curtis J. L., Veltmaat J. M., Del Moral P. M., Le L. T., Fairbanks T. J., Warburton D., Ford H., Wang K., Burns R. C., Bellusci S. (2006) Dev. Biol. 299, 373–385 [DOI] [PubMed] [Google Scholar]
- 48.Petiot A., Conti F. J., Grose R., Revest J. M., Hodivala-Dilke K. M., Dickson C. (2003) Development 130, 5493–5501 [DOI] [PubMed] [Google Scholar]
- 49.Lin Y., Chen L., Lin C., Luo Y., Tsai R. Y., Wang F. (2009) Dev. Biol. 329, 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Cell Metab. 2, 217–225 [DOI] [PubMed] [Google Scholar]
- 51.Ben-Dov I. Z., Galitzer H., Lavi-Moshayoff V., Goetz R., Kuro-o M., Mohammadi M., Sirkis R., Naveh-Many T., Silver J. (2007) J. Clin. Invest. 117, 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Razzaque M. S. (2009) Am. J. Physiol. Renal Physiol. 296, F470–F476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Estívariz C. F., Gu L. H., Scully S., Eli A., Jonas C. R., Farrell C. L., Ziegler T. R. (2000) Dig. Dis. Sci. 45, 736–743 [DOI] [PubMed] [Google Scholar]
- 54.Kage K., Fujita N., Oh-hara T., Ogata E., Fujita T., Tsuruo T. (1999) Biochem. Biophys. Res. Commun. 254, 259–263 [DOI] [PubMed] [Google Scholar]
- 55.Xu K., Chang C. M., Gao H., Shu H. K. (2009) Oncogene 28, 1410–1420 [DOI] [PubMed] [Google Scholar]
- 56.Pham H., Chong B., Vincenti R., Slice L. W. (2008) J. Cell. Physiol. 214, 96–109 [DOI] [PubMed] [Google Scholar]
- 57.Stoeltzing O., Liu W., Fan F., Wagner C., Stengel K., Somcio R. J., Reinmuth N., Parikh A. A., Hicklin D. J., Ellis L. M. (2007) Cancer Lett. 258, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Z., Liu L. Z., Dixon D. A., Zheng J. Z., Chandran B., Jiang B. H. (2007) Cell. Signal. 19, 1542–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsatsanis C., Androulidaki A., Venihaki M., Margioris A. N. (2006) Int. J. Biochem. Cell Biol. 38, 1654–1661 [DOI] [PubMed] [Google Scholar]
- 60.Anant S., Murmu N., Houchen C. W., Mukhopadhyay D., Riehl T. E., Young S. G., Morrison A. R., Stenson W. F., Davidson N. O. (2004) Gastroenterology 127, 1139–1149 [DOI] [PubMed] [Google Scholar]
- 61.Mason I. (2007) Nat. Rev. Neurosci. 8, 583–596 [DOI] [PubMed] [Google Scholar]
- 62.Ishikawa T. O., Jain N. K., Taketo M. M., Herschman H. R. (2006) Mol. Imaging Biol. 8, 171–187 [DOI] [PubMed] [Google Scholar]
- 63.Yaksh T. L., Dirig D. M., Conway C. M., Svensson C., Luo Z. D., Isakson P. C. (2001) J. Neurosci. 21, 5847–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmermann K. C., Sarbia M., Schrör K., Weber A. A. (1998) Mol. Pharmacol. 54, 536–540 [DOI] [PubMed] [Google Scholar]
- 65.Williams C. S., Mann M., DuBois R. N. (1999) Oncogene 18, 7908–7916 [DOI] [PubMed] [Google Scholar]
- 66.Loftin C. D., Tiano H. F., Langenbach R. (2002) Protaglandins Other Lipid Mediat. 177–185 [DOI] [PubMed] [Google Scholar]
- 67.Wang J., Hauer-Jensen M. (2007) Br. J. Radiol. 80, S41–S48 [DOI] [PubMed] [Google Scholar]
- 68.Kiba T. (2006) Digestion 74, 215–227 [DOI] [PubMed] [Google Scholar]
- 69.Ogawa K., Funaba M., Chen Y., Tsujimoto M. (2006) J. Immunol. 177, 6787–6794 [DOI] [PubMed] [Google Scholar]
- 70.Robson N. C., Phillips D. J., McAlpine T., Shin A., Svobodova S., Toy T., Pillay V., Kirkpatrick N., Zanker D., Wilson K., Helling I., Wei H., Chen W., Cebon J., Maraskovsky E. (2008) Blood 111, 2733–2743 [DOI] [PubMed] [Google Scholar]
- 71.Raskovalova T., Huang X., Sitkovsky M., Zacharia L. C., Jackson E. K., Gorelik E. (2005) J. Immunol. 175, 4383–4391 [DOI] [PubMed] [Google Scholar]
- 72.Grbic D. M., Degagné E., Langlois C., Dupuis A. A., Gendron F. P. (2008) J. Immunol. 180, 2659–2668 [DOI] [PubMed] [Google Scholar]