Abstract
The vesicular neurotransmitter transporter VMAT2 is responsible for the transport of monoamines into synaptic and storage vesicles. VMAT2 is the target of many psychoactive drugs and is essential for proper neurotransmission and survival. Here we describe a new expression system in Saccharomyces cerevisiae that takes advantage of the polyspecificity of VMAT2. Expression of rVMAT2 confers resistance to acriflavine and to the parkinsonian toxin 1-methyl-4-phenylpyridinium (MPP+) by their removal into the yeast vacuole. This expression system allowed identification of a new substrate, acriflavine, and isolation of mutants with modified affinity to tetrabenazine (TBZ), a non-competitive inhibitor of VMAT2 that is used in the treatment of various movement disorders including Tourette syndrome and Huntington chorea. Whereas one type of mutant obtained displayed decreased affinity to TBZ, a second type showed only a slight decrease in the affinity to TBZ, displayed a higher Km to the neurotransmitter serotonin, but conferred increased resistance to acriflavine and MPP+. A protein where both types of mutations were combined (with only three amino acid replacements) lost most of the properties of the neurotransmitter transporter (TBZ-insensitive, no transport of neurotransmitter) but displayed enhanced resistance to the above toxicants. The work described here shows that in the case of rVMAT2, loss of traits acquired in evolution of function (such as serotonin transport and TBZ binding) bring about an improvement in older functions such as resistance to toxic compounds. A process that has taken millions of years of evolution can be reversed by three mutations.
Keywords: Drug Resistance/Multidrug, Membrane/Proteins, Neurotransmitters, Subcellular Organelles/Vesicles, Transport, MPP, Reserpine, Tetrabenazine
Introduction
Neurotransporters play central roles in the overall process of neurotransmission. Sodium-coupled plasma membrane transporters actively remove the neurotransmitters from the synaptic cleft in a step essential for termination of the signal. The release of neurotransmitter by exocytosis is possible because of their storage inside synaptic vesicles in a process that depends on vesicular H+-coupled neurotransporters (VNT)2 (1). Three families of proteins responsible for the uptake of transmitter by secretory vesicles have been identified, one that includes the vesicular monoamine transporters (VMATs) and acetylcholine transporters (VAChT), a second that includes the vesicular GABA transporter (VGAT), and a third that includes the vesicular glutamate transporters (VGLUTs) (2–4).
Two monoamine transporters, VMAT1 and VMAT2, are responsible for the transport of dopamine, serotonin, adrenaline, and noradrenaline into the synaptic vesicles of neurons and the dense core vesicles of endocrine cells (2–4). VMAT2 is expressed by monoamine neurons in the central nervous system and selected peripheral endocrine populations. VMAT2 has a higher affinity for most monoamines than VMAT1, but only VMAT2 appears to recognize histamine (5, 6). Early studies using chromaffin granules from the adrenal medulla showed that the uptake of one protonated, and hence charged, cytoplasmic monoamine is coupled to the movement of two H+ in the opposite direction (1). The transport cycle results in net efflux of one charge. The H+ electrochemical gradient and the coupled stoichiometry allow for accumulation of monoamines at submolar concentrations in chromaffin granules (1).
The VMATs are the target of reserpine, which provided some of the first evidence for the monoamine hypothesis for depression (7, 8) and are involved in the function of amphetamines including methamphetamine, which acts primarily on dopamine neurons, and 3,4-methylenedioxymethamphetamine (MDMA), which promotes serotonin release (9). The VMATs also protect cells against the parkinsonian neurotoxin MPP+ (10, 11) by sequestering the compound in vesicles and away from its primary site of action in mitochondria.
Another clinically relevant drug is the non-competitive VMAT2 inhibitor TBZ that is used in the treatment of various movement disorders including Tourette syndrome and Huntington chorea (12). TBZ is a non-competitive inhibitor of transport and substrate binding (13). However, occupancy of the substrate binding site has little effect on TBZ binding (13, 14). The mode of interaction of the two binding sites and how TBZ brings about the inhibition are still unknown. The closely related VMAT1 isoform displays a decreased sensitivity to TBZ (6).
The structural basis for the function of VMAT remains unknown. VMAT belongs to the MFS superfamily of transporters and it displays a weak but distinct homology with bacterial multidrug transporters (1). In addition it became evident from its pharmacology that it is a multispecific transporter, and, when heterologously expressed, it confers resistance to toxic substrates because it removes them from their targets by transport into the vacuolar system (1, 15).
Here we describe the functional expression of rVMAT2 in Saccharomyces cerevisiae cells. rVMAT2 renders the cells resistant to MPP+ and acriflavine, a newly identified substrate, by a process that involves active removal of the drugs into the yeast vacuole. The development of this system allows us harnessing the power of yeast genetics to the study of the mechanism of rVMAT2. Screening of mutants with altered sensitivity to the non-competitive inhibitor TBZ brought about the isolation and characterization of proteins with a significantly modified pharmacological profile. A mutant rVMAT2 was constructed with three mutations obtained in the above screen. This mutant displayed properties of a multidrug transporter: it conferred a very robust resistance to acriflavine, MPP+ and other drugs but it lost the ability to transport neurotransmitter or binding the inhibitor TBZ.
EXPERIMENTAL PROCEDURES
Yeast Strains and Construction of Yeast Expression Vector
rVMAT2 cDNA with six histidine residues at the N terminus was cloned in the pAES426 yeast expression plasmid, under the control of the ADH1 (alcohol dehydrogenase) promoter. The plasmid contains the URA3 gene for selection in yeast, ampicillin-resistance marker and a 2-micron replication in yeast (35). Cloning was done using PCR, with HindIII and SpeI restriction enzymes. C-terminal GFP fusion was constructed using pFA6a-GFP(S65T)-kanMX6 according to Ref. 36. Site-directed mutagenesis was done with the QuikChange® II Site-directed Mutagenesis kit (Stratagene). Plasmid pAES426 with or without His rVMAT2 and derived mutants were routinely transformed into yeast strain ADU1-7 (US50–18C, yor1Δ, snq2D, pdr5Δ, pdr10Δ, pdr11Δ, ycf1Δ, pdr3Δ, ura3). The ura3 mutation in ADU1-7 was introduced to the original AD1-7 (16), kindly supplied by Dr. Stanislav Ulaszewski (Wroclav University), using 5-fluoroorotic acid (37).
Media
S. cerevisiae cells were grown in standard medium. Rich medium (YPD) contained 1% Bacto yeast extract, 2% Bacto peptone (both from Difco), and 2% glucose. Minimal medium (SD; 0.67% Bacto-yeast nitrogen base without amino acids and 2% glucose) was supplemented according to auxotrophic requirements as described in Ref. 38. Yeast cells were transformed by the method of Elble (39).
Phenotype Assay
Inhibition Zone Assay
S. cerevisiae strains were pregrown in liquid minimal medium to late log phase and inoculated on YPD plates. Sterile filter discs of 5-mm diameter were loaded with the desired amount of potential drugs and placed on the agar. The relative degree of drug resistance was determined by comparing the diameters of the inhibition zone after incubating the plates for 2–3 days at 30 °C.
Drug Sensitivity Assay
S. cerevisiae strains were pregrown in liquid minimal medium to late log phase. Cultures were diluted to a comparable density and then were decimal diluted. Dilutions (5 μl) were placed on YPD agar supplemented as indicated with the appropriate toxins or inhibitors: 40 μm acriflavine, 1.5 mm MPP+, 0.1–0.5 μm reserpine, or 2 μm TBZ. Plates were incubated for 2–3 days at 30 °C.
Phenotype in Liquid Medium
Overnight cultures in minimal medium were diluted in YPD or in the same medium (for MPP+ resistance) to A600 = 0.05 and supplemented with the corresponding toxic compounds and inhibitors. Growth (3 ml) was performed in 1.5 × 15 cm glass tubes or in 1.2 ml storage plates (ABgene), 200 μl per well, covered with gas permeable adhesive seal (Fig. 8, B and C). The absorbance at 600 nm was measured after overnight growth at 30 °C. Data analysis was performed using Origin 8.0 software (OriginLab, Northampton, MA). The lowest R2 value in the fits for all of the experiments was 0.95.
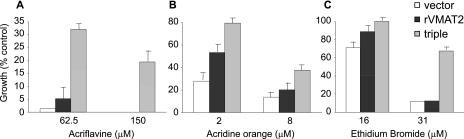
FIGURE 8.
rVMAT2-triple displays the phenotype of a multidrug transporter. Cells harboring empty pAES426 vector, rVMAT2, and rVMAT2-triple were tested for resistance on liquid medium with acriflavine (A), acridine orange (B), or ethidium bromide (C). Growth was analyzed after overnight incubation at 30 °C.
Scanning Laser Confocal Microscopy
Cells were scanned using the FV-1000 confocal microscope (Olympus, Japan), equipped with an IX81 inverted microscope. A 60×/1.35 oil immersion objective was used. GFP fluorescence was imaged using the 488-nm laser line for excitation and a 505–525-nm filter was used to collect the emission. Vacuolar staining with the dye N-(3-triethylammoniumpropyl)-4-(p-diethylamino-phenyl-hexatrienyl) pyridinium dibromide (FM 4-64) was as described in Ref. 17. The FM 4-64-stained sample was scanned using the 633-nm laser line for excitation, and a 655–755-nm filter was used to collect the emission. DIC images were taken at the same time.
Biochemical Characterization of rVMAT2
Membranes were prepared from yeast cells essentially as described (21). The binding of α-[2-3H]dihydrotetrabenazine (American Radiolabeled Chemicals, St. Louis, MO) was performed essentially as described in Ref. 22. The procedure for solubilization, purification, reconstitution, and uptake of [3H]serotonin (PerkinElmer Life Science) to proteoliposomes was performed as in Ref. 18.
Generation of Random Mutant Library
The library was generated using GeneMorph® II EZClone Domain Mutagenesis kit (Stratagene) according to the manufacturer's protocol.
Screening Procedures
The DNA library (1.5 μg) was transformed to ADU1-7 yeast cells by LiAc-heat shock transformation. At the end of the process, cells were plated on minimal medium at a concentration of 5 × 103-104 cells per plate. Replicas on selective plates containing 60 μm acriflavine with the addition of 1 or 2 μm TBZ were done with velvet. Enrichment trials were done in liquid medium, supplemented with 62.5 μm acriflavine and 5 μm TBZ. Cultures that grew in Erlenmeyer flask were plated on minimal medium without uracil (control for presence of the plasmid), single colonies were re-plated on plates containing 40 μm acriflavine ± 2 μm TBZ. rVMAT2 wild type was used as a control. DNA was purified from suspected colonies, propagated in Escherichia coli, re-purified, and transformed back to the S. cerevisiae ADU1-7 strain for the phenotype assay.
RESULTS
Functional Expression of rVMAT2 in S. cerevisiae Confers Resistance Phenotype
The pharmacological profile of VMAT (15) and its ability to transport toxic compounds into acidic organelles enabled the development of a system of functional expression in S. cerevisiae cells. After screening of yeast strains that are potentially sensitive to toxic compounds and can then serve as suitable hosts for phenotypic expression of VMAT2, we chose the strain ADU1-7 for routine use (16). ADU1-7 is a strain in which seven genes coding for ABC transporters were inactivated, and it is sensitive to several toxic compounds (16). Initial experiments using an inhibition zone assay allowed for a quick screen of several plasmids expressing rVMAT2 with various tags. We found that cells carrying a plasmid (pAES426), where rVMAT2 was expressed at relatively low levels from a constitutive promoter, were less sensitive to MPP+ and acriflavine than the host strain that contained the empty plasmid. MPP+ was already known as a substrate of VMAT and the ability of VMAT to confer resistance to CHO cells against the neurotoxin MPP+ was exploited for the cloning of VMAT1 (10). Acriflavine is a substrate of several multidrug transporters but was not previously known to interact with VMAT.
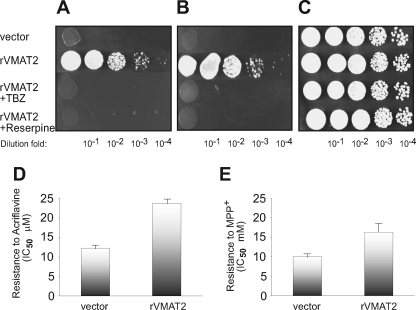
rVMAT2 fused to a His6 tag at either N′ or C′ terminal showed a similar phenotype as rVMAT2 with no tag (not shown). All the work described here was performed with rVMAT2 fused to a His6 tag at the N terminus of the protein. In the experiments shown in Fig. 1(A–C), the ability of cells to grow under various conditions was assessed in solid medium containing MPP+ (1.5 mm) or acriflavine (40 μm) in which 5 μl of logarithmic dilutions of an overnight culture were spotted. All cells tested grew well in the medium without toxicants at all of the dilutions tested (Fig. 1C). In the presence of either MPP+ (Fig. 1A) or acriflavine (Fig. 1B) at the above concentrations, cells carrying the vector alone did not grow at any of the dilutions. Under both conditions, cells carrying rVMAT2 displayed a very robust growth, only slightly lower than growth in the control plate. Full reversal of the resistance to both toxins was obtained upon addition of the VMAT2 inhibitors, reserpine (0.5 μm) or TBZ (2 μm) (Fig. 1, A and B). Neither one of the VMAT2 inhibitors had any effect on yeast growth in the absence of the toxin (Fig. 1C). To quantify the results described above, growth was also tested in liquid media and the IC50 for growth was determined (Fig. 1, D and E). Whereas growth of cells with vector alone was inhibited 50% at 12.3 ± 0.6 μm acriflavine and 10 ± 0.5 mm MPP+, the IC50 values for the strain expressing rVMAT2 were 23.8 ± 1.2 and 16.3 ± 2, respectively.
FIGURE 1.
rVMAT2 confers resistance against MPP+ and acriflavine, which is fully reversed in the presence of TBZ and reserpine. ADU1-7 cells harboring pAES426 with or without rVMAT2 were grown in minimal medium and diluted to comparable densities. After serial dilution, they were plated (5 μl) on rich solid medium (C) supplemented with 1.5 mm MPP+ (A), or 40 μm acriflavine (B). Where indicated, the plates contained 2 μm TBZ or 0.5 μm reserpine. Growth was analyzed after two overnights incubation at 30 °C. Growth in liquid media was performed as described under “Experimental Procedures” with increasing concentrations of acriflavine (D) or MPP+ (E), IC50 values were calculated from fits of the data obtained by Origin 8 software (the lowest R-square value is 0.98).
rVMAT2 Is Localized to the Vacuolar Membrane
To examine the intracellular localization of rVMAT2, we constructed a fusion of rVMAT2 with GFP. This construct is fully functional as judged by its ability to confer resistance against acriflavine (not shown). Vesicular transport activities are energized by a H+ electrochemical gradient (ΔμH+) (1). Various organelles may fulfill the acidic requirements necessary for activity, among them secretory vesicles, endosomes, and the vacuole.
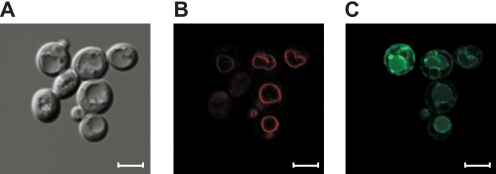
Because the fluorescence associated with this fusion protein suggested vacuolar localization, we stained the cells with the lipophilic styryl dye FM 4-64, used as a specific vacuolar membrane marker (17). The results shown in Fig. 2, B and C indicate that a large part of the fluorescence emitted by rVMAT2-GFP was coincident with that of the vacuolar membranes stained with FM 4-64. In addition, in some cells, expression was detected with a distribution that indicates that some of the protein is still in the endoplasmic reticulum. Therefore, it is possible to infer that the intracellular compartment to which rVMAT2 removes the toxicants is predominantly the cell vacuole. We assume that the vacuolar-type H+-ATPase (V-ATPase) that resides on membranes of the vacuole generates a pH gradient (acidic inside) that drives the accumulation of MPP+ and acriflavine inside the organelle.
FIGURE 2.
rVMAT2-GFP is localized mainly to the vacuolar membrane. ADU1-7 cells expressing rVMAT2-GFP were grown overnight in minimal medium, stained with 10 μm FM4–64, washed, and visualized in a confocal microscope as described under “Experimental Procedures.” Yeast cells in DIC (A), the vacuole can be noticed easily after exposure to hypotonic solution; FM 4-64, red staining, specific to the vacuole membrane (B). rVMAT2-GFP is visible mainly in the vacuole membrane. Some cells also show GFP fluorescence in the endoplasmic reticulum (C). Scale bars are 4 μm.
rVMAT2 Binds TBZ and Transports Serotonin
To estimate the amounts of functional protein in the cells and to further characterize its properties, we measured binding to the radiolabeled substrate [3H]TBZOH, an analog of TBZ, the high affinity non-competitive inhibitor of VMAT2 (13). The cell lysates displayed nanomolar affinity TBZOH binding, with a KD of 9.4 ± 2 nm. When using ∼100 μg of total membrane protein (obtained from about 8–10 ml culture) a Bmax value of ∼0.64 pmol binding sites was obtained. The expression levels estimated from this data are very low and correspond to only a few μg from 1 liter of culture. Efforts to improve expression using plasmids with stronger promoters reduced and impaired the ability of the cells to confer resistance to the toxic compounds, and membrane preparations from these cells showed little or no binding of [3H]TBZOH (data not shown).
The resistance of the transformed cells to MPP+ and acriflavine, together with [3H]TBZOH binding activity, suggests that the protein is functional. To further characterize the activity, we measured transport of serotonin in proteoliposomes. For this purpose we solubilized the membrane preparation with dodecylmaltoside (DDM), immobilized rVMAT2 onto Ni-nitrilotriacetic acid beads and reconstituted the transporter into proteoliposomes. The proton gradient (acidic inside) was generated by imposition of an ammonium gradient (high inside), a procedure that takes advantage of the higher permeability of the non-protonated species that rapidly leaks downhill (18). Proteoliposomes prepared from S. cerevisiae cells expressing rVMAT2 displayed time-dependent and proton-driven [3H]serotonin transport (supplemental Fig. S1A). The kinetic constants obtained from rate determination at various [3H]serotonin concentrations are shown in Table 1 (see also supplemental Fig. S1B). The Km value of 180 ± 27 nm is similar to the Km measured in vesicles isolated from mammalian cell lines expressing VMAT2 (6, 18) and ∼10 times lower than the value measured in native VMAT2 purified from bovine chromaffin granules and reconstituted in proteoliposomes (19).
TABLE 1.
Kinetic properties of rVMAT2 mutants
Growth inhibition by tetrabenazine (IC50) was determined in YPD medium supplemented with 100 μm acriflavine and increasing concentrations of the inhibitor. Membranes were prepared from yeast cells expressing the appropriate mutant and binding of α-[2-3H]dihydrotetrabenazine was performed as described under “Experimental Procedures.” Serotonin uptake in proteoliposomes was done as described under “Experimental Procedures” in the presence of 50 nm valinomycin and increasing concentrations of [3H]serotonin. To evaluate initial rates, the reaction was terminated after 1 min by rapid dilution followed by filtration, and the bound radioactivity was measured by scintillation counting. Vmax values were standardized for equivalent amounts of liposomes. Data analysis was performed using Origin 8.0 software (OriginLab, Northampton, MA). All experiments were done in duplicates and were repeated at least three times (the lowest R-square value is 0.95).
| Protein | Growth inhibition by TBZ IC50 | KDTBZ | Km | Vmax |
|---|---|---|---|---|
| nm | nm | nm | pmol/min | |
| rVMAT2 | 130 | 9.4 ± 2 | 180 ± 27 | 1.1 ± 0.07 |
| F136L | 6400 | ≫250 | 28 ± 7 | 0.38 ± 0.04 |
| I425F | 166 | 30.8 ± 5 | 1994 ± 708 | 1.2 ± 0.07 |
| V428A | 422 | 28.2 ± 2.3 | 325 ± 55 | 1.8 ± 0.12 |
The resistance to MPP+ and acriflavine together with the vacuolar localization and the biochemical characterization of rVMAT2 support the conclusion that the transporter expressed in S. cerevisiae is fully functional. rVMAT2 is targeted to acidic compartments in the yeast cell, mostly to the large vacuole, and confers resistance by actively removing the toxic compounds away from their target.
Random Mutagenesis
The functional expression provides a powerful tool to study rVMAT2. To take advantage of the yeast genetic power we screened for mutants impaired in their ability to bind TBZ, a high affinity non-competitive inhibitor of rVMAT2. A library of randomly mutagenized rVMAT2 was generated using error prone PCR with Mutazyme DNA polymerase. Random clones sequenced from this library contained on average 1.2 mutations per rVMAT2 reading frame. Selection was performed in yeast cells transformed with the library either in liquid non-permissive medium or by replica plating. After screening of a large number of independent clones, two types of mutants were identified, each one accounted for about 50% of the total number identified. The first ones (Type I) all were found to contain the same single base substitution: from thymine to cytosine that changes the codon for phenylalanine to that for leucine at position 136, within the second transmembrane domain. The same mutation was identified in mutants independently isolated by replica plating.
Type I: Phenylalanine 136 Contributes to High Affinity TBZ Binding
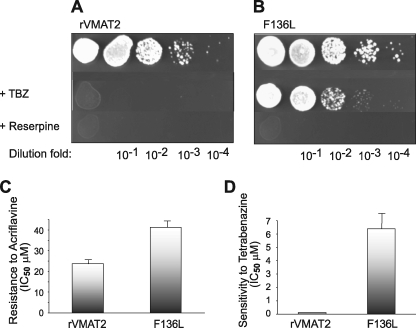
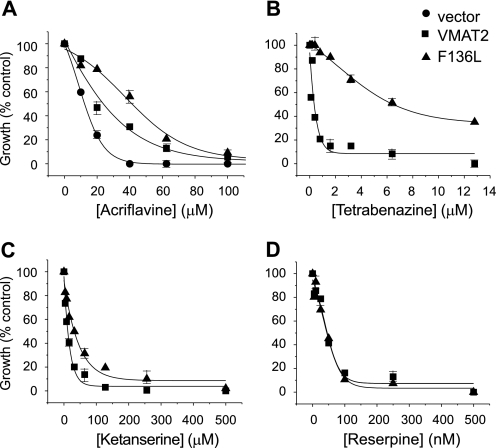
Yeast cells expressing rVMAT2-F136L maintained their resistance for growth in the presence of MPP+ but displayed a dramatic decrease in the sensitivity to the inhibitor TBZ in comparison to wild type (Fig. 3, A and B). Growth in the presence of the competitive inhibitor reserpine was not detected suggesting that the mutation affects differentially the inhibitor profile (Fig. 3). The mutant conferred a slightly better resistance to acriflavine than the wild type (Figs. 3C and 4A). Growth of ADU1-7 cells bearing mock plasmid was inhibited 50% at around 12.3 ± 0.6 μm acriflavine, the IC50 of the rVMAT plasmid-bearing cells is 23.8 ± 1.2 μm and that of rVMAT-F136L cells is 41.3 ± 2.1 μm (Fig. 4A). After overnight growth in liquid medium that contained acriflavine and increasing concentrations of TBZ, rVMAT-F136L cells were found to be significantly less sensitive toward the inhibitor with IC50 values of 0.13 ± 0.05 and 6.4 ± 1.2 μm, respectively for wild type and mutant (Figs. 3D and 4B). Importantly, both strains showed a small difference in sensitivity to ketanserine and no significant one in sensitivity toward reserpine (Fig. 4, C and D). Reserpine is a competitive inhibitor and, thereby most likely binds to a site different from the TBZ site. On the other hand, ketanserine is also a non-competitive inhibitor of VMAT transport, and its binding site was considered to overlap with that of TBZ. The results described here suggest that although the ketanserine and TBZ binding sites may overlap they do not necessarily share identical determinants.
FIGURE 3.
F136L alters rVMAT2 sensitivity toward TBZ in yeast cells. ADU1-7 cells harboring pAES426 rVMAT2 (A) or F136L (B) were grown in MPP+ and analyzed as in Fig. 1. Growth in liquid medium was performed with increasing concentrations of acriflavine. The calculated IC50 values are shown (C). Sensitivity toward TBZ was determined in the presence of 100 μm acriflavine and increasing concentrations of the inhibitor (D). The lowest R-square value is 0.98.
FIGURE 4.
The effect of the mutation F136L on rVMAT2-mediated resistance. ADU1-7 cells harboring empty pAE426 vector (circles), rVMAT2 (squares), or F136L (triangles) were grown overnight in minimal medium and diluted in rich medium containing increasing concentrations of acriflavine (A) or 100 μm acriflavine with increasing concentrations of TBZ (B), ketanserine (C), or reserpine (D). Absorbance was measured at 600 nm after overnight growth at 30 °C. Each curve represents a fit obtained by Origin 8 software (the lowest R-square is 0.96). A representative experiment of at least three replicas is shown.
Efforts to determine directly the affinity of TBZ to the mutant protein were carried out to no avail. Even at very high protein concentration, the binding was too low to allow accurate measurement of the KD. On the other hand, [3H]serotonin transport assayed in proteoliposomes reconstituted with the F136L mutant revealed transport activity with a Km to serotonin lower than that of the wild-type protein. Thus the Km was 28 ± 7 nm (Table 1) and a Vmax somewhat lower than of the wild-type protein (Table 1).
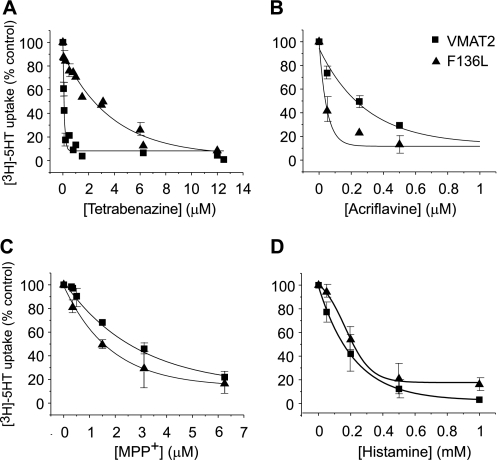
To further characterize the properties of the F136L mutant protein, we measured [3H]serotonin transport in the presence of different concentrations of substrates and inhibitors and used this dose-response analysis to estimate their apparent affinity (Fig. 5). Whereas F136L was about 30-fold less sensitive toward TBZ when compared with wild type (Fig. 5A, 2.4 ± 1 and 0.08 ± 0.005 μm, respectively), no major difference in IC50 was measured in response to MPP+ (Fig. 5C), histamine (Fig. 5D), and reserpine (not shown). These findings reinforce the hypothesis that F136L mutation affects mainly the affinity toward TBZ and has not significantly impaired other functions of the protein. Moreover, the apparently higher affinities to serotonin (Table 1) and to acriflavine (Fig. 5B) as measured in vitro and the increased resistance conferred to cells expressing the mutant F136L (Fig. 4A) suggest an enhanced recognition of some of the substrates by the F136L mutant. Because of its increased apparent affinity to acriflavine, the mutant protein can remove the toxin to concentrations lower than that achieved by wild-type rVMAT2.
FIGURE 5.
The F136L mutation modifies the pharmacological profile of rVMAT2. Liposomes (1.5 μl) reconstituted with rVMAT2 (squares) or F136L (triangles) were assayed for serotonin transport as described under “Experimental Procedures.” The reaction was terminated by filtration, and the bound radioactivity was measured by scintillation counting. [3H]Serotonin transport was assayed in the presence of increasing concentrations of TBZ (A), acriflavine (B), MPP+ (C), and histamine (D) to determine the concentration required to inhibit transport by 50% (IC50). IC50 were calculated with Origin 8 software. For each compound, a representative experiment of three replicas is shown.
An Aromatic Residue at Position 136 Contributes to the Sensitivity toward TBZ
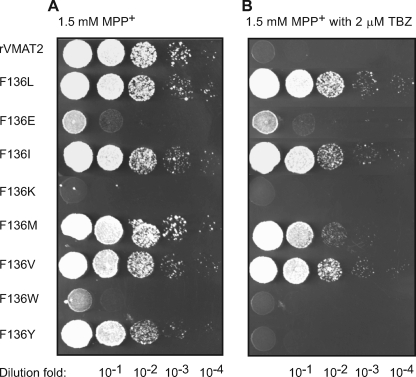
To further characterize the role of amino acids at position 136 in rVMAT2, we applied site directed mutagenesis and replaced Phe-136 with various amino acids. The mutants were tested for their ability to confer resistance against MPP+ and acriflavine and their sensitivity toward TBZ and reserpine. Similar results were obtained with both toxins, and the behavior in the presence of MPP+ is shown in Fig. 6. Substitution to Lys, Glu, and Trp impaired the ability of VMAT2 to confer resistance against the toxins. Whereas the effect of Lys and Trp substitutions resulted in full loss of activity, a Glu replacement showed some marginal resistance. Proteins with the aromatic amino acids Phe (in rVMAT2 wild type) and Tyr but not with the larger Trp at position 136, displayed full activity and exhibited similar susceptibility against TBZ. Mutant proteins with the hydrophobic amino acids Ile, Met, Val, and Leu at the same position exhibited essentially the same levels of activity but low sensitivity toward the inhibitor.
FIGURE 6.
Phenylalanine or tyrosine at position 136 confers high affinity binding of TBZ. ADU1-7 cells harboring pAES426 rVMAT2 with different substitutions at position 136 were grown overnight at 30 °C in minimal medium. 5-μl volumes of serial dilutions were spotted on rich solid medium supplemented with 1.5 mm MPP+ (A) and addition of 2 μm TBZ (B). Growth was analyzed after two overnight incubations at 30 °C.
All mutants were sensitive to reserpine as the wild type (data not shown). The results suggest that either one of the aromatic residues Phe and Tyr at position 136 may contribute to rVMAT2 sensitivity toward the inhibitor TBZ, and is most likely needed for an efficient binding of the inhibitor to the protein. The bulkier Trp has a deleterious effect on activity.
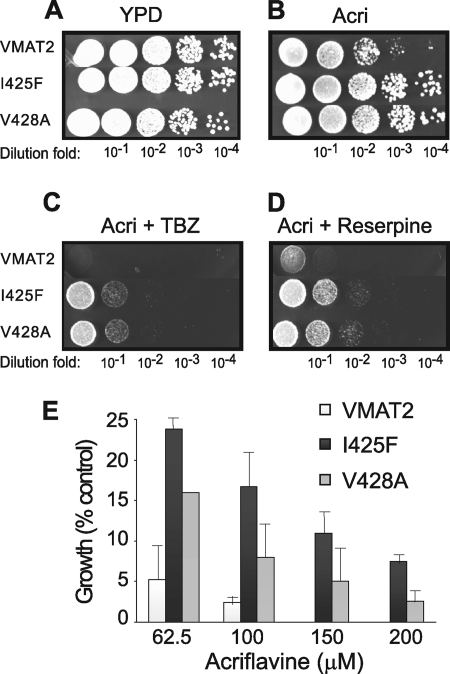
Type II Mutants Result from Mutations in TM11
In addition to the F136L mutant, two other mutants were isolated and further characterized. These mutants, I425F and V428A, displayed a phenotype different from the F136L even though they were isolated using the same strategy. Their growth on acriflavine in the presence of TBZ was substantial (Fig. 7C) but not as robust as the one displayed by F136L-rVMAT2. In addition, unlike F136L, growth was detectable also in the presence of reserpine (Fig. 7D). Whereas growth of rVMAT2-F136L cells was 50 times less sensitive toward TBZ, the difference in IC50 values for I425F and V428A mutants was only about 1.3- and 3.2-fold, respectively (Table 1).
FIGURE 7.
rVMAT2-I425F and V428A confer increased resistance to acriflavine. ADU1-7 cells harboring pAES426 rVMAT2, I425F, or V428A were grown on plates containing YPD (A), YPD with 40 μm acriflavine (B), YPD with 40 μm acriflavine and 2 μm TBZ (C), or YPD with 40 μm acriflavine and 100 nm reserpine (D). Growth in liquid media with different concentrations of acriflavine was analyzed after overnight incubation at 30 °C and is expressed as percent of growth of a culture without added acriflavine (E).
Notably, moreover, membranes prepared from cells expressing these two mutants bound [3H]TBZOH with an affinity only 2–3-fold lower than rVMAT2 (Table 1). Because it was surprising that such small differences would be sufficient for selection of these cells under the relatively stringent conditions used, we further investigated the phenotype. Remarkably, both mutants provided a more robust resistance to acriflavine than rVMAT2: as shown in Fig. 7E, growth of both mutants was evident even at concentrations as high as 200 μm acriflavine, a concentration well above that permissible for rVMAT2 cells. We suggest, therefore, that the reason that these mutants were selectable under the same stringent conditions used for isolation of F136L is due to a combination of two properties: superior growth on acriflavine and a small difference in the affinity to TBZ.
[3H]Serotonin transport assayed in proteoliposomes reconstituted with the I425F and V428A mutants revealed transport activity with a higher Km to serotonin. Thus the Km was 325 ± 55 and 1994 ± 708 nm for V428A and I425F, respectively, well above the value of 180 nm for rVMAT2 (Table 1).
To further characterize the role of amino acids at positions 425 and 428 in rVMAT2, we applied site-directed mutagenesis and replaced Ile-425 and Val-428 with various amino acids. The mutants were tested for their ability to confer resistance against MPP+ and acriflavine. Substitution of Val-428 to Lys or Asp impaired the ability of VMAT2 to confer resistance against the toxins. Mutant proteins with the hydrophobic amino acids Met (obtained also in the selection) and Leu at the same position conferred essentially the same levels of resistance as the V428A mutant and a mutant with Phe conferred slightly decreased resistance, essentially as wild-type rVMAT2. None of the replacements at position 425 conferred the robust resistance displayed by the Phe mutant. However, Ala, Leu, Met, and Trp supported the same level of resistance as rVMAT2, and Tyr generated a mutant with impaired function as judged by this assay (data not shown).
rVMAT2 with Three Mutations Lost the Ability to Transport Neurotransmitter but Confers Robust Resistance to a Variety of Toxic Compounds
A mutant rVMAT2 bearing the three mutations F136L, I425F, and V428A (rVMAT2-triple) was constructed and characterized. rVMAT2-triple conferred a considerably improved resistance to acriflavine (Fig. 8A) and to MPP+ (not shown) compared with wild type or to any of the single mutants (compare Figs. 8A and 7E). Moreover, it seems to have acquired novel specificities because it conferred a significant resistance to ethidium (Fig. 8C) and a marginal but noticeable resistance to acridine orange (Fig. 8B).
Despite the resistance conferred by the triple mutant, membranes prepared from cells expressing this mutant did not bind [3H]TBZOH (data not shown). [3H]Serotonin transport assayed in proteoliposomes reconstituted with the mutant revealed a very low transport activity that did not saturate in the concentration range tested (data not shown).
DISCUSSION
In this work we show that expression of rVMAT2 in S. cerevisiae confers a robust resistance to MPP+ and to acriflavine. MPP+ was already known as a substrate of VMATs but acriflavine, a substrate of many multidrug transporters, is identified here for the first time.
rVMAT2 is a protein specially suited to study in S. cerevisiae. It displays the pharmacological profile of a multidrug transporter (15) and provides resistance to MPP+ to CHO cells by removal into acidic compartments (10, 20). Previous efforts demonstrated the feasibility of the yeast system when using a bacterial multidrug transporter (21) but failed to provide a fully functional expression of bVMAT2 and rVMAT1 (22). The availability of a larger selection of genetically well-defined yeast strains with increased sensitivity to some of the substrates of VMAT led us to a renewed effort to screen for new substrates with a variety of plasmids.
The pharmacological properties of the protein expressed in yeast are unaltered as judged by the finding that reserpine, TBZ, and ketanserine, well-known inhibitors of rVMAT2, abolish the resistance. The biochemical characterization of the protein expressed in yeast suggests that its kinetic properties and specificity are indistinguishable from that expressed in mammalian cells (18) and very similar to the native bovine homologue previously studied mainly in chromaffin granules from bovine adrenal medulla (19). The localization, mainly in the vacuole of the yeast cell, supports the concept that drug transporters in eukaryotic cells can provide resistance not only by extrusion from the cell to the medium but also by compartmentalization into acidic compartments.
The strength of the yeast expression system stems from the ability to perform unbiased directed evolution experimentation based on the powerful phenotype of resistance in a relatively simple, fast growing unicellular organism that allows selection of specific mutants with desired properties. In the work described here we have generated two types of mutants of rVMAT2 that can grow in the presence of acriflavine even in the presence of high concentrations of TBZ. Type I mutants displayed a 50-fold decreased affinity to TBZ. The mutation resulted in replacement of Phe-136 to Leu, a site in TM2 that was not previously identified as involved in TBZ binding. The F136L mutation is an excellent example of the lack of bias in this technique: Phe-136 is conserved in several VNTs even in VMAT1 that displays a lower affinity to TBZ and VAChT that displays a very different substrate profile (supplemental Fig. S2A). Because of this, F136 was not a likely candidate for replacement using site directed mutagenesis. From the findings described here, we conclude that Phe-136 is one of the main, but certainly not the only, determinant of TBZ affinity. Analysis of the various replacements at this position revealed that replacement with hydrophobic amino acids such as Val, Met, Ile, and Leu generate a fully active protein that binds TBZ very poorly. Only a Tyr replacement generated a protein that binds TBZ, suggesting that an aromatic residue at position 136 is needed for efficient binding.
The interaction with competitive inhibitors such as reserpine or substrates such as serotonin, acriflavine, MPP+, and histamine is not impaired by the F136L mutation. Moreover, in the case of serotonin and acriflavine the mutant displays a higher apparent affinity. The higher apparent affinity to acriflavine of the F136L protein is reflected also in the more robust resistance conferred by its activity due to removal of the drug to lower concentrations than those achieved by the wild-type protein. The apparent affinity to ketanserine, an inhibitor that was known to interact at the same site as TBZ, is not affected to the same degree, suggesting that some of the binding interactions of both drugs must be different.
Type II mutants map at TM11 and maybe less directly involved in TBZ binding because their replacements have only a slightly decreased affinity to TBZ. The mutations resulted in replacement of Ile-425 and Val-428, sites that were not previously identified as involved in TBZ binding or substrate recognition. As the case for the F136L mutation, Ile-425 and Val-428 are conserved in several proteins throughout the vesicular amine transporter family (supplemental Fig. S2B). The mutation I425F displayed a lower Km toward serotonin and conferred a more robust resistance to acriflavine, suggesting a possible role in substrate recognition. When all the mutations were combined, the resulting protein, rVMAT2-triple, displayed a very interesting behavior: while it conferred an even higher resistance to acriflavine and MPP+ it lost practically completely the ability to transport serotonin and to bind TBZ. Moreover, it seems to have acquired novel specificities because it conferred a significant resistance to ethidium and a marginal but noticeable resistance to acridine orange. rVMAT2-triple resembles the properties of authentic multidrug transporters.
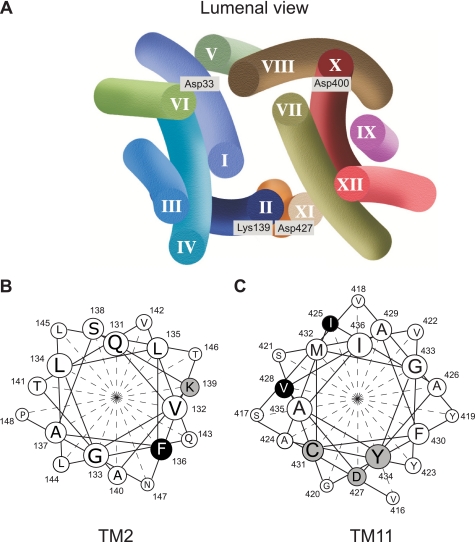
A structural model of rVMAT2 has been proposed, and, in this model, TM2 and TM11 are contiguous in space (Ref. 23 and Fig. 9). Several residues potentially involved in VMAT function and in TBZ binding have been identified in previous works (24–29). Along TM11 we find three of these residues (Fig. 9): Asp-427 was identified in the rVMAT1 isoform (Asp-431 in rVMAT1) as essential for transport. However, mutated proteins with Glu or Ser at the same position can perform partial reactions: they are unimpaired in ligand recognition and coupling of the electrochemical proton gradient to one of the protons transported (26). Cys-431 has been identified in human VMAT2 as a residue that confers sensitivity to thiol-reactive reagents, but the reaction is prevented by TBZ (27). Mutations in Tyr-434 reduce the affinity to serotonin and TBZ (28, 29). In TM2, Lys-139 has been identified as part of a charge pair with Asp-427 (24).
FIGURE 9.
Model of rVMAT2 and the cluster involved in TBZ binding. Charged residues facing the cavity and previously shown to play some role in function are highlighted (A). The helical projection of TM2 (B) and TM11 (C) is shown. Highlighted residues have been shown to modify rVMAT2 function or to provide targets for inhibitors.
Phe-136, Ile-425, and Val-428 identified in this work join now the above cluster (Fig. 9), and the finding strongly supports the suggestion that this section of the protein may be involved directly in TBZ binding and serotonin transport. The fact that only proteins with Phe and Tyr at position 136 display high affinity to TBZ led us to speculate that the residue at this position may be directly involved in aromatic interaction with TBZ. The mechanism of inhibition of transport by TBZ has been the subject of a large number of studies. TBZ is a potent non-competitive inhibitor of VMAT-mediated transport (13). It also inhibits substrate binding with a similar Ki. On the other hand, substrates inhibit TBZ binding only at concentrations at least hundred times higher than their affinities (13, 30). A corollary of this finding is that both sites (substrate and TBZ) can be occupied at the same time, provided the substrate binds first. We speculate that occupancy of the TBZ binding site prevents access to the substrate, probably by the formation of a steric barrier similar to what was suggested for the mode of inhibition of an antidepressant in the bacterial LeuT transporter (31, 32).
The fact that Phe-136, Ile-425, and Val-428 are conserved even in homologues that do not bind TBZ stresses the power of the approach described here that allows for an unbiased search of residues involved in one or other aspect of the catalytic cycle. Native vesicular neurotransmitters transporters have been extensively characterized at the biochemical level (1). However, as with many other mammalian membrane proteins, there is a lack of detailed high-resolution information. In addition, unlike the case of plasma membrane transporters, powerful analytical techniques such as electrophysiology do not seem to be easily applicable to transporters that are expressed in intracellular membranes. These facts prompted us to a renewed effort to develop new techniques for the study of VMAT. The yeast system combined with the approach of directed evolution provides us with a novel powerful tool to study a protein that plays a central role in the process of neurotransmission.
A variety of phylogenetic studies suggest that rVMAT2 and related vesicular neurotransporters have evolved from bacterial drug/H+ antiporters that have been implicated in bacterial resistance to antibiotics and other toxic compounds (1, 33, 34). From the work described here, it seems that in the case of rVMAT2, loss of traits acquired in evolution of function (such as serotonin transport and TBZ binding) bring about an improvement in older functions such as resistance to toxic compounds. A process that has taken millions of years of evolution can be reversed by three mutations.
Supplementary Material
Acknowledgments
We thank Dr. D. Engelberg and T. Ravid for advice in the work with yeast, Dr. Naomi Melamed-Book for confocal microscopy, Ofer Perl for performing part of the experiments in Table 1 and Fig. 5, and Yael Elbaz for constructs used in this work and for extensive discussions.
This work was supported, in whole or in part, by Grant NS16708 from the National Institutes of Health. This work was also supported by The Center for Innovation in Membrane Protein Production (P50 GM73210).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- VNT
- vesicular H+-coupled neurotransporter
- MDMA
- 3,4-methylenedioxymethamphetamine
- GFP
- green fluorescent protein
- MPP+
- 1-methyl-4-phenylpyridinium
- VMAT
- vesicular monoamine transporter
- TBZ
- tetrabenazine
- FM 4-64
- N-(3-triethylammoniumpropyl)-4-(p-diethylamino-phenyl-hexatrienyl) pyridinium dibromide
- TM
- transmembrane.
REFERENCES
- 1.Schuldiner S., Shirvan A., Linial M. (1995) Physiol. Rev. 75, 369–392 [DOI] [PubMed] [Google Scholar]
- 2.Eiden L. E. (2000) FASEB J. 14, 2396–2400 [DOI] [PubMed] [Google Scholar]
- 3.Erickson J. D., Varoqui H. (2000) FASEB J. 14, 2450–2458 [DOI] [PubMed] [Google Scholar]
- 4.Edwards R. H. (2007) Neuron 55, 835–858 [DOI] [PubMed] [Google Scholar]
- 5.Merickel A., Edwards R. H. (1995) Neuropharmacology 34, 1543–1547 [DOI] [PubMed] [Google Scholar]
- 6.Peter D., Jimenez J., Liu Y., Kim J., Edwards R. H. (1994) J. Biol. Chem. 269, 7231–7237 [PubMed] [Google Scholar]
- 7.Stitzel R. E. (1976) Pharm. Rev. 28, 179–208 [PubMed] [Google Scholar]
- 8.Freis E. (1954) N. Engl. J. Med. 251, 1006–1008 [DOI] [PubMed] [Google Scholar]
- 9.Schuldiner S., Steiner-Mordoch S., Yelin R., Wall S. C., Rudnick G. (1993) Mol. Pharmacol. 44, 1227–1231 [PubMed] [Google Scholar]
- 10.Liu Y., Peter D., Roghani A., Schuldiner S., Privé G. G., Eisenberg D., Brecha N., Edwards R. H. (1992) Cell 70, 539–551 [DOI] [PubMed] [Google Scholar]
- 11.Liu Y., Edwards R. H. (1997) Annu. Rev. Neurosci. 20, 125–156 [DOI] [PubMed] [Google Scholar]
- 12.Kenney C., Jankovic J. (2006) Expert. Rev. Neurother. 6, 7–17 [DOI] [PubMed] [Google Scholar]
- 13.Scherman D., Jaudon P., Henry J. P. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 584–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darchen F., Scherman D., Henry J. P. (1989) Biochemistry 28, 1692–1697 [DOI] [PubMed] [Google Scholar]
- 15.Yelin R., Schuldiner S. (1995) FEBS Lett. 377, 201–207 [DOI] [PubMed] [Google Scholar]
- 16.Rogers B., Decottignies A., Kolaczkowski M., Carvajal E., Balzi E., Goffeau A. (2001) J. Mol. Microbiol. Biotechnol. 3, 207–214 [PubMed] [Google Scholar]
- 17.Vida T. A., Emr S. D. (1995) J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adam Y., Edwards R. H., Schuldiner S. (2008) Am. J. Physiol. Cell Physiol. 294, 1004–1011 [DOI] [PubMed] [Google Scholar]
- 19.Stern-Bach Y., Greenberg-Ofrath N., Flechner I., Schuldiner S. (1990) J. Biol. Chem. 265, 3961–3966 [PubMed] [Google Scholar]
- 20.Liu Y., Roghani A., Edwards R. H. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 9074–9078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yelin R., Rotem D., Schuldiner S. (1999) J. Bacteriol. 181, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yelin R., Schuldiner S. (2001) Biochim. Biophys. Acta 1510, 426–441 [DOI] [PubMed] [Google Scholar]
- 23.Vardy E., Arkin I. T., Gottschalk K. E., Kaback H. R., Schuldiner S. (2004) Protein Sci. 13, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merickel A., Kaback H. R., Edwards R. H. (1997) J. Biol. Chem. 272, 5403–5408 [DOI] [PubMed] [Google Scholar]
- 25.Merickel A., Rosandich P., Peter D., Edwards R. H. (1995) J. Biol. Chem. 270, 25798–25804 [DOI] [PubMed] [Google Scholar]
- 26.Steiner-Mordoch S., Shirvan A., Schuldiner S. (1996) J. Biol. Chem. 271, 13048–13054 [DOI] [PubMed] [Google Scholar]
- 27.Thiriot D. S., Ruoho A. E. (2001) J. Biol. Chem. 276, 27304–27315 [DOI] [PubMed] [Google Scholar]
- 28.Finn J. P., 3rd, Edwards R. H. (1998) J. Biol. Chem. 273, 3943–3947 [DOI] [PubMed] [Google Scholar]
- 29.Finn J. P., 3rd, Edwards R. H. (1997) J. Biol. Chem. 272, 16301–16307 [DOI] [PubMed] [Google Scholar]
- 30.Scherman D., Henry J. P. (1984) Mol. Pharmacol. 25, 113–122 [PubMed] [Google Scholar]
- 31.Singh S. K., Yamashita A., Gouaux E. (2007) Nature 448, 952–956 [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z., Zhen J., Karpowich N. K., Goetz R. M., Law C. J., Reith M. E., Wang D. N. (2007) Science 317, 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vardy E., Steiner-Mordoch S., Schuldiner S. (2005) J. Bacteriol. 187, 7518–7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saier M. H., Jr., Paulsen I. T. (2001) Semin Cell Dev. Biol. 12, 205–213 [DOI] [PubMed] [Google Scholar]
- 35.Friedmann Y., Shriki A., Bennett E. R., Golos S., Diskin R., Marbach I., Bengal E., Engelberg D. (2006) Mol. Pharmacol. 70, 1395–1405 [DOI] [PubMed] [Google Scholar]
- 36.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 37.Boeke J. D., LaCroute F., Fink G. R. (1984) Mol. Gen. Genet. 197, 345–346 [DOI] [PubMed] [Google Scholar]
- 38.Guthrie C., Fink G. R. (1991) in Methods in Enzymology (Guthrie C., Fink G. R. eds), pp. xvii–xvii, Academic Press, New York [Google Scholar]
- 39.Elble R. (1992) BioTechniques 13, 18–20 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.