Abstract
Acquired resistance through genetic mutations is a common phenomenon in several cancer therapies using molecularly targeted drugs, best exemplified by the BCR-ABL inhibitor imatinib in treating chronic myelogenous leukemia (CML). Overcoming acquired resistance is a daunting therapeutic challenge, and little is known about how these mutations evolve. To facilitate understanding the resistance mechanisms, we developed a novel culture model for CML acquired resistance in which the CML cell line KCL-22, following initial response to imatinib, develops resistant T315I BCR-ABL mutation. We demonstrate that the emergence of BCR-ABL mutations do not require pre-existing BCR-ABL mutations derived from the original patient as the subclones of KCL-22 cells can form various BCR-ABL mutations upon imatinib treatment. BCR-ABL mutation rates vary from cell clone to clone and passages, in contrast to the relatively stable mutation rate of the hypoxanthine-guanine phosphoribosyltransferase gene. Strikingly, development of BCR-ABL mutations depends on its gene expression because BCR-ABL knockdown completely blocks KCL-22 cell relapse on imatinib and acquisition of mutations. We further show that the endogenous BCR-ABL locus has significantly higher mutagenesis potential than the transduced randomly integrated BCR-ABL cDNA. Our study suggests important roles of BCR-ABL gene expression and its native chromosomal locus for acquisition of BCR-ABL mutations and provides a new tool for further studying resistance mechanisms.
Keywords: Diseases/Cancer/Leukemia, Mutagenesis Mechanisms, Oncogene, Tyrosine Kinase, Reactive Oxygen Species (ROS), shRNA, BCR-ABL, Acquired Resistance, Chronic Myelogenous Leukemia, Imatinib
Introduction
Chronic myelogenous leukemia results from malignant transformation of a primitive hematopoietic cell by an oncogenic fusion gene BCR-ABL. Treatment with the potent ABL tyrosine kinase inhibitor imatinib mesylate (Gleevec®, STI-571, imatinib) results in complete cytogenetic responses in most chronic phase patients with infrequent relapse, but the treatment has poor responses and high frequency of relapse in advanced phases of the disease (1). Clinical resistance to imatinib is mediated primarily by genetic mutations of the BCR-ABL kinase domain and, to a lesser extent, by amplification of BCR-ABL gene (2). Numerous BCR-ABL mutations have been identified in relapsed CML3 patients, which confer various degrees of resistance to imatinib (2–4). Among them, the T315I mutation is the most resistant in that it does not respond to treatment with the more potent second generation of kinase inhibitors such as nilotinib (5, 6) and dasatinib (7).
In contrast to in vivo resistance, nearly all CML cell lines derived from blast crisis CML are sensitive to imatinib treatment in culture (8). Several resistant CML cell lines have been generated by exposing cells to gradually increasing concentrations of imatinib; however, the resulting resistant cells harbor BCR-ABL gene amplification but not mutations (9), in contrast to what is seen in patients. By expressing BCR-ABL cDNA in non-CML cell lines and/or random mutagenesis, multiple studies have demonstrated clinically relevant or novel BCR-ABL mutations that render resistance to imatinib in those cells (5–7, 10–15). Various BCR-ABL mutations can also occur in a short period of time when primary CML cells are cultured in growth factor-supplemented medium (16, 17). However, rapid generation of BCR-ABL mutations for acquired resistance in a CML cell line has not been reported and would be useful for our understanding of mechanisms of CML drug resistance and facilitate the development of new generations of BCR-ABL inhibitors.
In this study, we have developed a novel culture model for CML acquired resistance in which a blast crisis CML cell line, KCL-22, underwent initial apoptosis upon treatment with therapeutically effective doses of imatinib, but cells re-grew after 2 weeks with development of resistance through T315I BCR-ABL mutation. We have found that pre-existing BCR-ABL mutations were not required for the resistance. We have shown that the acquired resistance of KCL-22 cells on imatinib was dependent on the expression of BCR-ABL itself and that the native BCR-ABL translocation locus plays a role in promoting BCR-ABL mutations.
EXPERIMENTAL PROCEDURES
Cell Culture and Drugs
CML cell lines KCL-22 and K562 were purchased from German Collection of Cell Cultures, Braunschweig, Germany, and grown in RPMI 1640 medium with 10% fetal bovine serum (Hyclone, SH30071.03). The incoming cells were designated as passage 1. Imatinib (STI-571) was kindly provided by Novartis, Basel, Switzerland, and 6-thioguanine was purchased from Sigma.
Resistance Assay
One-half million KCL-22 cells were seeded in 1 ml of medium per well in 24-well plates and treated with different concentrations of STI-571. Cells were maintained in culture without changing medium. Aliquots of cells at specified time points were removed and counted on a hematocytometer. Cell viability was assessed by trypan blue exclusion. Typically, after 2 weeks in culture when the medium volume significantly decreased, fresh drug-free medium was supplied to the cells to restore that to the original volume for prolonged culture.
Soft Agar Colony Formation Assay
A standard two-layer soft agar culture was performed with a bottom layer of 0.6% agarose (Sigma, A9045) and a top layer of 0.35% agarose. For colony formation without drugs, 500 cells per well were seeded with warm top agar in 6-well plates and incubated for 3 weeks. Plates were then stained with 0.005% Crystal Violet for 1 h, and colonies were scored with the aid of a microscope. For drug resistance assay, 1,000,000 cells per well were seeded in 6-well plates in triplicate with STI-571 or 6-thioguanine added to both the top and bottom agar to their final concentrations. To clone or recover soft agar colonies for further analysis, individual colonies were plucked and expanded in liquid culture.
Cell Cycle, Cell Proliferation, and Apoptosis Analysis
For cell cycle analysis, cells were fixed in 70% ethanol at −20 °C overnight. After washing, cells were resuspended in phosphate-buffered saline containing 1 mg/ml RNase A, incubated for 30 min at 37 °C, and then stained with propidine iodine (50 μg/ml) for 30 min at room temperature before flow cytometry analysis. Cell proliferation was analyzed using an XTT cell proliferation kit (Roche Applied Science), and apoptosis was analyzed with annexin V (Pharmingen) as per the manufacturer's instruction.
Spectral Karyotyping and Fluorescent in Situ Hybridization (FISH) Analysis
For spectral karyotyping analysis, 10 mitotic cells of each sample were analyzed, 5 by GTG-band analysis and 5 by 24-color karyotyping. Triple-color FISH was performed with LSI ABL1/ASS (9q34.1)/BCR (22q11.2) probes (Vysis, Inc.) (18), and 200 cells of each sample were evaluated. These assays were done by City of Hope Cytogenetics Core Laboratory.
Sequencing Analysis
For sequencing cDNA, the ABL kinase domain was amplified by reverse transcription-PCR of total RNA with a high fidelity DNA polymerase (Stratagene) using a forward primer 5′-GCGCAACAAGCCCACTGTCTATGG and reverse primer 5′-GCCAGGCTCTCGGGTGCAGTCC that amplified the 579-bp kinase domain. To confirm mutations, the ABL kinase domain was also amplified by PCR using genomic DNA as templates with the intron primers 5′-GAGCCACGTGTTGAAGTCCT-3′ and 5′-TTTGTAAAAGGCTGCCCGGC-3′, which span ABL exon 6 for T315I mutation, and the intron primers 5′-GCCTGTCTCTGTGGGCTGAAG-3′ and 5′-TAATGCCAGCAGACGCCTTG-3′, which span ABL exon 5 for E255K and Y253H mutations. PCR products were cloned into the pCR2.1 vector using TA cloning kit (Invitrogen). At least 10 bacterial clones for each treatment group were sequenced by Sequencing Facility of Beckman Research Institute. Typically, about 30–50% of clones carried mutations because KCL-22 cells harbors one wild type allele of c-ABL gene that was also amplified for sequencing. For analysis of genomic DNA mutations of some clonal cells, purified PCR products from 1% agarose gel were directly sequenced without subcloning. We also carried out reverse transcription-PCR for sequencing BCR-ABL oligomerization and Src homology 3/2 (SH3/2) domains. For the oligomerization domain, we used forward primer 5′-GAGTGGGCGGGCATTGTTC and reverse primer 5′-GGGACTTTTTGCGCTCCATCT. For sequencing the BCR-ABL SH3/2 domain, we used primers described previously (13). For sequencing HPRT, the codon sequence was amplified by reverse transcription-PCR using a primer pair (forward, 5′-ACCGGCTTCCTCCTCCTGAG-3′; reverse, 5′-GATAATTTTACTGGCGATGT-3′) as described previously (19).
Cell Cloning by Limiting Dilution
Cells were counted and diluted to five cells per ml and seeded onto a 96-well plate with 100 μl (or 0.5 cell) per well. Individual cell seeding was then confirmed by microscopy, and single cell clones were grown and expanded for further analysis.
Immunoprecipitation and Western Blot Analysis
BCR-ABL expression and phosphorylation were analyzed by Western blot using anti-c-ABL monoclonal antibody (Pharmingen, 554148) and anti-phosphotyrosine antibody (Upstate Biotechnology, Inc., 05-321). To validate BCR-ABL phosphorylation, we pulled down BCR-ABL from 500 μg of total cell lysate of KCL-22 cells with 2 μg of anti-c-ABL and 100 μl of 50% slurry of protein G-agarose beads (Upstate Biotechnology, Inc.). The phosphorylation was detected by Western blot with tyrosine phosphorylation antibody.
Analysis of Reactive Oxygen Species (ROS) and DNA Damage
ROS was analyzed using redox-sensitive fluorochrome 2′,7′-dichlorofluorescein diacetate (Sigma) as described previously (16). When coupled with apoptosis analysis, cells were labeled with annexin V first and then 2′,7′-dichlorofluorescein diacetate. An H2AX phosphorylation assay kit (Millipore, 17-344) was used to analyze DNA damage by flow cytometry as per the manufacturer's suggestions.
shRNA Lentiviral Vectors and Gene Knockdown
Oligonucleotides for ABL shRNA (GTTGGTTCATCATCATTCA) were synthesized and cloned into the pSicoR vector (20) that contains a selection cassette for puromycin by a standard protocol. A scrambled shRNA was subcloned into the vector as a mock control. The VSV-G (G protein of vesicular stomatitis virus) pseudotyped lentiviral vectors were produced using a four-plasmid transfection system as described previously (21). For BCR-ABL knockdown, 1,000,000 cells were infected overnight with recombinant lentivirus by multiplicity of infection of 3 each in the presence of 8 μg/ml Polybrene. Under this condition, the transduction rate in these cells was typically about 99% (not shown).
BCR-ABL Overexpression Analysis
We produced amphotropic retroviral vectors for wild type p210 BCR-ABL, K1176R p210 mutant and the empty vector MIG R1, using Phoenix-Ampho packaging cells (ATCC) and carried out the transduction as described previously (22, 23). Briefly, KCL-22 cells were transduced with these vectors at a multiplicity of infection around 6. Cells were spin-infected by centrifugation at 1000 × g for 90 min and then returned to the incubator and cultured overnight. After removing viruses, cells were expanded in culture for 5 days, and green fluorescent protein (GFP)-expressing cells were isolated by fluorescent-activated cell sorting (FACS). For second round transduction, FACS-enriched cells were re-infected with the above vectors using the same conditions. We performed genomic DNA sequencing analysis of imatinib-resistant soft agar colonies using the intron primers for T315I mutation on the endogenous BCR-ABL locus and exon primers for 579-bp transduced BCR-ABL cDNA as described above. To increase PCR efficiency on the transduced BCR-ABL cDNA, we also used another reverse primer 5′-TAGTCCAGGAGGTTCCCGTAG to pair with the same exon forward primer used for 579-bp cDNA, which produced a 321-bp PCR product.
RESULTS
Novel Culture Model for CML Acquired Resistance Using Blast Crisis CML Cell KCL-22
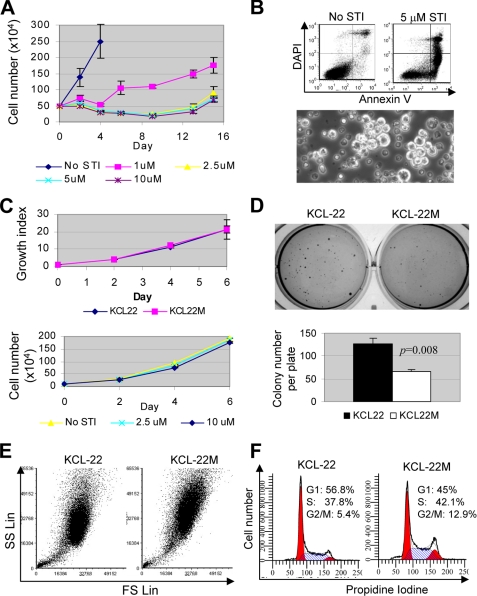
Imatinib at 1 μm selectively kills most cultured CML cells in a BCR-ABL-dependent manner with the exception of KCL-22 cells (8). In chronic phase CML patients, imatinib is given at 400 mg/day that produces the average peak plasma concentration at 4.4 μm and trough concentration at 2.0 μm (24). For blast crisis patients, imatinib dosage is increased to 600 mg/day. Therefore, we examined the effects of imatinib concentrations at 1, 2.5, 5, and 10 μm on the survival of KCL-22 cells during prolonged culture. We found that KCL-22 cells continued growing under 1 μm imatinib treatment but at a lower rate than in the absence of the drug. At 2.5 μm and higher concentrations, imatinib effectively suppressed cell growth and induced apoptosis over time (Fig. 1, A and B). Surprisingly, we found that small clusters of cells formed after 8–10 days in 2.5–10 μm treatment groups, and these cells appeared visibly larger with frequent irregular shapes (Fig. 1B), and after 2 weeks they repopulated the culture (Fig. 1A), indicating the relapse on the drug treatment. These emerging cells, named KCL-22M, were expanded and maintained in culture in the absence of imatinib. They grew equally well as KCL-22 cells and no longer responded to imatinib in the medium (Fig. 1C), but they formed fewer and smaller soft agar colonies (Fig. 1D). The abnormal size and shape of KCL-22M cells were confirmed by flow cytometric analysis, showing increase on both forward scatter (for size) and side scatter (for complexity) (Fig. 1E). KCL-22M cells exhibited a different cell cycle from KCL-22 cells by increasing S and G2/M fractions (p < 0.001, Fig. 1F).
FIGURE 1.
Novel model of CML acquired resistance. A, one-half million KCL-22 cells were treated with 1, 2.5, 5, and 10 μm imatinib (STI), and at days after the treatment as indicated, survival cells were counted. Relapse occurred on 2.5 μm and higher concentrations of imatinib 2 weeks post-treatment. B, top, apoptosis of KCL-22 cells after 6 days of treatment with 5 μm imatinib was analyzed by annexin V staining. DAPI, 4′,6-diamidino-2-phenylindole. Bottom, formation of clusters of resistant cells (bright) among scattered dead cells (dark) with 2.5 μm STI-571 treatment for 8 days. C, top, growth curves for resistant cells (KCL-22M) and KCL-22 cells analyzed by XTT. Growth indexes were relative XTT readings normalized to the initial XTT readings at day 0. Bottom, comparison of growth of KCL-22M cells in the absence and presence of imatinib. D, soft agar colony formation of KCL-22 and KCL-22M cells. Five hundred cells were seeded each well in 6-well plates in triplicate. E, comparison of cell size and complexity of KCL-22 and KCL-22M cells. KCL-22M cells exhibited increase at both forward scatter (FS) and side scatter (SC) parameters. SS Lin, side scatter in linear scale. F, comparison of cell cycle of KCL-22 and KCL-22M cells with propidium iodine staining.
Acquired Resistance of KCL-22 Cells Is Mediated by T315I BCR-ABL Mutation
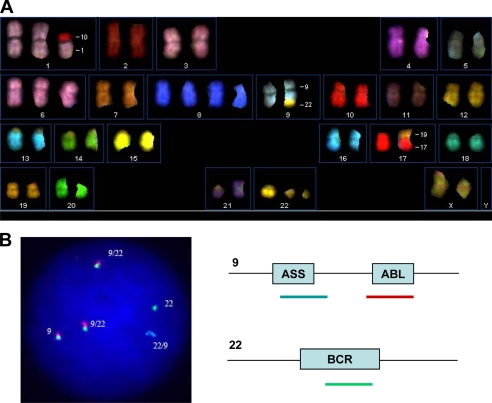
We characterized molecular alterations of KCL-22M cells. By spectral karyotyping and FISH analyses, we found that KCL-22M cells maintained the same cytogenetic profile as KCL-22 cells, i.e. 51,X,del(X)(p11.2p22.3),+der(1;10)(q10;p10),+6,+8,+8,t(9;22)(q34.1;q11.2),der(17;19)(q10;q10),+19,i(21)(q10),+der(22)t(9;22), and carried two Philadelphia chromosomes in all cells examined (Fig. 2 and data not shown). Our cytogenetic data were in line with previously reported karyotype for KCL-22 cells (25, 26) and showed no novel chromosomal rearrangements in KCL-22M cells, which was in contrast to another KCL-22 cell-derived imatinib-resistant cell line KCL-22R that has additional translocations (26). Using real time PCR, we found that the ABL DNA content and BCR-ABL RNA level in KCL-22M cells remained the same as in KCL-22 cells (supplemental Fig. 1), and consistently, the BCR-ABL protein level also remained the same in KCL-22M and KCL-22 cells (Fig. 3A). However, the BCR-ABL protein level in KCL-22 cells decreased after treatment with increasing concentrations of imatinib, and it stayed constant in KCL-22M cells (Fig. 3A). We found that the drug effectively inhibited tyrosine phosphorylation in KCL-22 cells; in contrast, tyrosine phosphorylation in KCL-22M cells remained relatively constant under imatinib treatment up to 5 μm (Fig. 3A). Slight decrease of phosphorylation under 10 μm imatinib was noticed, which was likely due to nonspecific activity of the drug under this concentration (8).
FIGURE 2.
Cytogenetic characteristics of KCL-22M cells. A, karyogram of 24-color spectral karyotyping. The composite karyotype is 51,X,del(X)(p11.2p22.3),+der(1;10)(q10;p10),+6,+8,+8,t(9;22) (q34.1;q11.2),der(17;19)(q10;q1),+19,i(21)(q10),+der(22)t(9;22)[10]. B, left, representative three-color FISH of BCR-ABL. Two Philadelphia chromosomes (9/22) were present in all cells. Right, the color code for FISH probes. ASS, argininosuccinate synthetase gene.
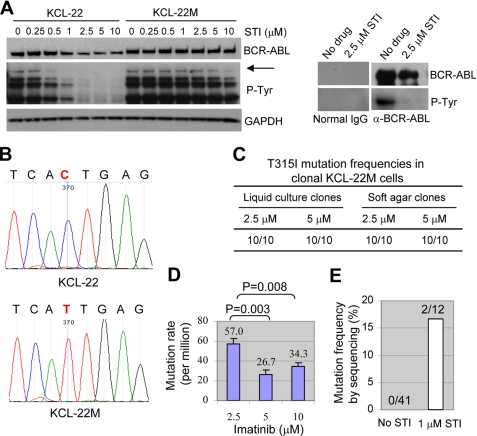
FIGURE 3.
Molecular characterizations of the new CML resistance model. A, left, Western blot analysis of BCR-ABL expression and tyrosine phosphorylation in KCL-22 and KCL-22M cells with and without imatinib treatment. The top band (arrow) detected by the tyrosine phosphorylation antibody corresponds to the position of BCR-ABL. Right, immunoprecipitation of KCL-22 cell lysate with BCR-ABL antibody or normal IgG followed by Western analysis for BCR-ABL expression and phosphorylation. P-Tyr, phosphotyrosine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, sequencing analysis of BCR-ABL kinase domain with cDNA or genomic DNA from KCL-22 and KCL-22M cells. Notice the point mutation of C to T (in bold and red) that causes T315I amino acid change. C, sequencing of BCR-ABL kinase domain mutations in clonal cells. KCL-22M cells derived from 2.5 and 5 μm imatinib treatment were subcloned by limiting dilution, and 10 clones each (liquid culture clones) were sequenced using genomic templates. Clonal cells were separately obtained by plating KCL-22 cells on soft agar with 2.5 or 5 μm imatinib, and 10 colonies each were sequenced. D, mutation frequencies of KCL-22 cells. One million KCL-22 cells were seeded on soft agar with the indicated concentrations of imatinib, and resistant colonies were scored after 3 weeks. E, mutation detection by conventional DNA sequencing. KCL-22 cells cultured without or with 1 μm imatinib for 2 weeks were analyzed. Forty one bacterial clones for untreated cells and 12 for treated cells were sequenced. Mutations were found in only two clones in the latter.
Given that BCR-ABL mutations account for most of clinical resistance, we next examined if BCR-ABL was mutated in KCL-22M cells. We sequenced both cDNA and genomic DNA for BCR-ABL kinase domain using the strategies described by Gorre et al. (2). In the 579-bp cDNA region of the BCR-ABL kinase domain covering the ATP-binding pocket and activation loop, we found only one C to T nucleotide change that resulted in amino acid T315I mutation at BCR-ABL in KCL-22M but not parental KCL-22 cells (Fig. 3B). Sequencing of genomic DNA further confirmed such a mutation (data not shown). To search for mutations on other BCR-ABL domains important for its kinase activity, we sequenced oligomerization and SH3/2 domains (27, 28), but no extra mutations were identified. It is worth mentioning that the SH3/2 sequencing fragment extended through most of the 579-bp kinase domain, and again, only T315I mutation was found.
To determine whether KCL-22M cells are a mixture of T315I and other mutants, we cloned individual mutant cells from KCL-22M cells relapsed after 2.5 and 5 μm imatinib treatment, respectively, and sequenced 10 clones each for BCR-ABL mutations using genomic templates. We found that all 20 clones carried the T315I mutation (Fig. 3C). To examine if the homogeneity of this mutation is a result of dominance of T315I mutant cells over cells bearing other mutations in liquid culture, we generated clonal resistant cells by plating KCL-22 cells on soft agar with 2.5 or 5 μm imatinib. We found that all 20 colonies, 10 from each concentration of imatinib, carried T315I mutation (Fig. 3C). Furthermore, we found that KCL-22 cells at various passages relapsed on imatinib, and all recurrent cells harbored T315I mutation (data not shown). Although all the above studies used a single dose imatinib treatment, we found that KCL-22 cells relapsed similarly with repeated doses of imatinib and developed T315I mutation only (supplemental Fig. 2 and data not shown). Our results suggest that KCL-22 cells developed acquired resistance predominantly, perhaps solely, through T315I BCR-ABL mutation.
We next measured the T315I mutation rate. By clonogenic assay described above, the average number of mutants per million cells was around 30 for 5 and 10 μm imatinib treatment and 57 for 2.5 μm imatinib treatment (Fig. 3D). If the plating efficiency of KCL-22M cells were considered (Fig. 1D), the mutation rates were about 2.5 × 10−4 and 4.8 × 10−4, respectively. The treatment with 2.5 μm imatinib resulted in a significantly higher mutation rate (Fig. 3D), given that KCL-22M derived from 2.5 μm imatinib did not respond to higher concentrations of the drug (data not shown).
To determine whether there is a pre-existing T315I mutation in KCL-22, we used two sensitive methods, a modified fluorescent allele-specific oligonucleotide-PCR assay (29) and a bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification assay (30), to detect T315I mutations in cDNA (by allele-specific oligonucleotide-PCR) or genomic DNA (by bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification) from untreated KCL-22 cells. We found that both methods reliably detected at least 1% mutant allele, but no T315I mutation was found in KCL-22 cells (supplemental Fig. 3). In our hands, we were unable to further increase the sensitivity of these methods without sacrificing the specificity, and therefore we could not use them to rule out the existence of rare mutant cells in KCL-22 cells. Intriguingly, however, after continuous culture of KCL-22 cells in a refractory dose of imatinib (1 μm) for 2 weeks, T315I mutation became readily detectable by conventional DNA sequencing (Fig. 3E). As T315I is the most resistant mutation for BCR-ABL inhibitors (5–7), this model recapitulates one key BCR-ABL mutagenesis process in clinical relapse. However, our approach of direct exposure of cells to therapeutically effective concentrations of imatinib did not work for CML cell lines K562 and KU812 that underwent rapid apoptosis (data not shown) as reported previously (8), probably before BCR-ABL mutations can be fully developed. It remains to be further tested whether this method can be applied to other CML cell lines. This may appear as a disadvantage when compared with the standard approach of generating imatinib resistance that can be applied to many cell lines. However, our model offers a unique advantage that BCR-ABL mutations can be rapidly produced within 2 weeks after one single exposure of cells to in vivo effective concentrations of imatinib, compared with the standard approach by multiple rounds of exposure of cells to gradually increasing concentrations of the drug for a period of several months.
Acquired Resistance of Clonal KCL-22 Cells through BCR-ABL Mutations
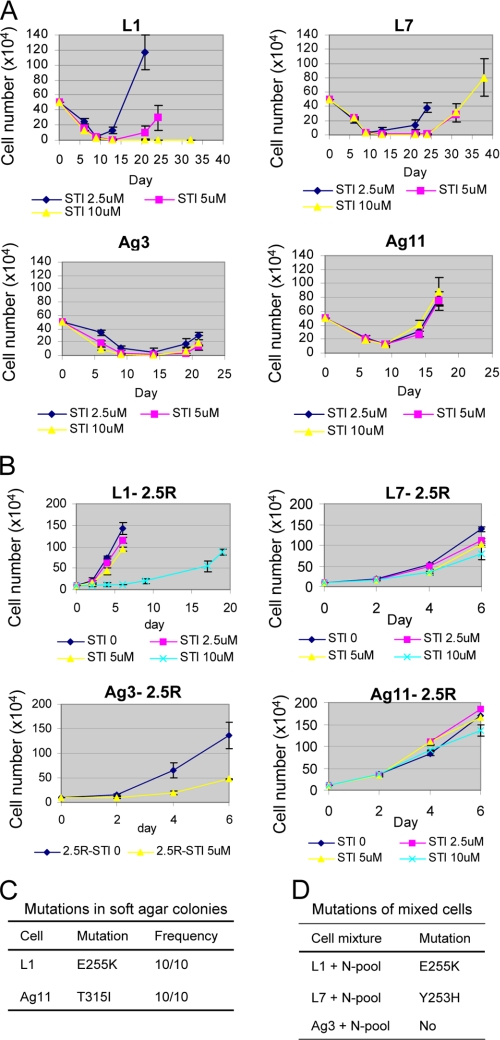
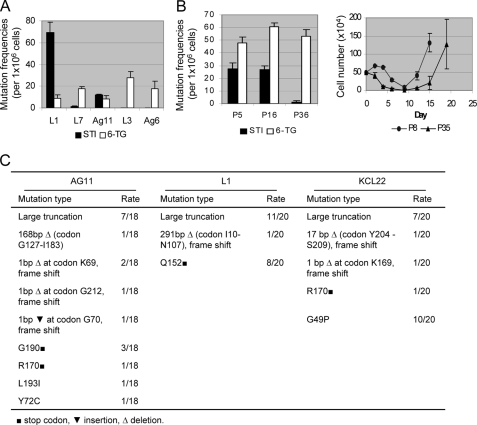
Because pre-existing T315I mutations are detected in some CML patients before imatinib treatment (3, 31), we set out to determine whether the rare pre-existing BCR-ABL mutant cells originated from the patient are required for development of resistance. Individual KCL-22 cell clones were isolated without drug treatment, 11 by the method of limiting dilution (clones L1–L11) and 13 by soft agar cloning (clones Ag1–Ag13), as shown in Table 1. We found that none of these clones carried BCR-ABL mutations and that imatinib induced apoptosis in all of them (Fig. 4A and data not shown). Most of clones failed to relapse, but four relapsed in 2 weeks with high frequency at different concentrations of imatinib, namely clones L1, L7, Ag 3, and Ag 11 (Table 1 and Fig. 4A). Intriguingly, clones L1, L7, and Ag11 relapsed with E255K, Y253H, and T315I BCR-ABL mutations, respectively, but clone Ag3 relapsed without BCR-ABL kinase domain mutations (Table 1). These BCR-ABL mutations conferred various degrees of clonal cell resistance to imatinib with T315I having the greatest protective effect (Fig. 4B). Compared with parental cells, more BCR-ABL mutations emerged from these clones, and all of them are the frequent mutations found in patients (3, 32). The ability of these clones to develop resistance through BCR-ABL kinase domain mutations suggests that the pre-existing rare mutant cells from the original patient are not required for the resistance.
TABLE 1.
Relapse and mutation analysis in clonal cells
No mutations have been detected in clones L1, L7, Ag3, and Ag11 before STI treatment. ND means not done.
| Clone | STI 2.5 μm |
STI 5 μm |
STI 10 μm |
|||
|---|---|---|---|---|---|---|
| Relapsed wells/seeded wells | Mutation | Relapsed wells/seeded wells | Mutation | Relapsed wells/seeded wells | Mutation | |
| L1 | 7/8 | E255K | 4/5 | E255K | 0/5 | |
| L2 | 0/6 | 0/3 | ND | |||
| L3 | 0/6 | 0/3 | ND | |||
| L4 | 0/6 | 1/3 | No | ND | ||
| L5 | 1/6 | No | 0/3 | ND | ||
| L6 | 0/6 | 0/3 | ND | |||
| L7 | 8/8 | Y253H | 4/5 | Y253H | 4/5 | Y253H |
| L8 | 0/6 | 0/3 | ND | |||
| L9 | 0/6 | 0/3 | ND | |||
| L10 | 0/6 | 0/3 | ND | |||
| L11 | 0/6 | 0/3 | ND | |||
| Ag1 | 0/6 | 0/3 | ND | |||
| Ag2 | 1/6a | ND | 0/3 | ND | ||
| Ag3 | 8/8 | No | 2/2 | No | 5/5 | No |
| Ag4 | 0/6 | 0/3 | ND | |||
| Ag5 | 0/6 | 0/3 | ND | |||
| Ag6 | 0/6 | 0/3 | ND | |||
| Ag7 | 1/6 | No | 0/3 | 0/3 | ||
| Ag8 | 5/6a | ND | 0/3 | 0/3 | ||
| Ag9 | 1/6 | No | 0/3 | 0/3 | ||
| Ag10 | 0/6 | 0/3 | ND | |||
| Ag11 | 8/8 | T315I | 2/2 | T315I | 5/5 | T315I |
| Ag12 | 0/6 | 0/3 | ND | |||
| Ag13 | 0/6 | 0/3 | ND | |||
a Wells relapsed after 50 days and remained sensitive to 2.5 μm STI 571.
FIGURE 4.
Acquired resistance of clonal CML cells on imatinib treatment. A, time courses of relapse for four KCL-22 clones as analyzed in Fig. 1A. Notice that clones L1 and Ag 11 relapsed on imatinib with time courses similar to parental cells, but clones L7 and Ag3 relapsed with slower time courses. B, resistance of recurrent clonal cells to higher concentrations of imatinib. Recurrent cells derived from 2.5 μm imatinib treatment for four clones were labeled with clone names followed by 2.5R. All recurrent cells were maintained in the medium without imatinib until analysis. Notice the different levels of resistance of L1–2.5R (E255K mutation), L7–2.5R (Y253H mutation), and Ag11–2.5R (T315I mutation) to the higher concentrations of imatinib. Growth of Ag3–2.5R (no mutation) was inhibited by 5 μm imatinib. C, cells from clones L1 and Ag11 were plated on soft agar with 2.5 μm imatinib for 3 weeks, and 10 colonies each were picked for sequencing analysis of BCR-ABL kinase domain mutations. D, mutations from mixed clonal cells. Equal numbers of eight never-relapsed clones were mixed to form a nonrelapse pool (N-pool). Clone L1, L7, or Ag3 was then mixed 1:1 with a nonrelapse pool for resistance analysis in liquid culture, and recurrent cells were analyzed for BCR-ABL mutations.
Emergence of different mutations in clones was surprising, and we further examined how these clones might develop different mutations from parental cells. We first examined the homogeneity of clonal mutations by plating L1 and Ag11 cells on soft agar with imatinib, and we found that all resistant L1 colonies carried the E255K mutation only, and all resistant Ag11 colonies carried the T315I mutation only (Fig. 4C), suggesting highly clone-specific mutation patterns for resistance. We then examined whether the parental mutation type (T315I) could be restored in clonal cells when they were placed in a culture environment with various clonal cells. We mixed L1, L7, or Ag3 cells, respectively, with an equal number of cells from a pool consisting of eight never-relapse clones. After treatment with 2.5 or 5 μm imatinib, we found that relapsed cells maintained the identical mutation phenotypes for L1, L7, and Ag3, respectively (Fig. 4D). These results suggest that each clone adapts to a stable resistance mechanism.
Most BCR-ABL Mutant Cells Are Not Derived from a Fixed Mutant Cell Pool
The limitation of BCR-ABL mutations to certain clonal cells prompted us to further examine whether the ability to form BCR-ABL mutations is restricted to a rare subpopulation of cells such as stably formed mutant cells or “pre-mutant” cells that are not yet mutated but destined to form mutations. We performed cell pool analysis by evenly dividing (by volume) 1 × 105 KCL-22 cells (calculated 25 mutants under 5 μm imatinib) into 24 wells, with an average of calculated 1 mutant cell per well, and we let the cells grow for 10 days in the absence of imatinib. If mutant cells were fully derived from a fixed mutant subpopulation, they would be randomly seeded into wells according to Poisson distribution, and thus 15 wells were expected to receive at least one mutant or pre-mutant cell. After 10 days, cells in all wells exhibited similar growth and reached an average of 1.8 × 106 cells per well. All cells in each individual well were then analyzed for imatinib-resistant clones by clonogenic assay with 5 μm imatinib. Strikingly, we found that only 3 wells produced resistant colonies as compared with the expected 15 wells (p = 0.0008) (Table 2). Even if we assumed that all mutants or pre-mutants happened to be seeded into these three wells, an unlikely event, we would anticipate enrichment of mutant or pre-mutant cells in these three wells. However, we did not observe the increased mutation frequency (Table 2).
TABLE 2.
Cell pool analysis of BCR-ABL mutant cells with pre-expansion
Expected number of wells receiving mutant cells was calculated by assuming stable “pre-existing” mutant cells were randomly distributed according to Poisson distribution.
| Starting KCL-22 cell number | 1 × 105 |
| Number of wells that cells were seeded | 24 |
| Calculated mutant cell number/well | 1 |
| Expected number of wells receiving mutant cells (total number of wells) | 15 (24)a |
| Average cell number/well after 10-day expansion | 1.8 × 106 |
| Actual number of resistant colonies under 5 μm imatinib detected for cells from each well (number of wells) | 1 (1) |
| 2 (1) | |
| 51 (1) | |
| 0 (21) | |
| Actual number of wells having mutant cells (total number of wells) | 3 (24)a |
| p value for expected versus actual number of wells having mutant cells | 0.0008 |
a The assumed mutant frequency under 5 μm imatinib was 2.5 × 10−4 as described in the text or 25 cells in 1 × 105 KCL-22 cells. p value was calculated with two-tailed Fisher exact test.
To validate this finding, we performed another cell pool assay by evenly dividing 1 × 105 KCL-22 cells (calculated 48 mutants under 2.5 μm imatinib) into 40 wells in the presence of 2.5 μm imatinib, an average of 1.2 mutant cell per well. Similarly, we evenly divided 20 KCL-22M cells into 20 wells in the presence of 2.5 μm imatinib. By Poisson distribution, 28 wells were expected to receive mutant or pre-mutant cells for KCL-22 plating, and 12 wells were expected to receive cells for KCL-22M plating. Strikingly, we found that only one well of KCL-22 cells relapsed and re-grew; in contrast, five wells of KCL-22M cells re-grew (p = 0.006) (Table 3). These data suggest that most BCR-ABL mutant cells found in KCL-22 cells upon imatinib treatment are unlikely derived from a fixed mutant or pre-mutant subpopulation, although we can not rule out the presence of a small portion of pre-existing mutant cells. Together with cell cloning analysis, our results indicate the emergence of BCR-ABL mutations may be a dynamic process influenced by culture and environmental conditions.
TABLE 3.
Cell pool analysis of BCR-ABL mutant cells without pre-expansion
Expected number of wells receiving mutant cells was calculated by the same assumption of Table 2. The assumed mutant frequency under 2.5 μm imatinib was 4.8 × 10−4 as described in the text or 48 cells in 1 × 105 KCL-22 cells. The re-growing cell numbers were counted 25 days after seeding of KCL-22 and 22 days for KCL-22M cells.
| Starting cell no. | Cell type |
|
|---|---|---|
| KCL-22 (1 × 105) | KCL-22M (20) | |
| Number of wells that cells were seeded | 40 | 20 |
| Calculated mutant cell number per well | 1.2 | 1 |
| Expected number of wells receiving mutant cells | 28 | 12 |
| Number of re-growing cells in each well with 2.5 μm imatinib (number of wells) | 1.3 × 106 (1) | 1 × 104 (2) |
| 0 (39) | 3.2 × 104 (1) | |
| 5 × 104 (1) | ||
| 1.2 × 106 (1) | ||
| 0 (15) | ||
| Actual number of wells with mutant cells re-growing (expected number of wells receiving mutant cells) | 1 (28)a | 5 (12)a |
| p value for actual/expected number of wells with mutant cells for KCL-22 versus KCL-22 m | 0.006 | |
a p value was two-tailed Fisher exact test.
BCR-ABL Versus HPRT Mutations
We next sought to determine whether BCR-ABL mutations in KCL-22 cells were correlated with high mutation rates or mutator phenotype in these cells. We compared the BCR-ABL mutation rate with the spontaneous mutation rate of the HPRT gene measured by cell resistance to 6-thioguanine. When analyzed side-by-side in three relapse-prone clones (L1, L7, and Ag11) and two never-relapse clones (L3 and Ag6), HPRT showed a relatively constant mutation rate among all clones regardless of their ability to relapse, whereas the BCR-ABL mutation rate was highly clone-dependent (Fig. 5A). Clone Ag3 developing resistance without BCR-ABL mutation also showed a similar HPRT mutation rate (data not shown). Besides, we found that the T315I mutation frequency in parental KCL-22 cells was relatively stable before 20 passages but declined significantly toward the late passage (p36), which delayed the relapse of late-passage KCL-22 cells (Fig. 5B). In contrast, the HPRT mutation rate remained relatively constant among various cell passages. Sequencing analysis confirmed HPRT mutations from both parental and clonal cells with a broad mutation spectrum (Fig. 5C) as reported previously (19). These results suggest that BCR-ABL mutations upon imatinib treatment are not correlated with mutator phenotype of parental or clonal KCL-22 cells.
FIGURE 5.
Comparison of BCR-ABL mutations with spontaneous HPRT mutations. A, measurement of BCR-ABL and HPRT mutation rates. One million cells of each clone were plated on soft agar with 2.5 μm imatinib (STI) or 2.5 μg/ml 6-thioguanine (6-TG) for side-by-side analysis of BCR-ABL and HPRT mutation frequencies, respectively. B, left, BCR-ABL and HPRT mutation rates of KCL-22 cells at passages 5, 16, and 36 measured by soft agar clonogenic assay. Imatinib, 5 μm; 6-thioguanine, 2.5 μg/ml. Right, time courses for relapse of KCL-22 cells on 2.5 μm imatinib at passages 8 and 35. Relapsed cells harbored T315I mutation in both cases. C, HPRT mutation spectrum. Ag11, L1, or KCL-22 cells were seeded on soft agar with 6-thioguanine, and 20 colonies each were picked for analysis of HPRT codon mutations. For Ag11, two colonies had sequencing failure and were excluded.
BCR-ABL Mutations Are Dependent on BCR-ABL Gene Expression
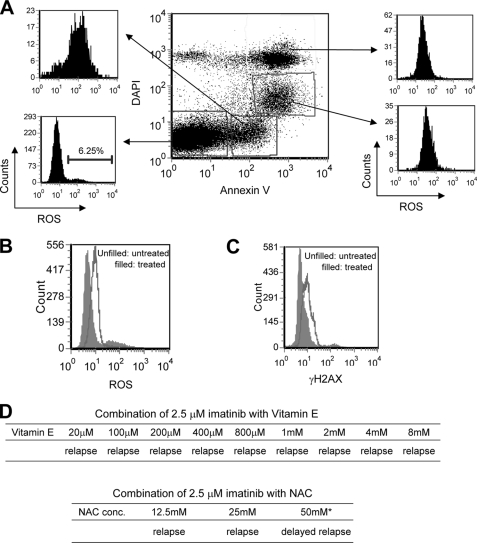
Koptyra et al. (16) demonstrate that BCR-ABL induces self-mutagenesis through increasing production of ROS, and imatinib or anti-oxidants reduce ROS production and BCR-ABL mutations. We examined the effects of imatinib treatment on ROS and DNA damage in our system. Following imatinib treatment, the bulk ROS level was reduced in the nonapoptotic fraction of KCL-22 cells similar to the findings of Koptyra et al. (16), although ROS increased in the apoptotic fractions (Fig. 6, A and B). Accordingly, the bulk DNA damage analyzed by γH2AX staining decreased in the nonapoptotic fraction of cells (Fig. 6C). Intriguingly, whereas 200 μm anti-oxidant vitamin E or N-acetylcysteine is able to reduce ROS and BCR-ABL mutations in the study of Koptyra et al. (16), in our studies even 100× higher concentration of NAC (25 mm) or 40× higher vitamin E (8 mm) failed to prevent imatinib-induced BCR-ABL mutations and relapse (Fig. 6D). These data suggest that BCR-ABL mutations in KCL-22 cells cannot be simply explained by ROS change.
FIGURE 6.
ROS and DNA damage in KCL-22 cells upon imatinib treatment. A, analysis of ROS in different apoptotic fractions of KCL-22 cells treated with 2.5 μm imatinib for 4 days. ROS increased in all apoptotic fractions of the cells with early apoptotic cells (top left square) having the highest level. Nonapoptotic fraction of the cells (bottom left square) had the lowest level of ROS. DAPI, 4′,6-diamidino-2-phenylindole. B, bulk ROS level decreased in nonapoptotic fraction of KCL-22 cells that were treated with 2.5 μm imatinib for 2 days. Similar results were seen for 1 day of drug treatment (not shown). C, bulk γH2AX level decreased in KCL-22 cells treated with 2.5 μm imatinib for 2 days. D, treatment with anti-oxidants. No effects of NAC or vitamin E were seen to prevent or delay KCL-22 cells from relapse on 2.5 μm imatinib except for 50 mm NAC that itself has significant cytotoxicity (data not shown).
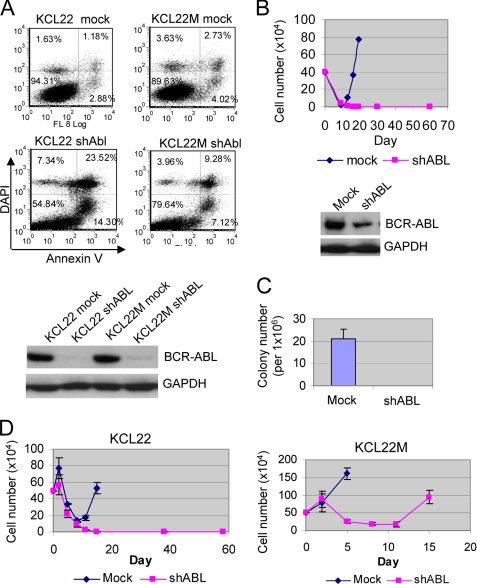
We then examined if BCR-ABL gene expression is required for its mutations. KCL-22 cells express e13a2 BCR-ABL fusion transcript, and the junction region sequence does not meet optimal shRNA design criteria (33). As reported previously (34), we found that shRNA targeting this region produced poor gene knockdown (data not shown). We therefore used an shRNA targeting ABL sequence described previously (35) to knock down BCR-ABL. We found that BCR-ABL knockdown rapidly inhibited proliferation and induced apoptosis of both KCL-22 and KCL-22M cells (Fig. 7A and data not shown), suggesting continued dependence of BCR-ABL for proliferation and survival of BCR-ABL mutant CML cells. Notably, a lower apoptosis rate was observed in KCL-22M cells (Fig. 7A), indicating that cells acquire additional survival advantage during development of resistance. No proliferation inhibition and apoptosis were observed in a BCR-ABL-negative leukemia cell line HL-60 when ABL was knocked down (data not shown), suggesting that effects in CML cells are BCR-ABL-specific.
FIGURE 7.
Induction of BCR-ABL mutations is dependent on BCR-ABL expression. A, top, analysis of apoptosis of KCL-22 and KCL-22M cells 6 days after BCR-ABL knockdown. Scrambled shRNA was used for mock knockdown. Less apoptotic cells were noticed in KCL-22M. Bottom, BCR-ABL protein levels after knockdown. DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, top, effects of BCR-ABL knockdown on acquired resistance on imatinib. After the knockdown, cells regrew, and they were selected for 5 days in puromycin and expanded in drug-free medium. These re-grown BCR-ABL knockdown KCL-22 cells and mock knockdown cells were treated with 5 μm imatinib, and cells were followed as described in Fig. 1. Bottom, BCR-ABL levels in the re-grown knockdown cells. C, effects of BCR-ABL knockdown on formation of imatinib-resistant colonies on soft agar. The re-grown BCR-ABL knockdown (shABL) or mock knockdown KCL-22 cells were plated on soft agar with 5 μm imatinib. The plating efficiency for mock and ABL knockdown was the same (data not shown). D, effects of combining BCR-ABL knockdown with imatinib treatment on acquired resistance. Cells were transduced overnight with lentiviral vectors, and imatinib was added right after the removal of viruses.
Both KCL-22 and KCL-22M cells with BCR-ABL knockdown re-grew after 2 weeks. We found that BCR-ABL expression was partially restored in re-grown knockdown KCL-22 cells but remained at a lower level than in mock knockdown cells (Fig. 7B). When treated with imatinib, re-grown BCR-ABL knockdown KCL-22 cells failed to relapse in liquid culture or form imatinib-resistant colonies on soft agar (Fig. 7, B and C). Similarly, BCR-ABL knockdown immediately followed by imatinib treatment also blocked relapse in KCL-22 cells but failed to prevent KCL-22M cells from regrowing (Fig. 7D). Although the shRNA knocks down both ABL and BCR-ABL, loss of DNA repair protein ABL is expected to further increase genetic mutation, which would contradict the above findings. Therefore, our results suggest that formation of BCR-ABL mutations is dependent on BCR-ABL gene expression and that combination of BCR-ABL knockdown and imatinib can block acquired resistance of KCL-22 cells on the drug.
Expression of Functional BCR-ABL from Endogenous BCR-ABL Locus for Acquisition of BCR-ABL Mutation
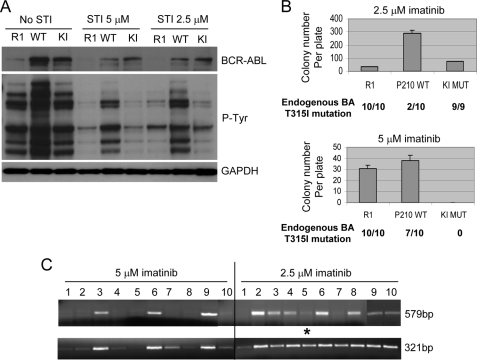
To determine roles of BCR-ABL kinase activity for its mutation, we overexpressed in KCL-22 cells wild type p210 BCR-ABL cDNA, K1176R p210 BCR-ABL mutant cDNA that abolishes its kinase activity (22, 36) or controls empty MIG retroviral vectors (23). The transduced cells were isolated by FACS for GFP expression. Overexpression of wild type BCR-ABL increased cellular phosphorylation as expected, but overexpression of the mutant BCR-ABL did not reduce cellular phosphorylation (Fig. 8A), suggesting K1176R BCR-ABL is not a dominant negative mutant in these cells. Neither wild type nor mutant BCR-ABL expression changed cell growth or soft agar cloning efficiency (supplemental Fig. 4).
FIGURE 8.
Effects of BCR-ABL overexpression on mutations. A, analysis of BCR-ABL expression and total cellular phosphorylation. KCL-22 cells were transduced with empty vector (R1), wild type p210 BCR-ABL (WT), or kinase-inactive p210 mutant (KI). Lysates were collected without or with 2.5 and 5 μm imatinib treatment. B, clonogenic assay of BCR-ABL-overexpressing cells for imatinib resistance with 1,000,000 cells plated each well in triplicate. Imatinib-resistant colonies were plucked and expanded for genomic sequencing of the endogenous BCR-ABL (BA) kinase domain. The number of colonies found with T315I mutation over the total number of colonies sequenced are listed below each category of graph bars. C, analysis of the transduced BCR-ABL cDNA in imatinib-resistant colonies overexpressing wild type BCR-ABL by PCR amplification of 579- and 321-bp fragments of BCR-ABL cDNA from genomic templates using exon primers. Both 579- and 321-bp fragments were sequenced. Only one colony, clone 5 from 2.5 μm imatinib treatment group marked by an asterisk, had BCR-ABL mutation (T315I). Colonies with T315I mutation of the endogenous BCR-ABL were as follows: clones 1, 2, 4, 5, 7, 8, and 10 from 5 μm imatinib group, and clones 1 and 7 from 2.5 μm imatinib group.
When treated with 2.5 μm imatinib, wild type BCR-ABL-overexpressing cells developed significantly higher numbers of imatinib-resistant colonies than empty vector or mutant BCR-ABL-transduced cells, and the majority of resistant colonies (8 out of 10) from wild type BCR-ABL-overexpressing cells did not harbor T315I mutation of the endogenous BCR-ABL (Fig. 8B), indicating that the transduced BCR-ABL cDNA may function as BCR-ABL gene amplification for resistance. In contrast, all resistant colonies in the empty vector or mutant BCR-ABL-transduced cells harbored T315I mutation of the endogenous BCR-ABL (Fig. 8B).
When treated with 5 μm imatinib, we found that resistance provided by wild type BCR-ABL cDNA was largely diminished, and the majority of resistant colonies (7 out of 10) from wild type BCR-ABL-overexpressing cells harbored T315I mutation of the endogenous BCR-ABL (Fig. 8B). Surprisingly, under this condition, mutant BCR-ABL completely blocked imatinib-resistant colony formation (Fig. 8B). Similar results were obtained when we carried out a second round of transduction with these vectors on the first round of GFP-enriched cells, aiming to further increase cDNA expression (data not shown). These results suggest a possible role of BCR-ABL kinase activity for its mutation when cells are under treatment with a high concentration of imatinib. However, we found that overexpression of mutant BCR-ABL did not enhance reduction of total cellular or a BCR-ABL substrate CRKL protein phosphorylation upon imatinib treatment (Fig. 8A and not shown). Therefore, functionally intact BCR-ABL may be important for generating mutations, but kinase activity may not be absolutely required.
BCR-ABL cDNA has been previously expressed in non-CML cells to generate random BCR-ABL mutations (5–7, 10–15) and is presumed to have similar mutagenesis capability. We examined mutations on the transduced wild type BCR-ABL cDNA in KCL-22 cells. The transduced gene was distinguished from the endogenous BCR-ABL by using exon primers that span multiple introns for genomic DNA PCR and sequencing. All 20 imatinib-resistant clones from 2.5 and 5 μm drug treatment had visible GFP expression (data not shown), suggesting that they all carried functional transduced BCR-ABL cDNA. We found that clones that did not harbor T315I mutation of the endogenous BCR-ABL tended to have a higher yield of PCR products, suggesting a higher copy number of the transduced BCR-ABL in these clones for resistance (Fig. 8C). Strikingly, we found that only one clone developed a BCR-ABL kinase domain mutation (T315I) on the transduced BCR-ABL among all 20 clones sequenced (Fig. 8C). Therefore, although the transduced BCR-ABL cDNA was capable of mutagenesis in KCL-22 cells, the mutation efficiency was significantly lower than that of the endogenous BCR-ABL (9 of 20 clones, p = 0.008). This difference is not a result of experimental artifacts. First, wild type BCR-ABL cDNA was transduced into KCL-22 cells using retroviral virions and would readily integrate into the genome of replicating KCL-22 cells. Nonintegrated viral DNA would be lost during the lengthy procedure for expanding transduced cells before and after FACS sorting, colony formation of imatinib-resistant cells on soft agar, followed by expanding clonal cells in liquid culture for assay. The stable integration of the cDNA was also evidenced by nearly homogeneous GFP expression of the expanded clonal cells in liquid culture (data not shown). Second, mutations were analyzed within the clonal population of wild type BCR-ABL-transduced cells, and both endogenous and transduced alleles for each clone were analyzed and compared. This is an internally controlled comparison, and therefore mutation efficiency is not affected by external factors such as retroviral transduction efficiency. Third, although KCL-22 cells have two translocation chromosomes, the lower mutation rate of transduced BCR-ABL cDNA is unlikely a result of lower copy number of the transduced BCR-ABL cDNA. The cDNA copy number may vary from clone to clone, but at least one copy of the cDNA is expected for each of 10 clones (clones 1, 2, 4, 5, 7, 8, and 10 under 5 μm and clones 1, 5, and 7 under 2.5 μm) that did not efficiently amplify the longer cDNA template (579 bp, Fig. 8C), whereas the other 10 clones would have higher copy numbers. However, the single clone that developed BCR-ABL cDNA mutation (clone 5 under 2.5 μm) carried low copy number of cDNA, suggesting that increasing cDNA copy number did not increase its mutation frequency. Together, our results suggest that the native BCR-ABL translocation locus in KCL-22 cells has inherently high mutagenesis potential, and expression of functional BCR-ABL from the locus promotes acquisition of BCR-ABL mutations.
DISCUSSION
In this study, we have developed a novel model for CML acquired resistance through BCR-ABL mutations using a naive blast crisis CML cell line KCL-22. In contrast to previous cell line models that are involved with gene amplification, altered BCR-ABL expression, and/or additional chromosomal rearrangements (9, 26, 37), genetic mutations of BCR-ABL primarily account for resistance in our model. Development of BCR-ABL mutations in this model is highly reproducible and occurs in 2 weeks with a single dose of imatinib treatment. T315I mutation has been shown in CML cell line KBM5 after several months of treatment with gradually increasing concentrations of imatinib (38), but the mutation induction time in KCL-22 cells is much shorter. Although a relatively short period of time (3–6 weeks) is also reported for developing BCR-ABL mutations in primary CML cells cultured in vitro (16, 17), our model is simpler and more tractable, using a commercially available cell line and routine culture conditions. Therefore, this model will be very useful for studying mechanisms of acquired resistance and developing strategies to prevent relapse.
Our finding that BCR-ABL mutations are dependent on BCR-ABL gene expression is in line with the previous finding that BCR-ABL can promote self-mutagenesis (16), which suggests that genome instability caused by BCR-ABL transformation is perhaps the driving force for its mutations. Consistent with this notion, we observe that clonal cells can derive mutations without pre-existing mutations from the original patient and that these clonal cells can develop distinct BCR-ABL mutations as well as resistance without mutations. Development of resistant BCR-ABL mutations may resemble adaptive mutations promoting survival and growth in bacteria under stressful conditions, which involves multiple DNA repair pathways (39, 40). BCR-ABL transformation alters regulation of multiple DNA repair pathways causing genome instability (41). It would be interesting to determine how altered DNA repair may influence mutations with this model in the future.
Although BCR-ABL cDNA has been widely used to study its mutagenesis, how the mutagenesis potential of randomly integrated BCR-ABL cDNA may compare with the native BCR-ABL locus has not been shown before. Our study now reveals that the BCR-ABL translocation locus itself may play a role in promoting mutations, as mutagenesis on the locus is far more efficient than on randomly integrated BCR-ABL cDNA in the same cells. Consistent with such locus-dependent influence, we show that HPRT mutation rate is relatively constant although the BCR-ABL mutation rate is more dynamic. Interestingly, the cell cloning process affects the mutagenesis process, resulting in distinct clonal mutations, which suggests a dynamic process for BCR-ABL mutagenesis on its native locus. We speculate that BCR-ABL mutations could be influenced by the epigenome structure of the translocation locus, and the environmental change or cloning process can result in subtle alteration of the local epigenome producing altered mutation hot spots. It would be interesting to test these possibilities in the future.
The specific inhibition of BCR-ABL kinase activity in KCL-22 cells turns out difficult, and the outcome is complex. Cellular phosphorylation in KCL-22 cells is not affected by overexpression of K1176R kinase-inactive BCR-ABL or by efficient delivery of BCR-ABL antibody (data not shown). Mutation phenotype in mutant BCR-ABL-overexpressing cells does not change with 2.5 μm imatinib treatment as anticipated, but surprisingly, mutant BCR-ABL blocks mutations at 5 μm imatinib. The precise mechanism for this difference is not yet clear. Because cells harboring mature tetramer BCR-ABL protein (28, 42) purely from mutant monomers may not be able to grow in culture, most cells for analysis should carry hybrid BCR-ABL protein with wild type and mutant monomers. The e13a2 BCR-ABL, expressed in KCL-22 cells, is known to have high tyrosine kinase activity (43). It is possible that the hybrid BCR-ABL retains normal phosphorylation ability on most substrates but may affect certain substrates regulating BCR-ABL mutations, and its effect would not be shown unless strong selection is applied. Alternatively, the mutant BCR-ABL may affect mutations independent of its kinase activity. K1176R BCR-ABL retains certain functions, such as the intact cellular localization and adhesion regulation (36), and may continue recruiting proteins necessary for mutagenesis but in a less efficient way.
Although ROS production is previously found to promote BCR-ABL mutations (16), inhibition of ROS by high concentrations of anti-oxidants was unable to prevent BCR-ABL mutations and relapse in our study. However, these two studies use quite different cell culture systems. KCL-22 cells have a basal level of ROS, compared with high levels of ROS in blast crisis CML cells previously studied, although low levels of ROS are also noticed in a subpopulation of CML cells (16). In addition, primary CML cell culture uses growth factor-supplemented medium (16, 17), whereas KCL-22 cells are growth factor-independent. Given that ABL kinase domain mutations can be detected even in normal human progenitor cells cultured in the growth factor medium (17), it is unclear if growth factor culture may affect ROS production and mutations.
How resistant mutations are generated upon cancer therapy remains a fundamental question for cancer biology, and overcoming acquired resistance is a daunting therapeutic challenge. The model developed in this study recapitulates key features of clinical resistance, namely rapid relapse through BCR-ABL mutations after direct exposure of CML cells to therapeutically effective concentrations of imatinib. This model will serve as an excellent tool for further uncovering resistance mechanisms that involve BCR-ABL gene expression and the native translocation locus. It will be also helpful for the design of strategies to prevent occurrence of additional mutations following drug treatment, and to help better manage or eliminate this devastating disease.
Supplementary Material
This work was supported by start-up funds from the City of Hope, grants from the United States Department of Defense, the STOPCANCER Foundation (to W. Y. C.), and a translational research grant from the V-Foundation (to W. Y. C. and R. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” Figs. 1–4, and additional references.
- CML
- chronic myelogenous leukemia
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase
- shRNA
- short hairpin RNA
- ROS
- reactive oxygen species
- GFP
- green fluorescent protein
- FACS
- fluorescent-activated cell sorting
- FISH
- fluorescent in situ hybridization
- SH
- Src homology
- XTT
- sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzene sulfonic acid hydrate.
REFERENCES
- 1.Deininger M. W., Druker B. J. (2003) Pharmacol. Rev. 55, 401–423 [DOI] [PubMed] [Google Scholar]
- 2.Gorre M. E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P. N., Sawyers C. L. (2001) Science 293, 876–880 [DOI] [PubMed] [Google Scholar]
- 3.Shah N. P., Nicoll J. M., Nagar B., Gorre M. E., Paquette R. L., Kuriyan J., Sawyers C. L. (2002) Cancer Cell 2, 117–125 [DOI] [PubMed] [Google Scholar]
- 4.Branford S., Rudzki Z., Walsh S., Grigg A., Arthur C., Taylor K., Herrmann R., Lynch K. P., Hughes T. P. (2002) Blood 99, 3472–3475 [DOI] [PubMed] [Google Scholar]
- 5.Weisberg E., Manley P. W., Breitenstein W., Brüggen J., Cowan-Jacob S. W., Ray A., Huntly B., Fabbro D., Fendrich G., Hall-Meyers E., Kung A. L., Mestan J., Daley G. Q., Callahan L., Catley L., Cavazza C., Azam M., Mohammed A., Neuberg D., Wright R. D., Gilliland D. G., Griffin J. D. (2005) Cancer Cell 7, 129–141 [DOI] [PubMed] [Google Scholar]
- 6.von Bubnoff N., Manley P. W., Mestan J., Sanger J., Peschel C., Duyster J. (2006) Blood 108, 1328–1333 [DOI] [PubMed] [Google Scholar]
- 7.Shah N. P., Tran C., Lee F. Y., Chen P., Norris D., Sawyers C. L. (2004) Science 305, 399–401 [DOI] [PubMed] [Google Scholar]
- 8.Deininger M. W., Goldman J. M., Lydon N., Melo J. V. (1997) Blood 90, 3691–3698 [PubMed] [Google Scholar]
- 9.Mahon F. X., Deininger M. W., Schultheis B., Chabrol J., Reiffers J., Goldman J. M., Melo J. V. (2000) Blood 96, 1070–1079 [PubMed] [Google Scholar]
- 10.von Bubnoff N., Veach D. R., van der Kuip H., Aulitzky W. E., Sänger J., Seipel P., Bornmann W. G., Peschel C., Clarkson B., Duyster J. (2005) Blood 105, 1652–1659 [DOI] [PubMed] [Google Scholar]
- 11.La Rosée P., Corbin A. S., Stoffregen E. P., Deininger M. W., Druker B. J. (2002) Cancer Res. 62, 7149–7153 [PubMed] [Google Scholar]
- 12.Bradeen H. A., Eide C. A., O'Hare T., Johnson K. J., Willis S. G., Lee F. Y., Druker B. J., Deininger M. W. (2006) Blood 108, 2332–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray A., Cowan-Jacob S. W., Manley P. W., Mestan J., Griffin J. D. (2007) Blood 109, 5011–5015 [DOI] [PubMed] [Google Scholar]
- 14.Azam M., Latek R. R., Daley G. Q. (2003) Cell 112, 831–843 [DOI] [PubMed] [Google Scholar]
- 15.Burgess M. R., Skaggs B. J., Shah N. P., Lee F. Y., Sawyers C. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3395–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koptyra M., Falinski R., Nowicki M. O., Stoklosa T., Majsterek I., Nieborowska-Skorska M., Blasiak J., Skorski T. (2006) Blood 108, 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X., Saw K. M., Eaves A., Eaves C. (2007) J. Natl. Cancer Inst. 99, 680–693 [DOI] [PubMed] [Google Scholar]
- 18.Huntly B. J., Bench A., Green A. R. (2003) Blood 102, 1160–1168 [DOI] [PubMed] [Google Scholar]
- 19.Osterholm A. M., Fält S., Lambert B., Hou S. M. (1995) Carcinogenesis 16, 1909–1912 [DOI] [PubMed] [Google Scholar]
- 20.Ventura A., Meissner A., Dillon C. P., McManus M., Sharp P. A., Van Parijs L., Jaenisch R., Jacks T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10380–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowolik C. M., Yam P., Yu Y., Yee J. K. (2003) Mol. Ther. 8, 324–331 [DOI] [PubMed] [Google Scholar]
- 22.Ramaraj P., Singh H., Niu N., Chu S., Holtz M., Yee J. K., Bhatia R. (2004) Cancer Res. 64, 5322–5331 [DOI] [PubMed] [Google Scholar]
- 23.Pear W. S., Miller J. P., Xu L., Pui J. C., Soffer B., Quackenbush R. C., Pendergast A. M., Bronson R., Aster J. C., Scott M. L., Baltimore D. (1998) Blood 92, 3780–3792 [PubMed] [Google Scholar]
- 24.Peng B., Lloyd P., Schran H. (2005) Clin. Pharmacokinet. 44, 879–894 [DOI] [PubMed] [Google Scholar]
- 25.Kubonishi I., Miyoshi I. (1983) Int. J. Cell Cloning 1, 105–117 [DOI] [PubMed] [Google Scholar]
- 26.Rosenhahn J., Weise A., Michel S., Hennig K., Hartmann I., Schiefner J., Schubert K., Liehr T., von Eggeling F., Loncarevic I. F. (2007) Int. J. Oncol. 31, 121–128 [PubMed] [Google Scholar]
- 27.Zhao X., Ghaffari S., Lodish H., Malashkevich V. N., Kim P. S. (2002) Nat. Struct. Biol. 9, 117–120 [DOI] [PubMed] [Google Scholar]
- 28.Smith K. M., Yacobi R., Van Etten R. A. (2003) Mol. Cell 12, 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willis S. G., Lange T., Demehri S., Otto S., Crossman L., Niederwieser D., Stoffregen E. P., McWeeney S., Kovacs I., Park B., Druker B. J., Deininger M. W. (2005) Blood 106, 2128–2137 [DOI] [PubMed] [Google Scholar]
- 30.Shi J., Liu Q., Sommer S. S. (2007) Hum. Mutat. 28, 131–136 [DOI] [PubMed] [Google Scholar]
- 31.Roche-Lestienne C., Soenen-Cornu V., Grardel-Duflos N., Laï J. L., Philippe N., Facon T., Fenaux P., Preudhomme C. (2002) Blood 100, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 32.Soverini S., Colarossi S., Gnani A., Rosti G., Castagnetti F., Poerio A., Iacobucci I., Amabile M., Abruzzese E., Orlandi E., Radaelli F., Ciccone F., Tiribelli M., di Lorenzo R., Caracciolo C., Izzo B., Pane F., Saglio G., Baccarani M., Martinelli G. (2006) Clin. Cancer Res. 12, 7374–7379 [DOI] [PubMed] [Google Scholar]
- 33.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. (2004) Nat. Biotechnol. 22, 326–330 [DOI] [PubMed] [Google Scholar]
- 34.Li M. J., McMahon R., Snyder D. S., Yee J. K., Rossi J. J. (2003) Oligonucleotides 13, 401–409 [DOI] [PubMed] [Google Scholar]
- 35.Zhelev Z., Bakalova R., Ohba H., Ewis A., Ishikawa M., Shinohara Y., Baba Y. (2004) FEBS Lett. 570, 195–204 [DOI] [PubMed] [Google Scholar]
- 36.Wertheim J. A., Forsythe K., Druker B. J., Hammer D., Boettiger D., Pear W. S. (2002) Blood 99, 4122–4130 [DOI] [PubMed] [Google Scholar]
- 37.Tipping A. J., Deininger M. W., Goldman J. M., Melo J. V. (2003) Exp. Hematol. 31, 1073–1080 [PubMed] [Google Scholar]
- 38.Ricci C., Scappini B., Divoky V., Gatto S., Onida F., Verstovsek S., Kantarjian H. M., Beran M. (2002) Cancer Res. 62, 5995–5998 [PubMed] [Google Scholar]
- 39.Rosenberg S. M. (2001) Nat. Rev. Genet. 2, 504–515 [DOI] [PubMed] [Google Scholar]
- 40.Karpinets T., Greenwood Dj., Pogribny I., Samatova N. (2006) Curr. Genomics 7, 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melo J. V., Barnes D. J. (2007) Nat. Rev. Cancer 7, 441–453 [DOI] [PubMed] [Google Scholar]
- 42.McWhirter J. R., Galasso D. L., Wang J. Y. (1993) Mol. Cell. Biol. 13, 7587–7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucas C. M., Harris R. J., Giannoudis A., Davies A., Knight K., Watmough S. J., Wang L., Clark R. E. (2009) Haematologica 94, 1362–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.