Abstract
During embryonic development, protein kinase A (PKA) plays a key role in cell fate specification by antagonizing the Hedgehog (Hh) signaling pathway. However, the mechanism by which PKA activity is regulated remains unknown. Here we show that the Misty somites (Mys) protein regulates the level of PKA activity during embryonic development in zebrafish. We isolate PKA regulatory type Iα subunit (Prkar1a) as a protein interacting with Mys by pulldown assay in HEK293 cells followed by mass spectrometry analysis. We show an interaction between endogenous Mys and Prkar1a in the zebrafish embryo. Mys binds to Prkar1a in its C terminus region, termed PRB domain, and activates PKA in vitro. Conversely, knockdown of Mys in zebrafish embryos results in reduction in PKA activity. We also show that knockdown of Mys induces ectopic activation of Hh target genes in the eyes, neural tube, and somites downstream of Smoothened, a protein essential for transduction of Hh signaling activity. The altered patterning of gene expression is rescued by activation of PKA. Together, our results reveal a molecular mechanism of regulation of PKA activity that is dependent on a protein-protein interaction and demonstrate that PKA activity regulated by Mys is indispensable for negative regulation of the Hh signaling pathway in Hh-responsive cells.
Keywords: Development Differentiation, Genetics, Protein/Protein-protein interactions, Signal Transduction, Cell Differentiation, Protein Kinase A (PKA), Hedgehog Signaling, Vertebrate Development, Zebrafish
Introduction
Protein kinase A (PKA)2 was first isolated from rabbit skeletal muscle as a protein kinase that catalyzes a cAMP-dependent phosphorylation (1). Since then, PKA has been found in all eukaryotes and has been demonstrated to regulate processes as diverse as growth, metabolism, gene expression, development, and memory. PKA forms an inactive holoenzyme containing a regulatory subunit dimer and two catalytic subunits. Classically, PKA is activated by cAMP binding to the regulatory subunit, which alters affinity of the regulatory subunit for the catalytic subunit and promotes dissociation into a dimer of regulatory subunits and two active monomeric catalytic subunits. The active catalytic subunits then phosphorylate protein substrates containing consensus phosphorylation motifs (2).
In invertebrate and vertebrate development, PKA antagonizes Hedgehog (Hh) signaling, which plays fundamental roles during pattern formation (3–11). For example, ectopic expression of Hh family members in zebrafish embryos leads to the expansion of proximal cell fate in the eyes, ventral cell fate in the neural tube, and adaxial cell fate in somites. Inhibition of PKA by a dominant negative form of PKA (dnPKA) mimics these effects, i.e. ectopic activation of Hh target genes in the eyes, neural tube, and somites, which leads to Hh-dependent cell differentiation. In contrast, activation of PKA by a constitutively active form of PKA blocks the effects of endogenous and ectopic Hh signaling (12–18). Results of these studies imply that the basal level of PKA activity during embryonic development is precisely regulated to properly specify cell fates. However, the molecular mechanism underlying the regulation of PKA activity in embryos remains unknown.
Zebrafish is an excellent model animal to identify developmental genes by forward genetics approaches. We previously identified misty somites (mys) as a gene essential for the somite boundary maintenance by transposon-mediated insertional mutagenesis in zebrafish (19, 20). Knockdown of mys resulted in defects not only in somites but also in the eyes, suggesting its roles in eye formation and differentiation as well as in somites (19). To elucidate the molecular basis of Mys protein function, we have screened for proteins that interact with Mys. In this study, we isolated PKA regulatory type Iα subunit (Prkar1a) as a protein interacting with Mys and characterized their interaction. Mys bound to Prkar1a, but not to the catalytic subunit (Prkac), dissociated Prkac from Prkar1a and activated PKA in vitro, indicating competition of Mys with Prkac for binding to Prkar1a. In contrast, knockdown of Mys in zebrafish embryos resulted in an increase in the amount of inactive PKA and a reduction in PKA activity, which induced ectopic activation of Hh target genes in the eyes, neural tube, and somites of embryos. Thus, our results provide a novel molecular mechanism of regulation of PKA activity that is dependent on the interaction between Mys and Prkar1a proteins, by which Hh signaling is negatively regulated during zebrafish development.
EXPERIMENTAL PROCEDURES
Fish and Extraction
Fish were maintained under standard laboratory conditions. TL was used as the wild-type fish. The syut4 line was obtained from the Zebrafish International Resource Center (Eugene, OR). Extracts from the embryos and distinct organs in adults were prepared as follows. One hundred embryos at 24-h post-fertilization (hpf) were dechorionated and transferred to phosphate-buffered saline and were dissociated into single cells by pipetting. The dissociated cells were washed three times with phosphate-buffered saline by centrifugation at 700 × g for 7 min. After removing excess phosphate-buffered saline, 100 μl of lysis buffer (LB: 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 50 mm sodium fluoride, 1 mm dithiothreitol, 100 μm (p-amidinophenyl)methanesulfonyl fluoride, 3 μg/ml of leupeptin) was added. The cells were sonicated for 10 s and centrifuged at 15,000 × g for 10 min at 4 °C. The supernatant was collected and used for immunoblotting, immunoprecipitation, and PKA activity assay. The brain, heart, and testis were homogenized with a pestle (Pellet Pestle, Kontes) in LB and centrifuged at 15,000 × g for 10 min at 4 °C. The ovary was centrifuged at 150,000 × g for 30 min at 4 °C in extraction buffer (100 mm β-glycerophosphate, 20 mm HEPES, pH 7.5, 15 mm MgCl2, 5 mm EGTA, 1 mm dithiothreitol, 100 μm (p-amidinophenyl)methanesulfonyl fluoride, 3 μg/ml of leupeptin). The supernatant was collected and used for immunoblotting.
Production of Mys and Prkar1a Antibodies
A part of the open reading frame (ORF) of the Mys protein (from 201 to 435 aa) was cloned into pGEX-KG to produce a fusion protein containing GST at the N terminus of Mys. GST-Mys-(201–435) protein was expressed in Escherichia coli, gel-purified, and injected into a rabbit as described previously (21). The full ORF of zebrafish Prkar1a protein was cloned into pGEX-KG. GST-Prkar1a protein was expressed in E. coli, gel-purified, and injected into mice to produce polyclonal antibodies. The obtained antiserums were affinity purified with the antigens.
Immunoblotting
Anti-GST-Mys polyclonal antibody, anti-Prkar1a polyclonal antibody, anti-Prkac monoclonal antibody (BD Biosciences), and anti-γ-tubulin monoclonal antibody (GTU-88, Sigma) were used to detect Mys, Prkar1a, Prkac, and γ-tubulin, respectively. Anti-FLAG monoclonal antibody (M2, Sigma), anti-GFP monoclonal antibody (Roche), and anti-Myc monoclonal antibody (MBL) were used to detect FLAG-tagged Mys, GFP-tagged Mys, and Myc-tagged Prkar1a, respectively. The samples were separated with SDS-PAGE gel, blotted onto an Immobilon membrane (Millipore), and probed with primary antibodies. The antigen-antibody complex was visualized by alkaline phosphatase-conjugated secondary antibodies.
Mass Spectometry Analysis
A pull-down assay with FLAG-tagged human Mys homolog in HEK293 cells followed by mass spectometry analysis was performed as described previously (22).
In Situ Hybridization and Immunostaining
In situ hybridization experiments were performed as described previously (23). For immunohistochemistry, the embryos were fixed at 22 hpf with 4% paraformaldehyde in phosphate-buffered saline, dehydrated, embedded in paraffin, and cut into 12-μm thick sections. Hoechst staining and immunostaining of the samples were performed as described previously (24). To analyze the subcellular localization of Mys, cells derived from embryos at 4 hpf were immunostained as described previously (19). Anti-GM130 monoclonal antibody (BD Biosciences) was used to detect the Golgi apparatus. The samples were observed under an LSM5LIVE confocal microscope (Zeiss).
Immunoprecipitation
Oligonucleotides encoding FLAG or Myc epitope tag or the ORF of GFP protein were cloned into pCS2+ (pCS2+FT, pCS2+MT, or pCS2+GFP). The full ORF or a fragment of Mys was cloned into pCS2+FT or pCS2+GFP. The full ORF or a fragment of Prkar1a was cloned into pCS2+MT. mys-flag and myc-prkar1a mRNAs were synthesized with an mMESSAGE mMACHINE SP6 Kit (Ambion Inc.). Six μg each of mys-flag and myc-prkar1a mRNAs were translated in rabbit reticulocyte lysate (Promega). Extracts from the embryos, rabbit reticulocyte lysates, or mixtures of PKA holoenzyme and Mys recombinant protein were 2-fold diluted with LB and incubated with affinity purified anti-Prkar1a polyclonal antibody, affinity purified anti-XMAP215 polyclonal antibody (25), anti-FLAG monoclonal antibody, anti-GFP monoclonal antibody, or anti-Prkac monoclonal antibody and 20 μl of protein G-Sepharose beads (GE Healthcare) at 4 °C for 1 h. The immunoprecipitates were washed 5 times with LB and used for immunoblotting.
PKA Activity Assay
The full ORFs of Mys and GST were cloned into pET21 ((Full)Mys-GST). Parts of the ORF of Mys (from 1 to 289 aa and 390 to 435 aa) were cloned into pET21 with the full ORF of GST ((ΔPRB)Mys-GST). The recombinant proteins were expressed in E. coli and purified with glutathione-Sepharose beads (GSH) (GE Healthcare). Five μl of 5 μg/ml of PKA holoenzyme (Sigma; B-Bridge International Inc.) was mixed with 10 μl of 0.2 μm (Full)Mys-GST, (ΔPRB)Mys-GST, GST, or cAMP. PKA activity was measured by using PepTag assays (Promega) according to the manufacturer's instructions. Briefly, 15 μl of the mixtures of PKA holoenzyme and recombinant proteins or 15 μl of embryonic extracts were incubated for 30 min at 30 °C with the PKA-specific peptide substrate, Leu-Arg-Arg-Ala-Ser-Leu-Gly (Kemptide). Samples containing 2 μg of PKA inhibitor (Sigma) were used as a control and the residual activity was considered as background. PKA activities in the embryos were normalized to equivalent amounts of Prkar1.
MO and mRNA Injection
One nl of a solution containing 8 mg/ml of mys antisense morpholino oligonucleotide (MO) targeted to the translational initiation site of the mys transcript or control 5mm MO containing 5-bp mismatches around the ATG codon (19) or 16 mg/ml of mys SD-MO targeted to the splice donor of the second exon of the mys gene (19) or prkar1a MO targeted to the translation initiation site of the prkar1a transcript (5′-CACTGCTCGTACTGCTGGACGCCAT-3′) (Gene Tools) was injected into one-cell stage embryos. To simultaneously knock down Mys and Prkar1a, 1 nl of a solution containing 8 mg/ml each of mys MO and prkar1a MO was injected into embryos. The full ORF of Sonic hedgehog (Shh) protein was cloned into pCS2+. shh and dnPKA (16) mRNAs were synthesized with an mMESSAGE mMACHINE SP6 Kit. One nl of a solution containing 400 μg/ml of shh mRNA or 100 μg/ml of dnPKA mRNA was injected into embryos.
Cyclopamine and Forskolin Treatment
Hh signaling induces different cell identities in a manner dependent on the dosage and hence on the levels of Smoothened (Smo) activity (26). The steroidal alkaloid cyclopamine specifically attenuates Smo activity in a dose-dependent manner (27, 28). To severely inhibit Smo function, embryos injected with mys MO or 5mm MO were dechorionated and incubated with 100 μm cyclopamine (Wako) at 10 hpf until fixation at 26 hpf. As a control, embryos injected with 5mm MO were dechorionated and incubated with 1% ethanol. To activate PKA, embryos injected with mys MO were dechorionated and incubated with 20 μm forskolin (Sigma) at 11 hpf until fixation at 20 and 26 hpf. As a control, embryos injected with mys MO or 5mm MO were dechorionated and incubated with 0.4% dimethyl sulfoxide.
RESULTS
Expression of Mys Protein
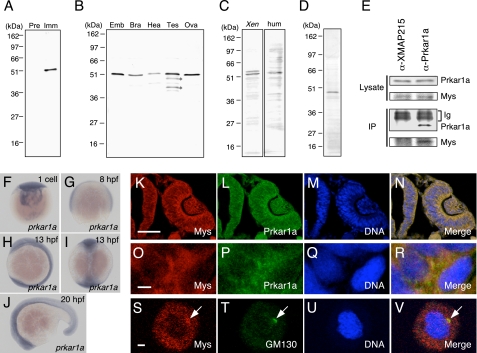
To investigate the mys gene product, we produced a rabbit polyclonal antibody raised against the recombinant protein encoded by the cDNA of a zebrafish mys gene. A single 52-kDa protein, the size of which is comparable with the predicted 435-amino acid Mys protein, was recognized specifically by immune, but not preimmune, serum in zebrafish embryos (Fig. 1A). Because the affinity-purified antibody with Mys recombinant protein recognized the same protein (Fig. 1B) and injection of the mys MO in embryos specifically reduced its expression (Fig. 4A), we conclude this to be zebrafish Mys. Mys was found to be expressed in the brain, heart, testis, and ovary in zebrafish adults (Fig. 1B). We then investigated whether the anti-Mys antibody recognizes Mys protein homologs that are found to be highly conserved in vertebrate genomes, because the C-terminal region of Mys used as the antigen retains high identity (19). As expected, Mys protein homologs were detected in Xenopus embryos and human embryonic kidney cells (HEK293 cells) (Fig. 1C). Taken together, these results show for the first time the expression of Mys and its protein homologs.
FIGURE 1.
Expression of Mys, prkar1a, and Prkar1 and an interaction between Mys and Prkar1a. A and B, identification of Mys by immunoblotting. A, extracts from zebrafish embryos immunostained with serum from a rabbit injected with recombinant Mys protein. Preimmune serum (Pre) and immune serum (Imm) were used. B, extracts from zebrafish embryos (Emb) and adult brain (Bra), heart (Hea), testis (Tes), and ovary (Ova) probed with affinity purified anti-Mys antibody. Asterisks show truncated forms of Mys. C, extracts from Xenopus laevis embryos (Xen) and HEK293 cells (hum) probed with affinity purified anti-Mys antibody. The existence of two types of proteins in extracts from Xenopus embryos may arise from whole genome duplication in X. laevis. D, characterization of anti-Prkar1a antibody. Extracts from zebrafish embryos immunostained with serum from a mouse injected with recombinant Prkar1a protein. E, interaction of endogenous Mys with Prkar1a. Extracts from zebrafish embryos (Lysate) and immunoprecipitations of the extracts (IP) with anti-XMAP215 mouse polyclonal antibody (α-XMAP215) and anti-Prkar1a mouse polyclonal antibody (α-Prkar1a) probed with anti-Prkar1a mouse antibody (Prkar1a) and anti-Mys rabbit antibody (Mys). Immunoglobulins (Ig) of the mouse polyclonal antibodies are shown. F–J, whole mount in situ hybridization with a prkar1a probe. Lateral views (F–H and J) and dorsal view (I) of zebrafish embryos at the one-cell stage (F), 8 hpf (G), 13 hpf (H and I), and 20 hpf (J). K–R, immunofluorescence of zebrafish embryos at 22 hpf with anti-Mys (Mys) and anti-Prkar1a (Prkar1a) antibodies. The embryo was simultaneously stained with Hoechst 33258 (DNA). Merged images are shown (Merge). K–N, transversal section of an eye and a neural tube, with dorsal at the top and medial at the left. O–R, enlarged images of K–N. S–V, immunofluorescence of cells derived from embryos at 4 hpf with anti-Mys (Mys) and anti-GM130 (GM130) antibodies. Arrows indicate the position of the Golgi apparatus. Bars represent 50 (K–N) and 2 μm (O–V).
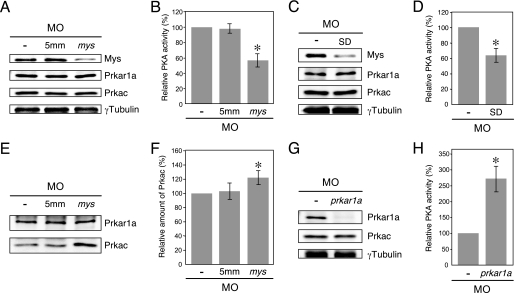
FIGURE 4.
Effects of Mys knockdown on PKA activity and amount of inactive PKA in zebrafish embryos. A, immunoblotting of extracts from wild-type embryos (−) and embryos injected with 5mm MO (5mm) and mys MO (mys). B, PKA activity in the embryos in A (mean ± S.D., n = 3; asterisk, p < 0.001, Student's t test). Knockdown of Mys by injection of mys MO resulted in reduction of PKA activity. C, immunoblotting of extracts from wild-type embryos (−) and embryos injected with mys SD-MO (SD). D, PKA activity in the embryos in C (mean ± S.D., n = 3; asterisk, p < 0.005, Student's t test). E, immunoprecipitations of extracts from wild-type embryos (−) and embryos injected with 5mm MO (5mm) and mys MO (mys) using anti-Prkar1a antibody. F, relative amount of Prkac coimmunoprecipitated with Prkar1a in the embryos in E (mean ± S.D., n = 3; asterisk, p < 0.01, Student's t test). Knockdown of Mys increased the amount of Prkac binding to Prkar1a. G, immunoblotting of extracts from wild-type embryos (−) and embryos injected with prkar1a MO (prkar1a). H, PKA activity in the embryos in G (mean ± S.D., n = 3; asterisk, p < 0.001, Student's t test). Knockdown of Prkar1a significantly increased PKA activity.
Interaction of Mys with Prkar1a
Our previous study showed that knockdown of mys in zebrafish embryos resulted in small eyes and severe somite defects (19). However, the molecular mechanisms of Mys function remain completely unknown. To elucidate the molecular basis of Mys function, we screened for Mys-interacting proteins by a pull-down assay with the human Mys homolog in HEK293 cells followed by mass spectrometry analysis (22). In this screening, 11 proteins were found to interact with the human Mys homolog. One of them was PKA regulatory type Iα subunit, Prkar1a. We focused on the interaction of Mys with Prkar1a in this study, and interactions with other proteins will be reported elsewhere.
Whole mount in situ hybridization analysis using a prkar1a probe showed ubiquitous expression of prkar1a transcripts in zebrafish embryos from the one-cell stage to 20 hpf (Fig. 1, F–J). The mys transcripts are also expressed ubiquitously during embryonic development (19). To analyze expression of Prkar1a protein, we produced a mouse polyclonal antibody specifically recognizing the Prkar1a protein (Fig. 1D). Immunostaining with anti-Mys and anti-Prkar1a antibodies showed co-expression of Mys and Prkar1a in almost all tissues, including the eyes, neural tube, and somites (Fig. 1, K–N, data not shown for somites). Mys and Prkar1a were co-localized in the cytoplasm within the cells (Fig. 1, O–R). To analyze the interaction between endogenous Mys and Prkar1a, we performed immunoprecipitation of Mys with anti-Mys antibody. Unfortunately, endogenous Mys was not precipitated with this antibody. We then performed immunoprecipitation of Prkar1a with anti-Prkar1a mouse polyclonal antibody followed by immunoblotting with anti-Mys rabbit antibody. Mys was coimmunoprecipitated with Prkar1a but not precipitated by a control anti-XMAP215 mouse polyclonal antibody (Fig. 1E), indicating interaction between endogenous Mys and Prkar1a in the zebrafish embryo.
Transiently expressed human Mys homolog, which is fused with GFP, is localized to Golgi apparatus in HeLa cells (29). The Mys homolog was recently named PRRC1 (proline-rich coiled coil 1) (GenBank number NP_570721). We then examined subcellular localization of endogenous Mys in cells derived from zebrafish embryos by immunostaining with anti-Mys antibody and anti-GM130 antibody, which recognizes a component of the Golgi apparatus. Endogenous Mys was found to be localized to the Golgi apparatus and distributed throughout the cytoplasm (Fig. 1, S–V).
Identification of Regions in Mys and Prkar1a That Are Responsible for the Interaction
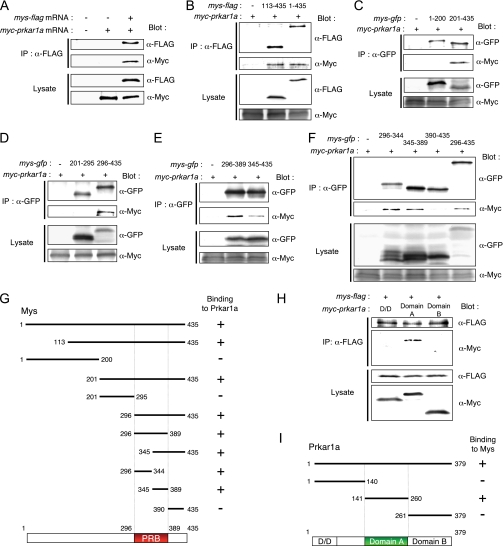
To further investigate the interaction between Mys and Prkar1a, Mys protein fused with the FLAG tag at its N or C terminus (FLAG-Mys or Mys-FLAG) and Myc-tagged Prkar1a (Myc-Prkar1a) were expressed in rabbit reticulocyte lysate and immunoprecipitated with anti-FLAG antibody. Myc-Prkar1a was coimmunoprecipitated with Mys-FLAG (Fig. 2A) but not with FLAG-Mys (data not shown). We then defined the region in Mys that binds to Prkar1a by expressing various FLAG-tagged parts of Mys. However, small fragments, less than 200 amino acids including the FLAG tag, were not synthesized in the lysate. Therefore, we replaced the FLAG tag with GFP. The results are summarized in Fig. 2G (for details, see Fig. 2, B–F). No binding of the N terminus (aa 1–200), middle part (aa 201–295), or C-terminal end (aa 390–435) of Mys to Myc-Prkar1a was detected, whereas all of the truncations containing aa 296–389 sequences bound to Myc-Prkar1a. These results indicate that Mys interacts with Prkar1a in its C-terminal region from aa 296 to 389. We refer to this 94- amino acid region as the PRB (Prkar1-binding) domain (Fig. 2G).
FIGURE 2.
Characterization of the interaction between Mys and Prkar1a. A, binding of Myc-Prkar1a to Mys-FLAG. Myc-Prkar1a was specifically coimmunoprecipitated with Mys-FLAG in lysate incubated with mys-flag mRNA (+) and myc-prkar1a mRNA (+) but not in lysate incubated with water (−) and myc-prkar1a mRNA (+). Expression of Mys-FLAG and Myc-Prkar1a in lysates is shown (Lysate). B–F, identification of the region in Mys that binds to Prkar1a. B, Myc-Prkar1a was coimmunoprecipitated with both Mys-(113–435)- and Mys-(1–435)-FLAG. C, Myc-Prkar1a was coimmunoprecipitated with Mys-(201–435)-GFP but not with Mys-(1–200)-GFP. D, Myc-Prkar1a was coimmunoprecipitated with Mys-(296–435)-GFP but not with Mys-(201–295)-GFP. E, Myc-Prkar1a was coimmunoprecipitated with both Mys-(296–389)- and Mys-(345–435)-GFP. F, Myc-Prkar1a was coimmunoprecipitated with Mys-(296–344)-, Mys-(345–389)-, and Mys-(296–435)-GFP but not with Mys-(390–435)-GFP. G, schematic diagrams showing the Mys fragments used in the experiments. The binding affinity of each molecule to Myc-Prkar1a is summarized on the right. PRB, Prkar1-binding domain. H, identification of the region in Prkar1a that binds to Mys. Myc-tagged domain A of Prkar1a, but not D/D or domain B, was coimmunoprecipitated with Mys-FLAG. D/D, dimerization/docking domain; domain A, cAMP-binding domain A; domain B, cAMP-binding domain B. I, schematic diagrams showing Prkar1a fragments used in the experiments. The binding affinity of each molecule to Mys-FLAG is summarized on the right.
To explore the role of Mys in the interaction with Prkar1a, we then examined the region in Prkar1a that binds to Mys. PKA regulatory subunits have a dimerization/docking (D/D) domain at the N terminus and two cAMP-binding domains: domain A at the middle and domain B at the C terminus (Fig. 2I) (2). Domain A interacts directly with the catalytic subunit (30). cAMP binds first to domain B, which induces a conformational change in domain A. The subsequent binding of cAMP to domain A causes dissociation of the catalytic subunits and activation of PKA (31). We expressed Myc-tagged D/D domain (aa 1–140), domain A (aa 141–260), and domain B (aa 261–379) of Prkar1a with Mys-FLAG followed by immunoprecipitation with anti-FLAG antibody. Myc-tagged domain A, but not the D/D domain or domain B, was coimmunoprecipitated with Mys-FLAG (Fig. 2, H and I), indicating that Mys interacts with domain A of Prkar1a.
Activation of PKA by PRB Domain-mediated Interaction of Mys with Prkar1a
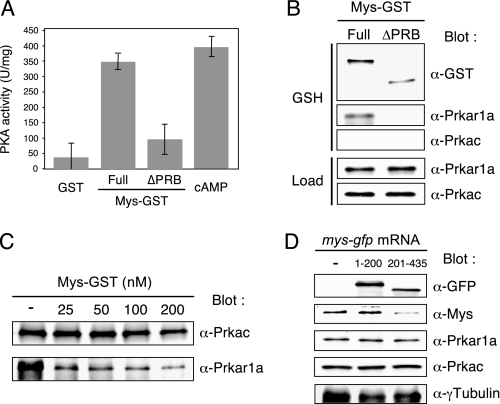
Because domain A of Prkar1a is a binding site of Mys, we hypothesized that Mys competes with the catalytic subunit for binding to domain A of Prkar1a, leading to release of the catalytic subunit and activation of the enzyme. To assess this hypothesis, inactive PKA holoenzyme was incubated with recombinant GST-tagged Mys ((Full)Mys-GST) or mutant Mys that lacks the PRB domain ((ΔPRB)Mys-GST), and PKA activity was assayed. Interaction of (Full)Mys-GST or (ΔPRB)Mys-GST with PKA was examined by a pull-down assay. PKA was activated by incubation with (Full)Mys-GST but not by incubation with GST and (ΔPRB)Mys-GST (Fig. 3A). The pull-down assay showed that (Full)Mys-GST, but not (ΔPRB)Mys-GST, interacted with Prkar1a, whereas neither of them interacted with the catalytic subunit (Prkac) (Fig. 3B). To confirm that binding of (Full)Mys-GST to Prkar1a dissociates Prkac from Prkar1a, we performed immunoprecipitation of Prkac followed by immunoblotting of Prkar1a. The amount of Prkar1a coprecipitated with Prkac was decreased in a manner dependent on the amount of (Full)Mys-GST (Fig. 3C). Taken together, these results indicate that binding of Mys to the Prkar1a via the PRB domain dissociates the Prkac from the Prkar1a, resulting in PKA activation.
FIGURE 3.
Effects of recombinant Mys on activity of purified PKA in vitro and effects of ectopic expression of Mys on protein expression in zebrafish embryos. A, effect of recombinant Mys on the activity of PKA holoenzyme. PKA activity of the holoenzyme incubated with GST, (Full)Mys-GST, (ΔPRB)Mys-GST, and cAMP (mean ± S.D., n = 3). B, pull-down assay of (Full)Mys-GST and (ΔPRB)Mys-GST incubated with PKA holoenzyme showing interaction of (Full)Mys-GST with Prkar1 but not with Prkac. Immunoblotting of loading control (Load) and precipitations with glutathione-Sepharose beads (GSH) of a mixture of Mys-GST and PKA holoenzyme. C, effects of (Full)Mys-GST on PKA holoenzyme formation. Immunoprecipitations with anti-Prkac antibody probed with anti-Prkac (α-Prkac) and anti-Prkar1a (α-Prkar1a) antibodies showing that increasing the amounts of (Full)Mys-GST decreases the amounts of Prkar1a binding to Prkac. D, effects of ectopic expression of mutant Mys-GFP on expression of endogenous Mys in zebrafish embryos. Immunoblotting of extracts from wild-type embryos (−) and embryos injected with mys-(1–200)-gfp mRNA or mys-(201–435)-gfp mRNA showing reduction of endogenous Mys in embryos expressing Mys-(201–435)-GFP.
We then examined the effects of ectopic expression of Mys on endogenous PKA activity by injection of mys-flag and truncated forms of mys-gfp mRNAs into zebrafish embryos. Unexpectedly, neither injection of mys-flag nor that of truncated forms of mys-gfp mRNAs affected PKA activity (data not shown). Immunoblot analysis showed that expression of endogenous Mys was reduced in embryos expressing Mys-FLAG and Mys-(201–435)-GFP but not in the embryos expressing Mys-(1–200)-GFP (Fig. 3D, data not shown for Mys-FLAG), suggesting the existence of a feedback mechanism that regulates the amount of Mys, in which the C-terminal region of Mys is involved. The results also suggest that embryos expressing exogenous Mys have a level of PKA activity similar to that in wild-type embryos because of reduction of endogenous Mys.
Reduction of PKA Activity by Knockdown of Mys in Zebrafish Embryos
To analyze the effects of Mys knockdown during vertebrate development, we first used mys mutant embryos carrying homozygous transposon insertions in the mys gene, which effectively but not completely disrupt transcription of the mys gene (19). However, immunoblot analysis showed that expression of the Mys protein was not effectively reduced in mutant embryos (data not shown), also suggesting the existence of a feedback mechanism at the post-transcriptional level. We then examined the effect of Mys knockdown on the activity of endogenous PKA by injection of mys MO targeted to the translation initiation site (19) in zebrafish embryos. 5mm MO containing 5-bp mismatches around the translation initiation site was used as a control. Injection of mys MO, but not 5mm MO, reduced Mys expression (∼50%), whereas neither of them affected Prkar1a and Prkac expression in the embryos at 24 hpf (Fig. 4A). PKA activity was reduced in embryos injected with mys MO but not in those injected with 5mm MO (Fig. 4B). To further confirm specificity of the mys MO effects, we injected mys SD-MO targeted to the splice donor of the second exon in embryos. Mys expression (∼60%) and PKA activity were reduced, whereas Prkar1a and Prkac expression was unaffected by injection of mys SD-MO (Fig. 4, C and D). Therefore, despite the expression of normal levels of Prkar1a and Prkac, knockdown of Mys leads to reduction of PKA activity, indicating that Mys ensures appropriate levels of PKA activity in the zebrafish embryo.
We further investigated the effects of Mys knockdown on PKA holoenzyme formation by immunoprecipitation of Prkar1a followed by immunoblotting of Prkac. As shown in Fig. 4E, Prkac was coimmunoprecipitated with Prkar1a, indicating the formation of inactive holoenzyme in embryos. The amount of Prkac precipitated with Prkar1a was unaffected by injection of 5mm MO (103 ± 12%, n = 3) but was increased by injection of mys MO (122 ± 9.6%, n = 3, p < 0.01) (Fig. 4, E and F). The fact that only 20% of inactive PKA was increased by knockdown of Mys suggested that the majority of PKA remains inactive in the embryos. To confirm this, prkar1a MO was injected into embryos and PKA activity was assayed. Immunoblot analysis showed that injection of prkar1a MO reduced the expression of Prkar1a (∼20%) but not that of Prkac (Fig. 4G). In these embryos, PKA activity was significantly increased (271 ± 39%, n = 3, p < 0.001) (Fig. 4H). Therefore, the majority of Prkacs form inactive holoenzyme with Prkar1a and a small fraction of them are activated by Mys binding to Prkar1a in the zebrafish embryo.
Ectopic Activation of Hh Signaling by Knockdown of Mys in Zebrafish Embryos
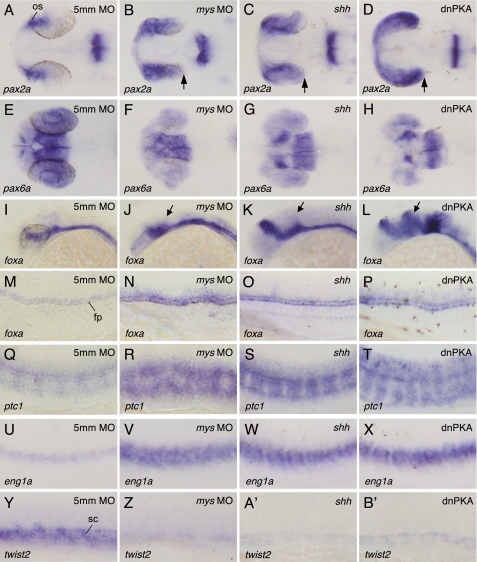
Because inhibition of PKA in zebrafish embryos by expressing dnPKA leads to ectopic activation of Hh target genes (14–16, 18), we then investigated whether the reduction of PKA activity by Mys knockdown alters Hh-mediated patterning of the eyes, neural tube, and somites. We examined pax2a, pax6a, foxa (also known as fkd4), ptc1, eng1a, and twist2 expression in mys MO-injected embryos. The expression of pax2a in the presumptive optic stalk (Fig. 5A), pax6a in distal regions of the eye primordia (Fig. 5E), foxa in floor plate (Fig. 5, I and M), ptc1 in cells adjacent to the notochord (Fig. 5Q), eng1a in slow muscle pioneer cells (Fig. 5U), and twist2 in the sclerotome (Fig. 5Y) was unaffected by injection of 5mm MO (n = 77, 79, 56, 54, 76, and 81, respectively). In contrast, embryos injected with mys MO exhibited an expansion of the pax2a expression domain and a reduction of pax6a expression in the eyes (Fig. 5, B and F, 79 of 80 and 70 of 75 embryos, respectively), an increase and expansion of foxa expression in the neural tube (Fig. 5, J and N, 50 of 55 embryos), an expansion of the expression domains of ptc1 and eng1a in somites (Fig. 5, R and V, 58 of 61 and 73 of 73 embryos, respectively), and a reduction of twist2 expression (Fig. 5Z, 86 of 86 embryos), consistent with the results of ectopic expression of Shh (Fig. 5, C, G, K, O, S, W, and A′) and dnPKA (Fig. 5, D, H, L, P, T, X, and B′) (12–16, 18). Because embryos injected with mys SD-MO exhibited altered patterning of gene expression similar to embryos injected with mys MO (supplemental Fig. S1) and the expression patterns of all genes were restored by injection of mys-(201–435)-gfp mRNA prior to the injection of mys MO (supplemental Fig. S2), we conclude that knockdown of Mys is responsible for the altered patterning of gene expression in the eyes, neural tube, and somites. Mys knockdown did not affect the expression of Wnt target genes, axin2 (32) and sp5l (33), and the expression of shh (data not shown), suggesting that Mys is specifically involved in Hh signaling and does not function upstream of shh.
FIGURE 5.
Effects of Mys knockdown and ectopic expression of Shh and dnPKA on patterning of gene expression in the eyes, neural tube, and somites. A and B′, whole mount in situ hybridization with probes for pax2a (A–D), pax6a (E–H), foxa (I–P), ptc1 (Q–T), eng1a (U–X), and twist2 (Y–B′). Dorsal (A–H) and lateral views (I–B′) of embryos injected with 5mm MO (A, E, I, M, Q, U, and Y), mys MO (B, F, J, N, R, V, and Z), shh mRNA (C, G, K, O, S, W, and A′), and dnPKA mRNA (D, H, L, P, T, X, and B′). Embryos fixed at 26 (A–X) and 20 hpf (Y–B′). Knockdown of Mys altered gene expression patterns in the eyes, neural tube, and somites (B, F, J, N, R, V, and Z), which is similar to those in embryos ectopically expressing Shh (C, G, K, O, S, W, and A′) and dnPKA (D, H, L, P, T, X, and B′). Arrows indicate the expanded region of pax2a expression (B–D) and foxa expression (J–L). os, optic stalk; fp, floor plate; sc, sclerotome.
Mys Antagonizes Hh Signaling Downstream of Shh and Smo
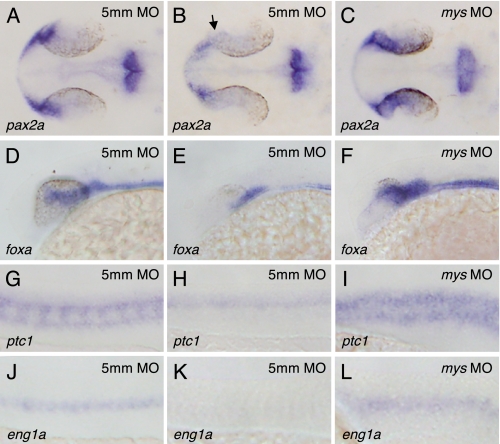
To confirm that ectopic gene expression is induced downstream of Shh, we injected mys MO into syut4 mutant embryos carrying a deletion in the shh gene (34) and examined the expression of pax2a, foxa, ptc1, and eng1a. Injection of 5mm MO did not affect shh homozygous mutant phenotypes, i.e. substantial reductions of expression domains of pax2a (Fig. 6B, arrow) (24% of the embryos derived from crosses between shh+/− fish, n = 25), foxa (Fig. 6E) (21% of the embryos, n = 24), ptc1 (Fig. 6H) (33% of the embryos, n = 24), and eng1a (Fig. 6K) (19% of the embryos, n = 21). In contrast, injection of mys MO resulted in increased expression of pax2a (Fig. 6C) (96% of the embryos, n = 28), foxa (Fig. 6F) (100% of the embryos, n = 29), ptc1 (Fig. 6I) (86% of the embryos, n = 28), and eng1a (Fig. 6L) (100% of the embryos, n = 27). These results provide evidence that Mys is involved in suppression of the expression of Hh target genes downstream of Shh.
FIGURE 6.
Involvement of Mys in suppressing expression of Hh target genes downstream of Shh. A–L, in situ hybridization with probes for pax2a (A–C), foxa (D–F), ptc1 (G–I), and eng1a (J–L). Dorsal (A–C) and lateral views (D–L) of the embryos derived from crosses between shh+/− fish and injected with 5mm MO (A, B, D, E, G, H, J, and K) and mys MO (C, F, I, and L). Embryos were fixed at 26 hpf. The shh mutant embryos injected with 5mm MO showed reduced expression of pax2a (B, arrow), foxa (E), ptc1 (H), and eng1a (K), whereas their wild-type siblings showed normal patterning of gene expression (A, D, G, and J). In contrast, the shh mutant embryos injected with mys MO showed increased expression of pax2a (C), foxa (F), ptc1 (I), and eng1a (L).
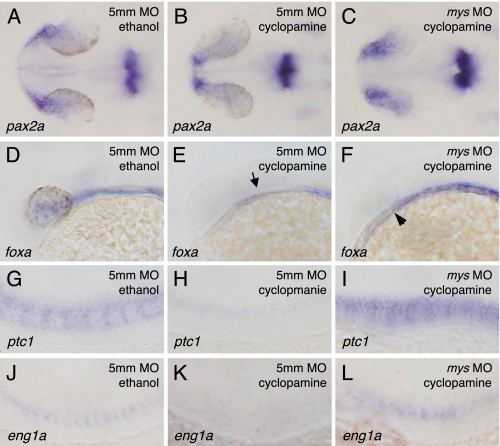
We next investigated whether Mys antagonizes the Hh signaling pathway downstream of Smoothened (Smo), a protein essential for transduction of all Hh signaling activity (35, 36), by inhibiting Smo function with cyclopamine (27). The embryos injected with 5mm MO and treated with cyclopamine showed significant reductions of expression domains of pax2a (Fig. 7B) (100%, n = 70), foxa (Fig. 7E) (100%, n = 40), ptc1 (Fig. 7H) (100%, n = 40), and eng1a (Fig. 7K) (100%, n = 78). In contrast, the embryos injected with mys MO and treated with cyclopamine showed increased expression of pax2a (Fig. 7C) (88%, n = 77) and ptc1 (Fig. 7I) (71%, n = 42), partial rescue of foxa expression (Fig. 7F) (86%, n = 42), and rescue of eng1a expression (Fig. 7L) (26%, n = 86). These results indicate that Mys is involved in antagonizing the Hh signaling pathway downstream of Smo, consistent with the epistatic relationship of Smo and PKA.
FIGURE 7.
Involvement of Mys in antagonizing Hh signaling pathway downstream of Smo. A–L, in situ hybridization with probes for pax2a (A–C), foxa (D–F), ptc1 (G–I), and eng1a (J–L). Dorsal (A–C) and lateral views (D–L) of embryos injected with 5mm MO (A, B, D, E, G, H, J, and K) and mys MO (C, F, I, and L) followed by ethanol (A, D, G, and J) or cyclopamine treatment (B, C, E, F, H, I, K, and L). Embryos were fixed at 26 hpf. Cyclopamine treatment induced significant reductions of pax2a expression in the eyes (B), foxa expression in the neural tube (E, arrow), ptc1 expression in cells adjacent to the notochord (H), and eng1a expression in somites (K). In contrast, knockdown of Mys resulted in increased expression of pax2a in the eyes (C) and ptc1 expression in somites (I), partial rescue of foxa expression in the neural tube (F, arrowhead) and rescue of eng1a expression in the somites (L) of the embryos incubated with cyclopamine.
Altered Patterning of Gene Expression Induced by Mys Knockdown Can Be Rescued by PKA Activation
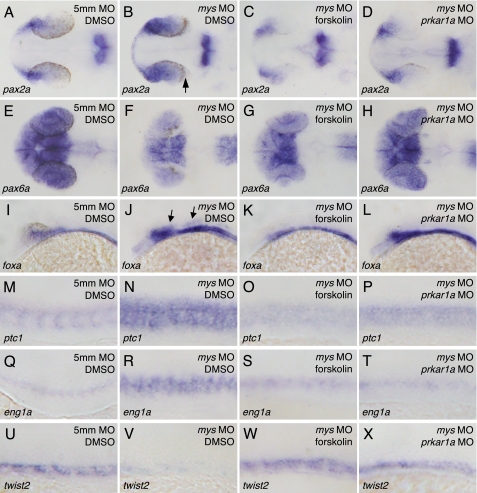
We finally examined whether activation of PKA is sufficient to rescue gene expression in mys MO-injected embryos. To recover the PKA activity, the mys MO-injected embryos were incubated with forskolin, which directly stimulates adenylyl cyclase to increase cellular cAMP levels and hence activates PKA. The PKA activity was also recovered by co-injection of prkar1a MO with mys MO in embryos. Incubation of the mys MO-injected embryos with dimethyl sulfoxide did not affect the altered patterning of gene expression (Fig. 8, B, F, J, N, R, and V). In contrast, treatment of mys MO-injected embryos with forskolin and co-injection of prkar1a MO with mys MO resulted in rescue of expression of pax2a (Fig. 8, C and D) (91%, n = 76 and 88%, n = 41, respectively), pax6a (Fig. 8, G and H) (93%, n = 74 and 71%, n = 41, respectively), foxa (Fig. 8, K and L) (100%, n = 74 and 88%, n = 41, respectively), ptc1 (Fig. 8, O and P) (77%, n = 35 and 67%, n = 51, respectively), eng1a (Fig. 8, S and T) (95%, n = 66 and 70%, n = 40, respectively), and twist2 (Fig. 8, W and X) (30%, n = 70 and 39%, n = 49, respectively). The results indicate that reduction of PKA activity is responsible for the altered patterning of gene expression in the eyes, neural tube, and somites in mys MO-injected embryos.
FIGURE 8.
Effects of PKA activation on the patterning of gene expression in the eyes, neural tube, and somites in mys MO-injected embryos. A–X, in situ hybridization with probes for pax2a (A–D), pax6a (E–H), foxa (I–L), ptc1 (M–P), eng1a (Q–T), and twist2 (U–X). Dorsal (A–H) and lateral views (I–X) of embryos fixed at 26 (A–T) and 20 hpf (U–X). Treatment with dimethyl sulfoxide (DMSO) did not affect gene expression patterning of control 5mm MO-injected embryos (A, E, I, M, Q, and U) and mys MO-injected embryos (B, F, J, N, R, and V). In contrast, treatment of the embryos with forskolin rescued gene expression patterns (C, G, K, O, S, and W). In addition, co-injection of prkar1a MO with mys MO resulted in rescue of gene expression patterns (D, H, L, P, T, and X). Arrows indicate the expanded region of pax2 (B) and foxa (J) expression.
DISCUSSION
A Molecular Mechanism of Regulation of PKA Activity by Mys
In this study, we identified the interaction between Mys and Prkar1a proteins by pull-down assay with FLAG-tagged human Mys homolog and showed the interaction of endogenous Mys with Prkar1a in zebrafish embryos (Fig. 1). In vitro assays showed that Mys interacts with domain A of Prkar1a via the PRB domain and this interaction activates PKA by dissociating Prkac from Prkar1a (Figs. 2 and 3). Results of Mys knockdown demonstrated that Mys ensures an appropriate level of PKA activity in the zebrafish embryo (Fig. 4). The basal PKA activity regulated by Mys is indispensable for negative regulation of the Hh signaling pathway in Hh-responsive cells during pattern formation of the eyes, neural tube, and somites of the zebrafish embryo (Figs. 5–8).
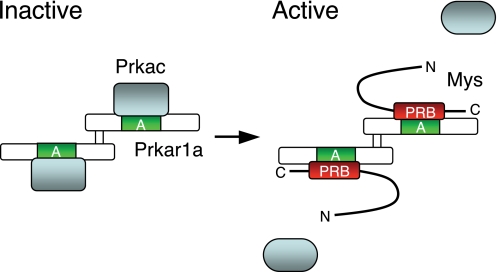
Based on our results and results of previous studies, we propose a molecular mechanism of regulation of PKA activity that is dependent on the interaction between Mys and Prkar1a proteins, by which Hh signaling is negatively regulated during vertebrate development (Fig. 9). PKA forms an inactive holoenzyme containing a Prkar1a dimer and two Prkacs. Prkac interacts directly with domain A of Prkar1a (31). PKA is activated by binding of Mys to Prkar1a domain A with its C-terminal PRB domain, resulting in release of Prkac from Prkar1a. One of the proteins known to be phosphorylated by PKA during embryonic development is Gli3, a bipotential transcription factor of Hh signaling. It has been demonstrated that PKA phosphorylates the consensus phosphorylation motifs of Gli3, which promotes truncation of Gli3 by limited proteolysis (37). The truncated Gli3 translocates to the nucleus and suppresses transcription of Hh target genes (11). A recent study has shown that Gli2 is also phosphorylated by PKA, leading to processing and degradation of Gli2 (38). The mutant Gli2, which is unable to be phosphorylated by PKA, is stable and expands expression regions of Hh target genes in the mouse embryo when inserted into the Gli2 locus, suggesting that phosphorylation of Gli2 by PKA is crucial for the Hh-regulated patterning of vertebrate embryos (39). Although no study has been reported in vertebrate, other Hh signaling components might be phosphorylated by PKA. In addition, PKA activity regulated by Mys might be implicated in phosphorylation of proteins playing important roles in physiological processes other than Hh signaling during embryonic development.
FIGURE 9.
Proposed model of the regulation of PKA activation by Mys. PKA forms an inactive holoenzyme containing a Prkar1a dimer and two Prkacs (left). Prkac binds to domain A of Prkar1a. Mys competes with Prkac for binding to domain A of Prkar1a via its C-terminal PRB domain, resulting in dissociation of Prkacs from a Prkar1a dimer and activation of PKA (right). The active Prkacs phosphorylate protein substrates. A, cAMP-binding domain A; PRB, Prkar1-binding domain.
Hh signaling has recently been demonstrated to be relevant to tissue homeostasis in adults (11, 40). Our finding that Mys is expressed in zebrafish adult tissues (Fig. 1B) suggests involvement of Mys in regulation of PKA activity and hence modulation of Hh signaling in the tissues.
PKA is known to antagonize Hh signaling in both invertebrates and vertebrates (11, 41). We previously found highly conserved Mys protein homologs in vertebrate genomes (19) and we revealed the expression of Xenopus and human Mys protein homologs in this study (Fig. 1C). The amino acid sequence of the PRB domain retains high identity in all homologs. These findings suggest that the role of Mys in regulation of PKA activity is conserved in vertebrates. In contrast to vertebrates, we could not detect any obvious counterpart of Mys in invertebrates in a previous study (19). However, genes containing the PRB domain identified in this study are present in invertebrate genomes: in Ciona intestinalis (GenBank number AK114792), Apis mellifera (XM_397072), and Anopheles gambiae (XM_001689177). Therefore, it is possible that these genes are similarly involved in regulation of PKA activity in invertebrates. Alternatively, it is possible that invertebrates ensure basal PKA activity by a mechanism different from that in vertebrates because an ortholog of mys has not been detected in the Drosophila genome.
Kamakari et al. (29) have reported the expression of transcripts of the gene encoding human Mys/PRRC. They revealed the existence of 6 splice variants of the transcripts, which encode four putative proteins of 311, 445, 462, and 464 aa. The transcript encoding the protein of 311 aa was shown to be expressed specifically in the liver and the other transcripts were shown to be expressed ubiquitously. By newly produced anti-human Mys/PRRC antibody, we detected low level expression of a 58-kDa protein in addition to the 54-kDa protein identified in this study (Fig. 1C) in extracts from HEK293 cells,3 suggesting that a protein of 445 aa corresponds to the 54-kDa protein and proteins of 462 and 464 aa correspond to the 58-kDa protein. The 445-aa protein fused with GFP was found to be localized to the Golgi apparatus with distribution in the cytoplasm as puncta (29), consistent with our observation of endogenous Mys (Fig. 1, S–V). In addition, a novel protein C04G6.4 was found to be the protein homolog in Caenorhabditis elegans (29). However, the function of C04G6.4 remains to be elucidated. They also showed that in the leukemia patient cell line the genomic region including the gene was deleted from the chromosomes, suggesting a putative involvement of the gene in leukemogenesis (29).
In this study, we showed that expression of Mys-(201–435)-GFP could rescue the altered patterning of gene expression induced by Mys knockdown, suggesting that the PRB domain, including the C-terminal region of Mys, is sufficient to exert its functions. We also showed that ectopic expression of the full-length and C-terminal region of Mys reduced expression of endogenous Mys but did not affect endogenous PKA activity. From these results, we hypothesize that PKA activity is maintained at a certain level by modulating the amount of Mys via an autoregulatory loop. An alternative explanation for the mechanism maintaining the basal level of PKA activity is that binding of Mys to Prkar1a is precisely regulated by modification of Mys in vivo, and therefore overexpression of Mys does not affect PKA activity. Further experiments will have to elucidate the molecular basis of the feedback mechanism by which the amount of Mys is regulated and have to clarify whether the amount or modification of Mys is involved in regulation of PKA activity.
In this study, we also found expression of the Mys protein in the mys mutant embryos. This is consistent with our previous observations that mys mutants show limited defects in somite boundaries at the early segmentation stage (12–13 hpf) and appear to be normal after this period (19). In addition, the mutant embryos survive to adulthood. These results suggest that Hh signaling is not severely altered in the mys mutant embryos.
The cAMP-dependent and -independent Activation Mechanisms of PKA
Levels of PKA activity are believed to in principle reflect intracellular cAMP levels in most cells. However, it has been suggested that a basal level of PKA activity that is independent of cAMP is both necessary and sufficient to suppress transcription of Hh target genes in Drosophila embryos, because low-level expression of the constitutively active form of PKA could substitute for the PKA catalytic subunit activity to confer normal Hh signal transduction (7, 42). In this regard, it is notable that a basic lipid, sphingosine, which acts as a second messenger, activates membrane-associated PKA type II by a mechanism independent of cAMP in cultured mammalian cells (43). Unlike cAMP, however, sphingosine activates PKA holoenzyme without dissociating catalytic subunits from a regulatory subunit dimer, indicating that the mechanism of PKA activation by sphingosine is distinct from that of PKA activation by Mys.
In mammals, four isoforms of PKA regulatory subunits, type Iα (Prkar1a), Iβ (Prkar1b), IIα (Prkar2a), and IIβ (Prkar2b), have been isolated. In general, the transcripts encoding Prkar1a and Prkar2a are expressed ubiquitously, whereas those encoding Prkar1b and Prkar2b have tissue-specific expression patterns in mouse embryos (44). In this study, we showed the ubiquitous expression of prkar1a transcripts and Prkar1a protein in the zebrafish embryo (Fig. 1, F–N). In addition, knockdown of Prkar1a significantly increased basal PKA activity in zebrafish embryos (Fig. 4, G and H). Although basal PKA activity is substantially increased in Prkar1a-deficient mouse embryos, which is consistent with our present results, a large amount of PKA responsive to cAMP is still retained and this PKA is thought to be inactivated by Prkar2a expressed in the mutant embryos (45). The results obtained in mice suggest that Prkar1a-deficient zebrafish embryos retain additional inactive PKA consisting of other regulatory subunits, such as Prkar2a. Mys seems to specifically interact with Prkar1a, because no interaction of Prkar1b, Prkar2a, and Prkar2b with the human Mys homolog was detected by pull-down assays despite the expression of all genes in HEK293 cells (data not shown), although we could not rule out the possibility that interactions of Mys with other regulatory subunits are weak and undetectable by our assays. In addition, Mys activates only a small part of PKA containing Prkar1a by dissociating Prkac from Prkar1a in zebrafish embryos (Fig. 4, A–F). The evidence demonstrates that PKA is dominantly inactivated by regulatory subunits and that only a small fraction of PKA is activated at least partially by Mys during embryonic development. However, we believe that this limited regulatory mechanism of PKA plays a key role in Hh-dependent cell differentiation.
Although Smo is known to be a seven-transmembrane protein related to G protein-coupled receptors, how Smo mediates Hh signaling activity remains obscure. In frog melanophores and cultured mammalian cells, exogenous Smo and stimulated Smo can activate members of the inhibitory G protein family, Gi proteins (46, 47). In zebrafish embryos, ectopic expression of a specific inhibitor of Gi proteins induces phenotypes similar to those caused by constitutive active PKA (48). In Drosophila embryos, a recent study has shown that a constitutively active Gi protein induces ectopic activation of Hh signaling and that mutations in the Gi protein result in reduction of Hh target gene expression (49). In addition, the basal level of cAMP was reduced in response to Hh through the Gi protein in cultured Cl8 cells, suggesting that Gi proteins may regulate PKA activity by reducing cAMP levels (49). However, it remains to be determined whether Gi proteins modulate Hh signaling through reducing cAMP levels or through other effectors in Drosophila embryos, because the low level of cAMP-independent PKA activity substitutes for endogenous PKA activity (50). In contrast to Drosophila embryos, activation and inhibition of Gi proteins have little effect on Hh signaling activity in chick embryos (51), suggesting that Gi proteins are not necessary for Hh signal transduction. The evidence suggests the contribution of more than one signaling pathway in transduction of Hh signaling, which may be different in distinct cell types, tissues, and organisms (52). Identification of Mys as a regulator of PKA activity would contribute to elucidation of the pathway through which Smo transduces Hh-signaling activity during early development in vertebrates. A detailed biochemical analysis is in progress to assess this hypothesis.
Additional Functions of Mys during Embryonic Development
The major role of PKA in early embryogenesis in fish is thought to be the modulation of Hh signaling, because zebrafish embryos expressing dnPKA exhibit no additional phenotypes compared with those ectopically expressing Hh family members (14). These embryos show small eyes and abnormal somite morphology. In contrast, mys MO-injected embryos display more broad defects, small eyes, abnormal somite morphology, curved body, and loss of somite boundaries in later stages of somitogenesis (19), suggesting additional functions of Mys during vertebrate development. To elucidate the molecular basis of Mys functions, including the mechanism by which somite boundaries are maintained, we are currently investigating the interactions of Mys with 10 other proteins that have been identified in our screening.
Supplementary Material
Acknowledgments
We thank H. Ochi, T. Yamamoto, and C. Sakai for technical advice, R. Ota for providing Xenopus embryos, and the Zebrafish International Resource Center for the zebrafish line.
This work was supported by grants from Inamori and Akiyama Foundations (to T. K.) and the New Energy and Industrial Technology Development Organization (NEDO) (to T. N.), a grant-in-aid for scientific research on Priority Areas “Systems Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to S. I. and K. K.), and the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN) from Bio-oriented Technology Research Advancement Institution (BRAIN) (to M. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
T. Kotani and M. Yamashita, unpublished data.
- PKA
- protein kinase A
- Hh
- Hedgehog
- dnPKA
- dominant negative form of PKA
- ORF
- open reading frame
- MO
- morpholino oligonucleotide
- Smo
- Smoothened
- hpf
- hours post-fertilization
- aa
- amino acid(s)
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- HEK
- human embryonic kidney
- D/D
- dimerization/docking.
REFERENCES
- 1.Walsh D. A., Perkins J. P., Krebs E. G. (1968) J. Biol. Chem. 243, 3763–3765 [PubMed] [Google Scholar]
- 2.Taylor S. S., Buechler J. A., Yonemoto W. (1990) Annu. Rev. Biochem. 59, 971–1005 [DOI] [PubMed] [Google Scholar]
- 3.Strutt D. I., Wiersdorff V., Mlodzik M. (1995) Nature 373, 705–709 [DOI] [PubMed] [Google Scholar]
- 4.Lepage T., Cohen S. M., Diaz-Benjumea F. J., Parkhurst S. M. (1995) Nature 373, 711–715 [DOI] [PubMed] [Google Scholar]
- 5.Li W., Ohlmeyer J. T., Lane M. E., Kalderon D. (1995) Cell 80, 553–562 [DOI] [PubMed] [Google Scholar]
- 6.Pan D., Rubin G. M. (1995) Cell 80, 543–552 [DOI] [PubMed] [Google Scholar]
- 7.Jiang J., Struhl G. (1995) Cell 80, 563–572 [DOI] [PubMed] [Google Scholar]
- 8.Fan C. M., Porter J. A., Chiang C., Chang D. T., Beachy P. A., Tessier-Lavigne M. (1995) Cell 81, 457–465 [DOI] [PubMed] [Google Scholar]
- 9.Hynes M., Porter J. A., Chiang C., Chang D., Tessier-Lavigne M., Beachy P. A., Rosenthal A. (1995) Neuron 15, 35–44 [DOI] [PubMed] [Google Scholar]
- 10.Epstein D. J., Marti E., Scott M. P., McMahon A. P. (1996) Development 122, 2885–2894 [DOI] [PubMed] [Google Scholar]
- 11.Ingham P. W., McMahon A. P. (2001) Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 12.Ekker S. C., Ungar A. R., Greenstein P., von Kessler D. P., Porter J. A., Moon R. T., Beachy P. A. (1995) Curr. Biol. 5, 944–955 [DOI] [PubMed] [Google Scholar]
- 13.Macdonald R., Barth K. A., Xu Q., Holder N., Mikkola I., Wilson S. W. (1995) Development 121, 3267–3278 [DOI] [PubMed] [Google Scholar]
- 14.Hammerschmidt M., Bitgood M. J., McMahon A. P. (1996) Genes Dev. 10, 647–658 [DOI] [PubMed] [Google Scholar]
- 15.Concordet J. P., Lewis K. E., Moore J. W., Goodrich L. V., Johnson R. L., Scott M. P., Ingham P. W. (1996) Development 122, 2835–2846 [DOI] [PubMed] [Google Scholar]
- 16.Ungar A. R., Moon R. T. (1996) Dev. Biol. 178, 186–191 [DOI] [PubMed] [Google Scholar]
- 17.Currie P. D., Ingham P. W. (1996) Nature 382, 452–455 [DOI] [PubMed] [Google Scholar]
- 18.Du S. J., Devoto S. H., Westerfield M., Moon R. T. (1997) J. Cell Biol. 139, 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotani T., Kawakami K. (2008) Dev. Biol. 316, 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotani T., Nagayoshi S., Urasaki A., Kawakami K. (2006) Methods 39, 199–206 [DOI] [PubMed] [Google Scholar]
- 21.Kotani T., Yoshida N., Mita K., Yamashita M. (2001) Mol. Reprod. Dev. 59, 199–208 [DOI] [PubMed] [Google Scholar]
- 22.Natsume T., Yamauchi Y., Nakayama H., Shinkawa T., Yanagida M., Takahashi N., Isobe T. (2002) Anal. Chem. 74, 4725–4733 [DOI] [PubMed] [Google Scholar]
- 23.Schulte-Merker S., Ho R. K., Herrmann B. G., Nüsslein-Volhard C. (1992) Development 116, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 24.Kotani T., Yamashita M. (2002) Dev. Biol. 252, 271–286 [DOI] [PubMed] [Google Scholar]
- 25.Kotani T., Yamashita M. (2005) Zygote 13, 219–226 [DOI] [PubMed] [Google Scholar]
- 26.Ingham P. W. (2008) Curr. Biol. 18, R238–241 [DOI] [PubMed] [Google Scholar]
- 27.Chen J. K., Taipale J., Cooper M. K., Beachy P. A. (2002) Genes Dev. 16, 2743–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolff C., Roy S., Ingham P. W. (2003) Curr. Biol. 13, 1169–1181 [DOI] [PubMed] [Google Scholar]
- 29.Kamakari S., Roussou A., Jefferson A., Ragoussis I., Anagnou N. P. (2005) Leuk. Res 29, 17–31 [DOI] [PubMed] [Google Scholar]
- 30.Saraswat L. D., Ringheim G. E., Bubis J., Taylor S. S. (1988) J. Biol. Chem. 263, 18241–18246 [PubMed] [Google Scholar]
- 31.Taylor S. S., Kim C., Cheng C. Y., Brown S. H., Wu J., Kannan N. (2008) Biochim. Biophys. Acta 1784, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. (2002) Mol. Cell. Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weidinger G., Thorpe C. J., Wuennenberg-Stapleton K., Ngai J., Moon R. T. (2005) Curr. Biol. 15, 489–500 [DOI] [PubMed] [Google Scholar]
- 34.Schauerte H. E., van Eeden F. J., Fricke C., Odenthal J., Strähle U., Haffter P. (1998) Development 125, 2983–2993 [DOI] [PubMed] [Google Scholar]
- 35.Chen W., Burgess S., Hopkins N. (2001) Development 128, 2385–2396 [DOI] [PubMed] [Google Scholar]
- 36.Varga Z. M., Amores A., Lewis K. E., Yan Y. L., Postlethwait J. H., Eisen J. S., Westerfield M. (2001) Development 128, 3497–3509 [DOI] [PubMed] [Google Scholar]
- 37.Wang B., Fallon J. F., Beachy P. A. (2000) Cell 100, 423–434 [DOI] [PubMed] [Google Scholar]
- 38.Pan Y., Bai C. B., Joyner A. L., Wang B. (2006) Mol. Cell. Biol. 26, 3365–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y., Wang C., Wang B. (2009) Dev. Biol. 326, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varjosalo M., Taipale J. (2008) Genes Dev. 22, 2454–2472 [DOI] [PubMed] [Google Scholar]
- 41.Huangfu D., Anderson K. V. (2006) Development 133, 3–14 [DOI] [PubMed] [Google Scholar]
- 42.Ohlmeyer J. T., Kalderon D. (1997) Genes Dev. 11, 2250–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Y., Pitson S., Hercus T., Murphy J., Lopez A., Woodcock J. (2005) J. Biol. Chem. 280, 26011–26017 [DOI] [PubMed] [Google Scholar]
- 44.Cadd G., McKnight G. S. (1989) Neuron 3, 71–79 [DOI] [PubMed] [Google Scholar]
- 45.Amieux P. S., Howe D. G., Knickerbocker H., Lee D. C., Su T., Laszlo G. S., Idzerda R. L., McKnight G. S. (2002) J. Biol. Chem. 277, 27294–27304 [DOI] [PubMed] [Google Scholar]
- 46.DeCamp D. L., Thompson T. M., de Sauvage F. J., Lerner M. R. (2000) J. Biol. Chem. 275, 26322–26327 [DOI] [PubMed] [Google Scholar]
- 47.Riobo N. A., Saucy B., Dilizio C., Manning D. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12607–12612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammerschmidt M., McMahon A. P. (1998) Dev. Biol. 194, 166–171 [DOI] [PubMed] [Google Scholar]
- 49.Ogden S. K., Fei D. L., Schilling N. S., Ahmed Y. F., Hwa J., Robbins D. J. (2008) Nature 456, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang J., Hui C. C. (2008) Dev. Cell 15, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Low W. C., Wang C., Pan Y., Huang X. Y., Chen J. K., Wang B. (2008) Dev. Biol. 321, 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Philipp M., Caron M. G. (2009) Curr. Biol. 19, R125–R127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.