Abstract
The human estrogen-related receptor α1 (ERRα1) is a member of an orphan receptor family closely related to the estrogen receptor. It has been demonstrated that estrogen modulates endothelial nitric oxide synthase (eNOS) expression through the estrogen receptor in endothelial cells. However, little is known about the relationship between ERRα1 and eNOS. In this study, we show that ERRα1 activates the estrogen response element (ERE) and eNOS promoter-dependent luciferase activity in COS-7 cells and bovine pulmonary artery endothelial cells. The endogenous ligand for ERRα1 has not been identified, but we show that these actions are dependent on serum constituents because ERRα1 fails to stimulate eNOS promoter-dependent luciferase activity in charcoal-treated serum. Furthermore, through the use of truncated eNOS promoter luciferase constructs, we demonstrate that the activation of eNOS transcription by ERRα1 is mediated via three regions: base pairs -1001 to -743, base pairs -743 to -265, and downstream from base pair -265 on the eNOS promoter. In addition, ERRα1 up-regulates eNOS mRNA and protein expression and stimulates eNOS activity in bovine pulmonary artery endothelial cells. These results suggest that ERRα1 has a potential role in the regulation of eNOS expression and may stimulate NO production by endothelial cells, which may in turn result in a protective effect against atherosclerosis.
Keywords: estrogen receptor, endothelium, atherosclerosis, ligand, transcription
Nitric oxide (NO) may attenuate the development of atherosclerosis by prevention of adhesion molecule expression and platelet aggregation and adhesion to endothelial cells (1, 2). NO is produced by two constitutive NOS isoforms [endothelial NO synthase (eNOS) and neuronal NOS (nNOS)] and by one inducible NOS isoform (iNOS) (3). In endothelial cells, eNOS expression and activation is proposed to be the most important factor governing vascular homeostasis and protection against atherosclerosis. This view is based on the observations that in vivo eNOS gene transfection stimulates endothelium-dependent relaxation (4) and the administration of the eNOS inhibitor N-nitro-l-arginine methyl ester (l-NAME) potentiates the development of atherosclerosis induced by a high-cholesterol diet in rabbits (5, 6).
Estrogen has been shown to have a protective effect on vascular endothelial cells in the prevention of atherosclerosis (7, 8) and has therefore been used for the prevention of cardiovascular disease in postmenopausal women (9). Estrogen stimulates NO production by endothelial cells through the phosphorylation and activation of eNOS (acute or immediate effect) (10) and also by up-regulation of eNOS expression (chronic or delayed effect) (11). Regarding eNOS up-regulation, estrogen was reported to stimulate eNOS transcription through the estrogen response element (ERE) (12) and Sp1-binding motif in the eNOS promoter (13). In addition, we have shown that estrogen prevents the eNOS mRNA destabilization that is induced by tumor necrosis factor α (14).
Estrogen modulates eNOS expression and activation through interaction with the estrogen receptors α (ERα) and β (ERβ) that are both expressed in endothelial cells (15, 16). ERs (ERα and ERβ) belong to a superfamily of nuclear receptors with highly conserved domain organization. A hormone-independent transactivation function (AF-1 region) is located in the N-terminal domain (A/B domain). The C domain is highly conserved in some receptors and has a binding capacity for specific regions of DNA. The D domain forms a bridge between the C and E domains. The E domain is the ligand-binding domain and includes the hormone-dependent transactivation function (AF-2 region). Although ERs are well known for their role in nuclear receptor-mediated gene expression, the acute or immediate response is mediated by ERs in the endothelial membrane (17). The important element for nuclear localization has been identified in the C and D domains of the ERα (18), and serine 522 plays a role in membrane association and translocation (19).
Two estrogen-related receptors (ERRα and ERRβ) were discovered by Giguère et al. (20), who used reduced stringency hybridization based on the DNA-binding domain (C domain) of the ERα (20). Recently, a third ERR has been identified and termed ERRγ (21). These ERRs are orphan receptors whose endogenous ligands are unknown. It was first reported that expression of the ERRα gene produced a protein of 63 kDa (20), but subsequent investigation revealed that transcription is actually initiated at the second Met in the ERRα gene sequence to produce a human ERRα1 of 53 kDa (22, 23). Human ERRα1 binds as a monomer (23) or homodimer (24), recognizes the SF-1 response element (SFRE; 5′-TCAAGGTCA-3′), and binds the ERE (5′-AGGTCANNNTGACCT-3′). ERRα1 binding is not activated by 17β-estradiol and is not inhibited by the ER antagonist, ICI164,384. Furthermore, ERRα1 transcriptional activity requires a serum-derived component that is removed by charcoal treatment (24).
Recently, Kraus et al. (25) showed that ERRα1 competes with ERα for binding to the ERE and inhibits ERE-dependent transactivation in ER-positive MCF-7 cells, an effect that does not occur in ER-negative HeLa cells, suggesting that initiation of ERE-dependent gene expression depends on the ratio of ERRα1 to activated ERα in the cells. In view of the knowledge that estrogen activates eNOS, as well as the cross-talk between the ERα and ERRα1, we investigated the relationship between ERRα1 and eNOS expression in endothelial cells. We report here that ERRα1 activates eNOS by means of up-regulation of eNOS mRNA and protein expression.
Materials and Methods
Plasmid Construction. The human ERRα1 was amplified from total RNA extracted from HeLa cells by RT-PCR. RNA was isolated by methods using TRIzol reagents (Invitrogen) and 1.0 μg was reverse-transcribed for 1 h at 42°C with the Advantage RT-for-PCR Kit (Clontech). Amplification of this cDNA was performed with the Advantage HF-2 PCR Kit (Clontech), using sense primer 5′-TTGGATCCGACCAGCGCCATGTCCAGCCAG-3′ and antisense primer 5′-GCCGAATTCCTTGCCTCAGTCCATCATGGC-3′(16). Amplified cDNA was cloned into pCR4-TOPO (Invitrogen). The sequenced ERRα1 cDNA was subcloned into pcDNA3.1(+) (Invitrogen) to construct ERRα1/pcDNA3.1(+) for high expression of ERRα1 in transfected cells. To confirm ERRα1 expression, we constructed ERRα1/pCMV-Tag4A cDNA to express an ERRα1-FLAG fusion protein. To construct ERRα1/pCMV-Tag4A cDNA, we used the XhoI restriction enzyme site in ERRα1 and pCMV-Tag4A (Stratagene). 3×ERE-TATA-luciferase cDNA was a kind gift from D. McDonnell (Duke University Medical Center, Durham, NC; ref. 26). Four eNOS promoter-fused luciferase cDNAs (pGL2 -1193/+109, pGL2 -1001/+109, pGL2 -743/ +109, and pGL2 -265/+109) were kind gifts from P. Marsden (University of Toronto, Toronto; ref. 27). To construct mutated eNOS promoter fused luciferase cDNAs (-803:A→C and -591:A→C) from pGL2 -1193/+109, we used the Gene-Tailor Site-Directed Mutagenesis System and Platinum Taq DNA Polymerase High Fidelity (Invitrogen). PCR was performed with the following primers: -803 sense primer 5′-CATGCTCCCACCAGGGCATCCAGCTCTTCCCT-3′ and antisense primer 5′-GATGCCCTGGTGGGAGCATGGGGGACTAGG-3′, and -591 sense primer 5′-GATACCCTAATGTCAGACTCCAGGACAAAAAG-3′ and antisense primer 5′-GAGTCTGACAT TAGGGTATCCCT TCCCCTC-3′. We prepared cDNA so that both sites were mutated with -803:A→C in the eNOS promoter fused with luciferase cDNA. These mutated cDNAs were confirmed by DNA sequencing.
Cell Culture. COS-7 cells were obtained from the American Type Culture Collection. Bovine pulmonary artery endothelial cells (BPAEC) were obtained from Cambrex (Santa Rosa, CA). BPAEC were used between passages 4 and 10. Cells were cultured at 37°C in a humidified atmosphere of 5% CO2 by using DMEM containing 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. For the serum test, BPAEC were maintained for 1 week in phenol red-free DMEM containing 10% charcoal-stripped FCS.
Transient Transfection and Luciferase Assay. To confirm ERRα1-FLAG fusion protein expression, ERRα1/pCMV-Tag4A cDNA was transfected into COS-7 cells and BPAEC. The day before transfection, 4 × 105 COS-7 cells or 5 × 105 BPAEC were seeded in six-well plates. Transfections were performed with Polyfect (Qiagen, Valencia, CA) for COS-7 cells and Effectene (Qiagen) for BPAEC according to the manufacturer's protocol. COS-7 cells were transfected with 0.5 and 1 μg of cDNA. BPAEC were transfected with 0.2 and 0.4 μg of cDNA. Cells were lysed 24 and 48 h after transfection in buffer (10 mM Tris, pH 7.4/1% SDS/1 mM sodium orthovanadate) for Western blotting.
To investigate ERRα1 transactivation activity, 2 × 105 COS-7 cells or 2.5 × 105 BPAEC were seeded in 12-well plates and incubated for 24 h. Transfections were performed as above. COS-7 cells were transfected with 1 μg of reporter cDNA, 0.1 μg of ERRα1 cDNA, and 10 ng of β-galactosidase (β-gal) cDNA. BPAEC were transfected with 0.3 μg of reporter cDNA, 30 ng of ERRα1 cDNA, and 30 ng of β-gal cDNA. Transfected cells were washed with PBS and lysed with luciferase assay system lysis buffer. Luciferase activity was measured in cellular extracts by using the luciferase assay system (Promega). To correct for transfection efficiency, luciferase activity was normalized to the corresponding β-gal activity, which was measured by the β-gal assay system (Promega).
Ribonuclease Protection Assay. Total RNA was isolated by methods using TRIzol reagents (Invitrogen). Twenty micrograms of RNA was dried by using a Speed-Vac (Savant) and stored at -80°C before use. Radiolabeled antisense RNA was synthesized by using the Riboprobe system (Promega) and [α-32P]CTP at 800 Ci/mmol (1 Ci = 37 GBq) (ICN). The antisense template for eNOS was from Cayman Chemical (Ann Arbor, MI). The template for β-actin was from Ambion (Austin, TX). Ribonuclease protection assay was performed by using the RPA-III Kit (Ambion), but RNase ONE (Promega) was used in place of the supplied RNase. Protected fragments were resolved on a 5% polyacrylamide:bisacrylamide (19:1) gel containing 25 mM Tris borate, 2.5 mM EDTA, and 8 M urea. Gels were dried and exposed to autoradiographic film. Band intensities were quantified by using NIH IMAGE software (http://rsb.info.nih.gov/nih-image), and levels of eNOS mRNA were normalized to β-actin band intensities.
Western Blotting. To detect ERRα1-FLAG fusion protein, 20 μg of protein were separated by SDS/10% PAGE. For eNOS protein expression, 10 μg of protein was separated by 7.5% SDS/7.5% PAGE. Gels were transferred to a PVDF-membrane and then placed in blocking solution [Tris-buffered saline (10 mM Tris, pH 8.0/150 mM NaCl), 0.05% Tween 20, and 5% nonfat milk] for 1 h. Blots were incubated for 1 h with anti-FLAG M2 monoclonal antibody (Sigma), anti-eNOS monoclonal antibody (BD Transduction Laboratories), or anti-actin monoclonal antibody (Chemicon), washed with Tris-buffered saline and 0.05% Tween 20, and incubated with horseradish peroxidase-conjugated secondary antibody. Bound IgG was visualized by using an enhanced chemiluminescence detection system (Pierce) according to the manufacturer's protocol. Band intensities were quantified with NIH IMAGE software.
eNOS Activity. eNOS activity was measured by monitoring the conversion of [3H]arginine to [3H]citrulline with the NOS Assay Kit (Cayman Chemical). Cells were washed with cold PBS, harvested by using a cell scraper, and collected by centrifugation. Cell pellets were resuspended in homogenization buffer (25 mM Tris, pH 7.4/1 mM EDTA/1 mM EGTA) and centrifuged at 12,000 rpm for 5 min at 4°C. The reactions were performed with 25 mM Tris (pH 7.4), 3 μM BH4, 1 μM FAD, 1 μM FMN, 1 mM NADPH (Sigma), 20 nCi/μl [3H]arginine, 600 μM CaCl2, and 0.1 μM calmodulin at room temperature for 30 min. The assays were terminated by the addition of stop buffer (50 mM Hepes, pH 5.5/5 mM EDTA) followed by the addition of 100 μl of equilibrated resin to remove the unconverted [3H]arginine. The eluates were transferred to a scintillation vial and scintillation fluid was added. Radioactivity was quantified by liquid scintillation spectrometry. Total protein concentrations were measured by Dc protein assay (Bio-Rad).
Statistical Analysis. Data were obtained from three or four separate experiments. Each value represents the mean ± SEM. Statistical significance was assessed by the Student t test, and differences between treatment groups were considered significant at P < 0.05.
Results and Discussion
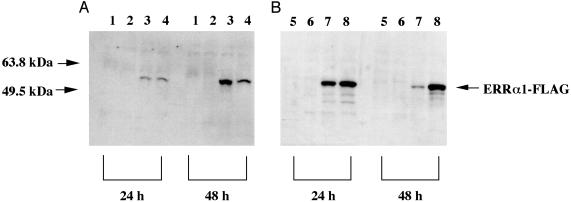
In studies designed to investigate the antiatherosclerotic effects of estrogen via the regulation of NO release from endothelial cells, it is well known that estrogen up-regulates eNOS expression (11). It has been reported that the ER is involved in this mechanism (11). ERRα1 was discovered by using a reduced-stringency hybridization based on the DNA-binding domain (C domain) of ERα (20). The sequence data show that ERRα1 shares high homology with ERα within the DNA-binding domain (C domain) (20). Based on these observations, our hypothesis is that ERRα1 binds to the eNOS promoter region and activates transcription, which results in increased eNOS expression and activity in endothelial cells. Confirmation of Expression of ERRα1 from ERRα1 cDNA. To confirm that the extracted ERRα1 cDNA gave rise to ERRα1 transcription and translation, and to discriminate exogenously expressed ERRα1 from endogenous ERRα1 in the cells, we transfected ERRα1/pCMV-Tag4A cDNA that can express ERRα1-FLAG fusion protein into COS-7 and BPAEC. Fig. 1 A (COS-7) and B (BPAEC) show that ERRα1-FLAG fusion protein transfection resulted in the expression a protein of the expected mass, visualized by using anti-FLAG monoclonal antibody in both cell types. This experiment confirmed that the transcription and translation of the ERRα1 cDNA resulted in expression of the ERRα1 protein.
Fig. 1.
ERRα1-FLAG fusion protein expression. Twenty micrograms of total protein was extracted 24 and 48 h after transfection of COS-7 cells (A) and BPAEC (B) and subjected to SDS/10% PAGE. Western blots were performed with anti-FLAG monoclonal antibody (10 μg/ml). Lane 1, 0.5 μg of pCMV-Tag4A-transfected; lane 2, 1.0 μg of pCMV-Tag4A-transfected; lane 3, 0.5 μgofERRα1/pCMV-Tag4A-transfected; lane 4, 1.0 μgofERRα1/pCMV-Tag4A-transfected; lane 5, 0.2 μg of pCMV-Tag4A-transfected; lane 6, 0.4 μg of pCMV-Tag4A-transfected; lane 7, 0.2 μg of ERRα1/pCMV-Tag4A-transfected; lane 8, 0.4 μg of ERRα1/pCMV-Tag4A-transfected.
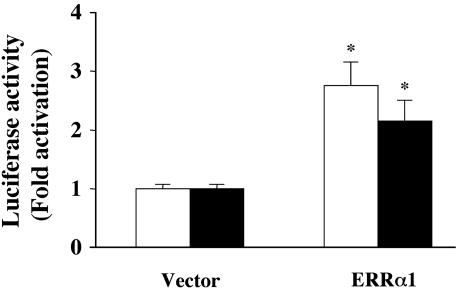
ERRα1 Activates ERE and eNOS Promoter-Dependent Luciferase Activity in COS-7 Cells. We examined whether the expression of ERRα1 activates ERE-dependent luciferase in COS-7 cells. As shown in Fig. 2, ERRα1 activated ERE-dependent luciferase 2.75-fold compared with vector [pcDNA3.1(+)] transfection. To investigate whether ERRα1 activates the eNOS promoter, we used a construct consisting of the eNOS promoter region (-1193/+109) fused to luciferase cDNA as a reporter gene. ERRα1 activated eNOS promoter-dependent luciferase (2.14-fold vs. vector transfection) in COS-7 cells.
Fig. 2.
Transcriptional activity of ERRα1 in COS-7 cells. ERRα1/pcDNA3.1(+) or pcDNA3.1(+) cDNA were transiently cotransfected with 3×ERE-luciferase (open bar) or eNOS promoter (-1193/+109) luciferase (filled bar) cDNA into COS-7 cells. Cells were cultured for 48 h. ERE and eNOS promoter-dependent luciferase activities were determined and plotted as fold activation over the individual luciferase activity with vector alone (*, P < 0.05 vs. vector).
With regard to eNOS and estrogen, it has been reported that estrogen up-regulates eNOS expression in fetal pulmonary artery endothelium (12), EA.hy926 cells (13), rat myocardium (28), and rat cerebral microvessels (29). In fetal pulmonary artery endothelium, estrogen stimulated ERE-dependent eNOS expression through the ER (12). In contrast, estrogen stimulated eNOS transcription through the transcription factor Sp1 in EA.hy926 cells (13). The present study represents the first demonstration that the ERRα1 activates eNOS promoter-dependent expression in COS-7 cells.
ERRα1-FLAG (ERRα1/pCMV-Tag4A transfection) did not activate ERE and eNOS promoter-dependent luciferase activity (data not shown). One explanation for this is that addition of the FLAG protein to the ERRα1 C-terminal may have inhibited ERRα1 transactivation. Another possibility is that an essential region was deleted to make the ERRα1-FLAG fusion protein or that the FLAG protein might disturb ERRα1 transactivation.
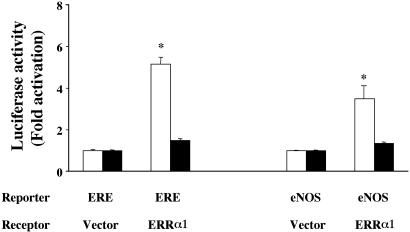
ERRα1 Activates the eNOS Promoter in BPAEC in a Serum-Dependent Manner. There are no previous reports that ERRα1 transactivates ERE and eNOS promoter-dependent luciferase in endothelial cells that express both ERα and ERβ. We transfected ERRα1/pcDNA3.1(+) cDNA and ERE luciferase or eNOS promoter-fused luciferase cDNA into BPAEC. As in the experiment using COS-7 cells, we observed that ERRα1 activates ERE (5.15-fold) and eNOS promoter-dependent luciferase (3.9-fold) (Fig. 3). The ER antagonist (ICI182, 780) did not inhibit this activation (data not shown). Kraus et al. (25) reported that ERRα1 down-regulates ERE-dependent luciferase activity in MCF-7 cells (ER-positive) and suggested that ERE-dependent gene expression depends on the ratio between ERRα1 expression and ligand-activated ERα in the cells. They showed that ERRα1 activates ERE-dependent luciferase activity in ER-negative HeLa cells. These data are consistent with our results in ER-negative COS-7 cells (Fig. 2). In endothelial cells, ERs are expressed, but their expression might be lower than that seen in MCF-7 cells. We believe that ERRα1 activates ERE-dependent luciferase activity in BPAEC because of the low ER expression in the cells. However, other explanations are possible.
Fig. 3.
Transcriptional activity of ERRα1 in BPAEC. ERRα1/pcDNA3.1(+) or pcDNA3.1(+) cDNA were transiently cotransfected with 3×ERE-luciferase or eNOS promoter (-1193/+109) luciferase cDNA into BPAEC. Transfected cells were cultured for 24 h in medium containing normal (open bars) or charcoal-stripped (filled bars) serum. ERE and eNOS promoter-dependent luciferase activities were determined and plotted as fold activation over the individual luciferase activity with vector alone (*, P < 0.05 vs. vector).
BPAEC were maintained in medium containing charcoal-stripped serum for 1 week to investigate whether ERRα1-dependent actions were mediated by unknown compounds in the serum. The ERE and eNOS promoter-dependent luciferase activities were not enhanced by ERRα1 transfection in BPAEC that had been treated with charcoal-stripped serum (Fig. 3). In addition, 17β-estradiol (10-8 M) did not activate luciferase in the charcoal-stripped serum (data not shown). Therefore, ERRα1 transactivates ERE and eNOS promoter-dependent luciferase in BPAEC and, as previously reported in ROS 17.2/8 cells (24), this activation is mediated by serum compounds that can be removed by charcoal treatment. The endogenous ligand for ERRα1, however, remains to be identified. Our results suggest that the ERRα1-mediated activation pathway in endothelial cells does not require estrogen or endogenous ER but does require serum factors that can be removed by charcoal treatment.
The ligand, either endogenous or synthetic, for ERRα1 has remained elusive. Furthermore, several investigators indicate that ERRα1 activates ERE-dependent luciferase activity in serum-free medium or charcoal-stripped serum (30, 31), suggesting that ERRα1 is constitutively active and does not require a ligand. However, ERRα1 shows little constitutive transactivation without cotransfection with the coactivator, proliferator-activated receptor γ coactivator 1. These observations suggest that ERRα1 requires this binding protein rather than a small lipophilic ligand for transactivation (32). Hong et al. (33) reported that ERR3 (ERRγ) transactivation does not require a ligand and that its activity depends on glucocorticoid receptor interacting protein (GRIP1) binding to ERRγ residues between 449 and 458 (murine), which are conserved hydrophobic residues (449-LF-450 and 453-ML-454). In the human ERRα1 amino acid sequence, these hydrophobic residues are highly conserved (412-KLFLEMLEAMMD-423).
The present data show that the ERRα1-FLAG fusion protein does not activate ERE and eNOS promoter-dependent luciferase activity compared with ERRα1 protein induced-transactivation (Figs. 2 and 3). Accordingly, we propose that GRIP1 may not be able to bind to the ERRα1 C-terminal region because the four-residue deletion (420-AMMD-423) leads to a failure to detect ERRα1-FLAG fusion protein-induced transactivational activity. However, we did not delete the highly conserved hydrophobic residues in the construction of the ERRα1-FLAG fusion protein. It appears that GRIP1 binding to the ERRα1 needs the additional residues around conserved sites, which we had deleted, or that binding is different between ERRα1 and ERRγ. GRIP1 binding to ERRγ leads to ligand-independent activation, as has been shown (33). Our data suggest that ERRα1-induced up-regulation of eNOS expression depends on serum compounds excluded by the charcoal.
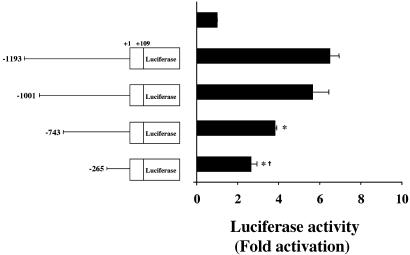
The eNOS Promoter Regions -1001+/-743, -743+/-265, and Downstream from -265 Are Necessary to Observe ERRα1-Dependent eNOS Promoter Activation. To determine the required domain for eNOS promoter activation by ERRα1, we used reporter genes consisting of four truncated eNOS promoters fused to luciferase cDNA. As shown in Fig. 4, -1001/+109 eNOS promoter-dependent luciferase activity did not show a significant decrease, but -743/+109 and -265/+109 eNOS promoter-dependent luciferase activities were decreased as compared with the -1193/ +109 eNOS promoter-dependent luciferase activity induced by ERRα1. In addition, the ERRα1-induced -265/+109 eNOS promoter-dependent luciferase activity was higher than transfection with vector only (2.5-fold activation). These results show that ERRα1-dependent eNOS promoter activation requires the regions -1001/-743, -743/-265, and downstream from -265 on the eNOS promoter.
Fig. 4.
ERRα1 transcriptional activity with truncated eNOS promoters in BPAEC. ERRα1/pcDNA3.1(+) cDNA was transiently cotransfected with four truncated eNOS promoter [-1193/+109, -1001/+109, -743/+109, or -265/+109] luciferase cDNAs into BPAEC. Cells were cultured for 24 h. eNOS promoter-dependent luciferase activities were determined and plotted as fold activation over the individual luciferase activity with vector alone (*, P < 0.05 vs. -1193/+109 eNOS promoter-dependent luciferase activity; †, P < 0.05 vs. -743/+109 eNOS promoter-dependent luciferase activity).
We propose that two steroidogenic factor 1 response element (SFRE)-like elements are necessary for the ERRα1 transactivation of the eNOS promoter, and these are located between -1001 and -743 (-805: 5′-TCAAGCTCT-3′: -796) and -743 and -265 (-593: 5′-TCAACCACA-3′: -585). Vanacker et al. (24) showed that ERRα1 could not bind to the mutated SFRE (-TCCAGGTCA-). Therefore, to confirm the exact region that is required for eNOS promoter activation, we mutated the predicted ERRα1-binding sites on the eNOS promoter (-803:A→C and -591:A→C; single and double mutants). By measuring luciferase activity, we could not detect a significant decrease by single or double mutations of the eNOS promoter (data not shown). These results suggest that ERRα1 binding may not require these two putative sites on the eNOS promoter. It remains to be determined where ERRα1 binds on the eNOS promoter. However, it is possible that ERRα1 activates the eNOS promoter through AP1 and Sp1. It has been shown that estrogen regulates insulin-like growth factor 1 gene transcription through the AP-1 motif (34) and cathepsin D gene expression through ER-Sp1 binding (35), and stimulates eNOS transcription through the transcription factor Sp1 in EA.hy926 cells (13).
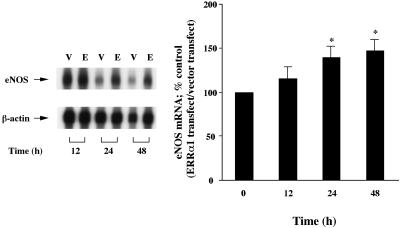
ERRα1 Up-Regulates Endogenous eNOS mRNA and Protein Expression in BPAEC. To investigate whether transfected ERRα1 can induce endogenous eNOS mRNA expression, we studied eNOS mRNA expression in ERRα1-transfected BPAEC by ribonuclease protection assay. As shown in Fig. 5, endogenous eNOS mRNA expression was up-regulated 24 h (1.39-fold) and 48 h (1.46-fold) after ERRα1 transfection.
Fig. 5.
ERRα1 up-regulates endogenous eNOS mRNA expression in BPAEC. ERRα1/pcDNA3.1(+) (E) or pcDNA3.1(+) (V) cDNA were transiently transfected into BPAEC. Transfected cells were cultured for 12, 24, and 48 h. Ribonuclease protection assay was performed with radiolabeled human eNOS antisense RNA. Band intensities were quantified by using nih image software and were normalized to β-actin band intensity. The percent control of eNOS mRNA was then calculated and further normalized to pcDNA3.1(+) (vector only control) [*, P < 0.05 vs. before transfection (0 h)].
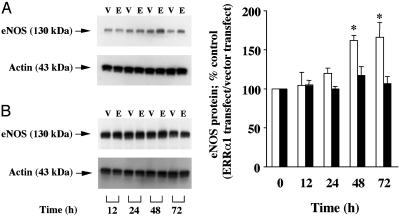
We also measured endogenous eNOS protein expression in ERRα1-transfected BPAEC. Fig. 6 shows that ERRα1 transfection significantly up-regulated eNOS protein expression 48 h (1.61-fold) and 72 h (1.65-fold) after ERRα1 transfection. We did not observe eNOS protein up-regulation induced by ERRα1 transfection in cells maintained in charcoal-stripped serum.
Fig. 6.
ERRα1 up-regulates endogenous eNOS protein expression in BPAEC. ERRα1/pcDNA3.1(+) (E) or pcDNA3.1(+) (V) cDNA were transiently transfected into BPAEC. Transfected cells were cultured for 12, 24, 48, and 72 h in medium containing normal serum (A and Right, open bars) or charcoal-stripped serum (B and Right, filled bars). Ten micrograms of total protein was extracted at each time point and separated by SDS/7.5% PAGE. Western blots were performed with anti-eNOS and anti-actin monoclonal antibodies. Each band intensity was quantified with nih image software and was normalized to the actin band intensity. The percent control of eNOS protein was then calculated and further normalized to pcDNA3.1(+) (vector only control) [*, P < 0.05 vs. before transfection (0 h)].
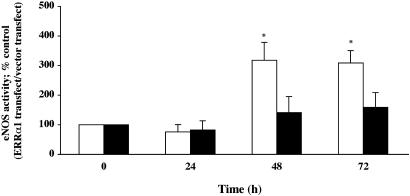
ERRα1 Activates Endogenous eNOS Activity in BPAEC. We measured eNOS activity in ERRα1-transfected BPAEC by using the [3H]arginine conversion assay. As shown in Fig. 7, eNOS activity was increased 48 h (3.17-fold) and 72 h (3.09-fold) after ERRα1 transfection in BPAEC. In a similar manner observed for eNOS protein expression, ERRα1 did not activate endogenous eNOS activity in BPAEC maintained in charcoal-stripped serum. Figs. 5, 6, 7 show that exogenously transfected ERRα1 regulates endogenous eNOS expression and activity. We observed that ERRα1 protein expression peaks ≈24 h after transfection. During this time period, expressed ERRα1 was found to bind to the eNOS promoter region, as was detected by luciferase assay. This may have resulted in the observed eNOS mRNA and protein up-regulation and activation.
Fig. 7.
ERRα1 increases eNOS activity in BPAEC. ERRα1/pcDNA3.1(+) or pcDNA3.1(+) cDNA was transiently transfected into BPAEC. Transfected cells were cultured for 24, 48, and 72 h in medium containing normal serum (open bars) or charcoal-stripped serum (filled bars). eNOS activity was measured by conversion of [3H]arginine to [3H]citrulline. The percent control of eNOS activity was calculated and normalized to pcDNA3.1(+) transfections [*, P < 0.05 vs. before transfection (0 h)].
It has been shown that ERRα1 can bind to DNA as a monomer (23) or homodimer (24) and that ERRα1 associates with ERα in vitro (23). It is proposed that ERRα1/ER heterodimers may exert different effects compared with ERRα1 monomers, ER homodimers, or ERα/ERβ heterodimers. In our BPAEC experiments, we have not examined the type of receptor complex that stimulates eNOS expression. However, there are many possibilities as to the components of this complex because BPAEC express ERα, ERβ, and ERRα1 (unpublished observations).
Until now, three human ERR subfamily members have been reported (ERRα, ERRβ, and ERRγ) that belong to the nuclear receptor superfamily. ERRα shares 63% and 61% amino acid identity in its ligand-binding domain and 91% and 93% identity in its DNA-binding domain, respectively, with ERRβ and ERRγ (20, 21). We propose that eNOS gene expression could be up-regulated not only by ERRα1 but also by ERRβ and ERRγ, owing to the close identity among the ERRα, ERRβ, and ERRγ DNA-binding domains.
Conclusions
Estrogen can affect endogenous ERα and ERβ expression in endothelial cells (16) and ERRα1 expression in the uterus (36). We have previously reported (11, 14) that estrogen up-regulates eNOS expression and stabilizes eNOS mRNA through the ER pathway in tumor necrosis factor α-stimulated bovine aortic endothelial cells. Adenovirus ERα transfection up-regulates endogenous eNOS expression and also inhibits basic fibroblast growth factor-induced endothelium migration, suggesting an atheroprotective effect (37). We have observed ERRα1 expression in endothelial cells (unpublished observations), and ERRα1 may perform a similar function to estrogen–ER modulation of eNOS expression in endothelial cells. Therefore, ERRα1 might also elicit antiatherosclerotic effects through the modulation of NO production.
In summary, ERRα1 transactivates ERE- and eNOS-dependent luciferase activity in COS-7 cells and BPAEC. This eNOS promoter activation by ERRα1 depends on three sites in the eNOS promoter region and the presence of serum compounds that are extracted by charcoal treatment. Furthermore, ERRα1 up-regulates endogenous eNOS mRNA and protein expression and stimulates eNOS activity. These results provide further evidence for regulation of endothelium-derived NO through the interaction of eNOS and nuclear receptors in the cardiovascular system. This pathway may account, at least in part, for the antiatherosclerotic actions of NO.
Acknowledgments
We thank Dr. D. McDonnell for kindly providing the 3×ERE-TATA-luciferase cDNA and Dr. P. Marsden for kindly providing the four eNOS promoter-fused luciferase cDNAs (pGL2 -1193/+109, pGL2 -1001/ +109, pGL2 -743/+109, and pGL2 -265/+109). We are especially grateful to Drs. Linda Connelly and Aaron Jacobs for technical assistance and helpful discussion and comments during the preparation of the manuscript. This work was supported by the Japan Foundation for Aging and Health and National Institute of Health Grants HL 58433 and HL 66999.
Abbreviations: BPAEC, bovine pulmonary artery endothelial cells; eNOS, endothelial NO synthase; ER, estrogen receptor; ERE, estrogen response element; ERR, estrogen-related receptor.
References
- 1.Ignarro, L. J., Buga, G. M., Wood, K. S., Byrns, R. E. & Chaudhuri, G. (1987) Proc. Natl. Acad. Sci. USA 84, 9265-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radomski, M. W., Palmer, R. M. & Moncada, S. (1990) Proc. Natl. Acad. Sci. USA 87, 5193-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith, O. W. & Stuehr, D. J. (1995) Annu. Rev. Physiol. 57, 707-736. [DOI] [PubMed] [Google Scholar]
- 4.Kullo, I. J., Mozes, G., Schwartz, R. S., Gloviczki, P., Crotty, T. B., Barber, D. A., Katusic, Z. S. & O'Brien, T. (1997) Circulation 96, 2254-2261. [DOI] [PubMed] [Google Scholar]
- 5.Cayatte, A. L., Palacino, J. J., Horten, K. & Cohen, R. A. (1994) Arterioscler. Thromb. 14, 753-759. [DOI] [PubMed] [Google Scholar]
- 6.Thakur, N. K., Hayashi, T., Sumi, D., Kano, H., Matsui-Hirai, H., Tsunekawa, T. & Iguchi, A. (2002) J. Cardiovasc. Pharmacol. 39, 298-309. [DOI] [PubMed] [Google Scholar]
- 7.Nathan, L., Pervin, S., Singh, R., Rosenfeld, M. & Chaudhuri, G. (1999) Circ. Res. 85, 377-385. [DOI] [PubMed] [Google Scholar]
- 8.Cid, M. C., Schnaper H. W. & Kleinman, H. K. (2002) Ann. N.Y. Acad. Sci. 966, 143-157. [DOI] [PubMed] [Google Scholar]
- 9.Gruchow, H. W., Anderson, A. J., Barboriak, J. J. & Sobocinski, K. A. (1988) Am. Heart J. 115, 954-963. [DOI] [PubMed] [Google Scholar]
- 10.Simoncini, T., Hafezi-Moghadam, A., Brazil, D. P., Ley, K., Chin, W. W. & Liao, J. K. (2000) Nature 407, 538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi, T., Yamada, K., Esaki, T., Kuzuya, M., Satake, S., Ishikawa, T., Hidaka, H. & Iguchi, A. (1995) Biochem. Biophys. Res. Commun. 214, 847-855. [DOI] [PubMed] [Google Scholar]
- 12.MacRitchie, A. N., Jun, S. S., Chen, Z., German, Z., Yuhanna, I. S., Sherman, T. S. & Shaul, P. W. (1997) Circ. Res. 81, 355-362. [DOI] [PubMed] [Google Scholar]
- 13.Kleinert, H., Wallerath, T., Euchenhofer, C., Ihrig-Biedert, I., Li, H. & Förstermann, U. (1998) Hypertension 31, 582-588. [DOI] [PubMed] [Google Scholar]
- 14.Sumi, D., Hayashi, T., Jayachandran, M. & Iguchi, A. (2001) Life Sci. 69, 1651-1660. [DOI] [PubMed] [Google Scholar]
- 15.Kim-Schulze, S., McGowan, K. A., Hubchak, S. C., Cid, M. C., Martin, M. B., Kleinman, H. K., Greene, G. L. & Schnaper, H. W. (1996) Circulation 94, 1402-1407. [DOI] [PubMed] [Google Scholar]
- 16.Ihionkhan, C. E., Chambliss, K. L., Gibson, L. L., Hahner, L. D., Mendelsohn, M. E. & Shaul, P. W. (2002) Circ. Res. 91, 814-820. [DOI] [PubMed] [Google Scholar]
- 17.Chen, Z., Yuhanna, I. S., Galcheva-Gargova, Z., Karas, R. H., Mendelsohn, M. E. & Shaul, P. W. (1999) J. Clin. Invest. 103, 401-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes, M. P., Li, L., Russell, K. S. & Bender, J. R. (2002) Vascul. Pharmacol. 38, 99-108. [DOI] [PubMed] [Google Scholar]
- 19.Razandi, M., Alton, G., Pedram, A., Ghonshani, S., Webb, P. & Levin, E. R. (2003) Mol. Cell. Biol. 23, 1633-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giguère, V., Yang, N., Segui, P. & Evans, R. M. (1988) Nature 331, 91-94. [DOI] [PubMed] [Google Scholar]
- 21.Heard, D. J., Norby, P. L., Holloway, J. & Vissing, H. (2000) Mol. Endocrinol. 14, 382-392. [DOI] [PubMed] [Google Scholar]
- 22.Yang, N., Shigeta, H., Shi, H. & Teng, C. T. (1996) J. Biol. Chem. 271, 5795-5804. [DOI] [PubMed] [Google Scholar]
- 23.Johnston, S. D., Liu, X., Zuo, F., Eisenbraun, T. L., Wiley, S. R., Kraus, R. J. & Mertz, J. E. (1997) Mol. Endocrinol. 11, 342-352. [DOI] [PubMed] [Google Scholar]
- 24.Vanacker, J. M., Bonnelye, E., Chopin-Delannoy, S., Delmarre, C., Cavaillčs, V. & Laudet, V. (1999) Mol. Endocrinol. 13, 764-773. [DOI] [PubMed] [Google Scholar]
- 25.Kraus, R. J., Ariazi, E. A., Farraell, M. L. & Mertz, J. E. (2002) J. Biol. Chem. 277, 24826-24834. [DOI] [PubMed] [Google Scholar]
- 26.Tzukerman, M. T., Esty, A., Santiso-Mere, D., Danielian, P., Parker, M. G., Stein, R. B., Pike, J. W. & McDonnell, D. P. (1994) Mol. Endocrinol. 8, 21-30. [DOI] [PubMed] [Google Scholar]
- 27.Karantzoulis-Fegaras, F., Antoniou, H., Lai, S. L. M., Kulkarni, G., D'Abreo, C., Wong, G. K. T., Miller, T. L., Chan, Y., Atkins, J., Wang, Y. & Marsden, P. A. (1999) J. Biol. Chem. 274, 3076-3093. [DOI] [PubMed] [Google Scholar]
- 28.Nuedling, S., Kahlert, S., Loebbert, K., Doevendans, P. A., Meyer, R., Vetter, H. & Grohè, C. (1999) Cardiovasc. Res. 43, 666-674. [DOI] [PubMed] [Google Scholar]
- 29.Mcneill, A. M., Kim, N., Duckles, S. P. & Krause, D. N. (1999) Stroke 30, 2186-2190. [DOI] [PubMed] [Google Scholar]
- 30.Xie, W., Hong, H., Yang, N. N., Lin, R. J., Simon, C. M., Stallcup, M. R. & Evans, R. M. (1999) Mol. Endocrinol. 13, 2151-2162. [DOI] [PubMed] [Google Scholar]
- 31.Tremblay, G. B., Kunath, T., Bergeron, D., Lapointe, L., Champigny, C., Bader, J. A., Rossant, J. & Giguère, V. (2001) Genes Dev. 15, 833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber, S. N., Knutti, D., Brogli, K., Uhlmann, T. & Kralli, A. (2003) J. Biol. Chem. 278, 9013-9018. [DOI] [PubMed] [Google Scholar]
- 33.Hong, H., Yang, L. & Stallcup, M. R. (1999) J. Biol. Chem. 274, 22618-22626. [DOI] [PubMed] [Google Scholar]
- 34.Umeyama, Y., Kawamori, R., Watada, H., Imano, E., Iwama, N., Morishima, T., Yamasaki, Y., Kajimoto, Y. & Kamada, T. (1994) J. Biol. Chem. 269, 16433-16442. [PubMed] [Google Scholar]
- 35.Krishnan, V., Wang, X. & Safe, S. (1994) J. Biol. Chem. 269, 15912-15917. [PubMed] [Google Scholar]
- 36.Shigeta, H., Zuo, W., Yang, N., DiAugustine, R. & Teng, C. T. (1997) J. Mol. Endocrinol. 19, 299-309. [DOI] [PubMed] [Google Scholar]
- 37.Tan, E., Gurjar, M. V., Sharma, R. V. & Bhalla, R. C. (1999) Cardiovasc. Res. 43, 788-797. [DOI] [PubMed] [Google Scholar]