“If there is any area in which a network thinking could trigger a revolution, I believe that biology is it.”
—Albert László Barabási1
Traditional biological and biochemical studies deal with relatively few components, allowing intuitive reasoning to guide hypotheses and experiments. For example, a popular experimental protocol in cardiovascular research is to examine gene functions through the use of transgenic or gene-targeted mice. Although clearly informative, it is becoming increasingly clear that such studies alone are not sufficient to explain complex processes such as arrhythmias, heart failure, and atherogenesis. Even a basic behavior, the action potential of a cardiomyocyte, requires the coordinated actions of >20 different ion transporters and channels.2 Perturbing proteins individually will help establish their functions, but it will not provide a full understanding of how they function together (quantitatively, temporally, and spatially). For this, a more global analysis, in which the activities of all of the relevant proteins are tracked over time and then integrated into a quantitative mathematical model, is required to provide a deeper level of understanding of cardiomyocyte dynamics.
A new branch of biology, called systems biology, seeks to identify the components of complex systems and to model their dynamic interactions.3 The approach arose in large part as a result of new technical and analytical developments. The Human Genome Project provided a biological “parts list,” and technologies such as massively parallel DNA sequencing, expression microarrays, and tandem mass spectrometric analyses of proteins and metabolites have made high-throughput analysis of biological systems feasible. To deal with the data explosion, novel statistical and other mathematical modeling approaches are being developed.
Basically, systems-based approaches involve 4 steps. The first is to define the system to be examined (eg, a cardiomyocyte). The second is to identify the components of the system (eg, the set of proteins regulating a property of interest). The third is to determine how the components interact with each other. This can be done experimentally or may be based on the published literature, and the set of components and their interactions is called a network. The fourth is to model the dynamics of the network mathematically (ie, how it changes over time or responds to various perturbations).
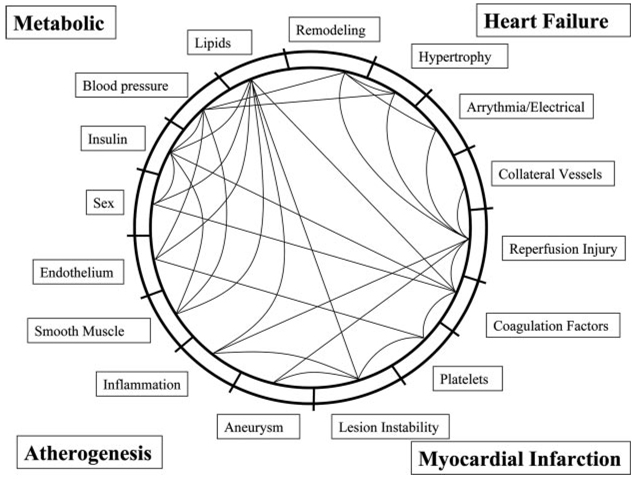
Given the complexity of the cardiovascular system and of cardiovascular diseases (Figure 1),4,5 systems-based approaches are likely to play an increasingly important role in elucidating the higher-order interactions underlying traits such as atherosclerosis, cardiac hypertrophy, heart failure, and arrhythmias. These approaches should have translational value in giving biological context to the multitude of genetic variants reported to be associated with disease and in providing a framework for the development of pharmacological treatments. Here, we review progress relevant to cardiovascular networks and their dynamics. We organized our review according to the different approaches used to model networks. In each section, we include examples relevant to cardiovascular biology and disease.
Figure 1.
Graph of the components (clinical traits) and their interactions (connecting lines) in cardiovascular disease. This crude network is based on the results of clinical studies and experimental models. Data derived from refs. 4, 5.
General Properties of Biological Networks
One of the central concepts in systems biology is that networks, rather than classic linear pathways, underlie biological processes. The concept of biological networks arose when classic metabolic pathways were represented as graphs in which the components (ie, metabolites) were called nodes and their interactions (ie, enzymatic steps converting one metabolite to another) were called links or edges. It then became clear that the overall structure of metabolic pathways was much more interconnected and redundant than previously recognized.6 Biological networks occur on many different levels such as genes, transcripts, proteins, metabolites, organelles, cells, organs, organisms, and social systems. In general, they appear to exhibit an architecture described mathematically as “scale free,” in which most nodes have few links but a small fraction of nodes (called hubs) are highly interconnected. This architecture arises naturally in systems that evolve under selective pressures where nodes randomly interact and form links with other nodes. The system grows over time by adding new nodes and links, and new links to already highly connected nodes are favored (preferential attachment, or the principle that “the richer get richer”).7 Scale-free architecture does not require but is fully compatible with a hierarchal modular organization of components,6,8 which is typical for biological systems (eg, the organization of metabolism into various modules as shown in Figure 2A).9–11 Thus, local groups of sparsely linked nodes form modules that are linked to other modules through their corresponding hub nodes. The challenge of systems biology is to construct detailed biological networks for each level, from the gene to the organism, and then to connect the levels by integrating orthogonal data sets.
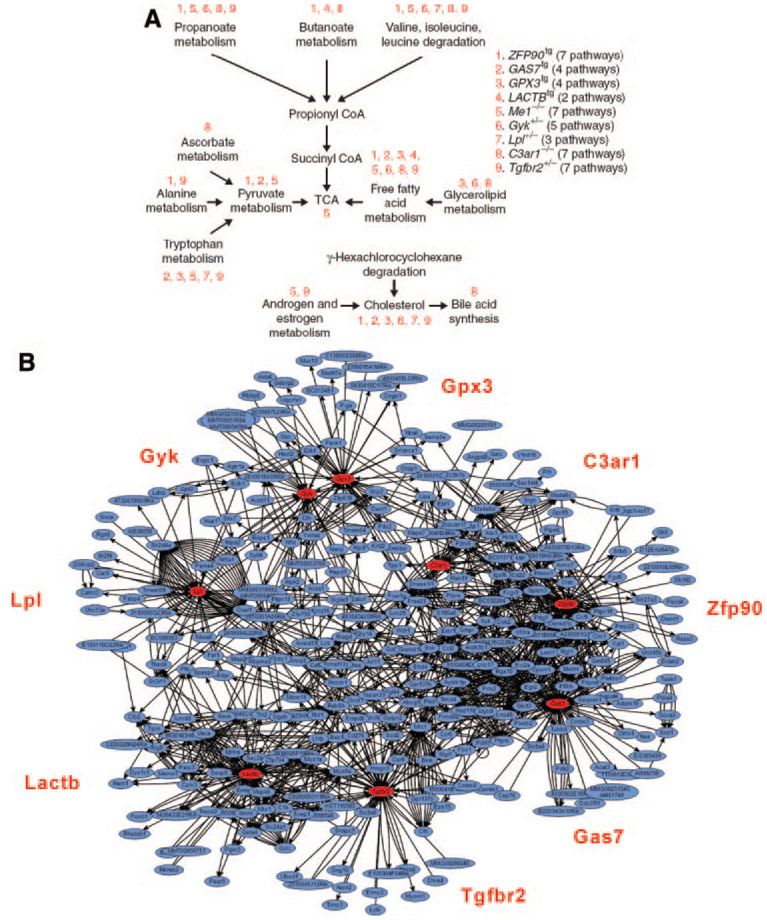
Figure 2.
Networks contributing to adiposity in mice. A, A set of curated pathways was shown to be enriched in genes differentially expressed between livers of fat and lean mice in a segregating mouse population.9 Note that these pathways tend to center on the tricarboxylic acid cycle, which is central in energy production. Through the use of systems genetics, a set of genes (labeled 1 through 9) were predicted to be causally related to adiposity in mice.10 Their effects on adiposity were validated with transgenic approaches, and expression array analyses of the transgenic mice showed that the differentially regulated genes were enriched in many of the same tricarboxylic acid–centered pathways (indicated by numbering above pathways).11 B, Systems genetics approaches were used to construct a directed “bayesian” coexpression network model based on global expression array analysis of segregating populations of mice. One subnetwork, or module, was significantly associated with adiposity. Notably, the genes causally involved in adiposity (above) correspond to hubs in the network. Data derived from refs. 9, 11.
Networks exhibit several key features that make them well suited for systems evolving in response to selective pressures such as biological systems operating through random mutation and natural selection. For self-organizing systems, properties such as adaptability and robustness are just as important as efficiency, and the redundancy of pathways in a network intuitively lends itself to adaptability and robustness more so than a purely linear pathway. Although redundancy might seem to compromise efficiency, networks overcome this limitation through their “small-world” effects.12 As an illustration, consider an Escherichia coli using glycolysis to metabolize glucose to pyruvate to generate ATP, which suddenly finds itself exposed to a different substrate such as alanine (Figure 2A). If the E coli metabolism had a linear architecture in which alanine was far removed from glucose, then it could be energetically costly (and hence costly to survival) to interconvert a whole array of intermediate metabolites to synthesize glucose to produce ATP. In a highly interconnected network, however, enzymatic pathways exist to convert the alanine to a hub metabolite, which in turn is converted to the hub metabolite (pyruvate) in the glycolysis module, which is then metabolized to generate ATP. Thus, the energetic cost to the E coli is minimized as a result of the “short-circuiting” effect of the highly interconnected hub nodes in a small-world network. This is analogous to airport networks in which airplanes making many stops to fly the geographically shortest route between 2 small towns may take much longer than flying from the small town to a large hub city (eg, Chicago) and then backtracking to the final destination, even though the total distance traversed is greater. Another example is the “6 degrees of separation” in social networks.12
Networks are inherently robust because there are multiple alternative pathways to get from one node to another; in a typical scale-free network, up to 80% of the links can be randomly destroyed before catastrophic network failure occurs. 13 The redundancy of pathways also makes networks adaptable to changing environmental conditions, as illustrated by the E coli example above. Most critical for evolutionary adaptability, however, is the feature of “emergent properties,” which arise when the interactions between the nodes in a network are nonlinear. Unlike linear systems, in which the whole is equal to the sum of the parts, nonlinear interactions can create a remarkable new set of collective behaviors that make the whole much greater than the sum of the parts. The most fundamental emergent property in biology, the self-oscillation underlying the cell cycle, could not exist without nonlinear interactions between subcellular components in the biological network of the cell. Other examples of emergent behaviors include developmental morphogenesis (pattern formation), circadian rhythms, excitability, and cardiac pacemaking.
From an evolutionary perspective, emergent behaviors provide a rich source of qualitatively new behaviors that can potentially confer a survival advantage to a biological system. Thus, it is not surprising that the logic of biological networks is typically nonlinear or that biological systems exhibit properties that may not be apparent until viewed as a whole. A key corollary is that pure reductionist approaches can provide us with a detailed parts list but not the whole view unless combined with integrative approaches.
With this general background, we now describe examples of different approaches being used to model biological networks relevant to cardiovascular biology and disease.
Networks Based on Prior Knowledge
The data in the published literature can be mined through the use of known associations (such as those with diseases or defined pathways) or simply correlations across data sets (such as cocitation, phylogenetic profiling, coexpression, and sequence similarity), allowing the modeling of functional networks. Numerous vast databases derived from high-throughput technologies such as the Gene Expression Omnibus, 14 the dbEST database at the National Center for Biotechnology Information, and GeneNetwork (www.genenetwork.org) are freely available. There are also systematic phenotyping projects such as the rat cardiovascular phenotyping project at the Medical College of Wisconsin.15 Such modeling can help reveal functions of genes about which little is known and can help identify genes underlying diseases.16 For example, Figure 1 summarizes published interactions in cardiovascular diseases derived from clinical and experimental databases.4,5 Our understanding of cardiovascular pathology is far from complete, so the network model is very imperfect, but the tremendous complexity of cardiovascular diseases is apparent. The goal of the studies described below is to create biological networks and to map them onto the disease network.
Systems-based approaches frequently rely on technologies capable of broadly interrogating the components of a system such as DNA sequencing (genetic variation), expression arrays (transcript levels), tandem mass spectrometry (protein and metabolite levels), or Chip-chip and Chip-seq (DNA–transcription factor interactions). The result is generally a long list of components, perhaps with some differences between samples, but generally lacking any unifying biological theme. The challenge is to extract meaning from such lists. A number of approaches have been developed to determine whether such lists are enriched for known pathways.16
For example, Ashley et al17 sought to identify networks involved in restenosis after a percutaneous coronary intervention. They examined 89 patients who underwent cardiac atherectomy for de novo atherosclerosis (n=55) or in-stent restenosis (n=34). Whole-genome gene expression profiling was then performed to examine the pathological samples, and the genes from the array were ordered in a rank list according to their differential expression between the classes. To construct an association network, the authors used text mining of Medline abstracts. Any 2 genes were considered an interaction verb if they appeared in the same sentence. In this way, certain highly connected genes, called nexus genes, were identified and proposed as candidates for involvement in restenosis.
A related method is gene set enrichment analysis. The gene sets are defined on the basis of prior biological knowledge, primarily published information. More than 1000 such lists have now been compiled and are publicly available.18 Using a simple statistical test such as the Kolmogorov-Smirnov test or Fisher exact test, one can ask if the list is enriched for any such gene sets. An early example of the use of this approach is a study by Mootha et al,19 who analyzed data obtained from muscle biopsies of diabetics compared with healthy control subjects. The results did not reveal any single genes that differed significantly in expression, but they did show that genes involved in oxidative phosphorylation exhibited reduced expression in diabetics when taken as a pathway or a gene set. A similar approach was used to identify pathways contributing to differences in obesity in randomized populations of mice.9 Progeny from an intercross between 2 different inbred strains, C57BL/6J and DBA/2J, were studied for obesity-related traits and for global expression in liver. With the gene set enrichment analysis, 13 annotated metabolic pathways were found to be significantly enriched among genes with an expression that was associated with obesity (Figure 2). Interestingly, all of these pathways centered on the tricarboxylic acid cycle, and recent transgenic studies have validated some of these findings.11
Networks Based on Physical Interactions
An intuitive criterion for analyzing network structure is that of physical proximity. Components situated near one another are more likely to exhibit functional connections than are distant components. For example, many proteins mediate their biological functions through protein interactions, including aspects such as signaling, regulation of gene expression, immunity, and molecular machines. Such networks can be based on literature such as the Human Protein Reference Database20 or on unbiased experimental studies. A variety of high-throughput methods have been used to construct protein interaction networks, including global yeast 2 hybrid analysis, tandem affinity purification/mass spectrometry, and protein arrays. Various methods for the prediction of protein-protein interactions have also been developed (eg, from coevolution events). Such methods have been applied in most detail to yeast, but a variety of organisms, including flies, worms, and mammalian cells, have been examined.21–23 The first draft of the human interaction map (interactome) comprises >70 000 predicted physical interactions between 6231 proteins.24 In interaction networks, the individual proteins are nodes, and the interactions connecting 2 proteins are links.
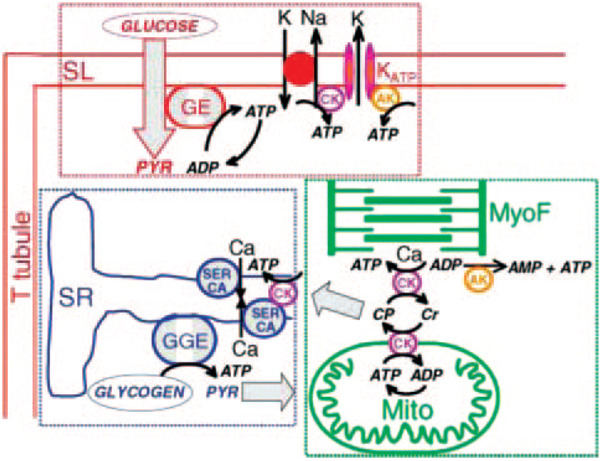
Another type of physical network can be constructed from functional interactions, eg, the modular compartmentalization of energy-generating systems in a cardiomyocyte (reviewed elsewhere25). These modules include glycolytic enzymes, glycogenolytic enzymes, and oxidative phosphorylation, which appear to be spatially distributed to optimize ATP delivery to specific ATPases (Figure 3). Glycolysis, which generates ATP through oxidation of glucose, preferentially serves energy channeling to the sarcolemma, where glucose is transported into the cell. Glycogenolysis appears to preferentially serve the sarcoplasmic reticulum to energize calcium cycling. Oxidative phosphorylation occurs in the mitochondria, channeling ATP to the myofilaments and throughout the cytoplasm. This is aided by the creatine kinase and adenylate kinase systems. Although the vast majority of energy is generated by oxidative phosphorylation, the glycolytic and glycogenolytic systems are low-capacity but high-specificity modules of the integrated metabolic network of a cardiomyocyte. There has been recent progress in identifying the protein components of mammalian organelles (eg, see the work by Foster et al26), and an important goal is to integrate proteomic networks with organelle networks.
Figure 3.
A spatially distributed, dynamic metabolic energy network in a cardiomyocyte. Energy modules include glycolytic enzymes (GE) associated with sarcolemma (SL) ATPases in red, glycogenolytic enzymes (GGE) associated with sarcoplasmic reticulum (SR) Ca ATPases (SERCA) in blue, and oxidative phosphorylation in mitochondria (Mito) associated with myofilament (MyoF) ATPases in green. Energy-distributing modules include creatine kinase (CK) in purple and adenylate kinase (AK) in orange. CP indicates creatine phosphate; Cr, creatine; KATP, ATP-sensitive K channel; and PRY, pyruvate.
Networks Based on Experimental Perturbations
Most biological studies involve some kind of perturbation such as a chemical treatment or genetic alteration, followed by analysis of the resulting effects. Conclusions about the causal interactions can then be drawn. For example, the function of a gene can be defined by “knockdown” in tissue culture using treatment with siRNA or by “knockout” in mice using gene targeting. In such experiments, whatever changes are observed are clearly the result of the single perturbation. But such experiments alone are not very useful for constructing gene networks because the components being analyzed have only 2 states (ie, wild type or knockout). Thus, to determine how the various components interact with each other, a series of single perturbations is required. For example, one could analyze a series of mouse knockouts or transgenics affecting overlapping pathways. The resulting changes in transcript levels or protein levels or activities could then be mathematically modeled as a network. Another kind of experiment involves the use of multiple perturbations. For example, natural populations exhibit functional variations (such as in gene expression) in hundreds or thousands of genes. Thus, one could examine the status of components in different individuals in the population and construct networks based on correlations between the components. In contrast to single perturbations, such studies have clear advantages for the construction of biological networks. Below, we discuss 2 examples of each.
Inflammation and macrophage activation are clearly implicated in atherogenesis, and a recent study by Ramsey and coworkers27 explored the macrophage transcriptional network mediated by Toll-like receptors (TLR) using a series of single gene perturbations. TLR recognize a variety of pathogen-associated molecules and certain endogenous ligands through adaptor molecules such as TRIF and Myd88 and then parallel crosstalking signaling pathways. In the case of macrophage TLR4, when stimulated with lipopolysaccharide, these activated pathways initiate a program leading to the differential expression of >1000 genes, including hundreds of transcription factors.28 Although these differentially expressed genes are known, the network of these interactions has proved difficult to address with traditional biochemistry. The authors combined 2 types of data to explain the network. First, they performed computational scanning of promoter sequences of clusters of coexpressed genes for known TF binding sites. Second, they used expression dynamics, modeling time course expression data to best fit the expression of a TF with potential target genes. The whole-genome expression array analyses were performed on primary bone marrow macrophages from 5 strains of mice (1 wild-type and 4 targeted strains) treated with 6 different TLR agonists at multiple time points between 0 and 48 hours. In all, 95 different combinations of strains, stimuli, and elapsed times were measured. The set of differentially expressed genes was clustered; promoter sequences of each gene were scanned for TF binding sites; and the temporal patterns of expression of TF and targeted genes were compared to identify potential causal influences. When integrated, the results provided a broad picture of the dynamic transcriptional program of the TLR network. The general approach seems applicable to other mammalian systems for which time course data are available.
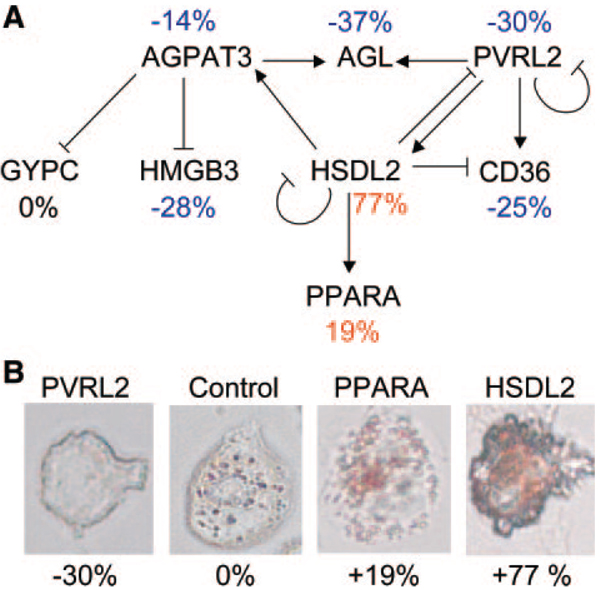
Skogsberg and colleagues29 used a series of single gene perturbations to create a network of genes mediating the development of advanced atherosclerotic lesions in mice. Briefly, they followed the development of atherosclerotic lesions in low-density lipoprotein receptor–null mice (a model of familial hypercholesterolemia) and observed a gradual initial growth phase, followed by an accelerated phase of rapid foam cell development and finally a plateau phase. Using a genetic switch that blocked secretion of lipoproteins to rapidly lower cholesterol, they observed that the switch blocked the development of advanced lesions if performed before the expansion phase and that this block was associated with altered expression of 37 genes, some of which had previously been associated with foam cell formation (CD36, PPARA). They then used siRNA knockdown of a subset of these genes to construct a network of cholesterol-responsive atherosclerosis target genes. For this, the authors examined expression patterns and cholesterol ester in a macrophage cell line (THP-1) after treatment with acetylated low-density lipoprotein in the presence or absence of siRNA to one of the candidate genes. Computational modeling of the expression data30 revealed a directed network of 8 genes involved in foam cell formation (Figure 4).
Figure 4.
A regulatory network for foam cell formation based on in vivo expression analyses in mice followed by perturbation in vitro. A, The above genes were found to be differentially regulated in the response to cholesterol lowering in atherosclerotic lesions in mice. They were then subjected to in vitro siRNA “knockdown” studies in a macrophage cell line. The predicted interactions (edges) are indicated, along with the percent change in cholesterol accumulation after cholesterol loading. Blue indicates an inhibition of cholesterol loading; red, an increase; and black, no change. B, Representative siRNA-treated cells are shown after 48 hours of incubation with acetylated low-density lipoprotein and staining for neutral lipids with Oil Red O. AGPAT3 indicates 1-acylglycerol-3-phosphate O-acyltransferase 3; AGL, amylo-1,6-glucosidase, 4-α-glucanotransferase; PVRL2, poliovirus receptor-related 2; GYPC, glycophorin C; HMGB3, high-mobility group box 3; HSDL2, hydroxysteroid dehydrogenase-like 2; CD36, CD36 molecule; PPARA, peroxisome proliferator-activated receptor-α. Reproduced from Skogsberg et al29 under the Creative Commons Attribution License. Copyright © 2008 Skogsberg et al.
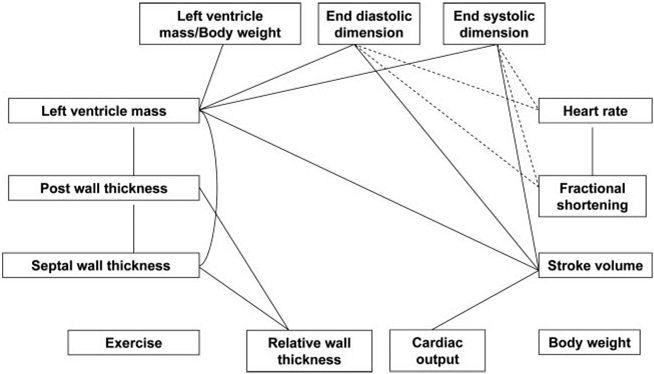
One of the first examples of the use of multiple, common genetic perturbations to construct functional networks for cardiovascular traits is described by Nadeau et al.31 These investigators examined a variety of cardiovascular traits, as well as exercise endurance and body weight, in a panel of genetically randomized mice (ie, a series of inbred strains differing in their genetic backgrounds as a result of mendelian segregation). Using echocardiographic and treadmill assays, the authors measured functions such as cardiac output, end-systolic dimensions, septal wall thickness, and heartbeats per minute. None of these traits represented adverse pathology but instead constituted genetically controlled differences in the normal range of variation. In addition, none of the traits showed mendelian inheritance but rather exhibited continuous variation that was consistent with multigenic control. A network was then constructed in which the traits (nodes) were assigned edges based on significant correlations (Figure 5). The assumption in such networks is that traits are correlated as a result of shared genetic determinants or causal interactions. The resulting network correctly identified known functional relationships based on physiological studies, and some interactions were confirmed through the use of single-gene mutant mice or treatment with pharmacological agents. Thus, this proof-of-principle study demonstrated that such networks are a powerful approach for characterizing functional relationships in complex biological systems.
Figure 5.
A network of cardiovascular traits based on trait correlations in genetically randomize mice. Solid lines indicate positive relationships between traits; broken lines, inverse relationships . The body weight and exercise nodes did not exhibit significant cosegregation with any other traits. Adapted from Nadeau et al31 with permission from Cold Spring Harbor Laboratory Press. Copyright © 2003 Cold Spring Harbor Laboratory Press.
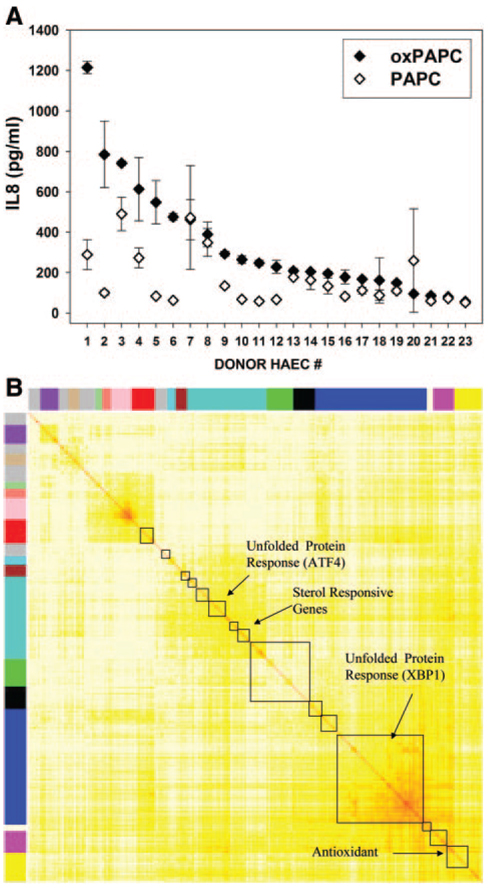
A similar strategy, using multiple genetic perturbations, was used to model an inflammatory network associated with atherosclerosis.32 Oxidized lipids are thought to promote atherosclerosis by stimulating endothelial cells to produce inflammatory cytokines such as interleukin-8, but the pathways involved are poorly understood. To examine this, transcript levels in the presence and absence of oxidized lipids were quantified in cultured endothelial cells derived from a series of random individuals (heart transplant donors). Altogether, >1000 genes were found to be significantly influenced by the oxidized lipids. In addition, between endothelial cells from different donors, there were striking differences in the responses of individual genes. This result was due to the fact that, in natural populations, there are many polymorphisms that perturb gene expression. These multiple common genetic variations were then used to group the genes according to the similarity of expression across individuals and thus create a “coexpression” network (Figure 6). In such networks, the nodes are genes and the edges represent correlations in the transcript levels between pairs of genes. A tutorial on the analysis of coexpression networks can be found at http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/. Altogether, 15 modules of highly connected genes were identified, and they were significantly enriched in genes for known pathways, including modules corresponding to 2 different arms of the unfolded protein response. In addition to identifying key pathways involved in the inflammatory response, the network was also useful for predicting regulatory mechanisms and identifying gene functions. For example, interleukin-8 and certain other cytokines thought to be involved in atherosclerosis were observed to occur in the XBP1 arm of the unfolded protein response, suggesting that these cytokines were regulated in part by the unfolded protein response. This prediction was validated through the use of siRNA and overexpression. The authors also were able to predict the functions of certain genes on the basis of their presence in modules. For example, a gene of unknown function (MGC4504) was a hub in the ATF 4 arm of the unfolded protein response, suggesting that it was an unfolded protein response gene regulated by ATF 4. This was confirmed, and subsequent studies have indicated that the function of the gene is related to apoptosis.33
Figure 6.
Coexpression analysis of the inflammatory responses of human endothelial cells based on common genetic variations in the population. Small pieces of human aortic arteries (obtained during the course of heart transplant surgery) were used to obtain pure, early-passage cultures of human endothelial cells. The endothelial cells from different donors were observed to exhibit significant differences in response to oxidized lipids. A, Production of interleukin-8 (IL8) by the cultures after treatment with biologically active oxidized phospholipids (solid symbols) vs control levels (open symbols). A total of 12 endothelial cell cultures isolated from individuals exhibiting various responses to oxidized lipids were subjected to microarray analysis, and >1000 genes were shown to be regulated by oxidized lipid treatment. Transcript levels of these genes were then used to model scale-free networks based on coexpression. A correlation matrix of all the genes was constructed, and their connectivities were used to generate a topographical overlap matrix (TOM) plot. B, Results shown as color-coded clusters (modules) of highly correlated genes in a symmetric plot. Correlations between genes are indicated by the color intensity (white corresponding to little or no correlation; red, strong correlation). The red diagonal represents correlations of each gene with itself, and offset from this are correlations with other genes. Some of the modules were highly enriched for curated pathways as shown. Adapted from Gargalovic et al32 according to the policy of Proceedings of the National Academy of Sciences. Copyright © 2006 by the National Academy of Sciences. HAECs indicates human aortic endothelial cells.
Systems Genetics
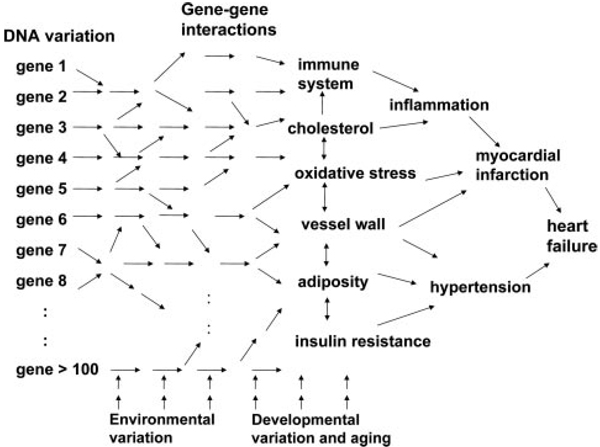
The endothelial example above shows how common genetic variations can be leveraged to model biological networks. This can be extended by simultaneously examining DNA variation and clinical phenotypes, as well as molecular phenotypes. This approach, called genetic genomics, integrative genetics, or systems genetics, is proving particularly powerful for the analysis of complex cardiovascular and metabolic traits.34–36 The basic concept is illustrated in Figure 7. Common forms of cardiovascular disease are due to multiple genetic factors, each contributing modestly to disease risk, and to environmental factors. These environmental and genetic factors perturb molecular phenotypes such as gene transcript levels, protein levels, and metabolite levels; genetic variation can also affect coding sequences and protein structure. These, in turn, perturb the cellular and physiological states contributing to the diseases. Classic genetics attempts to relate DNA variation directly to clinical phenotypes. Systems genetics, on the other hand, attempts to assess molecular phenotypes quantitatively and to identify patterns (networks) in them that are associated with clinical traits.
Figure 7.
“Molecular phenotypes” (such as transcript levels) can be used to identify networks contributing to complex cardiovascular disorders. Complex traits such as common forms of coronary artery disease and heart failure result from the interactions of multiple genetic variations, and environmental factors. Random populations of humans or experimental organisms can be examined for molecular phenotypes (such as transcript levels, protein levels, and metabolite levels) and for clinical traits to identify molecular patterns (networks) associated with the clinical traits. If DNA variants are typed, they can be associated with both molecular and clinical phenotypes.
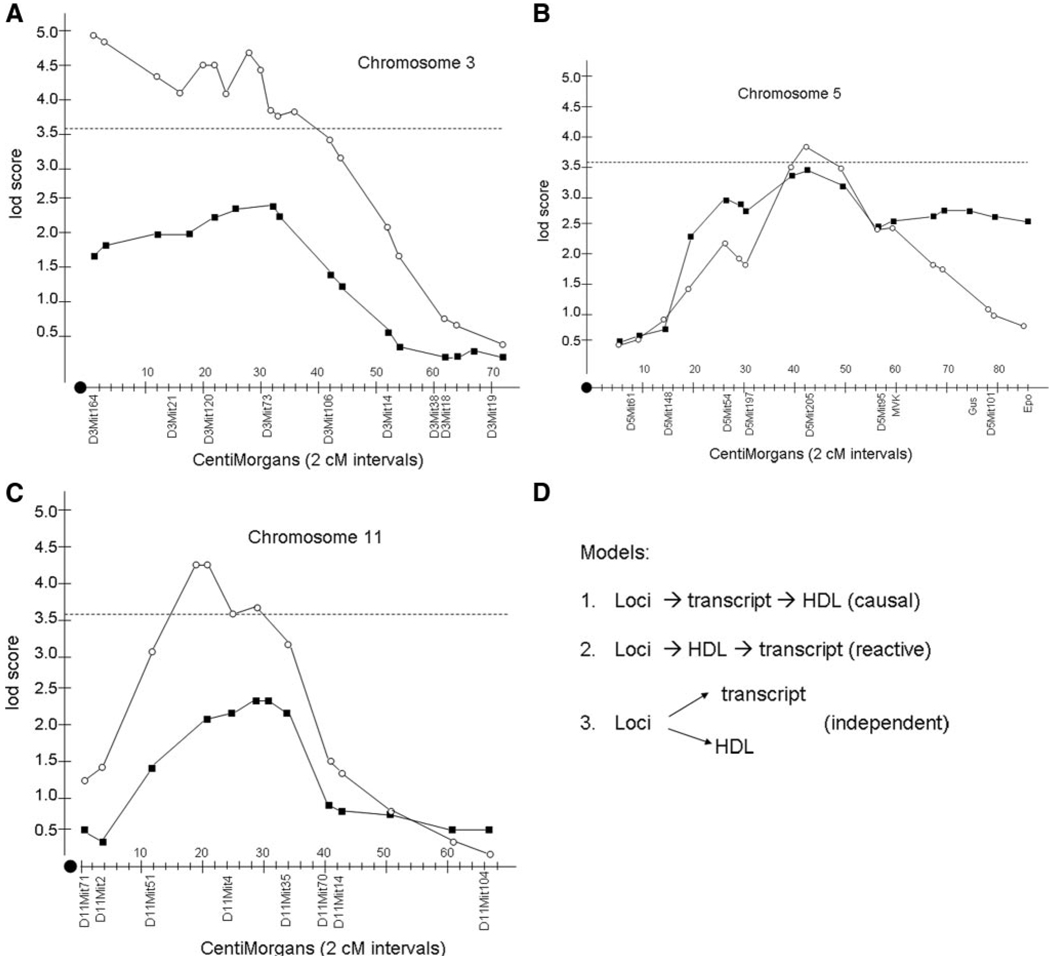
An early example of the approach is illustrated in Figure 8. A group of ≈300 genetically randomized mice (from a cross between 2 inbred strains) have been typed for DNA markers, transcript levels for an enzyme involved in cholesterol catabolism, and levels of plasma high-density lipoproteins (HDL). As in human populations, there are thousands of common variations that perturb gene expression among common laboratory strains of mice or rats. In this cross, genetic loci on chromosomes 3, 5, and 11 exhibited significant or suggestive association with HDL cholesterol levels. These same 3 loci also exhibited significant association for the transcript levels of the enzyme cholesterol 7α-hydroxylase (Cyp7a), which degrades cholesterol. The loci controlling HDL levels are called clinical quantitative trait loci; the loci controlling transcript levels are called expression quantitative trait loci. These results suggested 3 possible hypotheses: the loci control levels of the Cyp7a gene expression, which in turn influences HDL cholesterol levels; these loci control HDL cholesterol level, and this in turn perturbs Cyp7a transcript levels; and the 3 loci independently control levels of HDL cholesterol and Cyp7a transcripts. As discussed below, these possibilities can be modeled mathematically. These studies were carried out in the mid-1990s before the development of gene expression microarrays.37 Now, it is possible to carry out such analyses globally, examining relationships between DNA variation throughout the genome and levels of transcripts globally.
Figure 8.
Integration of DNA variation, transcript levels, and a clinical trait. A randomized population of mice (from an intercross between 2 strains of mice) was studied for DNA variation (genotyping), for levels of HDL cholesterol in plasma, and for transcript levels of a candidate gene, Cyp7a. A through C, A measure of statistical significance of linkage (lod score) for HDL cholesterol and Cyp7a transcript levels along the chromosomes, with the positions of typed genetic markers indicated. As shown, 3 overlapping loci (on chromosomes 3, 5, and 11) controlled both traits. D, There are 3 likely causal relationships between the components. Redrawn from Machleder et al37 according to the policy of The Journal of Clinical Investigation. Copyright © The American Society for Clinical Investigation.
An important concept for such analyses is that information flows from DNA to the molecular or clinical traits. Thus, in the example above, we can assume that DNA variation causes alterations in Cyp7a transcript levels and HDL levels but not the reverse. Thus, when a network is constructed, DNA can serve as a “causal anchor.” The relationships between DNA variation, transcript levels, and HDL levels can be modeled statistically. If Cyp7a transcript levels control HDL levels, then the relationship between the DNA variation and HDL would be expected to be considerably reduced or eliminated by mathematical conditioning on transcript levels using partial correlation coefficients. That is, because in this scenario the loci influence HDL only secondarily as a result of influencing transcript levels, transcript levels would be sufficient to explain HDL levels at those loci. Conversely, if HDL levels controlled transcript levels, conditioning on HDL would reduce or eliminate the DNA–transcript level associations. Schadt and colleagues36 developed a statistical procedure to distinguish between such possible relationships and to identify genes likely to be causally involved in complex traits. To validate the approach, they chose genes predicted to contribute to body fat in a segregating population of mice and tested the predictions by overexpressing the genes in transgenic mice or reducing expression in gene-targeted mice. To date, 9 genes have been tested; of these 9, all but 1 showed a significant impact on body fat.11 Such causal modeling is dependent on the concept of multiple perturbations discussed above. It would have little power to test for causal relationships between traits in studies with single perturbations such as a gene knockout study in mice because all traits would change as a result of the same perturbation and only environmental noise would distinguish primary from secondary effects.
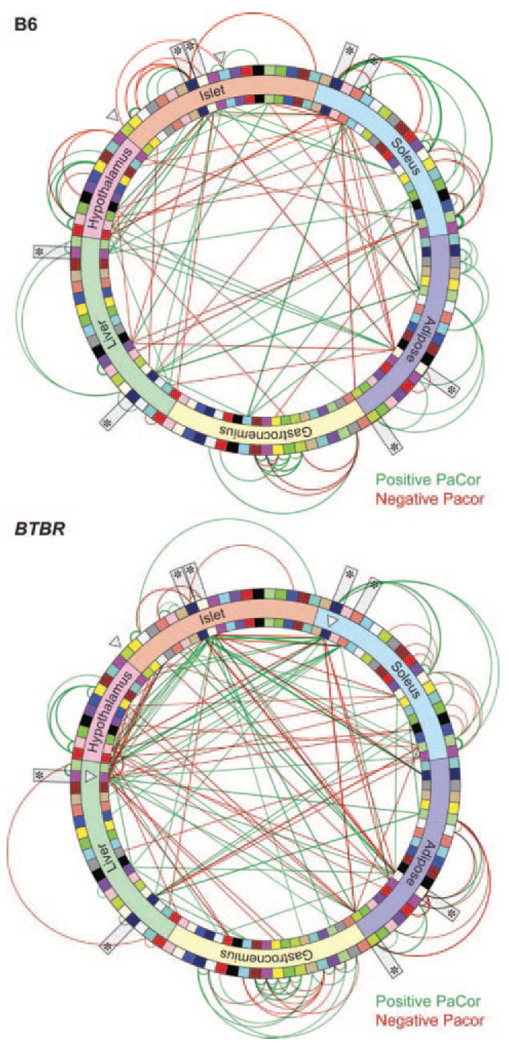
As discussed above for endothelial inflammation studies, data from such experiments can be modeled to generate coexpression networks. Such networks have exhibited several remarkable features. For example, some of the modules showed striking overall correlations with clinical traits. Ghazalpour et al38 identified liver modules that explained a significant fraction of adiposity or glucose levels in a segregating population of mice. Wang et al39 observed significant correlations between adipose gene modules and atherogenesis in a segregating population of mice on a hyperlipidemic genetic background. Keller et al40 also showed that the modules in different tissues can exhibit significant correlations, indicative of coordinated communication between different cells and organs (Figure 9). Striking sex effects have also been observed. For example, van Nas et al41 observed that modules in various tissues of a population of mice were generally conserved between sexes but that several showed dramatic differences in connectivity, including some that essentially broke into pieces in one sex compared with the other. Such emergent properties are likely to have important implications for cardiovascular and other complex diseases. Coexpression networks can also be significantly influenced by disease states and genetic backgrounds (Figure 9). In an elegant study in which pancreatic islet proliferation was measured with metabolic labeling, Keller et al40 observed differences in the overall networks of an obese, non–diabetes-prone strain of mouse (B6) compared with a diabetes-prone strain (BTBR), particularly in a cell-cycle module that is likely to be relevant to the development of type 2 diabetes mellitus (Figure 8). Conceptually, a powerful aspect of elucidating the topological structure of biological networks is the simplification of the system from a vast number of individual components (eg, 20 000 genes) to a smaller number of interacting modules (eg, <100 gene modules regulated by hub genes).
Figure 9.
Coexpression modules in different tissues are strongly correlated, and the connectivity of the networks is influenced by genetic background and disease status. Inbred C57BL/6 (B6) and BTBR mice, some carrying the ob leptin gene mutation resulting in massive obesity, were studied by global expression array analysis in 6 different tissues and at different ages, and the data were used to model coexpression networks in each tissue. The resulting modules are illustrated as colored bricks along the inside and outside of the network wheels. The edges, shown as arcs and lines, represent significant correlations of modules between tissues (inside the wheel) or within tissues (outside the wheel). A cell-cycle module with the pancreatic islets and those modules that form a direct connection to the cell cycle islet are highlighted with open arrows. Reproduced from Keller et al40 with permission from Cold Spring Harbor Laboratory Press. Copyright © 2008 Cold Spring Harbor Laboratory Press.
Although gene coexpression network analysis models the overall topological features of a network by portioning the transcriptome data into functional units (modules), how the genes interact and how information flows through the network are not explained. However, as discussed above, the relationships between genes in a network can be causally modeled, and bayesian methodologies that exploit the increased information of joint probabilistic mapping can be applied to produced “directed” graphs. Needham et al42 and Beaumont and Rannala43 provide reviews of bayesian analysis of complex systems. Chen and colleagues34 have used bayesian networks for liver and adipose gene expression in mouse crosses. Several subnetworks (coexpression modules) were observed to be significantly correlated with clinical traits, allowing prediction of genes likely to contribute importantly to these traits (Figure 2).
It is clear that systems genetics approaches can also be applied to humans, although access to tissues is an important limitation. Numerous studies have shown that loci contributing to transcript levels can be identified in human populations by linkage (expression quantitative trait loci) or association analyses (expression single nucleotide polymorphism, eSNP),44 and as discussed above, for human endothelial cells, it was feasible to construct coexpression networks. In fact, recent studies suggest that mouse and human coexpression networks show considerable overlap, and network analysis has been used to prioritize candidate genes identified in genome-wide association studies.45
Clearly, expression arrays provide a very incomplete picture of molecular phenotypes, and it is important that systems genetics be extended to proteins and metabolites. The most extensive of such analyses have been conducted in experimental organisms such as yeast and Arabidopsis.46 Recently, Chaibub Neto and colleagues47 integrated metabolic profiling with gene expression in a mouse cross-segregating for diabetes mellitus and validated some causal interactions.
Network Dynamics
The ultimate goal of systems biology is to apply knowledge about the structure of biological networks to understand their function, including how function changes over time (development, aging, diurnal cycles), between sexes,41 and in disease states. So far, such analyses have been performed in detail only in model organisms such as yeast and certain tissue culture cells (eg, Ramsey et al27). However, an understanding of network dynamics will be particularly important for the cardiovascular system, given the progressive nature of metabolic disease, atherosclerosis, and heart failure.
Perhaps the greatest challenge in elucidating network structure-function relationships relates to emergent properties. As discussed earlier, the nonlinear interactions in biological networks that drive evolution by generating novel adaptive collective behaviors cannot be understood solely by examining the properties of individual components of the network. Although reductionist approaches are essential to characterize components, integrative approaches that incorporate the dynamic interactions between components must be reintegrated back into the system to understand how these novel collective behaviors arise and how they are regulated.
Mathematical modeling and nonlinear dynamics provide the tools to analyze emergent properties and to define the system-level parameters controlling them. For this purpose, 2 modeling strategies can be used synergistically. Traditional detailed (high-dimensional) models are most valuable for directly linking biological networks across levels (eg, protein, organelle, cell, tissue, organism) in a biologically realistic manner. Conceptual (low-dimensional) models, on the other hand, are most valuable for analyzing the dynamics of novel behaviors at a given level.
Detailed modeling incorporates detailed molecular interactions into the model, with the advantage that physical biological entities can be represented explicitly, allowing corresponding biological experiments to be designed to evaluate the accuracy of the model. However, these models become very complex and difficult to analyze, especially with unknown parameters/rate constants. Once the topological structure of a network has been defined, however, computational techniques based on optimality assumptions can be used to test validity and predictive accuracy.48,49 For example, in metabolic networks, 2 commonly used optimality assumptions include flux balance analysis and minimization of metabolic adjustment. Flux balance analysis assumes that the goal of metabolism is to maximize growth (ie, maximizing the conversion of substrates into products that are essential for cell growth), whereas minimization of metabolic adjustment assumes that the metabolite network strives to minimize metabolic flux redistribution in response to perturbations. With these constraints, unknown parameter values in the network are explored computationally and assigned optimized values that maximize growth rate (flux balance analysis) or minimize metabolic flux redistribution (minimization of metabolic adjustment). With these techniques, knowledge of a restricted set of parameters, combined with the application of fundamental thermodynamic and evolutionary principles, can generate quantitative predictions and testable hypotheses. These approaches have successfully predicted experimental results in microbe responses to mutations and environmental changes, and similar approaches are being developed to model the adaptive responses of the mammalian myocardium to cardiac workload, acute ischemia, and heart failure.50–52
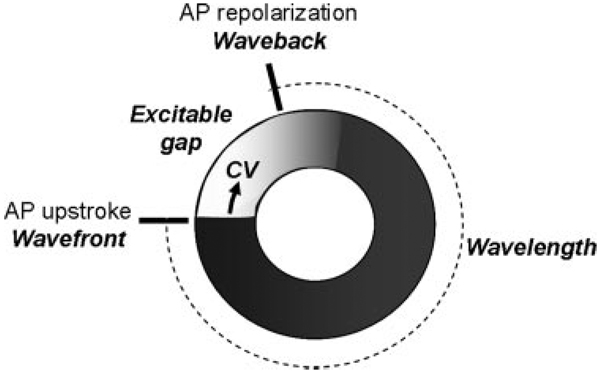
Conceptual modeling, on the other hand, ignores the explicit physical details and instead strives to capture dynamic principles underlying emergent behaviors by following Albert Einstein’s dictum that “things should be as simple as possible, but not too simple.” As an example, consider the emergent behavior of reentry, the most common cause of cardiac arrhythmias. Reentry does not exist at the level of the myocyte but emerges as a collective behavior at the level of tissue when individual myocytes are coupled diffusively by gap junctions. Conceptually, the simplest requirement for reentry is that the wavelength of the cardiac electric wave (the product of its conduction velocity and refractory period) be less than the available path length in the tissue so that an excitable gap exists between the wave front and the wave back (Figure 10). Thus, conduction velocity, refractoriness, wavelength, and excitable gap are system-level parameters that control the emergent behavior of reentry. It is important to note that these system-level parameters are phenomenological; ie, none is a discrete biological entity such as an ion channel protein. The goal of conceptual models is to identify such system-level phenomenological parameters that control the emergent behavior so that the physical entities such as ion channels (more specifically, the interactions between various ion channels) can be mapped onto the phenomenological parameters. Conceptual models can be very powerful, as illustrated by the example of reentry. The therapeutic concept of electric defibrillation, based on eliminating excitable gaps in reentrant circuits, preceded any detailed molecular or cellular knowledge of cardiac electrophysiology and yet remains the most effective therapy for preventing sudden cardiac death, despite 50 years of cellular/molecular investigation.
Figure 10.
Reentry as an emergent property of cardiac tissue. For reentry to occur, an excitable gap must be present between the wave front (action potential [AP] upstroke) and wave back (AP repolarization). CV indicates conduction velocity.
In summary, these 2 modeling approaches, although fundamentally different, are highly complementary.53 Conceptual models provide critical insight into how new properties emerge at a given level, but they do not directly relate phenomenological system parameters to physical biological entities. Detailed models link these phenomenological parameters to physical entities that can be experimentally manipulated. Combining both approaches provides a powerful strategy for generating hypotheses from conceptual models that can be related to specific biological parameters in detailed models, which can then be manipulated in biological experiments to test their validity. Thus, the interactions with biological experiments are direct and bidirectional. Excellent examples include quantitative modeling in electrophysiology in which iterated maps,54,55 simple 2-variable dynamical models,56 low-dimensional physiologically detailed models,57 complemented by very high-dimensional physiologically detailed models58–60 are used to elucidate the underlying dynamics of action potential excitation and wave propagation. A cardiovascular example relevant to morphogenesis includes calcification patterns in cultured vascular smooth muscle cells created by diffusible morphogens.61
Clinical Applications
Systems biology is providing the framework to analyze how the structures of biological networks relate to their functions, as required to understand human disease. Although clinical applications are still in their infancy, there have been a number of promising advances related to disease gene discovery, diagnosis, treatment, and general patterns of human disease.
One application that has already proven very useful is in providing a biological context to the many single nucleotide polymorphisms associated with disease in genome-wide association studies.45 Thus, if genes located near the single nucleotide polymorphisms are present in a network module, they can immediately be associated with annotated genes in that module. Baranzini62 have developed an interesting strategy for identifying subnetworks associated with common diseases, including cardiovascular disease, using genome-wide association study results. Thus, single-nucleotide polymorphisms with nominal evidence of association with the diseases were superimposed on the human protein interaction network, and subnetworks enriched for associated genes were identified.
A network perspective may prove valuable for diagnostic purposes. Recent genome-wide association studies have revealed that cardiovascular diseases, like most complex diseases, have a surprising complex genetic architecture and that there are no truly “major genes.” Thus, even highly heritable traits such as obesity and lipoprotein levels are determined by polymorphisms with effects that are, individually, very small (usually with the strongest genes exhibiting odds ratios <1.2). Moreover, only a small fraction of the genetic components of traits such as atherosclerosis have been explained by even very large genome-wide association studies involving thousands of individuals. Thus, the dream of individualized medicine using DNA analysis seems very distant. However, systems genetics studies, as discussed above, have shown that certain networks are highly correlated with clinical phenotypes, in some cases explaining up to 50% of the trait variance, much more than can be explained by individual DNA variations.38,40 This concept of using defined clusters of transcripts identified by global expression profiling has already proven useful in predicting outcomes in cancers and may be similarly effective in cardiovascular diseases.63
Although such networks can be readily constructed in experimental organisms, it will be challenging to obtain and integrate global data sets in humans in the context of cardiovascular disease. Molecular studies will be limited by tissue availability, but as discussed above, samples can be obtained from pathological sources such as endarterectomy and from accessible tissues such as blood and adipose. In a recent study, Lewis and colleagues64 took advantage of planned myocardial infarction procedures to create a metabolic profile related to myocardial injury. They applied mass spectrometry to quantify metabolites in blood of 36 patients undergoing alcohol septal ablation treatments for hypertrophic obstructive cardiomyopathy. A clear signature consisting of a set of altered metabolites associated with cardiac injury was thus obtained.
An important reason for elucidating the networks associated with a disease is to better understand how it can be targeted pharmacologically. If a given gene is not classically “targetable,” a network perspective may reveal connected targetable genes that regulate it. Alternatively, a network perspective could help avoid toxicities resulting from disruption of multiple edges of a node.10 It may also be useful for the development of “combination therapy” in which the desired effect is obtained by targeting multiple arms of the network. For example, the drug Vytorin reduces cholesterol by targeting both cholesterol synthesis with simvastatin (an inhibitor of HMG-CoA reductase) and dietary cholesterol uptake with ezetimibe (an inhibitor of Niemann-Pick type C1-like). It is also possible that some cardiovascular diseases are emergent properties of the network and thus are unlikely to be targetable by a single molecule.10
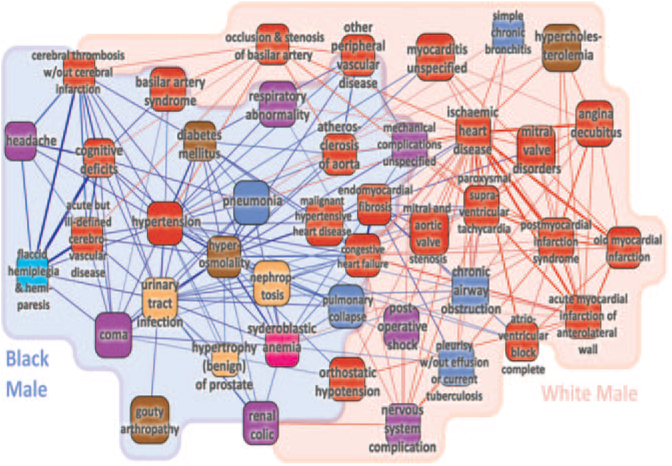
Network analyses are also revealing connections between diseases. Goh and coworkers65 constructed a bipartite graph in which one set of nodes corresponds to all known human genetic disorders and the second set to all known disease genes (the “diseaseome”). This could be transformed into a “human disease network” by connecting diseases sharing a gene or into a “disease gene network” by connecting genes sharing a disease. This network strategy offers the possibility of identifying general patterns of human disease not apparent from studies of individual disorders. For example, the disease gene network exhibited distinct functional modules in which various combinations of perturbed genes appeared to lead to diseases. The approach was also capable of predicting comorbidity patterns such as Alzheimer disease and myocardial infarction.66 In a related study, Hidalgo and colleagues67 constructed a network of pairwise comorbidity correlations for >10 000 diseases from >30 million medical records (a “phenotypic disease network”). They then used the results to study illness progression from a network dynamics perspective. For example, they found that the progression of disease occurred along the links of the network and differed among genders and ethnicities (Figure 11).
Figure 11.
A phenotypic database network based on comorbidities from >30 medical records.67 Shown are all diseases connected to hypertension and ischemic heart disease, emphasizing differences in links for black and white men. Comorbidities significantly stronger for black men are shown in blue; comorbidities stronger for white men are shown in red. Black edges were similar for black and white men. The node colors indicate different classes of diseases. Reproduced from Hidalgo et al67 under the Creative Commons Attribution License. Copyright © 2009 Hidalgo et al.
Finally, network analysis of disease offers a new, more specific, strategy for disease classifications.68 Thus, a systems-based approach allows one to quantitatively assess the molecular and environmental relationships that define a specific pathophenotype. From this perspective, disease is viewed as the result of interactions between a modular collection of genomic, proteomic, metabolomic, and environmental networks.
Summary
New technology is making it possible to see both the “forest” and the “trees” in biological systems. Systems biology might be best defined as the attempt to integrate new discovery-driven technologies that allow us to track biological networks holistically with classic hypothesis-driven approaches. Mathematical biology, in the form of bioinformatics to elucidate the structure of biological networks, as well as conceptual and detailed modeling to illuminate how network structure relates to function, will play an increasingly critical role in this effort. In this truly exciting era of cardiovascular biology, we can glimpse how the mysteries of human cardiovascular diseases might ultimately be solved.
Acknowledgment
We thank Xia Yang for assistance with Figure 2.
Sources of Funding
This work was supported by NIH/NHLBI 5PO1 HL28481 (Dr Lusis), NIH/NHLBI PO1 HL30568 (Dr Lusis), NIH/NHLBI RO1 HL094322 (Dr Lusis), NIH/NHLBI 1P01 HL078931 (Dr Weiss), NIH/NHLBI 1P01 HL08011 (Dr Weiss), and NIH/NHLBI 1R01 HL095663 (Dr Weiss).
Appendix
Glossary of Terms
Bayesian networks: Networks in which the connectivity of components is represented by joint probabilities. For example, in coexpression networks, the correlation of individual components can indicate functional associations, and causal interactions can be represented as joint probabilities.
Boolean networks: In these networks, each component is represented in 2 possible states (on/off). Boolean networks lead to relatively “coarse-grained” models lacking quantitative network properties.
Coexpression networks: Networks based on correlations between components in response to a variety of perturbations. In such networks, the connections are based on the degree of correlation, with the assumption that correlation suggests functional or causal interactions.
Complex traits: Traits that are due to multiple genetic, environmental, or developmental factors.
Connectivity (k): A measure of gene connectiveness. Genes with high k are all highly correlated with many other network genes.
Emergent properties: Properties that are not possessed by the individual components of a system but “emerge” in the assembled system.
Graphs: Mathematical representations of the pair-wise relationships between components in a system.
Hub: A node with many connections.
Module: A group of components tightly connected across a set of conditions or perturbations.
Network motifs: Subgraphs that occur in real networks relative to all possible links between a subset of nodes. For example, “feedback” or “feed-forward” loops occur commonly in biological regulatory systems.
Network models: Graphs containing multiple “components” or “nodes” connected by “edges.” For example, in a network of protein interactions, the nodes are the proteins and the edges are pair-wise interactions between the proteins. In “undirected’ graphs, causal relationships between nodes are not specified. In “directed” graphs, the edges, or “arcs,” indicate a direction of interaction.
Self-organizing system: A system that increases in complexity without being guided by an outside source. Examples in biology include the spontaneous folding of proteins and morphogenesis.
Small-world effects: Properties of biological networks resulting in short path lengths between nodes.
Systems-based approaches: Approaches that use computational and statistical methods to understand systems with a large number of components.
Weighted gene coexpression network analysis: A collection of algorithms designed to identify modules of coexpressed genes using microarray data. They are referred to as “weighted” because connection strengths (ie, correlations) range from 0 to 1. In an “unweighted” network, the connection strengths are binary, 0 or 1.
Footnotes
Reprints: Information about reprints can be found online at http://www.lww.com/reprints
Disclosures
None.
References
- 1.Barabási AL. Linked. London, UK: Penguin Books, Ltd; 2002. [Google Scholar]
- 2.Bondarenko VE, Szigeti GP, Bett GC, Kim SJ, Rasmusson RL. Computer model of action potential of mouse ventricular myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H1378–H1403. doi: 10.1152/ajpheart.00185.2003. [DOI] [PubMed] [Google Scholar]
- 3.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 4.Lusis AJ, Attie AD, Reue K. Metabolic syndrome: from epidemiology to systems biology. Nat Rev Genet. 2008;9:819–830. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topol E. Textbook of Cardiovascular Medicine. 3rd ed. Philadelphia, Pa: Lippincott Williams and Wilkins; 2007. [Google Scholar]
- 6.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 7.Barabasi AL, Bonabeau E. Scale-free networks. Sci Am. 2003;288:60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- 8.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 9.Ghazalpour A, Doss S, Sheth SS, Ingram-Drake LA, Schadt EE, Lusis AJ, Drake TA. Genomic analysis of metabolic pathway gene expression in mice. Genome Biol. 2005;6:R59. doi: 10.1186/gb-2005-6-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schadt EE, Lum PY. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders: reverse engineering gene networks to identify key drivers of complex disease phenotypes. J Lipid Res. 2006;47:2601–2613. doi: 10.1194/jlr.R600026-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, Karlsson J, Castellani LW, Mumick S, Wang K, Xie T, Coon M, Zhang C, Estrada-Smith D, Farber CR, Wang SS, van Nas A, Ghazalpour A, Zhang B, Macneil DJ, Lamb JR, Dipple KM, Reitman ML, Mehrabian M, Lum PY, Schadt EE, Lusis AJ, Drake TA. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strogatz SH. Sync: How Order Emerges From Chaos In the Universe, Nature, and Daily Life. New York, NY: Hyperion; 2003. [Google Scholar]
- 13.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 14.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoll M, Cowley AW, Jr, Tonellato PJ, Greene AS, Kaldunski ML, Roman RJ, Dumas P, Schork NJ, Wang Z, Jacob HJ. A genomic-systems biology map for cardiovascular function. Science. 2001;294:1723–1726. doi: 10.1126/science.1062117. [DOI] [PubMed] [Google Scholar]
- 16.Giallourakis C, Henson C, Reich M, Xie X, Mootha VK. Disease gene discovery through integrative genomics. Annu Rev Genomics Hum Genet. 2005;6:381–406. doi: 10.1146/annurev.genom.6.080604.162234. [DOI] [PubMed] [Google Scholar]
- 17.Ashley EA, Ferrara R, King JY, Vailaya A, Kuchinsky A, He X, Byers B, Gerckens U, Oblin S, Tsalenko A, Soito A, Spin JM, Tabibiazar R, Connolly AJ, Simpson JB, Grube E, Quertermous T. Network analysis of human in-stent restenosis. Circulation. 2006;114:2644–2654. doi: 10.1161/CIRCULATIONAHA.106.637025. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 20.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann WP, Constantinescu SN, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia JG, Pevsner J, Jensen ON, Roepstorff P, Deshpande KS, Chinnaiyan AM, Hamosh A, Chakravarti A, Pandey A. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chautard E, Thierry-Mieg N, Ricard-Blum S. Interaction networks: from protein functions to drug discovery: a review. Pathol Biol (Paris) 2009;57:324–333. doi: 10.1016/j.patbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 23.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 24.Lehner B, Fraser AG. A first-draft human protein-interaction map. Genome Biol. 2004;5:R63. doi: 10.1186/gb-2004-5-9-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss JN, Yang L, Qu Z. Systems biology approaches to metabolic and cardiovascular disorders: network perspectives of cardiovascular metabolism. J Lipid Res. 2006;47:2355–2366. doi: 10.1194/jlr.R600023-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Foster LJ, de Hoog CL, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Ramsey SA, Klemm SL, Zak DE, Kennedy KA, Thorsson V, Li B, Gilchrist M, Gold ES, Johnson CD, Litvak V, Navarro G, Roach JC, Rosenberger CM, Rust AG, Yudkovsky N, Aderem A, Shmulevich I. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput Biol. 2008;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stratford S, Hoehn KL, Liu F, Summers SA. Regulation of insulin action by ceramide: dual mechanisms linking ceramide accumulation to the inhibition of Akt/protein kinase B. J Biol Chem. 2004;279:36608–36615. doi: 10.1074/jbc.M406499200. [DOI] [PubMed] [Google Scholar]
- 29.Skogsberg J, Lundstrom J, Kovacs A, Nilsson R, Noori P, Maleki S, Kohler M, Hamsten A, Tegner J, Bjorkegren J. Transcriptional profiling uncovers a network of cholesterol-responsive atherosclerosis target genes. PLoS Genet. 2008;4:e1000036. doi: 10.1371/journal.pgen.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tegner J, Yeung MK, Hasty J, Collins JJ. Reverse engineering gene networks: integrating genetic perturbations with dynamical modeling. Proc Natl Acad Sci U S A. 2003;100:5944–5949. doi: 10.1073/pnas.0933416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadeau JH, Burrage LC, Restivo J, Pao YH, Churchill G, Hoit BD. Pleiotropy, homeostasis, and functional networks based on assays of cardiovascular traits in genetically randomized populations. Genome Res. 2003;13:2082–2091. doi: 10.1101/gr.1186603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Patel S, Nelson SF, Horvath S, Berliner JA, Kirchgessner TG, Lusis AJ. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci U S A. 2006;103:12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng H, Vera I, Che N, Wang X, Wang SS, Ingram-Drake L, Schadt EE, Drake TA, Lusis AJ. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc Natl Acad Sci U S A. 2007;104:4530–4535. doi: 10.1073/pnas.0607620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Machleder D, Ivandic B, Welch C, Castellani L, Reue K, Lusis AJ. Complex genetic control of HDL levels in mice in response to an atherogenic diet: coordinate regulation of HDL levels and bile acid metabolism. J Clin Invest. 1997;99:1406–1419. doi: 10.1172/JCI119300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, Horvath S. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang SS, Schadt EE, Wang H, Wang X, Ingram-Drake L, Shi W, Drake TA, Lusis AJ. Identification of pathways for atherosclerosis in mice: integration of quantitative trait locus analysis and global gene expression data. Circ Res. 2007;101:e11–e30. doi: 10.1161/CIRCRESAHA.107.152975. [DOI] [PubMed] [Google Scholar]
- 40.Keller MP, Choi Y, Wang P, Belt Davis D, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S, Steinberg HA, Chaibub Neto E, Kleinhanz R, Turner S, Hellerstein MK, Schadt EE, Yandell BS, Kendziorski C, Attie AD. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 2008;18:706–716. doi: 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, Arnold AP, Lusis AJ. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. A primer on learning in Bayesian networks for computational biology. PLoS Comput Biol. 2007;3:e129. doi: 10.1371/journal.pcbi.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beaumont MA, Rannala B. The Bayesian revolution in genetics. Nat Rev Genet. 2004;5:251–261. doi: 10.1038/nrg1318. [DOI] [PubMed] [Google Scholar]
- 44.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 45.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, Bumgarner RE, Schadt EE. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaibub Neto E, Ferrara CT, Attie AD, Yandell BS. Inferring causal phenotype networks from segregating populations. Genetics. 2008;179:1089–1100. doi: 10.1534/genetics.107.085167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahadevan R, Edwards JS, Doyle FJ., III Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J. 2002;83:1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cortassa S, O’Rourke B, Winslow RL, Aon MA. Control and regulation of mitochondrial energetics in an integrated model of cardiomyocyte function. Biophys J. 2009;96:2466–2478. doi: 10.1016/j.bpj.2008.12.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo RY, Liao S, Tao GY, Li YY, Zeng S, Li YX, Luo Q. Dynamic analysis of optimality in myocardial energy metabolism under normal and ischemic conditions. Mol Syst Biol. 2006;2:2006.0031. doi: 10.1038/msb4100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Zhang EY, Zhang J, Bache RJ, Beard DA. Phosphate metabolite concentrations and ATP hydrolysis potential in normal and ischaemic hearts. J Physiol. 2008;586:4193–4208. doi: 10.1113/jphysiol.2008.154732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss JN, Qu Z, Garfinkel A. Understanding biological complexity: lessons from the past. FASEB J. 2003;17:1–6. doi: 10.1096/fj.02-0408rev. [DOI] [PubMed] [Google Scholar]
- 54.Nolasco JB, Dahlen RW. A graphic method for the study of alternation in cardiac action potentials. J Appl Physiol. 1968;25:191–196. doi: 10.1152/jappl.1968.25.2.191. [DOI] [PubMed] [Google Scholar]
- 55.Weiss JN, Karma A, Shiferaw Y, Chen PS, Garfinkel A, Qu Z. From pulsus to pulseless: the saga of cardiac alternans. Circ Res. 2006;98:1244–1253. doi: 10.1161/01.RES.0000224540.97431.f0. [DOI] [PubMed] [Google Scholar]
- 56.Winfree AT. Electrical turbulence in three-dimensional heart muscle. Science. 1994;266:1003–1006. doi: 10.1126/science.7973648. [DOI] [PubMed] [Google Scholar]
- 57.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenstein JL, Winslow RL. An integrative model of the cardiac ventricular myocyte incorporating local control of Ca2+ release. Biophys J. 2002;83:2918–2945. doi: 10.1016/S0006-3495(02)75301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hund TJ, Rudy Y. Rate dependence and regulation of action potential and calcium transient in a canine cardiac ventricular cell model. Circulation. 2004;110:3168–3174. doi: 10.1161/01.CIR.0000147231.69595.D3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shannon TR, Wang F, Puglisi J, Weber C, Bers DM. A mathematical treatment of integrated Ca dynamics within the ventricular myocyte. Biophys J. 2004;87:3351–3371. doi: 10.1529/biophysj.104.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garfinkel A, Tintut Y, Petrasek D, Bostrom K, Demer LL. Pattern formation by vascular mesenchymal cells. Proc Natl Acad Sci U S A. 2004;101:9247–9250. doi: 10.1073/pnas.0308436101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, Uitdehaag BM, Kappos L, Polman CH, Matthews PM, Hauser SL, Gibson RA, Oksenberg JR, Barnes MR. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18:2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Itadani H, Mizuarai S, Kotani H. Can systems biology understand pathway activation? Gene expression signatures as surrogate markers for understanding the complexity of pathway activation. Curr Genomics. 2008;9:349–360. doi: 10.2174/138920208785133235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewis GD, Wei R, Liu E, Yang E, Shi X, Martinovic M, Farrell L, Asnani A, Cyrille M, Ramanathan A, Shaham O, Berriz G, Lowry PA, Palacios IF, Tasan M, Roth FP, Min J, Baumgartner C, Keshishian H, Addona T, Mootha VK, Rosenzweig A, Carr SA, Fifer MA, Sabatine MS, Gerszten RE. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J Clin Invest. 2008;118:3503–3512. doi: 10.1172/JCI35111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park J, Lee DS, Christakis NA, Barabasi AL. The impact of cellular networks on disease comorbidity. Mol Syst Biol. 2009;5:262. doi: 10.1038/msb.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hidalgo CA, Blumm N, Barabasi AL, Christakis NA. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol. 2009;5:e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]