Abstract
Chromosome 22q11.2 deletions are found in almost 90% of patients with DiGeorge/velocardiofacial syndrome (DGS/VCFS). Large, chromosome-specific low copy repeats (LCRs), flanking and within the deletion interval, are presumed to lead to misalignment and aberrant recombination in meiosis resulting in this frequent microdeletion syndrome. We traced the grandparental origin of regions flanking de novo 3 Mb deletions in 20 informative three-generation families. Haplotype reconstruction showed an unexpectedly high number of proximal interchromosomal exchanges between homologs, occurring in 19/20 families. Instead, the normal chromosome 22 in these probands showed interchromosomal exchanges in 2/15 informative meioses, a rate consistent with the genetic distance. Meiotic exchanges, visualized as MLH1 foci, localize to the distal long arm of chromosome 22 in 75% of human spermatocytes tested, also reflecting the genetic map. Additionally, we found no effect of proband gender or parental age on the crossover frequency. Parental origin studies in 65 de novo 3 Mb deletions (including these 20 patients) demonstrated no bias. Unlike Williams syndrome, we found no chromosomal inversions flanked by LCRs in 22 sets of parents of 22q11 deleted patients, or in eight non-deleted patients with a DGS/VCFS phenotype using FISH. Our data are consistent with significant aberrant interchromosomal exchange events during meiosis I in the proximal region of the affected chromosome 22 as the likely etiology for the deletion. This type of exchange occurs more often than is described for deletions of chromosomes 7q11, 15q11, 17p11 and 17q11, implying a difference in the meiotic behavior of chromosome 22.
INTRODUCTION
The 22q11.2 deletion syndrome is a genomic disorder encompassing the clinical entities of DiGeorge syndrome (DGS), velocardiofacial syndrome (VCFS), and conotruncal anomaly face syndrome (CAFS). Characteristic phenotypic features include conotruncal cardiac defects, cleft palate, thymus and parathyroid gland aplasia or hypoplasia with functional T-cell abnormalities and hypocalcemia. Facial dysmorphia includes a hooded appearance of the eyelids, hypertelorism, overfolded ears, bulbous nasal tip and micrognathia (1).
Most 22q11 deletions occur as de novo lesions, with 10% or less inherited from an affected parent (2). The vast majority of patients (80–90%) have the same approximately 3 Mb deletion that remains unchanged even when inherited, however, the phenotype can be widely variable within a family. Although smaller recurrent 1.5 Mb deletions occur, a smaller deletion does not indicate milder symptoms, making genotype–phenotype correlations difficult (3). Recent estimates indicate that the 22q11 deletion occurs in approximately 1:3000 live births (4), making this disorder the most common human microdeletion syndrome. The prevalence of these de novo 22q11.2 deletions indicates an extremely high mutation or rearrangement rate within this genomic region. The presence of large (200–500 kb) highly homologous chromosome-specific low copy repeats (LCRs) in 22q11 (5-7) is thought to predispose this region to aberrant recombination and subsequent deletion. Evidence of unequal meiotic exchanges in genomic regions containing such low copy repeats (8,9) has been provided for a number of human deletion syndromes including Williams–Beuren (chromosome 7q11.2) (10,11), Prader–Willi/Angelman (chromosome 15q11.2) (12,13), Smith–Magenis (chromosome 17p11.2) (14), neuro-fibromatosis type I (chromosome 17q11.2) (15) and for a total of 16 patients with de novo deletions of 22q11.2 (5,10,16). In attempts to determine deletion mechanisms, recent studies have demonstrated the presence of chromosomal inversions involving regions flanking the LCRs on 7q11.2 in a significant percentage (30%) of transmitting progenitors of Williams–Beuren syndrome. The inversions are thought to predispose that chromosome to rearrangement in meiosis (11,17).
In the work presented here, we are able to provide significant information about the meiotic behavior of chromosome 22 by studying 20 informative three-generation families of probands with de novo 3 Mb deletions of 22q11. We are able to demonstrate a statistically significant occurrence of interchromosomal meiotic exchanges associated with the deleted chromosome in these families. Other smaller studies have suggested a parent of origin bias for the 22q11.2 deletion (18-20). However, we find no evidence of such a bias in a much larger cohort. Further we find no evidence of an inversion of 22q11.2 in the chromosome that becomes deleted, or in the chromosomes of individuals with a DGS/VCFS phenotype who do not demonstrate evidence of a deletion. Thus we conclude that chromosome 22 has a pattern of meiotic behavior and structural rearrangement that is different from other chromosomes with apparently similar genomic architecture.
RESULTS
Size and parental origin of the deletion
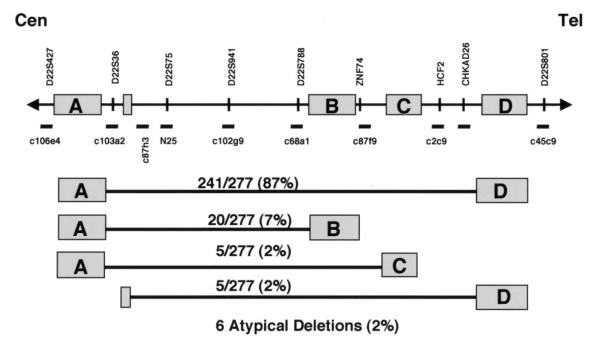
Families of patients with clinical features of DGS/VCFS and a deletion of 22q11.2 as determined by fluorescence in situ hybridization (FISH) with the N25 probe (Oncor) were recruited for this study (1). FISH (or microsatellite analysis in a few cases) was used to analyze peripheral blood samples from the parents of deleted probands in order to exclude familial deletions. Thus, all the deletions in this study occurred de novo in the proband. Figure 1 depicts the 22q11.2 region including the LCRs and relative location of markers and probes used for FISH. Probes adjacent to LCRs A and D (deletion endpoints of the standard 3 Mb deletion) and within that interval were used to determine the extent of the deletion. We found that in a group of 277 patients, 241 of the deletions (87%) span the same 3 Mb region from LCR A to LCR D. From the same cohort of 277, we found that 20 (7%) had a smaller approximately 1.5 Mb deletion extending from LCR A to LCR B (Fig. 1). Smaller numbers of individuals had other recurrent deletions (Fig. 1). Multiple microsatellite markers from within the region were used to determine the parental origin of the deletion (21) as shown in Figure 2. Sixty-five informative cases were examined. The results demonstrate that 35 deletions were of maternal origin and 30 were of paternal origin, indicating no bias in parental origin of the deletion.
Figure 1.
The 22q11.2 region including the locations of the LCR-22 segmental duplications A–D is depicted. Cosmid probes used for FISH are shown below the line. The names and relative locations of genes and STS markers are noted above the line. The figure is not drawn to scale. Results of deletion sizing experiments by FISH in a cohort of 277 patients is given in the lower part of the figure. The LCRs coinciding with the deletion endpoints are given with the relative percentage of patients that were found to be deleted for that given region.
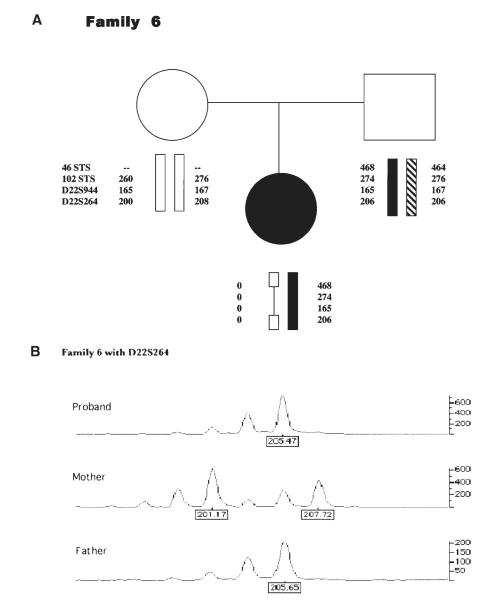
Figure 2.
(A) Four markers from within the 3 Mb deleted region that were used for parental origin determination are depicted in a sample pedigree. The names of the markers are listed to the left. Dashes represent a marker that did not amplify. The proband shows haploinsufficiency for alleles from her mother, noted by zeroes (0). (B) Genotyper data presented in the pedigree for marker D22S264 is given. Note the absence of commonly shared peaks, representing alleles, between mother and proband.
Haplotype analysis of deleted alleles
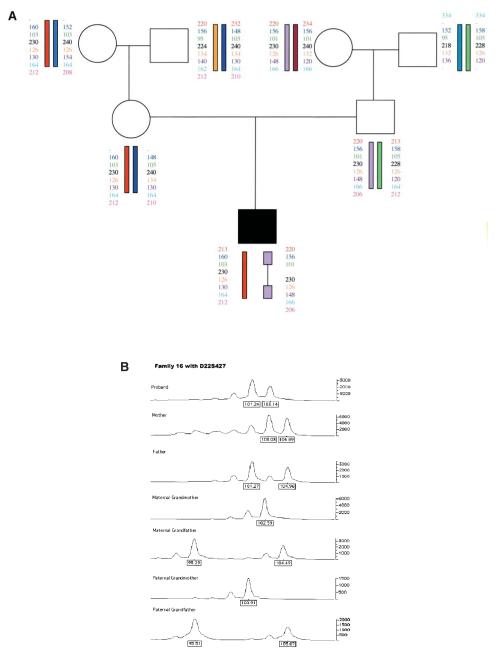
The meiotic exchange events that occurred in 20 informative de novo 3 Mb deletion cases were analyzed by examining their three-generation families for haplotype reconstruction. Grandparental alleles were used to establish the phase of marker transmission. A series of nine microsatellite markers flanking the deletion endpoints proximally (four markers) and distally (five markers) were used to genotype the families. Figures 3 and 4 provide examples of haplotype analysis showing evidence of the single intrachromosomal and one of the interchromosomal recombinations respectively, in two families from our cohort. The pedigrees and haplotype data of all 20 families are available as Supplementary Material. Table 1 summarizes the data obtained from these families. Family 19 has a set of affected identical twins. Haplotype reconstruction reveals that an interchromosomal mode of exchange between homologs predominates, occurring in 19/20 deleted chromosomes (ascertained by parental origin determination). Chisquare analysis based on expected numbers (10) demonstrates this finding to be statistically significant (P < 0.001). By contrast, when examining exchange events in the normal allele transmitted to these same individuals, we found only two crossovers in 15 informative meioses, consistent with the average genetic distance between the proximal and distal markers (10). In one of these two families (Family 11), the exchange on the normal chromosome was detected near a distal marker (TOP1P2), while the other family (Family 3), had a double crossover in the normal chromosome 22, one at a very proximal marker (F8VWFP2) and one detected by the most telomeric marker (D22S302). In our cohort, interchromosomal exchanges on the normal chromosome 22 did not occur in regions most closely flanking the deletion, as was observed for the deleted chromosome 22 haplotypes. In fact, the most distal marker, D22S302 (located in 22q13 and quite distant from the LCRs in the 22q11.2 region), was used primarily as a control to detect distal 22q exchanges that are consistent with normal meiotic recombination patterns. Interchromosomal exchanges involving this very distal marker were found on the normal chromosome 22 in a total of three families (Families 3, 18, 20; Supplementary Material).
Figure 3.
(A) Markers flanking the typical deletion endpoints that were used for haplotype reconstruction are shown in a three-generation pedigree consistent with an intrachromosomal exchange. Dashes depict a marker that did not amplify. (B) Sizes and intensity of allele peaks are shown from Genotyper analysis for a single marker (D22S427).
Figure 4.
(A) Haplotype reconstruction depicting an interchromosomal exchange. The regions flanking the deletion (shown by a thin bar) are derived from different grandparents, indicating the exchange. (B) Genotyper data from this family for a single proximal flanking marker.
Table 1.
High frequency of interchromosomal exchanges flanking de novo 3 Mb deletions
| Family no. | Parental origin | Crossover status | Deleted Proximal |
Distal | Normal Proximal |
Distal | Proband gender |
|
|---|---|---|---|---|---|---|---|---|
| 1 | Maternal | Crossover | MGM | MGF | PGF | PGF | Female | |
| 2 | Maternal | Crossover | MGM | MGF | PGM | PGM | Male | |
| 3 | Maternal | Crossover | MGF | MGM | PGM | PGF | Male | Normal has crossover |
| 4 | Maternal | Crossover | MGF | MGM | PGM | PGM | Male | |
| 5 | Maternal | Crossover | MGF | MGM | PGM | PGM | Female | |
| 6 | Maternal | Crossover | MGF | MGM | DEC | DEC | Female | |
| 7 | Paternal | Crossover | PGF | PGM | DEC | DEC | Male | |
| 8 | Paternal | Crossover | PGM | PGF | DEC | DEC | Female | |
| 9 | Paternal | Crossover | PGM | PGF | MGM | MGM | Male | |
| 10 | Maternal | Crossover | MGM | MGF | PGM | PGM | Male | |
| 11 | Maternal | Crossover | MGF | MGM | PGM | PGM | Female | Normal has crossover |
| 12 | Maternal | Crossover | MGF | MGM | NI | PGM | Female | |
| 13 | Maternal | Crossover | MGF | MGM | NI | NI | Female | |
| 14 | Paternal | Crossover | PGF | PGM | MGF | MGF | Female | |
| 15 | Paternal | Crossover | PGM | PGF | MGM | MGM | Male | |
| 16 | Paternal | No crossover | PGM | PGM | MGM | MGM | Male | |
| 17 | Maternal | Crossover | MGM | MGF | PGM | PGM | Female | |
| 18 | Maternal | No crossover | MGM | MGF | PGF | PGF | Male | |
| 19 | Maternal | Crossover | MGM | MGF | PGF | PGF | Male | Twin |
| Maternal | Crossover | MGM | MGF | PGF | PGF | Male | Twin | |
| 20 | Paternal | Crossover | PGF | PGM | MGM | MGM | Male |
DEC, deceased; NI, not informative.
We also determined that in the positions proximally flanking the deletion, in 20 informative meioses, there were 10 grandmaternally derived alleles, and 10 grandpaternally derived alleles. For the normal chromosome 22, there were 14 informative meioses. Eleven had grandmaternal origins of the proximal markers and three had grandpaternally-derived alleles. Further, when we compared parental age at conception in these 20 families, we found that among the parents whose chromosome 22 became deleted (parent of origin), the average maternal age was 30.8 years and average paternal age was 31.6 years. In the parents who transmitted the normal chromosome 22, the average maternal age was 30 years and average paternal age 32.3 years, showing no significant bias of parental age for either gender on meiotic exchanges or on the occurrence of a deletion.
Localization of meiotic exchange sites
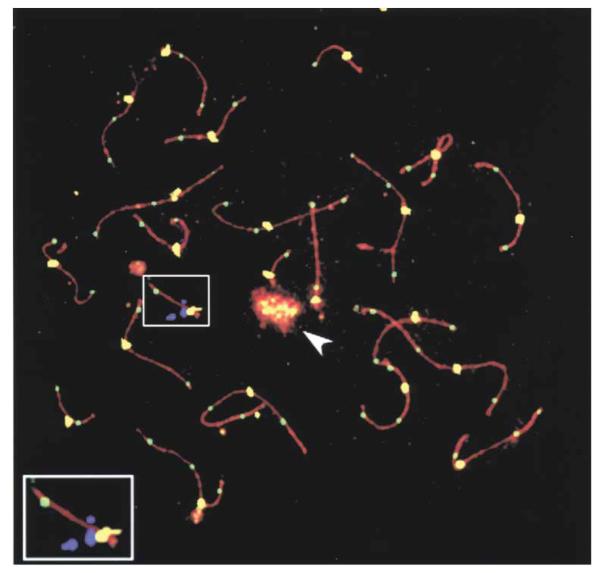
The DNA mismatch repair protein MLH1 has been shown to mark sites of crossovers (22-24). To determine if sequences involved in the deletion of chromosome 22 coincide with sites of meiotic crossover, the position of MLH1 foci relative to the position of cosmid 87f 9 (Fig. 1) were recorded using human spermatocytes (Fig. 5). Cosmid 87f 9 contains the ZNF74 locus and was used to mark the region of deletion. Initial data shows that in the first set of nuclei examined (n = 12), chromosome 22 had a single MLH1 focus. In 75% of these nuclei the focus was on the distal end of the synaptonemal complex. In 8.3% of nuclei the focus was immediately distal to c87f 9 and in 16.7% of nuclei the focus was between c87f 9 and the centromere.
Figure 5.
Immunostaining and FISH signals for a spermatocyte nucleus in pachynema. Anti-SCP3 antibodies (red), anti-MLH1 antibodies (green), CREST antisera (yellow) and c87f 9 (violet). The enlarged region is of chromosome 22. The chromosome 22 crossover site, as indicated by the anti-MLH1 focus (green), is at the distal end of the synaptonemal complex. The cosmid signal of c87f 9 (violet) is close to the centromere (yellow). The sex body containing the X and Y chromosomes is indicated by an arrowhead. In all cases, the CREST foci are significantly larger than the MLH1 foci.
Analysis for evidence of inversions
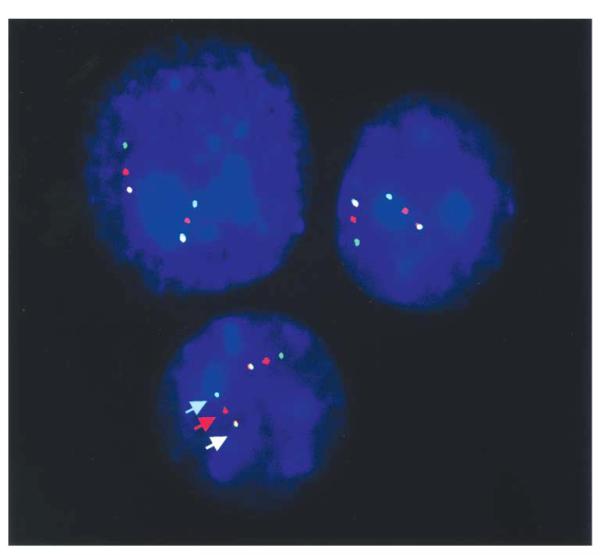
In order to identify whether submicroscopic inversions of 22q11.2 are responsible for its unusual meiotic behavior, FISH was performed using concurrent hybridization of three probes labeled in three colors (Fig. 6). The probes are located both within and flanking the typically deleted region (A–D in Fig. 1). To detect an inversion involving the standard 3 Mb region, the first probe set included: c106e4 labeled with Spectrum Green, c87h3 labeled with Spectrum Red and cHKAD25 labeled with both Spectrum Red and Green, producing a yellow signal. The second probe set was designed to detect an inversion involving the proximal 1.5 Mb region that is deleted in a smaller percentage of DGS/VCFS patients (A and B in Fig. 1). This probe set included: c87h3 labeled with Spectrum Red; c68a1 labeled with Spectrum Green; and c87f 9 labeled with both fluors. Using these methods we examined 22 parents whose chromosome 22 became deleted in their offspring. Samples available from 20 of the parents who had transmitted the normal chromosome to these same probands were also examined to serve as a population control. None of the 42 individuals tested carried an inversion of the typically deleted region in 22q11.2. Figure 6 shows interphase nuclei from one of these individuals as an example of our findings. In all cases the order of FISH signals observed in interphase nuclei was as predicted by the map and not indicative of an inversion. In addition, eight patients with a DGS/VCFS phenotype and no detectable deletion were also studied to determine whether inversion influences phenotype. None were found to harbor an inversion. Thus, we analyzed a total of 30 individuals in whom an inversion might be suspected, 22 were parents, eight were non-deleted probands, and none of the 30 were found to have large or small inversions. It appears then that, unlike what has been described for chromosome 7q11.2 in Williams–Beuren syndrome, inversions of material flanked by chromosome 22q LCRs are not a common predisposing configuration to the 22q11.2 deletion.
Figure 6.
FISH analysis of interphase nuclei to detect an inversion involving the standard 3 Mb region is shown. Probes were labeled in the order of the map (centromere to telomere) depicted in Figure 1, using a green, red, yellow order. All nuclei showed this color/probe order as demonstrated above, consistent with the map.
DISCUSSION
In the largest study to date, we have utilized microsatellite markers flanking the deletion endpoints to explore the deletion mechanism in 20 informative three-generation families of patients with a ‘typical’ 3 Mb de novo deletion of 22q11.2 as determined by FISH. Earlier reports have described a maternal parental origin bias in DGS/VCFS (18,19). Another study found instead, a preponderance of paternal progenitors (20). When these studies are combined, there are 33 maternal and 30 paternal chromosomes that become deleted. Here, we have examined 65 patients with 3 Mb de novo deletions and found no parental origin bias (35 maternal and 30 paternal). Taken together, these data (68 maternal : 60 paternal) do not support a deletion mechanism that is based on parental origin during meiosis.
Further, when we extended our studies back an additional generation to examine the organization of the deleted chromosome, we did not find an over-representation of grandmaternal alleles in the proximal flanking regions, which contrasts with earlier data that examined smaller numbers of deleted chromosomes 22 (10). Thus, we did not find any significant bias regarding the grandparental origin of the alleles flanking the deletion endpoints with respect to the parental origin of the deletion, the crossover frequency, or proband gender (Table 1). Additionally, there was no correlation of the severity of the proband’s cardiac or palate defect to the mode of exchange, grandparental allele location or parental origin of the deletion (S.C. Saitta, unpublished data).
A significant incidence of inversions among non-deleted transmitting progenitors of 7q11 deletions has been described in Williams syndrome (11,17), and is thought to contribute to the mechanism of the deletion. A recent study has also found evidence of inversions of 15q11–q13 in four of six mothers of patients with BP2/3 or class II deletions of chromosome 15q causing Angelman syndrome (25). This study also demonstrated the inversion in normal controls estimating its presence in 4.5% of the general population. These findings led us to further examine whether inversions were playing a role on 22q11. We examined 22 parents whose chromosome became deleted in the probands, as well as the non-transmitting parent as a control, and found no chromosomal inversions flanked by LCRs A and D (Fig. 6), the LCRs that mediate ~90% of the deletions (Fig. 1). In addition, we analyzed the smaller region encompassed by LCRs A and B and found no inversions. This is consistent with a recent study that examined 18 unselected parents of patients with 22q11.2 deletions for inversions around LCRs A and B and found no evidence of such rearrangements (26). We then examined eight non-deleted DGS/VCFS patients to assess whether an inversion might explain their phenotype by virtue of a position effect, but no inversions were detected. Thus, it does not appear that inversions facilitated by the presence of LCRs are a major predisposing factor to the 22q11.2 deletion or to the phenotype associated with it.
By tracing the transmission of the deleted chromosome 22 in three-generation families, we found that 19/20 had interchromosomal meiotic exchanges in proximal 22q, flanking the deletion. When reviewing previous reports of aberrant exchanges in comparable, apparent 3 Mb de novo 22q11 deletions, 8/10 were found to be interchromosomal in one study (10), 3/3 in another (16), and 0/2 or both intrachromosomal in another (5), for a total of 11/15. Although the study of Trost (16) examined five patients, one patient had a familial deletion transmitted from an affected parent, and another patient appeared by microsatellite analysis to have a smaller 1.5 Mb deletion. Thus, we did not include the two cases for comparison with our patients. We now add 19/20 to the 11/15, for a total of 30/35 aberrant interchromosomal exchanges leading to a deletion of 22q11.2. The current study used proximal and distal flanking markers covering the same physical distance as an earlier study (10). In that report, the number of meiotic crossovers expected by random analysis of 10 individuals (based on sex-averaged distances from Genethon and GDB linkage maps and taking interference into consideration) was estimated to be 0.98 (or 1) of 10. Based on this calculation, we would have expected four exchanges in 35 events rather than the 30/35 that have been detected. This number is highly significant (P < 0.0001) and most likely relates to differences in the 22q11.2 region that affect its behavior during meiosis. In fact, the pericentromeric region of 22q has a genomic architecture that appears to predispose it to rearrangements such as deletions, duplications, and translocations. This ‘instability’ is related to the large, highly homologous (98–99% identity at the sequence level) modular, LCRs or segmental duplications that allow for misalignment prior to meiotic exchange. The aim of this study was to examine whether the meiotic behavior of 22q11 is similar to other chromosomes that contain LCRs predisposing them to rearrangement. The data presented here suggest that it is not.
When reviewing aberrant deletion-producing exchanges found on other LCR-containing chromosomes that are associated with microdeletion syndromes, a striking pattern of meiotic behavior does not emerge. Rather, intrachromosomal or meiotic exchange between sister chromatids occurs as often as interchromosomal crossovers. For Williams syndrome and chromosome 7q11, in the only study where deletion size and inversion status were known, 3/7 interchromosomal and 4/7 intrachromosomal exchanges in non-inverted deleted chromosomes were found (11). An earlier study of Williams syndrome noted 12/12 interchromosomal exchanges on deleted chromosomes 7 (27). Two other studies found a total of 13/22 interchromosomal exchanges in cases thought to have a uniform deletion size (10,28). If these four studies are grouped together, there could potentially be as many as 28/41 interchromosomal exchanges (68%), a number that begins to approach what is now described for chromosome 22q (86%). However, it is likely that as many as one-third of the chromosome 7q11 deletions in the latter three reports, or 11/34, occurred on an inverted chromosome, making direct comparisons with our findings on 22q difficult. In Prader–Willi syndrome caused by a deletion of paternal 15q11, two of six informative meioses showed interchromosomal exchanges, while Angelman syndrome, caused by a deletion of maternal 15q11, found all three informative cases to have interchromosomal exchanges (29). Here, small numbers and the phenomenon of imprinting on 15q11 make comparisons with 22q difficult. For the LCR-mediated deletions that cause Smith–Magenis syndrome on 17p11, the data show 5/13 interchromosomal and 8/13 intrachromosomal exchanges (14). For the microdeletions of 17q11 that lead to NF1, five of six informative meioses demonstrated an interchromosomal mechanism (15), however, the numbers are small and all five of the interchromosomal cases were maternal in origin, implying other factors that might bias the outcome.
Our study examined not only the aberrant exchanges that lead to a deletion, but also transmission of the normal 22q homolog in the same families. In contrast to the deleted homolog, we observed that the normal chromosome 22 did not have frequent interchromosomal exchanges in the region flanking the putative deletion endpoints. We found two in 15 informative meioses, consistent with that expected for the average genetic distance. Further, these two interchromosomal exchanges detected in the normal chromosomes 22 did not appear to occur in regions immediately flanking the deletion on proximal 22q, but were in more distal regions, consistent with recombination maps of chromosome 22q. It is still possible that the normal chromosome may be undergoing significant intrachromosomal exchanges in the regions flanking the LCRs; however, this would not be detectable by our methods of tracing the inheritance of informative polymorphic markers. These findings suggest that the genomic structure or sequence of the chromosome 22 that becomes the deleted homolog may be different from the one that remains intact, a difference that makes it susceptible to misalignment, double-stranded breaks, and deletion. Molecular studies of the modular arrangements within the segmental duplications, especially LCRs A and D, in the parents of deleted individuals may help elucidate these mechanisms. Such a task would be arduous however, given the high level of sequence identity among the multiple segmental duplications on this chromosome, and the ‘unclonable’ gaps still present in the region (30). Nonetheless, further work toward an explanation is necessary to better understand not only how 22q11.2 becomes deleted, but also why it is the most frequently encountered microdeletion even amongst the group of chromosomes with similar genomic architecture.
The deletions and duplications that give rise to these genomic diseases are often referred to as meiotic recombination ‘hot spots’. If this were indeed the case, 22q11 deletions should constitute an exchange subset within a region already exhibiting a high rate of recombination. Our meiotic data indicate that this is not the case as the vast majority of MLH1 foci and genetic recombination events on normal chromosomes 22 appear to take place in the distal region of 22q. Further, our results are consistent with the data from a significantly larger study (23) where more than 200 spermatocytes were examined for the location of MLH1 foci. An alternative possibility to recombination ‘hot spots’ is that the exchange occurs during the synaptic process when a sequence-based check for homology occurs.
Recent experiments that examined exchange rates and synaptonemal complex length in meiosis (23) may provide some insight into the meiotic behavior of chromosome 22. Using antibodies against MLH1 and FISH to identify sites of meiotic exchange in human and mouse spermatocytes, these investigators were able to physically assess crossover frequency and locations of the exchanges. Their findings indicate that synaptonemal complexes better reflect the genetic distance than the physical distance of a chromosome. They were able to compare data from structurally normal human chromosomes 1, 16, 21 and 22. The data demonstrate that, similar to what is suggested by recombination studies, exchanges on chromosome 22q occur most frequently in the telomeric region, at a rate apparently twice that for pericentromeric 22q. If relative comparisons of the numbers of exchanges are made, approximately 25% of the exchanges occur in the regions near the centromere [Supplementary Material from Lynn et al. (23)] and if inter- and intrachromosomal mechanisms are equally likely, one would expect that 12.5% of these exchanges would occur by an interchromosomal mechanism. This is consistent with what we have found for the haplotype analysis of the normal 22q in our families, that is, 2/15 meioses (13.3%). However, the deleted chromosomes 22 do not appear to conform to these ‘rules’. While it is logical to infer that the majority of crossovers in the Lynn et al. (23) study are quite distal on chromosome 22 and distant from the commonly deleted region, their studies did not use FISH probes that would allow correction for differential compaction of the chromosomes. The use of the cosmid probe c87f 9 in the present study allows us to take this factor into consideration. Thus, our meiotic exchange localization results are consistent with those inferred from the Lynn et al. (23) study and differ from our genetic analysis of deleted chromosomes 22.
One possible reason for this variance may be what the above study (23) also considered: that synaptonemal complexes, and exchange locations might form in response to double-strand breaks. Unstable genomic regions or those more prone to double-strand breaks would thus be more likely to have a crossover. Since the pericentromeric region of chromosome 22q shows an unusual degree of genomic instability, it is presumably susceptible to double strand breaks. The presence of palindromic sequences on 22q, for example, may play a major role in this susceptibility (30). Palindromic sequences have already been implicated in double strand breaks that mediate translocations of 22q11, including the t(11;22), which is the most frequently occurring recurrent constitutional translocation (30). The 22q11 deletion itself is unusually frequent as well, detected 10 times more often (1:3000 incidence), than other, less commonly occurring microdeletion syndromes. We also know now that the frequent aberrant exchanges resulting in deletion occur primarily by interchromosomal exchange in regions that flank or are within LCR copies in proximal 22q, during meiosis I.
Although at first glance the genomic architecture of 22q11 appears similar to the other LCR-containing chromosomes prone to deletion, significant differences can also be noted by more extensive analysis. The risk factors for LCR-mediated aberrant recombination include size, number, complexity and homology of the segmental duplications. The 22q11 region has an extreme combination of these factors. The segmental duplications are quite large (LCRs A and D are 250–500 kb) and have a complex modular structure with sequence identity of 98–100% (7). In fact, on chromosome 22q11, there are four large segmental duplications densely packed into the pericentromeric region of a small, acrocentric chromosome. It is perhaps this combination of factors that sets the striking pattern of meiotic behavior of 22q apart from other segmental duplication-containing chromosomes.
MATERIALS AND METHODS
Clinical evaluation of the patients
Patients were referred to the Clinical Genetics Center at The Children’s Hospital of Philadelphia (CHOP) for evaluation of dysmorphia, cardiac or organ malformation, or developmental delay. Several patients who had already been diagnosed with the 22q11.2 deletion were referred from outside hospitals to CHOP for comprehensive genetics and subspecialty evaluation. The cohort reported here were enrolled in ongoing research studies after informed consent was obtained under an IRB-approved protocol. The patients were examined and evaluated by the same clinical geneticist (E.H.Z.).
Molecular haplotype analysis
DNA was extracted from buccal brushes of the subjects using published methods (31). In some cases, DNA isolated from the peripheral blood or from transformed lymphoblasts of a proband was used. Six short tandem repeat polymorphisms (STRPs) or microsatellite markers from within the deleted region (46STS, D22S941, 102STS, D22S944 115STS, D22S264) were chosen from those previously used for determining the parental origin of the deletion (21,32). Nine markers flanking the standard 3 Mb deletion were chosen from those reported in the literature (10,33,34) or from the Genome Database. Four map proximal (centromeric) to LCR A, (F8VWFP1, F8VWFP2, D22S420, D22S427) and five are distal (telomeric) to LCR D (D22S257, D22S303, D22S1683, D22S1709, TOP1P2). LCRs A and D delineate the boundaries of the common 3 Mb deletion (3). Another marker (D22S302) located near the 22q telomere was added as a control to assess for recombination in this region. Fluoresceinated PCR primers for the STRP markers were designed using TET, HEX and 6-FAM fluors conjugated to the 5′ ends of the forward primers (IDT; Coralville, IA, USA). PCR was performed in a final volume of 25 μl, with 25–50 ng of genomic DNA, 10× buffer, dNTPs (2.5 mM each), 10 pmol of primers, and 2.5 U of AmpliTaq (all reagents were purchased from Roche). Samples using DNA extracted from buccal samples required a 5 min denaturation at 95°C prior to adding polymerase for optimal results. PCR was then performed using conditions published for the primers. Products were analyzed on an ABI Prism 377 sequencer in the Nucleic Acid/Protein core facility at CHOP. Genescan and Genotyper software (ABI) was used to analyze the size and relative intensity of each product. Haplotypes were reconstructed by following segregation of the markers and using grandparental alleles to set meiotic phase. The most parsimonious interpretation was used for genotypes observed in families where only one grandparent was available, and assumed no recombination.
Deletion detection and sizing
All probands were determined to have a deletion of the 22q11.2 region by absence of one copy of either the N25 or TUPLE probe (Oncor/Vysis) when FISH was performed in a clinical cytogenetics laboratory. Parents of the probands were also analyzed by FISH studies of peripheral blood lymphocytes. Those that showed a deletion were excluded from this study, and were referred to Clinical Genetics for follow-up as per the approved protocol. Subsequent FISH was performed on metaphase spreads from the deleted probands to assess the extent of the deletion using probes and methods described previously (7).
Fluorescent antibody localization (FAL) and FISH
Spermatocytes isolated from testicular biopsies from normal males were prepared for immunostaining using the same method described for the preparation of mouse spermatocytes (35). In addition, immunostaining of synaptonemal complexes was performed with antibodies to SCP3 (23) and meiotic exchange sites were localized using antibodies against MLH1 (24). CREST antiserum (1 : 2000) was applied to visualize centromeres (23). Four separate images were captured for each nucleus (CREST, Cy5; SCP3, rhodamine, MLH1, FITC and DAPI). The location of each nucleus on the slide was recorded.
After image acquisition of the FAL signals, the spermatocyte preparation underwent a single round of denaturation and FISH. The antifade solution was removed by washing the slides in 4× SSC/0.2% Tween 20. The denaturation time was 15 min and the hybridization time 120 h. The probes used were cosmid c87f 9 (Fig. 1) and a commercially available chromosome paint for the long arm of chromosome 22. The cosmid was labeled with biotin by standard nick translation methods, and the chromosome 22 paint was supplied already labeled with Spectrum Orange. Anti-avidin FITC was applied during the post-hybridization washes. Each nucleus imaged in the previous FAL experiment was located and two additional separate images captured (c87f 9, FITC; chromosome paint, rhodamine). The images were treated uniformly to remove background using Adobe Photoshop software (Adobe Systems Inc., San Jose, CA, USA) and merged. The number of MLH1 foci for chromosome 22 was recorded. The position of the c87f 9 signal was observed relative to the MLH1 foci and centromere of chromosome 22. The q-arm of chromosome 22 was divided into three equal regions (proximal, medial and distal to the centromere) as previously defined [Supplementary Material in Lynn et al. (23)]. The number of MLH1 foci for chromosome 22 was recorded and the relative position scored within the defined regions. The position of the c87f 9 signal was also scored within the defined regions.
Inversion detection
In order to detect an inversion of chromosomal material surrounding the LCR-22s, FISH was performed using two sets of three cosmid probes labeled in three different colors. The first probe set included: c106e4 labeled with Spectrum Green; c87h3 labeled with Spectrum Red; and cHKAD25 labeled with both Spectrum Red and Green, producing a yellow signal. The second probe set included: c87h3 labeled with Spectrum Red; c68a1 labeled with Spectrum Green; and c87f 9 labeled with both. Cosmids were labeled directly with Spectrum Red or Green, and FISH was performed using standard conditions (6). Fifty interphase nuclei were examined for each individual. Only interphase nuclei that clearly showed the requisite six signals in an unambiguous position for each set of three probes were scored.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Danielle Conforto for technical support, Dr Eric Rappaport of the Nucleic Acid/Protein Core at CHOP for helpful suggestions for genotyping using buccal swab DNA, the staff of the 22q and You Center at CHOP, and the patients, parents and grandparents who participated in this study. This work was supported by K08-HL04487, and the Children’s Health Research Center (P30-HD28815) at Children’s Hospital of Philadelphia to S.C.S.; P01-DC02027, R01-CA39926, and the Charles E.H. Upham Chair in Pediatrics to B.S.E.; and R01-HD39384, and the Robert Leet and Clara Gutherie Patterson Award to T.A.
Footnotes
SUPPLEMENTARY MATERIAL Supplementary Material is available at HMG Online.
REFERENCES
- 1.McDonald-McGinn DM, Kirschner R, Goldmuntz E, Sullivan K, Eicher P, Gerdes M, Moss E, Solot C, Wang P, Jacobs I, et al. The Philadelphia Story: the 22q11.2 deletion: report on 250 patients. Genet. Couns. 1999;10:11–24. [PubMed] [Google Scholar]
- 2.McDonald-McGinn DM, Tonnesen MK, Laufer-Cahana A, Finucane B, Driscoll DA, Emanuel BS, Zackai EH. Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: cast a wide FISHing net! Genet Med. 2001;3:23–29. doi: 10.1097/00125817-200101000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Emanuel BS, McDonald-McGinn D, Saitta SC, Zackai EH. The 22q11.2 deletion syndrome. Adv. Pediatr. 2001;48:39–73. [PubMed] [Google Scholar]
- 4.Burn J, Goodship J. Developmental genetics of the heart. Curr Opin Genet Dev. 1996;6:322–325. doi: 10.1016/s0959-437x(96)80009-8. [DOI] [PubMed] [Google Scholar]
- 5.Edelmann L, Pandita RK, Morrow BE. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 1999;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 7.Shaikh TH, Kurahashi H, Saitta SC, O’Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 8.Emanuel BS, Shaikh TH. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 9.Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. A. Rev. Genom. Hum. Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- 10.Baumer A, Dutly F, Balmer D, Riegel M, Tukel T, Krajewska-Walasek M, Schinzel AA. High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum. Mol. Genet. 1998;7:887–894. doi: 10.1093/hmg/7.5.887. [DOI] [PubMed] [Google Scholar]
- 11.Bayes M, Magano LF, Rivera N, Flores R, Jurado L.A. Perez. Mutational mechanisms of Williams-Beuren syndrome deletions. Am. J. Hum. Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrozzo R, Rossi E, Christian SL, Kittikamron K, Livieri C, Corrias A, Pucci L, Fois A, Simi P, Bosio L, et al. Inter- and intrachromosomal rearrangements are both involved in the origin of 15q11–q13 deletions in Prader–Willi syndrome. Am. J. Hum. Genet. 1997;61:228–231. doi: 10.1086/513907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Chromosome breakage in the Prader–Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am. J. Hum. Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw CJ, Bi W, Lupski JR. Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am. J. Hum. Genet. 2002;71:1072–1081. doi: 10.1086/344346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Correa C. Lopez, Brems H, Lazaro C, Marynen P, Legius E. Unequal meiotic crossover: a frequent cause of NF1 microdeletions. Am. J. Hum. Genet. 2000;66:969–974. doi: 10.1086/302920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trost D, Wiebe W, Uhlhaas S, Schwindt P, Schwanitz G. Investigation of meiotic rearrangements in DGS/VCFS patients with a microdeletion 22q11.2. J. Med. Genet. 2000;37:452–454. doi: 10.1136/jmg.37.6.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW. A 1.5 million-base pair inversion polymorphism in families with Williams–Beuren syndrome. Nat. Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seaver LH, Pierpont JW, Erickson RP, Donnerstein RL, Cassidy SB. Pulmonary atresia associated with maternal 22q11.2 deletion: possible parent of origin effect in the conotruncal anomaly face syndrome. J. Med. Genet. 1994;31:830–834. doi: 10.1136/jmg.31.11.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demczuk S, Levy A, Aubry M, Croquette MF, Philip N, Prieur M, Sauer U, Bouvagnet P, Rouleau GA, Thomas G, et al. Excess of deletions of maternal origin in the DiGeorge/velo-cardio-facial syndromes. A study of 22 new patients and review of the literature. Hum. Genet. 1995;96:9–13. doi: 10.1007/BF00214179. [DOI] [PubMed] [Google Scholar]
- 20.Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J. Med. Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitta SC, Harris SE, McDonald-McGinn DM, Emanuel BS, Tonnesen MK, Zackai EH, Seitz SC, Driscoll DA. Independent de novo 22q11.2 deletions in first cousins with DiGeorge/velocardiofacial syndrome. Am. J. Med. Genet. 2004;124A:313–317. doi: 10.1002/ajmg.a.20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie D-M, Monell C, Arnheim N, Bradley A, Ashley T, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 23.Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 24.Froenicke L, Anderson LK, Wienberg J, Ashley T. Integrating genetic recombination and meiotic chromosome structure in male mice. Am. J. Hum. Genet. 2002;71:1353–1368. doi: 10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gimelli G, Pujana MA, Patricelli MG, Russo S, Giardino D, Larizza L, Cheung J, Armengol L, Schinzel A, Estivill X, Zuffardi O. Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum. Mol. Genet. 2003;12:849–858. doi: 10.1093/hmg/ddg101. [DOI] [PubMed] [Google Scholar]
- 26.Gebhardt GS, Devriendt K, Thoelen R, Swillen A, Pijkels E, Gewillig M, Fryns JP, Vermeesch JR. No evidence for a parental inversion polymorphism predisposing to rearrangements at 22q11.2 in the DiGeorge/Velocardiofacial syndrome. Eur. J. Hum. Genet. 2003;11:109–111. doi: 10.1038/sj.ejhg.5200930. [DOI] [PubMed] [Google Scholar]
- 27.Urban Z, Helms C, Fekete G, Csiszar K, Bonnet D, Munnich A, Donis-Keller H, Boyd CD. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am. J. Hum. Genet. 1996;59:958–962. [PMC free article] [PubMed] [Google Scholar]
- 28.Dutly F, Schinzel A. Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams–Beuren syndrome. Hum. Mol. Genet. 1996;5:1893–1898. doi: 10.1093/hmg/5.12.1893. [DOI] [PubMed] [Google Scholar]
- 29.Robinson WP, Dutly F, Nicholls RD, Bernasconi F, Penaherrera M, Michaelis RC, Abeliovich D, Schinzel AA. The mechanisms involved in formation of deletions and duplications of 15q11–q13. J. Med. Genet. 1998;35:130–136. doi: 10.1136/jmg.35.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel B, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 31.Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ. Health Perspect. 1999;107:517–520. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Driscoll DA, Emanuel BS, Mitchell LE, Budarf ML. PCR assay for screening patients at risk for 22q11.2 deletion. Genet. Test. 1997;1:109–113. doi: 10.1089/gte.1997.1.109. [DOI] [PubMed] [Google Scholar]
- 33.Buetow KH, Ludwigsen S, Scherpbier-Heddema T, Quillen J, Murray JC, Sheffield VC, Duyk GM, Weber JL, Weissenbach J, Gyapay G, et al. Human genetic map. Genome maps V. Science. 1994;265:2055–2070. doi: 10.1126/science.8091228. [DOI] [PubMed] [Google Scholar]
- 34.Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane–Yeboa K, Warburton D, et al. Molecular definition of 22q11.deletions in 151 velo-cardio-facial syndrome patients. Am. J. Hum. Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.