Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor, which controls adipocyte differentiation. We targeted with homologous recombination the PPARγ2-specific exon B, resulting in a white adipose tissue knockdown of PPARγ. Although homozygous (PPARγhyp/hyp) mice are born with similar weight as the WT mice, the PPARγhyp/hyp animals become growth retarded and develop severe lipodystrophy and hyperlipidemia. Almost half of these PPARγhyp/hyp mice die before adulthood, whereas the surviving PPARγhyp/hyp animals overcome the growth retardation, yet remain lipodystrophic. In contrast to most lipodystrophic models, the adult PPARγhyp/hyp mice only have mild glucose intolerance and do not have a fatty liver. These metabolic consequences of the lipodystrophy are relatively benign because of the induction of a compensatory gene expression program in the muscle that enables efficient oxidation of excess lipids. The PPARγhyp/hyp mice unequivocally demonstrate that PPARγ is the master regulator of adipogenesis in vivo and establish that lipid and glucose homeostasis can be relatively well maintained in the absence of white adipose tissue.
The peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor that acts as a lipid sensor, integrating the control of energy, lipid, and glucose homeostasis (1). The actions of PPARγ are mediated by two protein isoforms, the widely expressed PPARγ1 and adipose tissue-restricted PPARγ2 with an additional 28 aa in the NH2 terminus (2–4). PPARγ is the master regulator of differentiation and energy storage by adipocytes (5–8). Despite undisputed arguments that support a pivotal role of PPARγ in adipocyte differentiation in vitro, the PPARγ field has been slowed by the absence of good animal models for PPARγ deficiency, because homozygous PPARγ-deficient animals are embryonic lethal (8). This has had a restrictive impact on studies aimed at unraveling the pleiotropic roles of PPARγ in adult homeostasis. We therefore generated, by homologous recombination, mice that carry a hypomorphic mutation at the PPARγ2 locus and characterized the molecular and metabolic phenotype of these mice.
Methods
Homologous Recombination. The main features of our targeting strategy are shown in Fig. 1A. The loxP sites were inserted in reverse orientation at position -45 of the PPARγ2 gene, 445 bp downstream of the exon B splice site and at the 3′ end of the frt-PGKneo-frt cassette. The Pro-12–Ala mutation that was introduced in the B exon was flanked by an EcoRI site. Chimeric animals were generated from two independently targeted embryonic stem (ES) cell clones (nos. 84 and 73). Heterozygous animals, derived from the two ES cell clones, were backcrossed for seven generations to mice with either a SV129 or a C5S7BL/6J background and then intercrossed to generate PPARγhyp/hyp mice for analysis. The Pro-12–Ala knock-in PPARγAla12Ala animals were generated by intercrossing PPARγhyp/hyp mice with mice that expressed the FLP recombinase under the control of a cytomegalovirus promoter to remove the neomycin cassette.
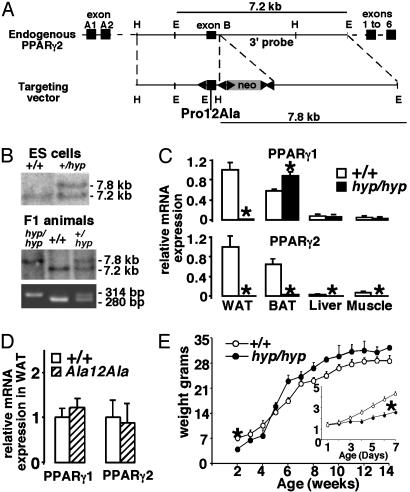
Fig. 1.
Targeting of the PPARγ2 gene. (A) Schematic representation of the mouse PPARγ2 gene (Upper) and targeting vector (Lower). EcoRI (E), HindIII (H), loxP (left arrow), neomycin cassette (gray box), frt sites (right arrow), and exons (dark boxes) are indicated. (B) Southern blot and PCR analysis of ES cell clones and mice. (C) Quantitative RT-PCR analysis of PPARγ1 and PPARγ2 mRNA in WAT, BAT, liver, and muscle. (D) Quantitative RT-PCR analysis of PPARγ1 and PPARγ2 mRNA in WAT of WT and PPARγAla12Ala mice. (E) Body weight gain in males (n > 10). (Inset) Weight gain in the postnatal period (n > 17) is shown.
Animal Experiments. Age- and gender-matched mice with a 50%/50% C57BL/6J/129Sv or pure C57BL/6J or pure 129Sv background were used. Most experimental animals were derived from C57BL/6J mice originating from ES cell clone 84, although crucial experiments were repeated in mice on a different genetic background and/or derived from ES cell clone 73. Some mice were gavaged with 30 mg/kg rosiglitazone for 2 weeks. Blood and tissue analysis and clinical biochemistry were as described (9, 10).
RNA Analysis. RNA preparation was as described (9). cDNA was synthesized by using the SuperScript System (Invitrogen) and random hexamer primers. Quantitative RT-PCR was performed by using LightCycler FastStart DNA Master SYBR Green I from Roche Diagnostics according to the manufacturer's protocol. The sequences of primers used are available at www-igbmc.u-strasbg.fr/Departments/Dep_V/Dep_VA/Publi/Paper.html. GAPDH mRNA or 18S rRNA was used as the invariant control.
Data Analysis. Data are presented as means ± SEM. Differences analyzed with Student's t test were considered statistically significant at P < 0.05 and are indicated by an asterisk in the figures.
Results
Targeting the PPARγ2 Locus. The proline residue at position 12 in the PPARγ2 gene was replaced with alanine (Pro-12–Ala) by homologous recombination in ES cells (Fig. 1A). This strategy also introduced three loxP sites for the removal of selection marker or exon B. The male chimera that originated from two independent ES cell clones (Fig. 1B) transmitted the mutant PPARγ2 allele to their offspring (Fig. 1B). Unexpectedly, both PPARγ2 and PPARγ1 transcripts were significantly reduced in the white adipose tissue (WAT) of the homozygous (PPARγhyp/hyp) animals (Fig. 1C). In the brown adipose tissue (BAT), liver, and muscle, the PPARγ2 mRNA expression is also virtually undetectable. The PPARγ1 mRNA expression remained unchanged in liver and muscle. PPARγ1 levels were, however, found to be increased in the BAT. Sequencing of the PPARγ2 RT-PCR products confirmed that the gene was correctly targeted and spliced (data not shown). These data demonstrate that targeting of the PPARγ2 locus disrupted PPARγ2 mRNA expression and in addition altered PPARγ1 mRNA expression in WAT. Importantly, the virtual absence of PPARγ2 mRNA negated any possible effect the Pro-12–Ala mutation may have on the phenotype of the PPARγhyp/hyp animals. When we excised the neomycin cassette by breeding homozygous mice with animals expressing FLP recombinase, the resulting PPARγAla12Ala (neomycin excised) mice have normal mRNA levels of PPARγ1 and PPARγ2 in WAT (Fig. 1D). This finding demonstrates that the absence of PPARγ expression in WAT and the phenotype of the PPARγhyp/hyp mice (described below) is a consequence of the neomycin cassette interfering with PPARγ expression.
PPARγ2 Gene Targeting Produces Lipodystrophic Mice. Heterozygous animals were intercrossed, and pups were born at expected Mendelian ratio in SV129, C57BL/6J, and mixed SV129 (50%)/C57BL/6J (50%) backgrounds (data not shown). The homozygous animals were indistinguishable from their littermates at birth. During the first week, these mice became severely growth retarded (Fig. 1E Inset) and 24% died, although they were suckling and nursed actively; the mortality increased to >40% by the time of weaning. By week 5 the surviving homozygous animals had a similar body weight as their littermates (Fig. 1E).
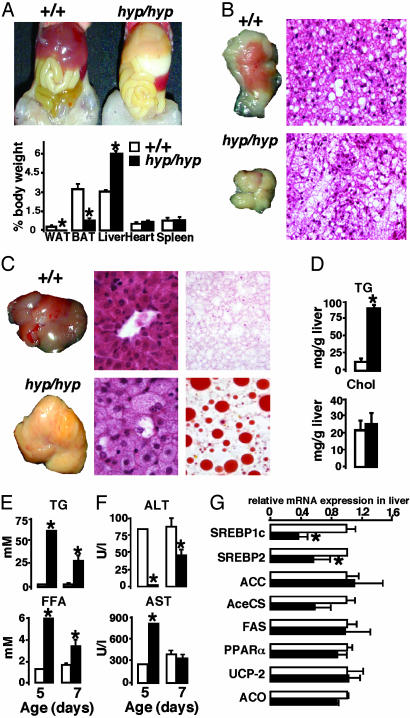
Analysis at 1 week showed that homozygous animals had no WAT (Fig. 2A), whereas the BAT was smaller, paler in color, and infused with lipids (Fig. 2B). The body temperature of the PPARγhyp/hyp animals was diminished relative to WT mice (31.5 ± 0.2°C versus 33.9 ± 0.3°C). PPARγhyp/hyp animals had massive hepatomegaly (Fig. 2 A and C) caused by macrovesicular steatosis, subsequent to an increase in liver triglycerides (TGs; Fig. 2 C and D). Serum TG and free fatty acid (FFA) levels were higher in the homozygous animals at day 5 (Fig. 2E) and abnormal alanine aminotransferase (ALT) and aspartate transaminase (AST) levels indicated severe liver damage (Fig. 2F). Between days 5 and 7 liver function was recovering in the surviving homozygous animals, because serum ALT, AST, TG, and FFA were normalizing. Gene expression in the liver of homozygous mice at 7 days showed reduced levels of sterol regulatory element-binding protein-1c and 2 (SREBP1c and SREBP2) mRNA. Other genes involved in energy metabolism [FA synthase (FAS), acetyl-CoA synthetase, acetyl-CoA carboxylase (ACC), PPARα, acyl-CoA oxidase (ACO), and uncoupling protein 2 (UCP-2)] did not differ between both genotypes (Fig. 2G), indicating that the elevated FFA plasma concentrations were not caused by excessive FA production or decreased catabolism in the liver. Heart, skeletal muscle, and kidney also accumulated lipids (data not shown).
Fig. 2.
Lipodystrophy in young PPARγhyp/hyp mice. (A) Exposed ventral view of a 7-day-old PPARγ+/+ and PPARγhyp/hyp mouse and percentage of organ over body weights (n > 10). (B) Gross morphology and histology of interscapular BAT (×4,000). (C) Gross morphology, histology (×6,000), and Oil red O staining of the liver. (D) Hepatic TG and cholesterol content in both genotypes (n = 4). (E) Serum TG and FFA levels at 5 and 7 days of age (n = 6–8). (F) Serum ALT and AST levels at 5 and 7 days of age. (G) Liver mRNA levels of SREBP1c, SREBP2, FAS, ACC, acetyl-CoA synthetase (AceCS), PPARγ, UCP-2, and ACO were determined by quantitative RT-PCR.
Consequences of Lipodystrophy in the Homozygous Mice. The homozygous animals remained lipodystrophic throughout life (examined at 4, 8, 12, and 20 weeks) with an absence of visceral WAT and very sparse s.c. WAT depots (Fig. 3 A and B). In the rare cases where s.c. WAT was detected in homozygous mice, adipocytes were hypertrophic, reflecting the intensified demand for TG storage (Fig. 3C). In the fed state, serum FFAs were elevated in PPARγhyp/hyp mice, but they did not increase upon fasting, like in the WT animals, reflecting the absence of WAT stores (Fig. 3D). TG (Fig. 3E), but not cholesterol, levels were reduced in homozygous mice (data not shown).
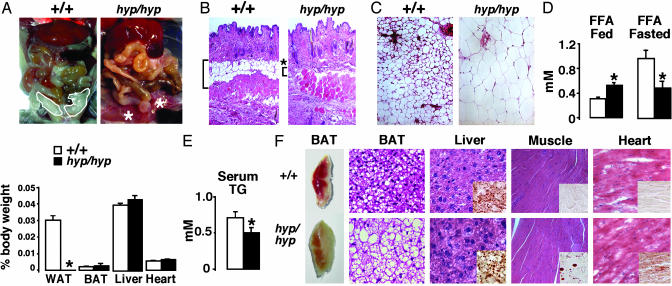
Fig. 3.
Lipodystrophy and lack of liver steatosis in adults. (A) Exposed ventral view of a 20-week-old mouse and percentage of organ over body weights (n > 8). (B) Skin histology (×4,000). The white adipocytes in the hypodermis are shown in brackets. (C) Histology of the s.c. WAT (×4,000). (D) Serum FFA in the fed and fasted state. (E) Serum TG in the fasted mice. (F) Morphology of interscapular BAT, histological sections of BAT, liver, skeletal muscle, and heart of a representative PPARγ+/+ (Upper) and a PPARγhyp/hyp mouse (Lower) (hematoxylin/eosin stain, Oil red O staining in the Inset; ×4,000).
The physiologic consequences of a near absence of WAT were then assessed. All organs were similar in size between the genotypes (Fig. 3A). The BAT of PPARγhyp/hyp mice had a different morphology because it was thickly veiled by WAT and very pale, presumably because of an increase in WAT-like unicellular adipocytes (Fig. 3F). Importantly, the livers of adult homozygous mice were normal in size, morphology, and function (normal ALT and AST levels) and showed no signs of inflammation or fibrosis (Fig. 3 A and F). TG levels of the adult homozygous livers were also similar to the WT (data not shown). The skeletal muscle and the heart, however, both contained more lipids upon Oil red O staining (Fig. 3F).
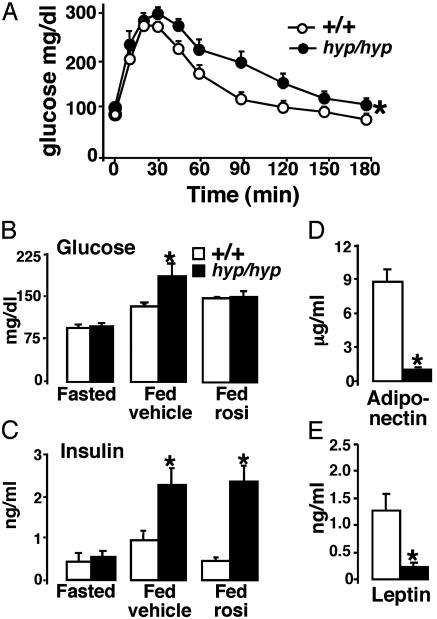
Because muscle lipid accumulation is associated with glucose intolerance, we performed an i.p. glucose tolerance test (IPGTT). During the IPGTT glucose levels were consistently higher in PPARγhyp/hyp mice (Fig. 4A). Serum glucose and insulin concentrations were normal in fasted homozygous mice, whereas in the fed state both levels were elevated, indicative of a mild insulin resistance (Fig. 4 B and C). Treatment with the PPARγ agonist rosiglitazone alleviated the glucose intolerance, but not the insulin resistance in homozygous mice (Fig. 4 B and C).
Fig. 4.
Metabolic consequences of PPARγ targeting. (A) Serum glucose levels after i.p. glucose tolerance test with PPARγ+/+ (○) and PPARγhyp/hyp (•) mice (n = 8). (B) Serum glucose in fasted and fed state in vehicle-treated or rosiglitazone (30 mg/kg per day for 2 weeks)-treated mice after meal tolerance test (n = 4–8). (C) Serum insulin in fasted and fed state (treated with either vehicle or rosiglitazone) after a meal tolerance test (n = 4–8). (D) Serum adiponectin (n > 8). (E) Serum leptin (n > 8).
WAT-derived signaling factors, adiponectin and leptin, were reduced (Fig. 4 D and E). Leptin has been shown to be important in sexual maturation (11). The homozygous mice were fertile, but when homozygous animals were crossed their litter size was reduced, which was in contrast to the litters obtained from intercrosses between heterozygotes. The homozygous mice did not demonstrate the normal relationship between plasma leptin and food intake (12), because the quantity of food consumed was comparable between the genotypes. The homozygous mice showed, however, mild polydypsia, and polyuria (data not shown), secondary to urinary water loss, a hallmark of glucose intolerance.
Metabolic Compensation for Lipodystrophy Occurs Mainly in the Muscle. The essential requirement for PPARγ in WAT was illustrated by the aberrant gene expression in the homozygous animals. Adipogenic markers and PPARγ target genes were decreased [leptin, adiponectin, and lipoprotein lipase (LPL)], whereas genes involved in FAS and β-oxidation and energy dissipation (ACO, UCP-2, and UCP-3) were reduced (Fig. 5A).
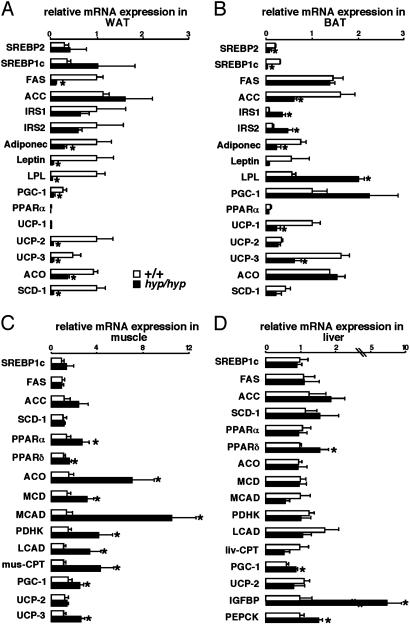
Fig. 5.
Muscle compensation of lipodystrophy in adult PPARγhyp/hyp mice. Quantitative RT-PCR of mRNA levels in WAT (A), BAT (B), muscle (C), and liver (D) of WT (empty bars) and homozygous (filled bars) mice. MCD, malonyl-CoA decarboxylase; MCAD, medium chain acyl-CoA dehydrogenase; PDHK, pyruvate dehydrogenase kinase 4; LCAD, long chain acyl-CoA dehydrogenase; CPT, carnitine acyltransferase; PEPCK, phosphoenolpyruvate carboxykinase.
The BAT morphology and gene responses reflected the increased demand for FFA storage caused by the absence of WAT. Although PPARγ2 mRNA was virtually absent in BAT of homozygous mice, PPARγ1 mRNA was increased (Fig. 1C). Correspondingly, there was an increase in the expression of PPARγ target genes like LPL, explaining the artial transdifferentiation of brown into white adipocytes in the homozygous mice (Figs. 1C and 5B). The increased FFA uptake subsequent to the increase in LPL may have altered BAT function because UCP-1 and UCP-3 mRNA levels decreased, insulin receptor substrate 1 (IRS1) and IRS2 mRNA expression increased, and FA synthesis seemed down-regulated (SREBP1c and ACC) in homozygous mice (Fig. 5B).
Fed homozygous mice had a higher level of FFA than WT mice, reflecting the absence of WAT (Fig. 3D), which consequently increases the availability of natural PPAR ligands. The increased availability of PPAR ligands was reflected by the induction of genes that control FFA catabolism [PPARα, PPARδ, ACO, malonyl-CoA decarboxylase, medium chain acyl-CoA dehydrogenase, pyruvate dehydrogenase kinase 4, long chain acyl-CoA dehydrogenase, muscle carnitine acyltransferase-1, PPARγ coactivator 1 (PGC-1), and UCP-3] in the muscle of homozygous mice (Fig. 5C). In comparison, expression of genes involved in FA synthesis remained unchanged [PPARγ1, SREBP1c, FAS, ACC, and stearoyl-CoA desaturase 1 (SCD-1)]. Distinguishing between the contribution of PPARα and PPARβ/δ to explain the induction of genes involved in FFA catabolism is difficult, because both receptors regulate similar genes (13, 14). Traditionally, PPARα induces FA oxidation in response to stresses, such as fasting and exercise, and is not required to maintain constitutive activity of FA oxidation enzymes in the skeletal muscle (13). Conversely, PPARβ/δ is the most abundant PPAR in this tissue and is induced in response to FFA. Moreover, UCP-3, which was strongly enhanced in the homozygous mice, has been shown to be increased by PPARβ/δ, particularly in the absence of PPARα (13–15). This finding suggests that activation of signaling through PPARα and PPARβ/δ plays a prominent role in compensating for the absence of WAT by up-regulating FA oxidation in the skeletal muscle in response to elevated FFA levels.
Lipid metabolism and, most notoriously, β-oxidation genes in the liver remained unchanged, reflecting the capacity of the muscle to compensate for the absence of WAT (Fig. 5D). This was further supported by the similar levels of β-oxidative enzymatic activity within the liver in the PPARγhyp/hyp and WT mice (Table 1, which is published as supporting information on the PNAS web site). Gluconeogenesis seems induced in the liver of fasted homozygous animals as reflected by the induction of phosphoenolpyruvate carboxykinase mRNA, which is most likely secondary to the increase in PGC-1 mRNA expression (Fig. 5D) (16, 17). Like in SREBP1c-deficient lipodystrophic mice (18), insulin-like growth factor binding protein mRNA, a marker of insulin resistance, was increased in homozygous animals. Taken together, these data indicate that the liver contributes to the mild glucose intolerance and suggest that the muscle is the major compensatory organ for lipid metabolism (Fig. 5 C and D).
Discussion
Because PPARγ knockout mice are embryonic lethal (8), the effects of the absence of PPARγ have not been extensively characterized in vivo. Our homologous recombination strategy resulted in a WAT-specific PPARγ knockdown, as a consequence of the presence of the neomycin cassette in the proximity of the B exon. The introduced genomic modifications encompass a region ≈500 bp downstream of the PPARγ2-specific B exon and are distant [>33 kb (2)] from the PPARγ1 A2 exon, underscoring the importance of the PPARγ2 locus, which seems to control the expression of PPARγ1 in WAT, but not in other tissues. This regulatory cascade between the two PPARγ isoforms during early adipocyte differentiation, where PPARγ2 expression precedes that of PPARγ1, has been described (7, 19). Because homozygous mice are normal at birth, the PPARγ2 isoform may not be required for placental or cardiac development (6, 8, 20). Targeting of the PPARγ2 locus severely compromised WAT development postnatally, underscoring that PPARγ2 is fundamental for adipogenesis. WAT is crucial for development, which became evident by the early mortality of homozygous mice. PPARγ2, however, appears dispensable for the establishment of BAT. Although adult homozygous animals lack visceral WAT, some WAT was present around the BAT and in certain s.c. depots. Altogether, these observations indicate that PPARγ isoforms direct depot-specific regulation of adipose tissue development.
Human studies have associated PPARγ mutations with autosomal dominant familial partial lipodystrophy (21–23). The lipodystrophic neonatal phenotype in the PPARγhyp/hyp mice resembles human congenital generalized lipodystrophy (CGL). In CGL, normally the adipose deficiency is accompanied by severe insulin resistance, hyperinsulinemia, hyperglycemia, hypertriglyceridemia, and fatty liver, which persists throughout life (24). Although the neonatal phenotype mirrors CGL, adult mice overcome the fatty liver and hyperlipidemia and are only mildly glucose intolerant, which is distinct from most other lipodystrophy models (25–29).
The FFA levels in adult PPARγhyp/hyp mice remain rather low considering the absence of WAT. We hypothesized that these mice achieve this steady-state FFA level by increasing FA catabolism in the skeletal muscle (Fig. 5C). This increase is likely caused by enhanced expression and activity (subsequent to increased availability of their FA ligands) of PPARα and PPARβ/δ to prevent lipid imbalance. The catabolism of excess FA by the muscle of the PPARγhyp/hyp mice results in a decrease of serum and liver lipids and an improvement in liver function, ultimately, converting the dysfunctional neonatal fatty liver into a rather normal adult liver. Similarly, the lipoatrophic fld (lipin) mouse overcomes liver steatosis after weaning by a combination of genetic makeup and developmental induction of both an increased capacity for FA oxidation and TG secretion (30–32). Overall, this demonstrates the coordinated management of lipid homeostasis between the three PPAR subtypes within the predominant FA oxidizing organs muscle (PPARα and PPARβ/δ), liver (PPARα), and fat storage adipose tissues (PPARγ).
The comparison of PPARγhyp/hyp mice and PPARγ+/- mice provides insights into how PPARγ coordinates energy homeostasis in metabolic tissues. Ubiquitous reduction of PPARγ in all tissues in the PPARγ+/- mice results in normal body weight and fat depots. These mice are insulin sensitive and resistant to diet-induced obesity (20, 33); similar results have also been observed in mice in which PPARγ activity was inhibited pharmacologically (34, 35). In comparison, PPARγhyp/hyp mice with reduced PPARγ levels in WAT are lipodystrophic and mildly glucose intolerant, thus implicating WAT as the fundamental target tissue for PPARγ to maintain glucose tolerance. In the absence of WAT, such as the case in adult PPARγhyp/hyp mice, glucose tolerance is compromised mainly by the increased gluconeogenesis subsequent to the induction of PGC-1 and phosphoenolpyruvate carboxykinase in the liver. Treatment with PPARγ agonists can overcome the glucose intolerance, although insulin resistance is not corrected. The results obtained with the PPARγ agonist in our WAT PPARγhyp/hyp mice together with recent data in liver- and muscle-specific PPARγ knockout mice indicate that primarily the WAT and not the liver and muscle are crucial for the protective effects of PPARγ agonist on insulin resistance (36–38).
Few treatments have proven effective for lipodystrophic syndromes. The PPARγ agonist troglitazone has been the only treatment for lipodystrophy that induces adipocyte differentiation (39). Thiazolidinediones hence increase total body fat and also improve metabolic control in patients with lipoatrophic diabetes (40). In comparison, leptin only improves the metabolic consequences of the absence of WAT (27, 41–43). Lipodystrophy in the PPARγhyp/hyp mice provides a mechanistic rational to support the use of PPARγ agonists in human lipodystrophic syndrome.
In conclusion, the PPARγhyp/hyp mouse model reveals that the PPARγ2 locus is the critical regulator of adipogenesis in vivo, because PPARγhyp/hyp mice have a severe lipodystrophic syndrome with a significant neonatal mortality. Surviving PPARγhyp/hyp mice have only limited metabolic consequences of the lipodystrophy because of an efficient compensation by other organs, particularly the muscle. These data also have implications for the treatment of patients because they lend support to the hypothesis that selective PPARγ modulators with a partial agonist (9) or even antagonist profile (34, 35), rather than full agonists, could be the preferred therapeutic strategy to treat metabolic disorders (reviewed in ref. 1). PPARγ modulators with a reduced adipogenic drive, but with agonist activity in other tissues, might be ideally positioned to maintain glucose homeostasis in these metabolic disorders. The genetic evidence obtained in the PPARγhyp/hyp mice also provides an argument that PPARα and PPARβ/δ in the skeletal muscle may be effective targets to treat the metabolic syndrome.
Supplementary Material
Acknowledgments
We thank C. Haby, O. Wendling, M. van Werkhoven, and the staff of the Institut Clinique de la Souris for technical assistance and P. Chambon, R. Reiter, R. Wanders, and the Auwerx laboratory for support and discussion. This work was supported by grants from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Hopitaux Universitaires de Strasbourg, the European Union, the European Molecular Biology Organization, and the National Institutes of Health.
Abbreviations: PPARγ, peroxisome proliferator-activated receptor γ; ES, embryonic stem; WAT, white adipose tissue; BAT, brown adipose tissue; TG, triglyceride; FA, fatty acid; FFA, free FA; FAS, FA synthase; ALT, alanine aminotransferase; AST, aspartate transaminase; SREBP, sterol regulatory element-binding protein; ACC, acetyl-CoA carboxylase; ACO, acyl-CoA oxidase; UCP, uncoupling protein; LPL, lipoprotein lipase; PGC, PPARγ coactivator; SCD-1, stearoyl-CoA desaturase; IRS, insulin receptor substrate.
References
- 1.Picard, F. & Auwerx, J. (2002) Annu. Rev. Nutr. 22, 167-197. [DOI] [PubMed] [Google Scholar]
- 2.Zhu, Y., Qi, C., Korenberg, J. R., Chen, X.-N., Noya, D., Rao, M. S. & Reddy, J. K. (1995) Proc. Natl. Acad. Sci. USA 92, 7921-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fajas, L., Auboeuf, D., Raspe, E., Schoonjans, K., Lefebvre, A. M., Saladin, R., Najib, J., Laville, M., Fruchart, J. C., Deeb, S., et al. (1997) J. Biol. Chem. 272, 18779-18789. [DOI] [PubMed] [Google Scholar]
- 4.Kliewer, S. A., Forman, B. M., Blumberg, B., Ong, E. S., Borgmeyer, U., Mangelsdorf, D. J., Umesono, K. & Evans, R. M. (1994) Proc. Natl. Acad. Sci. USA 91, 7355-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tontonoz, P., Hu, E. & Spiegelman, B. M. (1994) Cell 79, 1147-1156. [DOI] [PubMed] [Google Scholar]
- 6.Rosen, E. D., Sarraf, P., Troy, A. E., Bradwin, G., Moore, K., Milstone, D. S., Spiegelman, B. M. & Mortensen, R. M. (1999) Mol. Cell 4, 611-617. [DOI] [PubMed] [Google Scholar]
- 7.Ren, D., Collingwood, T. N., Rebar, E. J., Wolffe, A. P. & Camp, H. S. (2002) Genes Dev. 16, 27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barak, Y., Nelson, M. C., Ong, E. S., Jones, Y. Z., Ruiz-Lozano, P., Chien, K. R., Koder, A. & Evans, R. M. (1999) Mol. Cell 4, 585-595. [DOI] [PubMed] [Google Scholar]
- 9.Rocchi, S., Picard, F., Vamecq, J., Gelman, L., Potier, N., Zeyer, D., Dubuquoy, L., Bac, P., Champy, M. F., Plunket, K., et al. (2001) Mol. Cell 8, 737-747. [DOI] [PubMed] [Google Scholar]
- 10.Picard, F., Gehin, M., Annicotte, J. S., Rocchi, S., Champy, M. F., O'Malley, B., Chambon, P. & Auwerx, J. (2002) Cell 111, 931-941. [DOI] [PubMed] [Google Scholar]
- 11.Chehab, F. F., Lim, M. E. & Lu, R. (1996) Nat. Genet. 12, 318-320. [DOI] [PubMed] [Google Scholar]
- 12.Saladin, R., De Vos, P., Guerre-Millo, M., Leturque, A., Girard, J., Staels, B. & Auwerx, J. (1995) Nature 377, 527-529. [DOI] [PubMed] [Google Scholar]
- 13.Muoio, D. M., MacLean, P. S., Lang, D. B., Li, S., Houmard, J. A., Way, J. M., Winegar, D. A., Corton, J. C., Dohm, G. L., Kraus, W., et al. (2002) J. Biol. Chem. 277, 26089-26097. [DOI] [PubMed] [Google Scholar]
- 14.Wang, Y. X., Lee, C. H., Tiep, S., Yu, R. T., Ham, J., Kang, H. & Evans, R. M. (2003) Cell 113, 159-170. [DOI] [PubMed] [Google Scholar]
- 15.Beckstead, R., Ortiz, J. A., Sanchez, C., Prokopenko, S. N., Chambon, P., Losson, R. & Bellen, H. J. (2001) Mol. Cell 7, 753-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randle, P. J. (1998) Diabetes Metab. Rev. 14, 263-283. [DOI] [PubMed] [Google Scholar]
- 17.Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., et al. (2001) Nature 413, 179-183. [DOI] [PubMed] [Google Scholar]
- 18.Liang, G., Yang, J., Horton, J. D., Hammer, R. E., Goldstein, J. L. & Brown, M. S. (2002) J. Biol. Chem. 277, 9520-9528. [DOI] [PubMed] [Google Scholar]
- 19.Saladin, R., Fajas, L., Dana, S., Halvorsen, Y. D., Auwerx, J. & Briggs, M. (1999) Cell Growth Differ. 10, 43-48. [PubMed] [Google Scholar]
- 20.Kubota, N., Terauchi, Y., Miki, H., Tamemoto, H., Yamauchi, T., Komeda, K., Satoh, S., Nakano, R., Ishii, C., Sugiyama, T., et al. (1999) Mol. Cell 4, 597-609. [DOI] [PubMed] [Google Scholar]
- 21.Hegele, R. A., Cao, H., Frankowski, C., Mathews, S. T. & Leff, T. (2002) Diabetes 51, 3586-3590. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal, A. K. & Garg, A. (2002) J. Clin. Endocrinol. Metab. 87, 408-411. [DOI] [PubMed] [Google Scholar]
- 23.Savage, D. B., Tan, G. D., Acerini, C. L., Jebb, S. A., Agostini, M., Gurnell, M., Williams, R. L., Umpleby, A. M., Thomas, E. L., Bell, J., et al. (2003) Diabetes 52, 910-917. [DOI] [PubMed] [Google Scholar]
- 24.Garg, A. (2000) Am. J. Med. 108, 143-152. [DOI] [PubMed] [Google Scholar]
- 25.Moitra, J., Mason, M. M., Olive, M., Krylov, D., Gavrilova, O., Marcus-Samuels, B., Feigenbaum, L., Lee, E., Aoyama, T., Eckhaus, M., et al. (1998) Genes Dev. 12, 3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J. K., Gavrilova, O., Chen, Y., Reitman, M. L. & Shulman, G. I. (2000) J. Biol. Chem. 275, 8456-8460. [DOI] [PubMed] [Google Scholar]
- 27.Shimomura, I., Hammer, R. E., Ikemoto, S., Brown, M. S. & Goldstein, J. L. (1999) Nature 401, 73-76. [DOI] [PubMed] [Google Scholar]
- 28.Shimomura, I., Hammer, R. E., Richardson, J. A., Ikemoto, S., Bashmakov, Y., Goldstein, J. L. & Brown, M. S. (1998) Genes Dev. 12, 3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montenegro, R. M., Jr., Montenegro, A. P., Fernandes, M. I., de Moraes, R. R., Elias, J., Jr., Gouveia, L. M., Muglia, V. F., Foss, M. C., Moreira, A. C., Martinelli, C., et al. (2002) J. Pediatr. Endocrinol. Metab. 15, 441-447. [DOI] [PubMed] [Google Scholar]
- 30.Langner, C., Birkenmeier, E., Roth, K., Bronson, R. & Gordon, J. (1991) J. Biol. Chem. 266, 11955-11964. [PubMed] [Google Scholar]
- 31.Peterfy, M., Phan, J., Xu, P. & Reue, K. (2001) Nat. Genet. 27, 121-124. [DOI] [PubMed] [Google Scholar]
- 32.Rehnmark, S., Giometti, C. S., Slavin, B. G., Doolittle, M. H. & Reue, K. (1998) J. Lipid Res. 39, 2209-2217. [PubMed] [Google Scholar]
- 33.Miles, P. D., Barak, Y., He, W., Evans, R. M. & Olefsky, J. M. (2000) J. Clin. Invest. 105, 287-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rieusset, J., Touri, F., Michalik, L., Escher, P., Desvergne, B., Niesor, E. & Wahli, W. (2002) Mol. Endocrinol. 16, 2628-2644. [DOI] [PubMed] [Google Scholar]
- 35.Yamauchi, T., Waki, H., Kamon, J., Murakami, K., Motojima, K., Komeda, K., Miki, H., Kubota, N., Terauchi, Y., Tsuchida, A., et al. (2001) J. Clin. Invest. 108, 1001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsusue, K., Haluzik, M., Lambert, G., Yim, S. H., Gavrilova, O., Ward, J. M., Brewer, B., Jr., Reitman, M. L. & Gonzalez, F. J. (2003) J. Clin. Invest. 111, 737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavrilova, O., Haluzik, M., Matsusue, K., Cutson, J. J., Johnson, L., Dietz, K. R., Nicol, C. J., Vinson, C., Gonzalez, F. J., Reitmann, M., et al. (2003) J. Biol. Chem. 278, 34268-34276. [DOI] [PubMed] [Google Scholar]
- 38.Norris, A. W., Chen, L., Fisher, S. J., Szanto, I., Ristow, M., Jozsi, A. C., Hirshman, M. F., Rosen, E. D., Goodyear, L. J., Spielgelmann, B., et al. (2003) J. Clin. Invest. 112, 608-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer, P., Moller, P., Bindl, L., Melzner, I., Tornqvist, H., Debatin, K. M. & Wabitsch, M. (2002) J. Clin. Endocrinol. Metab. 87, 2384-2390. [DOI] [PubMed] [Google Scholar]
- 40.Arioglu, E., Duncan-Morin, J., Sebring, N., Rother, K. I., Gottlieb, N., Lieberman, J., Herion, D., Kleiner, D. E., Reynolds, J., Prekumar, A., et al. (2000) Ann. Intern. Med. 133, 263-274. [DOI] [PubMed] [Google Scholar]
- 41.Petersen, K. F., Oral, E. A., Dufour, S., Befroy, D., Ariyan, C., Yu, C., Cline, G. W., DePaoli, A. M., Taylor, S. I., Gorden, P., et al. (2002) J. Clin. Invest. 109, 1345-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gavrilova, O., Marcus-Samuels, B., Leon, L. R., Vinson, C. & Reitman, M. L. (2000) Nature 403, 850-851. [DOI] [PubMed] [Google Scholar]
- 43.Colombo, C., Cutson, J. J., Yamauchi, T., Vinson, C., Kadowaki, T., Gavrilova, O. & Reitman, M. L. (2002) Diabetes 51, 2727-2733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.