Summary
It is well established that protein sequence determination may be achieved by mass spectrometric analysis of protonated tryptic peptides subjected to collisional activation. When separated by nanoflow HPLC, a high percentage of peptides from complex mixtures of proteins can usually be identified.
Recently alternative, radical-driven fragmentation approaches of electron capture dissociation and the more common electron transfer dissociation (ETD) have been introduced and made widely available. In order to utilize these techniques in large scale proteomics studies it is important to characterize the performance of these fragmentation processes on peptides formed by a range of enzymatic cleavages.
In this study we present a statistical analysis of the ion types that are observed from peptides produced by different enzymes, and highlight the different characteristics of ETD spectra of doubly-charged precursors in comparison to precursors of higher charge states.
Introduction
Protein and peptide identification using gas phase fragmentation analysis is the cornerstone of most proteomic studies, and has allowed in-depth studies to be performed of complex biological samples1. Practically all of these studies have been performed using collision induced dissociation (CID) to fragment peptides, most commonly produced by proteolytic cleavage using the enzyme trypsin. Tryptic peptides are particularly appropriate for CID analysis, as the presence of a basic amino acid on the C-terminus and the N-terminal amino group produce ladder sequences of residues from both peptide termini2. However, it is generally the case that as peptides become longer in length, the effectiveness of CID for peptide identification diminishes. This is partly due to the sequence-dependent fragmentation produced in CID, where cleavages next to, or between certain amino acids are energetically preferred, leading to a decreased number of peptide backbone cleavages3, 4. It is also influenced by the less efficient fragmentation in CID when there are basic residues in the middle of the peptide sequence, due to the higher energy threshold for charge-directed fragmentation5; fragmentation of doubly-protonated precursors in CID are significantly more likely to lead to successful peptide identifications.
Alternative fragmentation techniques to CID have recently become more widely available. These include the radical-based fragmentation approaches of electron capture dissociation (ECD)6 and electron transfer dissociation (ETD)7. Both of these techniques induce peptide fragmentation through mechanisms that are not driven by bond strength, and thus they produce spectra with different characteristics: no strong cleavage preferences based on peptide sequence; the presence of basic internal residues does not adversely affect the distribution of peptide backbone cleavages; and retention of labile post-translational modifications. As a result, several researchers have begun to investigate whether enzymes with different protein cleavage specificities may yield better results than trypsin when ETD analysis is applied 8, 9.
The types of fragment ions observed in ECD and ETD spectra of peptides have been described by a number of groups 10-12. Unlike CID fragmentation, the most common cleavage is of the N-Cα bond to form c ions and radical z. ions. It is also possible to observe a hydrogen transfer reaction between these two products to form c-1. (also referred to as c.) and z+1 ions (or z′). Smaller numbers of b and y ions may also be present, especially if supplemental activation energy is applied to the charge-reduced molecular species to assist in breaking non-covalent interactions between fragments13. Supplemental activation is routinely used on the most common mass spectrometers equipped for ETD analysis; those from Thermo and Agilent. Lastly, side-chain neutral losses can be observed; although in ETD these are generally only detected from the charge-reduced molecular species rather than from fragment ions.
Knowledge of the frequency of occurrence of the ion types formed as a result of ETD processes would be very important for optimizing search engines for identifying peptides from fragmentation spectra in proteomic analysis. There are several search engines available for ETD fragmentation analysis. Amongst these, there is currently no consensus about which ion types to consider, let alone an appropriate weighting between the different ion types. For example, the search engine OMSSA, which has been used by a number of researchers for ETD analysis, only considers c and z. ions14. At the other extreme is the search engine Protein Prospector, which considers all of the sequence ion types that are present in ETD spectra (i.e. b, c-1, c, y, z. and z+1), with different weighting factors for each ion type. Other search engines consider different subsets of these ion types. As a result of this disparity of ion types considered, when presented with ETD data, the peptide identifications can differ significantly among search engines, as highlighted in a recent comparison15.
In this study we report the frequency of matching the fragment ion types observed in ETD analyses. Studies were performed using a selection of different enzymes to assess peptide-specific differences.
Experimental Section
Mass Spectrometry
All data were acquired on an LTQ-Orbitrap (Thermo) with ETD ion introduction through the rear of the instrument. Peptides were separated using a Nanoacquity LC system (Waters), with a BEH130 C18 75 μM ID × 150 mm (Waters) column using either a ninety minute or two hour gradient and operating at a flow rate of roughly 400 nl/min.
Precursor ion masses were measured in the Orbitrap, while ETD data were acquired in the linear ion trap. Automated gain control (AGC) was set to 10 000 for the linear trap in MS/MS experiments, while the AGC target value was 100 000 for the fluoranthene ions. The minimal signal required for precursor ion selection was set to 10 000, the isolation window was set to 3 Th, and the activation time was 200 msec. All ETD spectra were acquired with supplemental activation on.
The tryptic datasets were generated from several mouse post-synaptic density (PSD) preparations, and some of these data were used in a recent publication16. The data for the other cleavage types was generated from preparations of mouse synaptosomes and nuclei from a mouse cell line. In some of these experiments CID and ETD data were acquired, but for the purposes of the results in this manuscript only the ETD data were further analyzed.
Data Analysis
Peaklists of ETD spectra were extracted using in-house software PAVA17, which creates separate peaklists for CID and ETD data, and then searched using Batch-Tag in Protein Prospector version 5.318, 19. For each spectrum Protein Prospector splits the mass range of fragment ions observed in half and then searches against the twenty most intense ions in each half of the fragment ion spectrum (i.e. searching with a total of 40 ions, providing there are at least 20 ions in each half of the mass peaklist)18. For ETD data, Batch-Tag considers b, c-1., c, y, z. and z+1 ions, with the score weighting of each ion type as in Table 1. These weightings were derived from an in-house small-scale analysis prior to this present study.
Table 1.
Scores assigned by Batch-Tag to different fragment ion types in ETD.
| Ion Type | Score |
|---|---|
| b | 0.3 |
| c-1 | 0.3 |
| c | 2.1 |
| y | 1.2 |
| z. | 3.2 |
| z+1 | 1.9 |
For ion trap ETD spectra, like most search engines, Batch-Tag assumes an inability to determine fragment ion charge state, and thus does not attempt to de-isotope the peaklists. In this analysis results from a number of different experiments were combined together even though individual datasets were searched with different database versions. All data were searched against databases of SwissProt where a randomized version of the database was concatenated onto the normal database. For PSD preparations, only rodent entries were considered, and all data was searched allowing for a precursor mass tolerance of 15 ppm and a fragment mass tolerance of 0.6 Da. Results were then filtered using the program SearchCompare19. For tryptic data the acceptance criteria for identifications were requiring at least one peptide with an expectation value of 0.01 or better to identify a protein, then requiring other peptides identified to these proteins to have expectation values of 0.05 or better. For data from other proteolytic cleavage specificities, a threshold of 0.05 was used at both peptide and protein level. In the results for the tryptic data, a total of four matches to the decoy part of the database were obtained, giving an estimated peptide false discovery rate of 0.2%. Similarly, estimated false discovery rates were 0.5%, 1.7% and 0%, for endoproteases LysC, LysN digests, and CNBr cleavages respectively, according to decoy database hits.
For all peptides reported in the searches described above, a script was written in-house that ran MS-Product20, extracting all ion matches to each identified spectrum. This script was written to consider all possible sequence ion types observed in ETD spectra (b, c-1., c, y, z. and z+1) and also considered x ions, which are not formed in ETD analysis, so could be used as a measure of random peak matching. In addition, instead of only considering singly- or doubly-charged fragments, MS-Product considers ions as being up to one charge less than the precursor ion, providing there were enough basic residues in the fragment sequence to explain the presence of such a charge state; i.e. for quadruply-charged precursors it considers some fragment ions as being potentially up to triply-charged, providing the fragment contained at least three basic residues (or two basic residues and the peptide N-terminus). The combined results for all spectra in the dataset/s were then analyzed using Excel.
Results
Ion statistics for tryptic peptides were obtained from a large number of LC-MS runs of peptides derived from repeated analyses of mouse post-synaptic density preparations. Some of these data were used in a recent publication16. All the data was acquired on an LTQ-Orbitrap with ETD. Precursor ions were measured in the Orbitrap, but all fragment ions were measured in the linear ion trap. Together, around 70,000 spectra were acquired, from which 2149 unique peptide sequences were identified (over 10,000 spectra were identified, but many peptides were identified multiple times). An in-house script was used to extract the ion types that were matched in all these 2149 spectra. The cumulative number of each ion type matched was determined and these values were normalized to determine the relative frequency of observation of different ion types. These results are reported in Table 2. In the list of ion types considered ‘x’ ions are included. Such fragments are not generated by ETD, so these represent the frequency of random peak matches.
Table 2.
Frequency of observation of different ion types matched in tryptic ETD spectra. Numbers represent the fraction of all matched peaks in a given spectrum that match to each ion type. Included in the list of ions considered is ‘x’ ions, which are not formed in ETD, so represent a measure of the random peak matching frequency. The first column reports average results, which are then split between those from doubly-charged precursors and those of higher charge state in the next two columns.
| Ion Type | All | 2+ Precursor | 3+ or higher precursor |
|---|---|---|---|
| b | 0.03 | 0.02 | 0.03 |
| c | 0.29 | 0.11 | 0.31 |
| c-1 | 0.05 | 0.09 | 0.04 |
| x | 0.03 | 0.02 | 0.03 |
| y | 0.11 | 0.13 | 0.10 |
| z | 0.30 | 0.30 | 0.28 |
| z+1 | 0.19 | 0.31 | 0.15 |
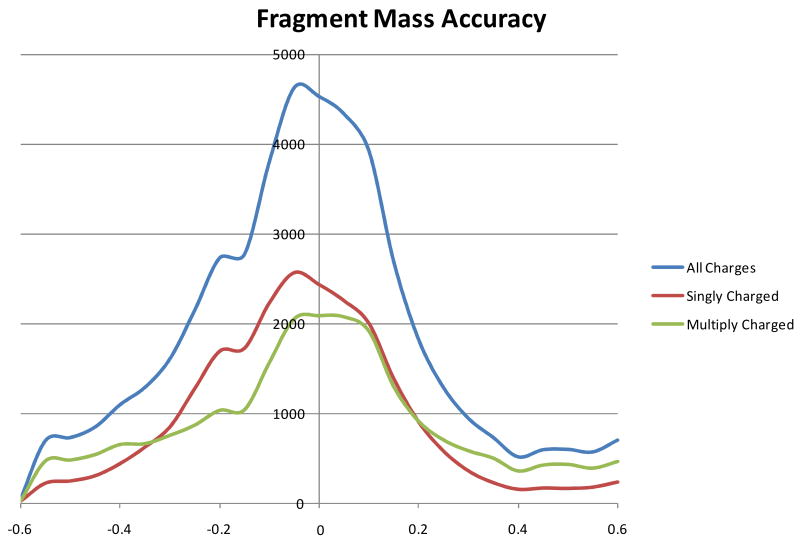
The fragment peak lists were searched with a mass tolerance of +/- 0.6 Th, and using this value some peaks were matched to multiple fragment ion types; e.g. z. and z+1. Figure 1 shows a plot of the mass errors between theoretical and observed fragment ion mass values. The likelihood of randomly matching a multiply-charged fragment ion is higher than a singly charged; e.g. if you consider a triply charged fragment ion while allowing an error of up to 0.6 Th in mass measurement, this value actually corresponds to +/- 1.8 Da in deconvolved mass tolerance. To assess this effect, the results for singly-charged fragment ions and multiply-charged fragment ions were separated. As can be seen by comparing the red and green lines in Figure 1, the frequency of matching multiply-charged fragment ions with mass errors of greater than +/- 0.3 Th was significantly higher than singly-charged. The mass accuracy of multiply-charged fragments should be identical. Hence, these results demonstrate that there were a measurable amount of incorrect matches to peaks considered as multiply-charged, and thus matches to fragments considered as multiply-charged are of lower reliability (when the fragment charge-state is not discernable).
Figure 1.
Histogram of the frequency of matching ions in ETD spectra as a function of the mass error between theoretical and observed m/z. Data was binned into 0.05 Da intervals. The blue line shows results for all assignments, whereas the red and green lines separate results by charge state of assignment.
We have previously commented that the characteristics of ETD fragmentation spectra of doubly-charged precursors differ from those of higher charge states21. To systematically describe this observation, we separated the fragmentation results of 2+ precursor ions from those of higher charge-states, and the separated results are the final two columns of Table 2. These results illustrate that there are significant differences in the frequency of observation of certain ion types in spectra of doubly-charged precursors compared to higher charge states. The frequency of occurrence of z. and z+1 ions is essentially identical in doubly-charged spectra and nearly as many c-1. as c ions are observed. This observation contrasts with results from spectra of higher-charged precursors, where z+1 ions are less frequently observed than z. and c-1. ions are rarely detected. In spectra of doubly-charged precursors of tryptic peptides the frequency of observation of C-terminally derived ions (y and z type) is roughly three times higher than those N-terminally derived. For spectra derived from higher charge state precursors, the frequency of c and z. ions observed is similar. About 3% of ions matched were to x ions, suggesting that about 3% of matches to other ions may be random. Hence, the results in Table 2 suggest that the frequency of observation of genuine b ions in ETD spectra is extremely low.
The bias described in the previous paragraph toward C-terminal ions for tryptic peptides is not surprising, due to the presence of the basic C-terminal residue, and for doubly-charged precursors the C-terminal residue is likely to be the only basic residue in the sequence. However, these results show that as soon as another proton is added this preference is lost.
The data discussed above was acquired on a Thermo instrument. In addition, we have analyzed some of the data acquired in a recent study comparing search engine performance at analyzing ETD data, where the data was acquired on an Agilent ion trap employing supplemental activation15. The ion statistic results observed from this different instrument were very similar (data not shown), suggesting the results presented in this present study should be broadly applicable to ETD data acquired on a variety of instruments.
To get an impression of the intensity of the peaks for the different ion types, the tryptic peptide spectra were re-analyzed considering differing numbers of peaks per spectrum. These results are shown in Table 3. In each case the spectral mass range was still split in half, and for the twenty m/z peak list the ten most intense from each half of the spectrum were used, and for sixty peaks the thirty most intense (or all the peaks if there were less than thirty) were used. Those fragments that are typically more intense would expect to represent a higher percentage of peaks matched when a lower number of peaks are considered. Hence, interpretation of the data shown in Table 3 indicates that peaks for z. and z+1 ions are generally more intense and y ion peaks are typically of lower intensity in all spectra. It also indicates that c ion peaks are not intense in doubly-charged spectra, but are in spectra of higher charge states.
Table 3.
Percentage of peaks matched to different ion types using different thresholds for the number of ions considered in each spectrum. For selecting peaks, Protein Prospector splits the spectral mass range in half and takes the ‘n’ most intense peaks in each half of the spectrum to reach the total listed at the top of the table; e.g. the 20 peaks represent the 10 most intense in each half of the spectrum. For the final columns; some spectra will not contain 60 peaks, in which case the full peak list is used.
| #Peaks Prec. Charge | 20 2+ | 20 >2+ | 40 2+ | 40 >2+ | 60 2+ | 60 >2+ |
|---|---|---|---|---|---|---|
| b | 0.02 | 0.02 | 0.02 | 0.03 | 0.03 | 0.04 |
| c | 0.11 | 0.36 | 0.11 | 0.31 | 0.11 | 0.31 |
| c-1 | 0.09 | 0.04 | 0.09 | 0.04 | 0.10 | 0.05 |
| x | 0.01 | 0.02 | 0.02 | 0.03 | 0.02 | 0.03 |
| y | 0.09 | 0.07 | 0.13 | 0.10 | 0.15 | 0.13 |
| z | 0.34 | 0.34 | 0.30 | 0.28 | 0.28 | 0.28 |
| z+1 | 0.35 | 0.15 | 0.31 | 0.15 | 0.30 | 0.16 |
To investigate whether the observed ion type trends are in common with results obtained using other enzymes, spectra acquired on preparations digested with either the enzyme Lys-N, Lys-C or the chemical cleavage agent CNBr were analyzed. The fraction of each ion type identified with each cleavage specificity, both for doubly-charged and higher charge state precursors is shown in Table 4. As can be seen, Lys-C produced 50% more z. ions than c ions in doubly-charged precursor spectra but, similarly to trypsin, showed little bias for higher charge-state precursors. For the enzyme Lys-N an inverse bias to trypsin was observed; i.e. c ions are roughly three times more likely to be observed than z. ions in spectra of doubly-charged precursors. For spectra of higher precursor charge state there is still a bias toward detecting c ions, but it is less pronounced, and the summed number of z. or z+1 ions is nearly as high as the total of c or c-1.. For CNBr, similar numbers of N-terminal and C-terminal ions are observed, and these ratios were not altered significantly by the charge state.
Table 4.
Frequency of matching different ion types in ETD spectra of peptides produced by different cleavages. Numbers represent the fraction of all matched peaks in a given spectrum that match to each ion type. Results are separated into those from doubly-charged precursor ions and those of a higher charge state. (Note: tryptic numbers differ slightly from Table 3 due to the lack of consideration of ‘x’ ions).
| Enzyme Prec. Charge | LysC 2+ | LysC >2+ | LysN 2+ | LysN >2+ | CNBr 2+ | CNBr >2+ | Trypsin 2+ | Trypsin >2+ |
|---|---|---|---|---|---|---|---|---|
| b | 0.02 | 0.03 | 0.06 | 0.04 | 0.04 | 0.05 | 0.03 | 0.03 |
| c-1 | 0.11 | 0.05 | 0.17 | 0.06 | 0.14 | 0.08 | 0.10 | 0.04 |
| c | 0.20 | 0.33 | 0.49 | 0.42 | 0.31 | 0.36 | 0.11 | 0.34 |
| y | 0.13 | 0.09 | 0.05 | 0.06 | 0.06 | 0.06 | 0.14 | 0.11 |
| z | 0.30 | 0.29 | 0.17 | 0.27 | 0.27 | 0.25 | 0.31 | 0.31 |
| z+1 | 0.23 | 0.20 | 0.07 | 0.15 | 0.18 | 0.21 | 0.32 | 0.17 |
We have presented normalized results depending on the charge state. However, the frequency of matching precursors at a given charge state differs depending on the enzyme/cleavage used and also the performance of the database search engine. A breakdown of the precursor charge states of the peptides identified using Protein Prospector version 5.3 with each enzyme is presented in Table 5. When a particular peptide could be identified from two different charge states (meeting the quoted acceptance criteria) then both of them were counted, unlike in the ion frequency analysis above, where only the most confident matching charge state of a given peptide was used to create the ion statistics. These results show that for all cleavage specificities considered, triply-charged precursors are the most commonly identified in ETD analysis. However, enzymes that produce larger peptides lead to identification of more precursors of higher charge state (4+ or higher) than trypsin, which produces the smallest peptides of those compared in this study.
Table 5.
Fraction of identified spectra with each protein cleavage specificity broken down by precursor charge state.
| LysC | LysN | CNBr | Trypsin | |
|---|---|---|---|---|
| 2+ | 0.31 | 0.22 | 0.21 | 0.32 |
| 3+ | 0.43 | 0.49 | 0.34 | 0.55 |
| 4+ | 0.20 | 0.23 | 0.27 | 0.12 |
| 5+ | 0.05 | 0.04 | 0.14 | 0.01 |
| >5+ | 0.01 | 0.01 | 0.04 | 0.00 |
Discussion
The presented results show that for most enzymes the characteristics of doubly-protonated precursor ion spectra differ significantly from those of higher charge states, with the location of the basic residue in the peptide sequence being the prime determinant of the sequence ion series actually observed. Multiply-charged ions usually feature basic sites not only at one terminus. Thus, for higher charge states this bias almost completely disappears to yield similar ion frequencies for all enzymes. The observation that the location of basic residues dictates the fragments observed is not unique to ETD data and is well documented for singly protonated CID spectra of peptides22. Indeed, a similarity in behavior between singly-charged CID spectra and doubly-charged ETD spectra makes sense, as after electron transfer the radical species that undergoes fragmentation is now singly-charged.
Another broad observation is that y ions are significantly more frequently present in ETD spectra than b ions, and in doubly-charged spectra from trypsin and Lys-C they are fairly common. The statistics suggest that b ion matching is at a similar level as random peak matching (x ions). However, we have observed spectra where we are confident that a genuine b ion is present; these results show this is rare occurrence.
Across all enzymes, the frequency of observation of hydrogen transfer products (c-1. and z+1 ions) in relation to the more conventional c and z. ions was significantly higher in spectra of doubly-charged ions compared to spectra from higher charged precursors, and z+1 ions are considerably more common than c-1. fragments. These results suggest that search engines that do not consider the hydrogen transfer products of c-1. and z+1 are going to perform sub-optimally, particularly on doubly-charged precursor ion spectra.
For all cleavage specificities studied triply-charged spectra were the most often identified. However, these results do not simply represent the frequency of formation of peptides of a particular charge state, but rather include a bias based on the subsequent ability to identify the peptide using ETD. The fragmentation of doubly-charged precursors by ETD leads to the formation of fewer fragmentation products than the activation of higher charge states. Hence, a lower percentage of doubly-charged precursors are identified in ETD than higher charge state precursors. Other problems are encountered with precursors of much higher charge state (5+ or higher). The low resolution ion trap fragmentation spectra do not allow fragment ion charge states to be determined. Most search engines consider that fragment ions could be singly or doubly charged, but usually not higher charge states. This is because the frequency of observation of higher charge states in most peptide fragmentation data is low (partly because the majority of data is tryptic, so higher charge state precursors are not common), and so by considering, for example, triply-charged products, a higher percentage of peaks are matched at random than genuine peak identifications, so it becomes counterproductive to consider them. However, for precursors of 5+ or higher there will be triply-charged fragment ions observed and search engines generally will not match them. Another issue with the low resolution spectra is the inability to identify the monoisotopic mass for multiply-charged fragments. Peaklist generation software will sometimes report an average rather than monoisotopic peak mass value, and this average mass for larger fragments may be outside the mass tolerance allowed for matching fragment ions. This effect may cause search engines to report some z. ions as z+1 ions, so the actual frequency of z+1 ion formation, especially in spectra from long peptides, may be slightly lower than reported in the analysis presented here. Some researchers / software search for average masses of multiply-charged fragments to counteract this difficulty in labeling the monoisotopic peak. However, this is not a perfect solution either, as for many fragments (especially in the lower mass range) the monoisotopic mass is correctly identified. It should be noted that most of these problems would be solved if fragments were measured, for example, in an orbitrap, as this would provide much higher mass accuracy and resolution, consistently producing monoisotopic rather than average masses, and the higher mass accuracy would allow searching that would only match the correct isotope peak. However, most researchers currently performing ETD do not have access to an orbitrap detector, and even for those that do, in the first generation orbitrap instruments there is a significant sensitivity loss in transferring ions to the orbitrap for detection, meaning measurement in the ion trap identifies more components. The newly announced LTQ-Orbitrap Velos reportedly exhibits more efficient ion transfer to the orbitrap, which may make the higher quality measurement in the orbitrap preferable.
Conclusions
The results presented here are the first statistical analysis of ion types observed in ETD data measured in an ion trap. They report the frequency of matching different ion types in peptides produced by cleavage with a range of different enzymes or chemical cleavage. Having a more comprehensive understanding of the ion types to expect provides important information required to optimize database searching strategies for robust matching of ETD spectra. Of particular interest, the results reveal noticeable differences in the fragmentation spectra of doubly-charged precursors compared to precursors of higher charge states. In general, doubly-charged ETD spectra contain fewer fragment ions, partly due to shorter peptide lengths and partly due to less efficient fragmentation13. Also, doubly-charged fragmentation spectra contain a higher percentage of hydrogen transfer fragment products (c-1. and z+1 ions). These two factors combined mean that sensitive identification of doubly-charged precursors using ETD requires slightly different considerations than precursors of higher charge states (i.e. higher weighting toward hydrogen transfer products and increased weighting for y ions when trypsin or Lys-C have been used) and if this is not taken into account, then search engines may perform poorly on this data type. This prediction is borne out by the results in a recent comparison of different search engines analyzing a common dataset, where the major differences between search engines were due to differences in doubly-charged precursor identification15.
We believe the information highlighted in this study is important for the further development of software tools that will facilitate interpretation of ETD fragmentation spectra of peptides and proteins.
Acknowledgments
This work was funded by the NIH National Center for Research Resources grant P41 RR001614 and the Vincent J. Coates Foundation. We thank Agnes Thalhammer and Ralf Schoepfer for the PSD and synaptosomal samples and Samuel Myers for the mouse cell nuclei.
References
- 1.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Medzihradszky KF. Methods Enzymol. 2005;402:209–244. doi: 10.1016/S0076-6879(05)02007-0. [DOI] [PubMed] [Google Scholar]
- 3.Kapp EA, Schutz F, Reid GE, Eddes JS, Moritz RL, O'Hair RA, Speed TP, Simpson RJ. Anal Chem. 2003;75:6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH. Anal Chem. 2005;77:5800–5813. doi: 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. J Mass Spectrom. 2000;35:1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 6.Zubarev RA. Curr Opin Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Coon JJ, Shabanowitz J, Hunt DF, Syka JE. J Am Soc Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Proc Natl Acad Sci U S A. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taouatas N, Drugan MM, Heck AJ, Mohammed S. Nat Methods. 2008;5:405–407. doi: 10.1038/nmeth.1204. [DOI] [PubMed] [Google Scholar]
- 10.Zubarev RA, Kelleher NL, McLafferty FW. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 11.Cooper HJ, Hudgins RR, Hakansson K, Marshall AG. J Am Soc Mass Spectrom. 2002;13:241–249. doi: 10.1016/S1044-0305(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 12.Fung YM, Chan TW. J Am Soc Mass Spectrom. 2005;16:1523–1535. doi: 10.1016/j.jasms.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ. Anal Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi A, Bai DL, Geer LY, Shabanowitz J, Hunt DF. Int J Mass Spectrom. 2007;259:197–203. doi: 10.1016/j.ijms.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandasamy K, Pandey A, Molina H. Anal Chem. 2009;81:7170–7180. doi: 10.1021/ac9006107. [DOI] [PubMed] [Google Scholar]
- 16.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Proc Natl Acad Sci U S A. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynn AJ, Chalkley RJ, Baker PR, Medzhiradszky KF, Guan S, Burlingame AL, Denver CO. American Society for Mass Spectrometry. Santa Fe, NM; 2008. [Google Scholar]
- 18.Chalkley RJ, Baker PR, Huang L, Hansen KC, Allen NP, Rexach M, Burlingame AL. Mol Cell Proteomics. 2005;4:1194–1204. doi: 10.1074/mcp.D500002-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. Mol Cell Proteomics. 2008;7:2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalkley RJ, Hansen KC, Baldwin MA. Methods Enzymol. 2005;402:289–312. doi: 10.1016/S0076-6879(05)02009-4. [DOI] [PubMed] [Google Scholar]
- 21.Chalkley RJ, Baker PR, Lynn AJ, Burlingame AL, Memphis TN. 2009 [Google Scholar]
- 22.Paizs B, Suhai S. Mass Spectrom Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]