Summary
We describe a cell-based kinetic profiling approach using impedance readout for monitoring the effect of small molecule compounds. This non-invasive readout allows continuous sampling of cellular responses to biologically active compounds and the ensuing kinetic profile provides information regarding the temporal interaction of compounds with cells. The utility of this approach was tested by screening a library containing FDA approved drugs, experimental compounds and nature compounds. Compounds with similar activity produced similar impedance-based time-dependent cell response profiles (TCRP). The compounds were clustered based on TCRP similarity. We identified novel mechanisms for existing drugs, confirmed previously reported calcium modulating activity for COX-2 inhibitor, celecoxib and identified an additional mechanism for the experimental compound, monastrol. We also identified and characterized a new anti-mitotic agent. Our findings indicate TCRP approach provides predictive mechanistic information for small molecule compounds.
Introduction
Drug discovery and development is both time and resource-intensive endeavor which is not always guaranteed to result in safe and efficacious drugs. This problem was recently highlighted by issuance of safety alert by the FDA for the diabetes drug Avandia and also the withdrawal of the pain medication, Vioxx from the market (Nathan, 2007; Slordal and Spigset, 2006; Topol, 2004). At least part of the explanation centers on the fact that modern drug discovery has mostly undertaken a target-centric approach while the ultimate effect of drugs are the culmination of on-target and less well understood off-target interactions (Abraham, 2006). The potential interaction of drugs with unintended targets or pathways could either prove fatal leading to toxicity and unwanted side effects or could prove to be beneficial. It is imperative to come up with strategies to identify and understand the specific biochemical nature of the on target activity as well as off-target activity of lead compounds earlier in the drug discovery process in order to optimize structural features leading to desirable outcomes, while avoiding or minimizing less desirable attributes. To address this issue a number of multi-dimensional high throughput phenotypic approaches have been proposed to assess the global responses to specific perturbations in cells.
Multi-dimensional phenotypic profiling approaches including gene expression profiling, proteomic profiling, protein-fragment complementation profiling, high content microscopy-based profiling and cell line cytotoxicity based profiling has the capacity to measure both the interaction of compounds on intended targets and also generate testable hypotheses concerning mechanism of action and off-target effects (Gunther et al., 2003; Hieronymus et al., 2006; Leung et al., 2003; MacDonald et al., 2006; Marton et al., 1998; Perlman et al., 2004; Scherf et al., 2000; Wei et al., 2006; Weinstein et al., 1997; Young et al., 2008). In all cases a very large information-rich data set is generated which can be used to cluster compounds based on profiles and search for specific patterns of activity. All the profiling approaches have had various degrees of success not only in elucidating mechanism of action of unknown compounds, but also unraveling new and novel targets for existing drugs.
The extent to which protein targets are modulated by drugs or small molecule compounds are dependent on a number of factors, including expression levels of the target, the effective concentration of the compound and importantly the time needed for the compound to perturb the target. Some compounds can be fast acting, such as agonists of certain GPCRS or ion channels, leading to immediate biochemical and cellular changes, while others can have a more prolonged effect. One of the limitations of current multi-dimensional phenotypic profiling approaches is that typically a single time point after compound addition is chosen to assess the effect of the compound and therefore the conclusion regarding mechanism of action is based upon the time point at which the samples are processed. It can be envisioned that if the effect of the compound is manifested at any other time, or if a compound has multiple and kinetically distinct effects, it can be easily overlooked. To address this limitation, we have devised a live cell morphological profiling approach for dynamic monitoring of the effect of small molecules compounds that is based on impedance measurement of cells growing on microelectronic sensors integrated in wells of microtiter plates (Atienza et al., 2006). Microelectronic-based monitoring of cells using impedance was first described by Giaever and Keese (1984) and the key feature of this approach is that it is a non-invasive readout and cellular responses to treatments can be continually sampled inside the well throughout the length of the assay(Giaever and Keese, 1993). The ensuing kinetic profile can provide information regarding the temporal interaction of the compounds with the cells. In this report we tested the utility of impedance-based profiling approach by screening and profiling a small compound library containing 2000 compounds of FDA approved drugs, experimental biological compounds and nature compounds (Kocisko et al., 2003). We found that small molecule compounds with similar biological activity produced similar impedance-based time-dependent cell response profiles (TCRP). Due to the kinetic nature of the profiling approach both short term and long term compound activity can be measured, allowing for detection of temporally isolated but distinct activities of small molecules and potentially off-target effects. These findings indicate that using impedance-based monitoring and profiling of cellular response upon exposure to biologically active compounds can provide incisive and quantitative information and novel mechanisms for existing drugs as well as experimental biological compounds and can be used to complement other profiling approaches.
Results
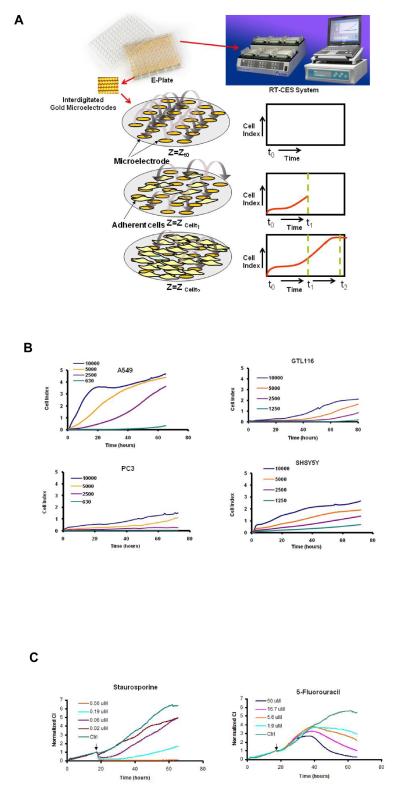
We have designed a Real-Time Cell Electronic Sensing (RT-CES) system (Figure 1A) that utilizes microelectronic plates (E-Plates) integrated with gold microelectrode arrays on glass substrate in the bottom of the wells to measure cellular status in real-time (Solly et al., 2004). In the presence of media, application of low AC voltage (20 mV) produces a small electric field between the electrodes which can be impeded by the presence of adherent mammalian cells leading to large changes in measured impedance (Figure 1A). The extent of impedance change is proportional to the number of cells inside the well and the inherent morphological and adhesive characteristics of the cells. Figure 1B shows the time and density dependent growth and proliferation of 4 different cell lines in the E-Plates. Each cell line produces its own distinct growth kinetic profiles due to unique morphological, adhesive and growth rate characteristics. Since it is well established that cellular properties at the level of cell number, morphology and adhesive interactions can be differentially modulated by biologically active compounds (Abraham et al., 2008; Wang et al., 2008; Young et al., 2008) we sought to determine how modulation of these parameters by small molecule compounds affect the impedance readout. Figure 1C shows the time-dependent effect of two cytotoxic compounds, the general kinase inhibitor staurosporine and the anti-metabolite 5-fluorouracil on A549 cells. Even though both of these compounds are well known apoptotic inducers, they produced unique dose and time-dependent cell response profiles (TCRP). Interestingly, staurosporine appears to have an immediate effect on the cells, while 5-FU takes longer time to manifest its effect. It was of interest to determine if the TCRP can be predictive of biological mechanism; for example, do compounds with similar mechanism produce similar TCRP?
Figure 1.
Real-Time monitoring of adherent cells by the RT-CES system (A) The RT-CES System is composed of a plate station accommodating up to six 96 well E-Plates, an electronic analyzer and a computer which runs the software for automatic and real-time data acquisition and display. The E-Plates are integrated with interdigitated gold microelectrodes. A Schematic representation of microelectrodes and the principle of utilizing cell electrode impedance to non-invasively measure adherent cells are shown. In the absence of cells, the baseline impedance (Z0) at time t0 represents the impedance of the gold microelectrodes. Addition of cells to the sensor microelectrodes leads to changes in impedance signal at time t1 (Zcellt1) that is directly proportional to the number of cells seeded on the sensors and is displayed as Cell Index (CI). The CI value changes with time (Zcellt2) and reflects the morphology, adhesion and number of cells inside the well. (B) Real-time monitoring of the density-dependent growth and proliferation of four cell lines on the RT-CES platform. The cells suspensions were transferred to E-Plates and placed on the RT-CES reader for real-time monitoring every 30 minutes for the duration of the assay. (C) Time-dependent cell response profiles (TCRP) of two different cytotoxic agents using A549 cells. The arrow indicates the point of compound addition.
To test the hypothesis that TCRP can be predictive of mechanism of compound action, a 2000 compound library consisting of FDA approved drugs (50%), natural products (29%), bioactive compounds (18%), herbicides and insecticides (3%) were screened against A549 non-small lung cancer cells and PC3 prostate cancer cell lines using the RT-CES platform. While TCRPs can be generated from both cell types, for the purposes of this report we will primarily show the data from A549 cells and only discuss the data from PC3 cell lines for comparative purposes. We measured both short and long term compound effects. For short term, the compound effect was monitored every 2 minutes after compound addition for a total duration of one hour. The high sampling frequency ensures the acquisition of immediate and transient compound effects on the cell. The short term monitoring is automatically proceeded by long term monitoring protocol collecting the data every 30 minutes from 1 hour to 48 hours after compound addition. This time duration was chosen for the long term response because it would allow sufficient time for the compounds to interact and modulate their target(s) and also result in sufficiently distinguishable TCRPs. Two complementary approaches were undertaken to select the hits from the screen. First, compounds were divided between those resulting in short term response and long term response. For short term response, any compound resulting in normalized cell index change equivalent to 25% or greater of the mean of the control (DMSO) at any time point between compound addition and 1 hour later were chosen as a “hit”. For long term responses, the criterion for hit selection was a change in normalized cell index of 40% or greater of control at any time point between 1 hour after compound addition until the end of the screen. However, if a compound resulted in normalized CI values higher than control for the entire duration of the long term response, then the hit selection criteria was set at 25% of control. The above hit selection criteria of 25% or 40% over control for short-term and long-term hits are chosen based of the consideration that a false positive hit rate should be controlled at about 10%, taking into account the assay variation in RT-CES platform. These criteria not only allow for the identification of compounds with biological activity but also sufficiently distinguishable TCRPs to be used for clustering analysis. The 1 hour selection for short-term response is mainly based on consideration for identifying fast responding cellular events and is used as a demonstration of the concept of separating time series data into different segments for analysis. For other cells or other data from the RT-CES platform, it may be possible to use different time windows for classifying the cell response types. Based on the hit selection criteria described we identified a total of 190 short term and 261 long term hits using A549 cells. Performing the screen in PC3 cells and applying the same criteria for hit selection resulted in a total 160 short term and 440 long term hits, with 75 common hits between A549 in the early response and 104 common hits with A549 cells in the late response. These results indicate that by expanding the cell type for screening and profiling, it may be possible to cover a larger subset of intracellular targets.
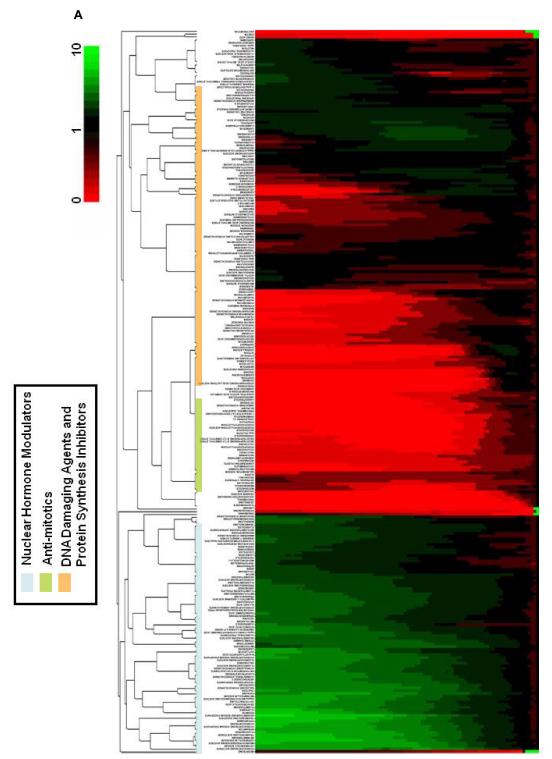
The hits from short term response was treated as one group and the hits from long term response was subjected to an agglomerative hierarchical clustering analysis as described in the methods section (Figure 2A). A complete list of the clustered compounds is shown in Supplementary Figure 2. If TCRPs are indicative of biological activity then compounds with similar biological activity would be grouped together. Indeed, we could identify at least five clusters and subclusters based on comparison with compounds with known mechanism which can be associated with different mechanistic classes such as calcium level and pathway modulators, steroidal nuclear receptor modulators, anti-mitotics, DNA damaging agents and protein synthesis inhibitors (Figure 2A and Supplementary Figures 1-5).
Figure 2.
Clustering analysis and TCRPs of compounds associated different mechanistic groups (A) Agglomerative, hierarchical clustering analysis of TCRPs in the long-term response. The descriptor is the change in CI as a function of time. A complete list of the compounds is shown in Supplementary Figure 2. (B) Typical A549 TCRPs of short-term response group together with compound classes. Most compounds in this sub-group appear to be antagonists of GPCRs and modulators of calcium. The light blue arrow on the x-axis in this graph and proceeding graphs indicate that the short term data was collected every 2 min. (C) Typical A549 TCRP associated with compounds and compound classes which modulate the nuclear hormone receptors. (D) Typical A549 TCRP for tubulin modulating compounds and compound classes. (E) Typical A549 TCRP for compounds which affect nucleotide and DNA synthesis. (F) Typical A549 TCRP for compounds which affect protein synthesis.
The hits that were selected for their short term response was treated as one group and displayed two related impedance-based TCRP (Figure 2B). In both instances there was an immediate drop in cell index (within 5 min) followed by either partial or complete recovery of the cell index curve similar to control curve or alternatively, the other group continued to maintain very low cell indices after the initial decline most likely due to cytotoxicity. Analysis of the compounds with known mechanisms in these groups revealed that the majority of these compounds are antagonists of various GPCR signaling pathways including that of histamine, dopamine and serotonin receptors (Figure 2B and Supplementary Figure 2). Interestingly, most of these compounds displayed similar TCRP in PC3 cells as well (data not shown), indicating their mode of action and consequent morphological cellular response is similar in both cell lines. Based on structural considerations, the hits with known mechanisms in this category can be divided into several structurally-related groups of compounds, including thioxanthenes, phenothiazines, dibenzazepines, dibenzoxazepines, selective estrogen receptor modulators (SERM), dihydropyridine and estrogenic compounds (Figure 2B). Closer examination of compounds in this category revealed another common underlying mechanistic feature, which is modulation of intracellular calcium levels or pathways within the cells either directly or indirectly. For example, the compound amlodipine besylate is a well know L-Type voltage-gated calcium channel inhibitor, while triflupromazine and other phenothiazines are known to interfere with the pleiotrophic effect of calcium/calmodulin action on intracellular pathways (Epstein et al., 2007; Motohashi, 1991). Interestingly, SERM such as tamoxifen, a well known estrogen receptor antagonist also resulted as a hit in this category. Indeed, tamoxifen can cause an increase in intracellular calcium concentrations in a number of cell types (Greenberg et al., 1987; Zhang et al., 2000). Furthermore, there is also evidence that tamoxifen can directly bind and inhibit calmodulin (Lam, 1984).
A number of nuclear hormone receptor agonists, including those that modulate glucocorticoid receptor, mineralcorticoid receptor, progesterone receptor and androgen receptor clustered together (Figure 2C and Supplementary Figure 3). The unique TCRP for this category is characterized by an increase in cell index above the control curve throughout the length of the assay. We have also observed similar TCRP for agonists of peroxisome proliferator activated receptor (PPAR) and retinoic acid receptor (RAR) (data not shown). The fact that modulators of all of these nuclear hormone receptors display similar TCRPs suggests in addition to their specific effects, they share common mechanisms leading to similar cellular effects. One of the common features of nuclear hormone receptors is that they can induce G1 arrest in certain cell types (Wu et al., 2006), possibly through upregulation of cyclin-dependent kinase inhibitors (Addeo et al., 2004). Interestingly, using a high content microscopy profiling approach, Young et al (2008) also observed that most corticosteroid compounds clustered as a group which they identified as G1 arrest phenotype. However, the nuclear receptor like responses using impedance measurement appears to be cell type specific as we did not observe similar response profiles for these groups of compounds in PC3 cells (data not shown).
Another group of compounds whose TCRP co-clustered are well characterized modulators of tubulin polymerization (Figure 2D and Supplementary Figure 4). The impedance-based TCRP for these compounds are characterized by an initial steady decline in cell index followed by recovery after about 14-16 hours post-treatment. This category includes compounds such as paclitaxel, colchicine, nocadazole and nocadazole related compounds and podophyllotoxins. Our screening assay does not seem to distinguish between agents stabilizing microtubules such as paclitaxel and agents that disrupt or prevent tubulin polymerization such as nocadazole (Jordan and Wilson, 2004). It is likely that the TCRP we observe here is reflective of a common underlying global cellular response to these classes of compounds such as cell cycle arrest at mitosis, characterized by unique morphological features such as cell rounding and transient detachment of the cells (Jordan and Wilson, 2004). To better understand this pattern, we treated cancer cells with paclitaxel and fixed and stained the cells with anti-tubulin antibodies at different time points after treatment (Supplementary Figure 6). The initial decline in cell index correlates with morphological changes due to cell rounding as a result of mitotic arrest. At 16 hours, which is coincident with the lowest point of cell index in the TCRP, the majority of the cells (85-90%) are arrested at mitosis determined by intense staining of the spindle apparatus by anti-tubulin antibody. The recovery of the cell index 24 hours after treatment correlates with a subpopulation of cells escaping the arrest and are larger cells that have failed to undergone cytokinesis. This phenomenon has been described as mitotic slippage and is observed in cell types lacking a robust mitotic checkpoint(Weaver and Cleveland, 2005). In addition, non-tubulin targeting compounds, such as monastrol which inhibits the mitotic kinesin Eg5 also displays similar TCRP as those of tubulin targeting agents in our assays (Figure 4). Collectively, the data demonstrate the impedance-based TCRPs are reflection of global cellular morphological responses to anti-mitotic agents and represent a unique signature for mitotic arrest. Exposure of a panel of 6 other cell types to paclitaxel and other tubulin targeting agents resulted in overall similar TCRPs (Supplementary Figure 6B).
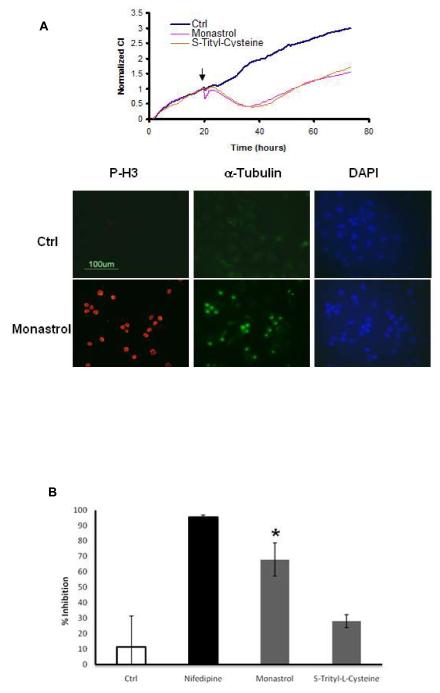
Figure 4.
Using the time resolution offered by TCRP to identify off-target effect for compounds with established activity. (A) Monastrol and S-trityl-cysteine result in a long term TCRP indicative of mitotic arrest. Monastrol (100 μM final concentration) but not S-trityl-cysteine (11 μM final concentration) leads to a short-term response profile indicative of modulation of calcium. The right panel shows staining of A549 cells with phospho-histone H3 antibody after treatment with monastrol for 16 hours. (B) Treatment HEK 293 cells stably expressing the human Cav1.2 voltage-gated L type calcium channel with monastrol (100μM) leads to inhibition of calcium uptake (* ρ value < 0.001 compared to control), while S-trityl-L-Cysteine (20 μM) does not have a significant effect. The error bars represent standard deviations (N of at least 4 samples) and the data is representative of at least 3 independent experiments.
Another major subcluster in our analysis is composed of compounds interfering with DNA synthesis, unwinding and replication, transcription and translation. The TCRP for these compounds is characterized by an initial and sustained increase in cell index above that of the control followed by a decrease in cell index below control levels, reflecting an ultimate cytotoxic response (Figure 2E). These compounds primarily include topoisomerase inhibitors and antimetabolites (Figure 2E and Supplementary Figure 5). Within this cluster, we also observed a subcluster containing TCRPs for compounds inhibiting protein translation such as cyclohehimide and emetine (Figure 2F). The compounds in this category are also known to induce cell-cycle arrest primarily at G1, S or G1/S transition followed by induction of cell death.
The data thus far support the hypothesis that TCRP can be used to cluster compounds with similar mechanism of action. However, the utility of such an approach is best determined by the predictive value it may provide as a compound screening or profiling tool. We next analyzed the TCRP within each cluster to determine its predictive capacity for compounds with unknown mechanism or other reported mechanisms.
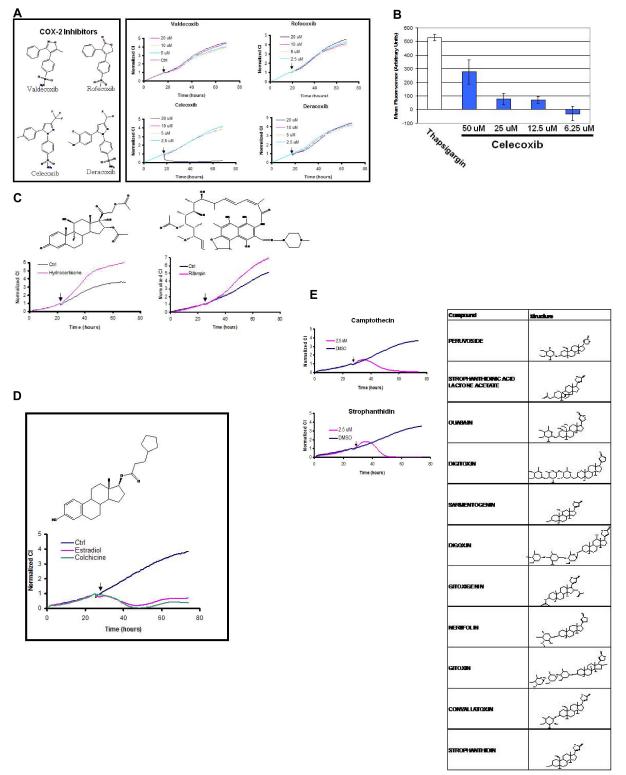
We analyzed the compounds in the calcium level and pathway modulators group and noticed that the TCRP for celecoxib, a cyclooxygenase-2 (COX-2) inhibitor, was present in this group. COX-2 inhibitors, such as rofecoxib, valdecoxib, deracoxib and celecoxib have received considerable attention recently due to cardiotoxicity side effects and all these compounds were present in the library (Figure 3A). Interestingly, at the concentrations the screen was conducted only celecoxib resulted as a hit amongst the four structurally related compounds. Based on the immediate TCRP for celecoxib, the prediction is that celecoxib may be modulating calcium levels within the cell. Indeed, it has been shown previously that celecoxib treatment leads to an immediate increase in internal calcium levels within the cell (Pyrko et al., 2007; Tanaka et al., 2005). To confirm this observation, we measured calcium levels in A549 cells after treating with different doses of celecoxib and thapsigargin as positive control. Celecoxib treatment led to a dose-dependent increase in calcium, as shown previously (Figure 3B).Testing rofecoxib at the same doses did not lead to any detectable calcium release (data not shown). These results support the argument that in addition to inhibiting COX-2, celecoxib may function by alternative mechanisms, especially at doses ultimately resulting in cytotoxicity (Kardosh et al., 2008; Pyrko et al., 2007). From a structure activity relationship, it is interesting that all the COX-2 inhibitors shown in Figure 3A share a common backbone structure, yet the unique side group modifications of celecoxib affords it the ability to modulate intracellular calcium levels. This observation also underscores the challenge in target-driven drug discovery where it is easy to introduce and overlook activities unrelated to the target during lead optimization process.
Figure 3.
TCRPs can be predictive of mechanism of action. (A) COX-2 inhibitors, valdecoxib, rofecoxib, deracoxib and celecoxib were tested in A549 cells at different doses. Only celecoxib resulted in a short-term response TCRP, indicating it might affect calcium levels inside the cell. (B) Direct measurement of calcium levels in the cell using fura-2 demonstrated that celecoxib can induce an increase in calcium levels inside the cell. (C) The TCRP for rifampin co-clustered with well known modulators of nuclear hormone receptors, such as hydrocortisone. (D) The TCRP for estradiol co-clustered with those of other tubulin modulating agents such as colchicine. (E) Compounds with similar backbone structure as digoxin resulted in TCRP similar to camptothecin, indicating that they may affect DNA synthesis or repair as predicted by the TCRP.
In the nuclear hormone modulator cluster we identified the compound rifampin, an antibiotic used for the treatment of tuberculosis(Zhang and Amzel, 2002)(Figure 3C and Supplementary Figure 2). Rifampin acts directly on messenger RNA synthesis in prokaryotes by inhibiting DNA-primed RNA polymerases, especially those of grampositive bacteria such as Mycobacterium tuberculosis. Rifampin has been shown to act as an agonist for glucocorticoid receptors (Calleja et al., 1998).
Close examination of the compounds resulting in mitotic arrest TCRP revealed there are other compounds and drugs not readily recognized for their anti-mitotic activity (Supplementary Figure 4). One of these is estradiol, estrogen receptor agonist (Figure 3D). It has been shown that estradiol and some of its analogues disrupt microtubule polymerization by directly binding to tubulin (Aizu-Yokota et al., 1994; Bogatcheva et al., 2007; Tinley et al., 2003). As a matter of fact, one of the breakdown products of estradiol with anti-proliferative and tubulin binding properties, 2-methoxyestradiol is currently in clinical trials for metastatic breast cancer (James et al., 2007).
In a separate screen in A549 cells we also identified the compound dibenzyltrisulfide (DBTS), a bioactive natural compound found in a subtropical shrub, Petiveria Alliacea (Rosner et al., 2001) displaying similar TCRP as that of epothilone B (Supplementary Figure 6C). Based on the TCRP pattern, we hypothesized DBTS may induce mitotic arrest by targeting tubulin polymerization. Indeed, treatment of MCF-7 cells with DBTS disrupts the microtubule cytoskeleton compared to cells treated with DMSO (Supplementary Figure 6D). To determine if DBTS directly target tubulin polymerization, microtubule assembly assay was performed in the presence of increasing concentrations of DBTS (Supplementary Figure 6E). DBTS dose-dependently inhibited tubulin assembly into microtubules, suggesting that it induces mitotic arrest by directly targeting tubulin polymerization.
In the DNA-damaging subcluster a group of compounds known as cardiac glycosides were identified. These compounds are members of the digoxin family widely used for treatment of heart conditions (De Mey and Snoeck, 1980). They act by inhibiting the sodium/potassium (Na/K) ATPase pump in heart cells, slowing down the extrusion of calcium and causing increased contractility. In our screen, digoxin and related compounds co-clustered with compounds inducing DNA damage such as topoisomerase inhibitors (Figure 3F). These results are corroborated by recent publications demonstrating certain cardiac glycosides can inhibit topoisomerase I and II, providing an explanation for the similarity in their TCRP with DNA damaging agents (Bielawski et al., 2006; Lopez-Lazaro et al., 2005). Interestingly, Young et al (2008) using high content microscopy also demonstrated cardiac glycoside family co-clustered with DNA damaging agents.
One of the major obstacles in target-centric approach to drug discovery is lack of a methodology for identifying and understanding potential off-target effects. We wanted to determine if the TCRP approach can be used to identify off-target activity of compounds. Testing the compound monastrol in A549 cells resulted in a biphasic, short and long term TCRPs (Figure 4A). The long term TCRP is similar to anti-mitotic agents as expected based on the reported mechanism of action of monastrol (Mayer et al., 1999) and was confirmed by phospho-histone H3 staining (Figure 4A right panel). The short term response, while unexpected, resembles the profile of compounds modulating intracellular calcium levels (Figure 2B) and also leads to a transient change in cell morphology as judged by staining of the actin cytoskeleton (data not shown). The compound S-trityl-Cysteine, another Eg5 inhibitor (DeBonis et al., 2004), also led to mitotic arrest TCRP but did not induce the early phase response (Figure 4A). This observation indicated the calcium modulator-like response maybe an off-target effect or additional mechanism of monastrol, rather than relating to inhibition of Eg5. Searching the literature using the chemical name for the core structure of monastrol, 6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate yielded at least one reference indicating compounds resembling monastrol contain calcium channel antagonistic activity (Kumar et al., 2002). We sought to test the possibility that monastrol treatment may modulate the activity of voltage-gated L-type calcium channels. This was tested by measuring calcium uptake through voltage-gated L-type calcium channel Cav1.2 stably expressed in HEK 293 cells. The data in Figure 4B shows that at concentrations which monastrol induces mitotic arrest TCRP (Figure 4A), it significantly blocks calcium uptake (about 70% inhibition) through Cav1.2 channels, while S-Trityl-Cysteine did not have any significant effects. Nifedipine, a known inhibitor of voltage-gated L-type channels, potently inhibited (94.7%) calcium uptake in this assay. While we can’t rule out other mechanisms, the data is consistent with the prediction that monastrol can modulate L-type voltage gated calcium channel activity in addition to targeting Eg5.
Discussion
The approach described in this paper brings to bear two important features of impedance-based monitoring of cellular response upon compound treatment which are important for generating mechanism-specific profiles. First, the interaction of adherent cells with the microelectrodes and generation of impedance response is dependent on three cellular parameters, namely cell number, morphology and attachment quality. All three parameters are intricately linked to signaling pathways regulating various facets of cellular physiology and therefore amenable to modulation by biologically active small molecule compounds in a manner that is dependent on mechanism and concentration of the compound being tested. Rather than measuring compound activity against a single target or pathway, our approach allows for expansion of the biological space at which a compound is screened and provide ample opportunity to identify biological activity of small molecule compounds in an unbiased manner. Because the impedance readout is non-invasive, it allows for the other critical feature of our approach which is inclusion of time resolution in the screening assay. Time dependent compound interaction with cells and unique modulation of cell number, adhesion and overall morphology ultimately cumulates in TCRP which can be used to distinguish between compounds with different activities. The time factor is critical in assessing compound activity(s) as the modulation of the target by the compound or consequent manifestation of the cellular response can be time-dependent. Therefore, the results obtained by other screening and profiling approaches are dependent on the time point at which the assay is processed and any activity that may manifest itself at other than the end point can be missed.
We have demonstrated the importance of the temporal resolution in the assay by using the TCRP to predict a calcium modulating activity for celecoxib, a COX-2 inhibitor (Figure 3A and 3B). Our observation is supported by recent publications, indicating some aspects of celecoxib activity may be COX-2 independent (Kardosh et al., 2008; Pyrko et al., 2007). The time-dependent dual activity of monastrol also highlights the predictive aspect of our current approach. The TCRP allowed us to identify a potential calcium channel inhibitory activity of monastrol not described previously (Figure 4).
While the data presented here support the hypothesis that TCRP can be predictive of mechanism of action and potential off-target drug interactions, we feel that the overall predictive value of the system can be improved by performing the screen and clustering taking into account the dose-response of the compounds and by expanding the pool of cells. Similar to the NCI 60 cell line panel, the TCRP of compounds across multiple cell lines would allow a greater coverage of the expanse of targets being screened and can improve the predictive capacity of the assay. Because the assay is not limited to cell lines or genetically modified cells, primary or disease relevant cell types can also be used making the predictive value of the assay more physiologically relevant. Finally, the TCRP approach can be an important complement to previously described compound profiling approaches such as high content microscopy. The time resolution of impedance-based TCRP can be combined with the multiplexing power of high content microscopy to focus in on specific cellular responses to drugs or compounds and further resolve the on or off targets drug interactions.
Significance
The extent to which protein targets are modulated by drugs or small molecule compounds are dependent on a number of factors, including expression levels of the target, the effective concentration of the compound and the time needed for the compound to perturb the target. One of the limitations of current multi-dimensional phenotypic profiling approaches is that typically a single time point is chosen to assess the effect of compounds and the conclusion regarding the mechanism of action of the compound is based upon the time point at which the samples are processed. It can be envisioned that if the effect of the compound is manifested at any other time, or if a compound has multiple and kinetically distinct activities, it can be overlooked. The TCRP approach described in this work addresses this limitation because the profile generated is time-dependent and in combination with measurement of cell number, morphology and adhesion allows greater expansion of the “biological space” at which compounds are screened and provides ample opportunity detect and identify biological activity associated with small molecules. The utility of the TCRP approach was demonstrated by identifying novel activities and “off-target effects” for certain drugs currently in use in the clinic and experimental compounds. The time resolution provided by the TCRP approach can be used in conjunction with phenotypic profiling approaches to obtain additional mechanistic data associated with small molecule compounds.
Experimental Procedure
Cell culture
All the cells used in this study were purchased from ATCC. Cells are cultured in a standard humidified incubator at 37 °C with 5% CO2 according to optimal media and growth conditions specified by ATCC.
Reagents
All the reagents were purchased form Sigma unless indicated otherwise. The Spectrum 2000 compound library was purchased from MS Discovery. Phospho-histone H3 (Ser 10) antibody was purchased from Cell Signaling Technologies.
Time-dependent cell response profiling using the Real-time Cell Electronic Sensing (RT-CES)
The RT-CES system has been described previously (Atienza et al., 2006; Solly et al., 2004). For time-dependent cell response profiling, 90 μl of media was added to wells 96 well electronic plates (E-Plates) to obtain background readings followed by the addition of 100 μl of cell suspension. The E-plates containing the cells were allowed to incubate at room temperature for 30 min and placed on the reader in the incubator for continuous recording of impedance as reflected by cell index or CI. After 20-24 hours the cells were treated with the compounds from the library. To prepare the compounds for screening, 2 μL of compounds (10 mM) dissolved in DMSO were transferred from the compound plate to 96 well round bottom microtiter plates and diluted with 50 μL of cell growth media. 10 μL of the diluted compounds were then transferred to the E-Plate containing the cells (20 μM final). Each compound dilution plate also contained DMSO only diluted wells as well as media only wells in addition to wells containing 25 nM paclitaxel and 1 μM MG132. The cells were monitored every two minutes for duration of 1 hour after compound addition to capture the short term response and for every 30 min from 1 hour after compound addition to about 48 hours to capture the long term response.
Impedance and Cell Index (CI) measurements by RT-CES system
To quantify cell status based on the measured cell-electrode impedance, a parameter termed Cell Index is derived, according to
where Rb(f) and Rcell(f) are the frequency dependent electrode resistances (a component of impedance) without cells or with cell present, respectively. N is the number of the frequency points at which the impedance is measured. Thus, Cell Index is a quantitative measure of the status of the cells in an electrode-containing well. Under the same physiological conditions, more cells attached on to the electrodes leads to larger Rcell(f) value, leading to a larger value for Cell Index. Furthermore, for the same number of cells present in the well, a change in the cell status such as morphology will lead to a change in the Cell Index. A “Normalized Cell Index” at a given time point is calculated by dividing the Cell Index at the time point by the Cell Index at a reference time point. Thus, the Normalized Cell Index is 1 at the reference time point.
Fluorescence microscopy
A549 cells were seeded in 16-well Lab-tec chamber slides and allowed to attach and spread for 24 h. The cells were treated with compounds at the indicated final concentrations and fixed with 100% ice cold methanol. The cells were permeabilized in PBS containing 0.2% Triton X 100 washed and blocked with PBS containing 0.5% BSA followed by staining with either FITC-conjugated anti-tubulin antibody and anti-phospho-histone H3 antibody (S-10). The cells were washed 3 times with PBS, treated with anti-fade reagent containing DAPI (Invitrogen, Carlsbad, CA) and visualized and imaged using a Nikon E400 epifluorescence microscope and Nikon ACT software.
Calcium Measurements
A549 cells and HeK293 cells stably expressing hCav1.2 a and b cDNAs (Chantest) were seeded in 96 well clear bottom black wall plastic culture plates and transferred to recording chamber of Molecular Devices FLIPR imaging system. Cellular fluorescence was excited by 488 nm illumination through the bottom of 96 well plate by an argon laser and dye fluorescence imaged with a cooled CCD camera. Fluorescence was displayed on line and saved in a file for offline analysis with FLIPRControl software.
For calcium measurements, the cells were loaded with 4 μM Fluo-3AM ester dye and 0.4% pleuronic acid in Hanks Buffer-phosphate solution (HB-PS) for one hour at room temperature. For celecoxib mediated calcium release, A549 cells were washed twice in HB-PS assay buffer and the assay was performed in this assay. For monastrol experiments in Hek-293-Cav1.2 cells, cells were washed twice with Ca-free HB-PS assay buffer and the assay was performed in this buffer. Fluorescence from each plate was monitored for stability of signal. After a uniform response was obtained from the plate in a mock experiment without fluid addition, the measurement with fluid addition was started. The recording consisted of a 30 sec period of baseline data followed by addition of compound plate solutions to each well. The effect of the compounds were monitored for 13.5 min, the first two min sampled at a frequency of 1 sample every second and the last 12 min at a frequency of 1 sample every 5 sec. Control data were subtracted from the test article data to remove background changes in the signal. For monastrol experiment, the compounds were prepared in Na-free HB-PS to depolarize cells with a final concentration of 72 mM KCl and an increase extracellular calcium from 0 to 5 mM at the time of fluid addition.
Microtubule Assembly Assays
Microtublule assembly assays were performed using the fluorescence-based Tubulin Polymerization Kit from Cytoskeleton, Inc. (Denver, CO). Briefly, 2 mg/mL tubulin (Cytoskeleton) was mixed with DBTS at different final concentration (30 μM, 10 μM, 3 μM, 1 μM and 0.5 μM), vincristine (1.5 μM), or DMSO, and the polymerization reaction was performed at 37°C in 20% glycerol, 80 mM PIPES, 2.0 mM MgCl2, 0.5 mM EGTA, pH6.9 and 1 mM GTP and monitored every minute for 45 min in a temperature controlled plate reader (Beckman) at excitation and emission of 360 nm and 450 nm, respectively.
Clustering Analysis
The TCRP for the hits were subjected to an agglomerative, hierarchical clustering analysis (http://en.wikipedia.org/wiki/Cluster_analysis) classified into different clusters so that the TCRP within a cluster shared same or similar patterns. The algorithm treats each response curve as a separate cluster and merges them into successively larger clusters by continuously calculating distances between TCRP curves, searching for minimum distances and grouping minimum-distance curves into a cluster. Following approaches were taken in the curve clustering algorithm: (1) TCRP curves were normalized at the last measurement time point prior to compound addition; (2) the normalized CI curves were projected to a common time coordinate by using interpolation so that the TCRP curves from different experiments with different impedance-measurement time points can be directly compared and the distances between these curves can be calculated; (3) The Euclidean distance in an n-space was used as the distance between two single curves each having n data points; (4) searching for minimum distance between two curves by fixing one curve and moving another one horizontally (time) within a small and given region (4 minutes) to correct the effects of experimental variations in compound addition time for different wells on the calculation of the distance between different response profiles. In this work, the straight forward Euclidean distance based hierarchical clustering algorithm is used for demonstrating the profiling of the cell responses based a time-dependent series data. It is recognized that one may use other approaches for defining the distances between curves or other clustering methods for analyzing the TCRPs.
Supplementary Material
Figure S1 List of and TCRP of hits with short-term responses.
Figure S2 List of clustered long-term hits.
Figure S3 List compounds whose TCRP co-clustered with nuclear hormone receptors
Figure S4 List compounds whose TCRP co-clustered with compounds that modulate microtubule polymerization.
Figure S5 List compounds whose TCRP co-clustered with compounds affecting DNA synthesis and repair and compounds inhibiting protein synthesis.
Figure S6 The TCRP for compounds that modulate microtubule polymerization reflects cellular processes associated with mitotic arrest. (A) HT1080 cells were seeded in 16 well chamber slides, 18 hours later treated with 12.5 nM paclitaxel followed by fixation with methanol 14 hours later. The cells were stained with rhodmaine-conjugated anti-tubulin antibody and visualized and photographed using a CCD camera connected to an eppifluorescence microscope. The antibody intensely stains the mitotic spindle marking cells that are actively undergoing mitosis or are arrested at mitosis. The extent of mitotic arrest is quantified by calculating the mitotic index. (B) The indicated cell panels were seeded in the wells of the E-Plate and 24 hours later treated with 12.5 nM paclitaxel and continuously monitored by the RT-CES system. (C) Treatment of A549 cells growing on E-Plates with 10 μM DBTS or 20 nM Epothilone B, results in similar TCRP (D) Treatment of MCF-7 cells with 10 μM DBTS or DMSO as a control followed by fixation and staining using anti-tubulin antibody conjugated to fluorescine. (E) DBTS inhibits the extent of tubulin polymerization into microtubule using purified and fluorescently-tagged porcine tubulin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham RT. Signalomic signatures enlighten drug profiling. Nat Chem Biol. 2006;2:295–296. doi: 10.1038/nchembio0606-295. [DOI] [PubMed] [Google Scholar]
- Abraham VC, Towne DL, Waring JF, Warrior U, Burns DJ. Application of a high-content multiparameter cytotoxicity assay to prioritize compounds based on toxicity potential in humans. J Biomol Screen. 2008;13:527–537. doi: 10.1177/1087057108318428. [DOI] [PubMed] [Google Scholar]
- Addeo R, Casale F, Caraglia M, D’Angelo V, Crisci S, Abbruzzese A, Di Tullio MT, Indolfi P. Glucocorticoids induce G1 arrest of lymphoblastic cells through retinoblastoma protein Rb1 dephosphorylation in childhood acute lymphoblastic leukemia in vivo. Cancer Biol Ther. 2004;3:470–476. doi: 10.4161/cbt.3.5.838. [DOI] [PubMed] [Google Scholar]
- Aizu-Yokota E, Ichinoseki K, Sato Y. Microtubule disruption induced by estradiol in estrogen receptor-positive and -negative human breast cancer cell lines. Carcinogenesis. 1994;15:1875–1879. doi: 10.1093/carcin/15.9.1875. [DOI] [PubMed] [Google Scholar]
- Atienza JM, Yu N, Kirstein SL, Xi B, Wang X, Xu X, Abassi YA. Dynamic and label-free cell-based assays using the real-time cell electronic sensing system. Assay Drug Dev Technol. 2006;4:597–607. doi: 10.1089/adt.2006.4.597. [DOI] [PubMed] [Google Scholar]
- Bielawski K, Winnicka K, Bielawska A. Inhibition of DNA topoisomerases I and II, and growth inhibition of breast cancer MCF-7 cells by ouabain, digoxin and proscillaridin A. Biol Pharm Bull. 2006;29:1493–1497. doi: 10.1248/bpb.29.1493. [DOI] [PubMed] [Google Scholar]
- Bogatcheva NV, Adyshev D, Mambetsariev B, Moldobaeva N, Verin AD. Involvement of microtubules, p38, and Rho kinases pathway in 2-methoxyestradiol-induced lung vascular barrier dysfunction. Am J Physiol Lung Cell Mol Physiol. 2007;292:L487–499. doi: 10.1152/ajplung.00217.2006. [DOI] [PubMed] [Google Scholar]
- Calleja C, Pascussi JM, Mani JC, Maurel P, Vilarem MJ. The antibiotic rifampicin is a nonsteroidal ligand and activator of the human glucocorticoid receptor. Nat Med. 1998;4:92–96. doi: 10.1038/nm0198-092. [DOI] [PubMed] [Google Scholar]
- De Mey C, Snoeck J. Review of the use of digitalis glycosides in ventricular dysrhythmia. Acta Cardiol. 1980;35:153–165. [PubMed] [Google Scholar]
- DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, Wade RH, Kozielski F. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol Cancer Ther. 2004;3:1079–1090. [PubMed] [Google Scholar]
- Epstein BJ, Vogel K, Palmer BF. Dihydropyridine calcium channel antagonists in the management of hypertension. Drugs. 2007;67:1309–1327. doi: 10.2165/00003495-200767090-00005. [DOI] [PubMed] [Google Scholar]
- Giaever I, Keese CR. Monitoring fibroblast behavior in tissue culture with an applied electric field. Proc Natl Acad Sci U S A. 1984;81:3761–3764. doi: 10.1073/pnas.81.12.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever I, Keese CR. A morphological biosensor for mammalian cells. Nature. 1993;366:591–592. doi: 10.1038/366591a0. [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Carpenter CL, Messing RO. Calcium channel antagonist properties of the antineoplastic antiestrogen tamoxifen in the PC12 neurosecretory cell line. Cancer Res. 1987;47:70–74. [PubMed] [Google Scholar]
- Gunther EC, Stone DJ, Gerwien RW, Bento P, Heyes MP. Prediction of clinical drug efficacy by classification of drug-induced genomic expression profiles in vitro. Proc Natl Acad Sci U S A. 2003;100:9608–9613. doi: 10.1073/pnas.1632587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- James J, Murry DJ, Treston AM, Storniolo AM, Sledge GW, Sidor C, Miller KD. Phase I safety, pharmacokinetic and pharmacodynamic studies of 2-methoxyestradiol alone or in combination with docetaxel in patients with locally recurrent or metastatic breast cancer. Invest New Drugs. 2007;25:41–48. doi: 10.1007/s10637-006-9008-5. [DOI] [PubMed] [Google Scholar]
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- Kardosh A, Golden EB, Pyrko P, Uddin J, Hofman FM, Chen TC, Louie SG, Petasis NA, Schonthal AH. Aggravated endoplasmic reticulum stress as a basis for enhanced glioblastoma cell killing by bortezomib in combination with celecoxib or its non-coxib analogue, 2,5-dimethyl-celecoxib. Cancer Res. 2008;68:843–851. doi: 10.1158/0008-5472.CAN-07-5555. [DOI] [PubMed] [Google Scholar]
- Kocisko DA, Baron GS, Rubenstein R, Chen J, Kuizon S, Caughey B. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J Virol. 2003;77:10288–10294. doi: 10.1128/JVI.77.19.10288-10294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B, Kaur B, Kaur J, Parmar A, Anand RD, Kumar H. Thermal/Microwave assisted synthesis of substituted tetrahydropyrrimidines as potent calcium channel blockers. Indian Journal of Chemistry. 2002;41:1526–1530. [Google Scholar]
- Lam HY. Tamoxifen is a calmodulin antagonist in the activation of cAMP phosphodiesterase. Biochem Biophys Res Commun. 1984;118:27–32. doi: 10.1016/0006-291x(84)91062-3. [DOI] [PubMed] [Google Scholar]
- Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- Lopez-Lazaro M, Pastor N, Azrak SS, Ayuso MJ, Austin CA, Cortes F. Digitoxin inhibits the growth of cancer cell lines at concentrations commonly found in cardiac patients. J Nat Prod. 2005;68:1642–1645. doi: 10.1021/np050226l. [DOI] [PubMed] [Google Scholar]
- MacDonald ML, Lamerdin J, Owens S, Keon BH, Bilter GK, Shang Z, Huang Z, Yu H, Dias J, Minami T, et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat Chem Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, Stoughton R, Burchard J, Slade D, Dai H, et al. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Motohashi N. Phenothiazines and calmodulin (review) Anticancer Res. 1991;11:1125–1164. [PubMed] [Google Scholar]
- Nathan DM. Rosiglitazone and cardiotoxicity--weighing the evidence. N Engl J Med. 2007;357:64–66. doi: 10.1056/NEJMe078117. [DOI] [PubMed] [Google Scholar]
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- Pyrko P, Kardosh A, Liu YT, Soriano N, Xiong W, Chow RH, Uddin J, Petasis NA, Mircheff AK, Farley RA, et al. Calcium-activated endoplasmic reticulum stress as a major component of tumor cell death induced by 2,5-dimethyl-celecoxib, a non-coxib analogue of celecoxib. Mol Cancer Ther. 2007;6:1262–1275. doi: 10.1158/1535-7163.MCT-06-0629. [DOI] [PubMed] [Google Scholar]
- Rosner H, Williams LA, Jung A, Kraus W. Disassembly of microtubules and inhibition of neurite outgrowth, neuroblastoma cell proliferation, and MAP kinase tyrosine dephosphorylation by dibenzyl trisulphide. Biochim Biophys Acta. 2001;1540:166–177. doi: 10.1016/s0167-4889(01)00129-x. [DOI] [PubMed] [Google Scholar]
- Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- Slordal L, Spigset O. Heart failure induced by non-cardiac drugs. Drug Saf. 2006;29:567–586. doi: 10.2165/00002018-200629070-00003. [DOI] [PubMed] [Google Scholar]
- Solly K, Wang X, Xu X, Strulovici B, Zheng W. Application of realtime cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev Technol. 2004;2:363–372. doi: 10.1089/adt.2004.2.363. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Tomisato W, Hoshino T, Ishihara T, Namba T, Aburaya M, Katsu T, Suzuki K, Tsutsumi S, Mizushima T. Involvement of intracellular Ca2+ levels in nonsteroidal anti-inflammatory drug-induced apoptosis. J Biol Chem. 2005;280:31059–31067. doi: 10.1074/jbc.M502956200. [DOI] [PubMed] [Google Scholar]
- Tinley TL, Leal RM, Randall-Hlubek DA, Cessac JW, Wilkens LR, Rao PN, Mooberry SL. Novel 2-methoxyestradiol analogues with antitumor activity. Cancer Res. 2003;63:1538–1549. [PubMed] [Google Scholar]
- Topol EJ. Failing the public health--rofecoxib, Merck, and the FDA. N Engl J Med. 2004;351:1707–1709. doi: 10.1056/NEJMp048286. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou X, Bradley PL, Chang SF, Perrimon N, Wong ST. Cellular phenotype recognition for high-content RNA interference genome-wide screening. J Biomol Screen. 2008;13:29–39. doi: 10.1177/1087057107311223. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, Opferman JT, Sallan SE, den Boer ML, Pieters R, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ, Jr., Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Wu K, DuPre E, Kim H, Tin UC, Bissonnette RP, Lamph WW, Brown PH. Receptor-selective retinoids inhibit the growth of normal and malignant breast cells by inducing G1 cell cycle blockade. Breast Cancer Res Treat. 2006;96:147–157. doi: 10.1007/s10549-005-9071-1. [DOI] [PubMed] [Google Scholar]
- Young DW, Bender A, Hoyt J, McWhinnie E, Chirn GW, Tao CY, Tallarico JA, Labow M, Jenkins JL, Mitchison TJ, Feng Y. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat Chem Biol. 2008;4:59–68. doi: 10.1038/nchembio.2007.53. [DOI] [PubMed] [Google Scholar]
- Zhang W, Couldwell WT, Song H, Takano T, Lin JH, Nedergaard M. Tamoxifen-induced enhancement of calcium signaling in glioma and MCF-7 breast cancer cells. Cancer Res. 2000;60:5395–5400. [PubMed] [Google Scholar]
- Zhang Y, Amzel LM. Tuberculosis drug targets. Curr Drug Targets. 2002;3:131–154. doi: 10.2174/1389450024605391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 List of and TCRP of hits with short-term responses.
Figure S2 List of clustered long-term hits.
Figure S3 List compounds whose TCRP co-clustered with nuclear hormone receptors
Figure S4 List compounds whose TCRP co-clustered with compounds that modulate microtubule polymerization.
Figure S5 List compounds whose TCRP co-clustered with compounds affecting DNA synthesis and repair and compounds inhibiting protein synthesis.
Figure S6 The TCRP for compounds that modulate microtubule polymerization reflects cellular processes associated with mitotic arrest. (A) HT1080 cells were seeded in 16 well chamber slides, 18 hours later treated with 12.5 nM paclitaxel followed by fixation with methanol 14 hours later. The cells were stained with rhodmaine-conjugated anti-tubulin antibody and visualized and photographed using a CCD camera connected to an eppifluorescence microscope. The antibody intensely stains the mitotic spindle marking cells that are actively undergoing mitosis or are arrested at mitosis. The extent of mitotic arrest is quantified by calculating the mitotic index. (B) The indicated cell panels were seeded in the wells of the E-Plate and 24 hours later treated with 12.5 nM paclitaxel and continuously monitored by the RT-CES system. (C) Treatment of A549 cells growing on E-Plates with 10 μM DBTS or 20 nM Epothilone B, results in similar TCRP (D) Treatment of MCF-7 cells with 10 μM DBTS or DMSO as a control followed by fixation and staining using anti-tubulin antibody conjugated to fluorescine. (E) DBTS inhibits the extent of tubulin polymerization into microtubule using purified and fluorescently-tagged porcine tubulin.